Background: Important details of the structures of lipomannan and lipoarabinomannan remain unknown.
Results: New details on the branching structure of LM and the number of arabinan chains in LAM are here provided.
Conclusion: Mature LAM carries a single arabinan chain attached near the middle of the mannan core.
Significance: Results allow for a working model of the biosynthetic pathway of LM and LAM.
Keywords: Glycolipid Structure, Glycosyltransferase, Mass Spectrometry (MS), Mycobacterium tuberculosis, Polysaccharide, Lipoarabinomannan, Lipomannan, Phosphatidylinositol Mannosides
Abstract
Lipoarabinomannan (LAM) is composed of a phosphatidylinositol anchor followed by a mannan followed by an arabinan that may be capped with various motifs including oligosaccharides of mannose. A related polymer, lipomannan (LM), is composed of only the phosphatidylinositol and mannan core. Both the structure and the biosynthesis of LAM have been studied extensively. However, fundamental questions about the branching structure of LM and the number of arabinan chains on the mannan backbone in LAM remain. LM and LAM molecules produced by three different glycosyltransferase mutants of Mycobacterium smegmatis were used here to investigate these questions. Using an MSMEG_4241 mutant that lacks the α-(1,6)-mannosyltransferase used late in LM elongation, we showed that the reducing end region of the mannan that is attached to inositol has 5–7 unbranched α-6-linked-mannosyl residues followed by two or three α-6-linked mannosyl residues branched with single α-mannopyranose residues at O-2. After these branched mannosyl residues, the α-6-linked mannan chain is terminated with an α-mannopyranose at O-2 rather than O-6 of the penultimate residue. Analysis of the number of arabinans attached to the mannan core of LM in two other mutants (ΔembC and ΔMSMEG_4247) demonstrated exactly one arabinosyl substitution of the mannan core suggestive of the arabinosylation of a linear LM precursor with ∼10–12 mannosyl residues followed by additional mannosylation of the core and arabinosylation of a single arabinosyl “primer.” Thus, these studies suggest that only a single arabinan chain attached near the middle of the mannan core is present in mature LAM and allow for an updated working model of the biosynthetic pathway of LAM and LM.
Introduction
Lipoarabinomannan (LAM)4 is a high molecular weight amphipathic lipoglycan that makes up one of the major components of the cell envelope of mycobacteria. Emerging data point to a critical role of LAM and biosynthetic precursors in the integrity of the mycobacterial cell envelope and cell division (1). Consistently, many of the enzymes involved in the biosynthesis of these glycoconjugates are essential for Mycobacterium tuberculosis growth, providing exciting opportunities for drug development (1). The importance of these molecules in promoting the entry of M. tuberculosis inside phagocytic cells, regulating phagosome maturation, and modulating the host immune response has also been well documented (1–3). The fact that even subtle variations in their structures, including their degree of acylation and mannan branching, the length of their mannan and arabinan domains, and the nature of the substituents capping the non-reducing end of the arabinan domain, dramatically impact their biological activities adds further support to the hypothesis that these molecules serve as important modulators of host-pathogen interactions in the course of infection.
LAM was first recognized as a lipidated carbohydrate after isolation from Mycobacterium leprae (4). The presence of an anchoring phosphatidylinositol mannosyl unit was recognized thereafter as was the basic structure of the α-6-linked mannan core with terminal mannosyl residues attached to the 2-position of some of the mannosyl residues, although the arrangement of the branching was not determined (5). The structure of LAM is complex and heterogeneous with four distinct structural domains, including a phosphatidylinositol anchor (5–7), a branched mannan (5), a branched arabinan (8), and capping of the β-terminal arabinofuranosyl residues. The phosphatidylinositol anchor is composed of a myo-inositol phosphoryl diacylglycerol substituted at the 2-position with a single mannopyranose (Manp) and at the 6-position with the mannan (6); this structure is identical to those found in the mycobacterial phosphatidylinositol mannosides (PIM) and lipomannan (LM), which are thought to be biosynthetic precursors of LAM. To date, our cumulative structural data are consistent with the mannan core in both LAM and LM being based on an α-(1,6)-linked backbone, substituted to varying degrees at position 2 with single α-Manp residues and containing ∼35 α-Manp residues in total (1, 2), although the length of the mannan core of LM and degree of branching may vary depending on the Mycobacterium species.
The arabinan portion of LAM contains about 60 arabinofuranosyl (Araf) units (1) that are present as an unknown number of arabinan chains each attached to the mannan (i.e. one arabinan chain of 60 units or two chains of 30 units or 3 chains of 20 units). The arabinan region is conveniently visualized as an internal region and non-reducing termini. The innermost region consists of a linear α-(1,5)-linked Araf backbone of unknown and likely variable length that is attached at its reducing end to the mannan core. This linear region is then followed by a branched region the details of which are poorly understood, although an interesting hypothesis suggesting elongation and further branching based on the structure of the more fully understood arabinan present in arabinogalactan has been put forth (9). The non-reducing termini are better understood and consist of branched hexaarabinofuranosides (Ara6) and linear tetraarabinofuranosides (Ara4). In both these cases the arabinan ends with a β-Araf-(1,2)-α-Araf unit (two of these in the case of Ara6 and one in the case of Ara4). In Mycobacterium smegmatis and Mycobacterium fortuitum the β-Araf units can be capped with an inositol phosphate residue; more commonly, in slow-growing species such as M. tuberculosis and M. leprae, they are capped with α-Manp, α-Manp-(1,2)-α-Manp, and α-Manp-(1,2)-α-Manp-(1,2)-α-Manp (1, 2, 10). The mannose caps can themselves be capped in M. tuberculosis with an unusual α-linked sugar, 5-deoxy-5-methylthio-xylo-furanose (11–13).
Nothing was known until recently about the enzymes, intermediates, and steps involved in the biosynthesis of the mannan backbone and arabinan domain of LM/LAM. We and others recently characterized the M. tuberculosis gene product of Rv2174 as an α-(1,6)-mannosyltransferase (ManT) involved in the latter stages of the elongation of mannan in mycobacteria and corynebacteria (14–15). The knock-out of the orthologous M. smegmatis gene (MSMEG_4241) yielded a mutant that accumulated a precursor LM (designated as pLM) considerably shorter in size than the wild-type (WT) LM (14) but still containing α-(1,6) Manp residues and thus revealing that more than one α-1,6-ManT is involved in LAM biosynthesis with MSMEG_4241 participating in latter stages of mannan elongation. The α-1,6-ManT(s) responsible for the presence of the α-(1,6) mannosyl residues at the reducing end of LAM has not been identified but will be designated ManT-E for earlier in the pathway. Rv2174 (and MSMEG_4241) are designated α-1,6-ManT-L for later in the pathway. We identified Rv2181 as the α-1,2-ManT responsible for the synthesis of the α-(1,2) Manp-linked branches characteristic of the mannan backbone of LM and LAM (16). Inactivation of the M. smegmatis orthologous gene (MSMEG_4247) by allelic exchange blocked the synthesis of mature, branched LM and resulted in the accumulation of a LAM-like product shown by MALDI-TOF/MS to be lower in molecular weight than WT LAM and devoid of 2,6-linked Manp and the corresponding terminal Manp residues. This product had little alteration in the known arabinan motifs, suggesting that a linear α-(1,6) mannan devoid of any α-(1,2) Manp branching is capable of being arabinosylated in a normal fashion (16). In a similar vein, inactivation of MSMEG_6387 (embC) (17), the gene product of which is the major α-(1,5)-arabinosyltransferase (AraT) used in LAM biosynthesis, yielded an M. smegmatis mutant devoid of LAM, as expected, but producing an LM-like molecule (17).
Herein we used these recombinant strains (see Table 1) to determine how many arabinans are present on the mannan backbone of LAM and to elucidate the structural details of the mannan chain itself and where on the mannan chain the arabinan is attached. The novel structural knowledge gained from these studies allow for an updated working model of the biosynthetic pathway of LAM and LM.
TABLE 1.
Strains used in these studies
| Mutant strain | Disrupted gene product | Polymer made | Previously used names | Mannan branching | Size of mannan (no. of Manp residues) |
|---|---|---|---|---|---|
| ΔMSMEG_4241a (14) | α-1,6-ManT | LM-like; no LAM or LAM-like | MutLM | Yes | 13–17 (18–20 minor) |
| ΔMSMEG_6387 (17) | α-1,5-AraT (EmbC) | LM-like; no LAM or LAM-like | Yes | 17–29 | |
| ΔMSMEG_4247b (16) | α-1,2-ManT | LAM-like; no LM or LM-like | MutLAM | No | 8–18 |
EXPERIMENTAL PROCEDURES
Culture Conditions
M. smegmatis mc2155 was grown in 7H9-OADC broth (Difco) with 0.05% Tween 80 or LB broth. The ΔMSMEG_4247, ΔMSMEG_4241, and ΔembC knock-out mutants of M. smegmatis were cultured in the presence of kanamycin (50 μg/ml).
Extraction of Glycosylated Phosphotidylinositol Polymers
M. smegmatis strains were harvested in late logarithmic phase from liquid medium and lyophilized. Dried cells were delipidated with organic solvents and freeze-thawed before disruption by sonication. The resulting suspension was refluxed in 50% ethanol several times; extracts were combined, evaporated, and digested with proteinase K followed by dialysis and subjection to a two-step purification protocol involving hydrophobic and gel exclusion chromatography. The resulting LAM-like and LM-like-containing fractions were monitored on Tricine gels stained with periodic acid/Schiff reagent, pooled, and dialyzed.
MALDI-TOF/MS
The matrix used was 2,5-dihydroxybenzoic acid at a concentration of 10 mg/ml in a mixture of water/acetonitrile (1:1 (v/v)) containing 0.1% TFA and 10 μg of lipoglycan mixed with 1.0 μl of the matrix solution. Analyses were performed on an Ultraflex MALDI-TOF/TOF instrument (Bruker, Bremen, Germany) using reflector mode detection. Mass spectra were recorded in the negative mode for the underivatized sample on a 30-ns time delay with a grid voltage of 94% and full accelerating voltage (25 kV). The mass spectra were mass-assigned through external calibration.
Methylation of the Lipoglycan
Samples were permethylated using the NaOH/dimethyl sulfoxide slurry method as described (18). Permethylated samples as recovered by chloroform/water partitioning were analyzed directly or further loaded onto a reverse phase C18 Sep-Pak cartridge (Waters) for fractionation into stepwise acetonitrile- and methanol-eluted fractions. They were then used for MALDI-TOF analysis and/or a portion was hydrolyzed, reduced, and acetylated as described (19) for linkage analysis.
Mass Spectrometry/Mass Spectrometry (MS/MS) Analyses
MALDI-MS profiling and collision-induced dissociation (CID) MS/MS sequencing were performed on either a quadrupole-time of flight (Q/TOF) Ultima MALDI (Micromass) or a 4700 Proteomics Analyzer (Applied Biosystems), both operated in reflectron positive ion mode. For acquisition on the Q/TOF instrument, the permethylated samples in acetonitrile were mixed 1:1 with α-cyano-4-hydroxycinnamic acid (CHCA) matrix (5 mg/ml in 50% acetonitrile, 0.1% TFA) for spotting onto the target plate. MS survey and CID MS/MS data were manually acquired. For data acquisition on the TOF/TOF instrument (4700 Proteomics Analyzer), the permethyl derivatives in acetonitrile were mixed 1:1 with a 2,5-dihydroxybenzoic acid (DHB) matrix (10 mg/ml in water), whereas underivatized samples in water were mixed 1:1 with 10 mg/ml DHB matrix in acetonitrile for spotting onto the target plate. For high energy CID MS/MS on the TOF/TOF, the potential difference between the source acceleration voltage and the collision cell was set at 2 or 3 kV to obtain optimum fragmentation. The indicated collision cell pressure was normally increased from 3 × 10−8 torr (no collision gas) to 5 × 10−7 torr or up to a maximum of 2 × 10−6 torr (argon).
RESULTS
Analysis of Truncated LM Present in M. smegmatis ΔMSMEG_4241
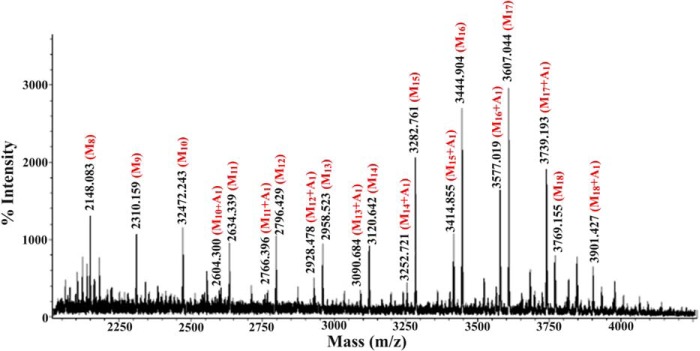
M. smegmatis ΔMSMEG_4241 is missing α-1,6-ManT-L that is used in the later elongation of the mannan present in LM and LAM. This mutant mostly produces pLM of 12–18 mannosyl residues compared with 21–34 mannosyl residues in WT LM (14). No arabinosyl-containing LM or LAM-like molecules are found suggesting that α-1,6-ManT-L is required for arabinosylation to occur. The LM-like molecule produced by M. smegmatis ΔMSMEG_4241 was purified and yielded the expected truncated LM with 12–17 mannosyl residues (with quantitatively minor species containing 5–12 and 18–20 residues), as determined by the value of their M+Na+ ions in MALDI-TOF/MS as previously reported.
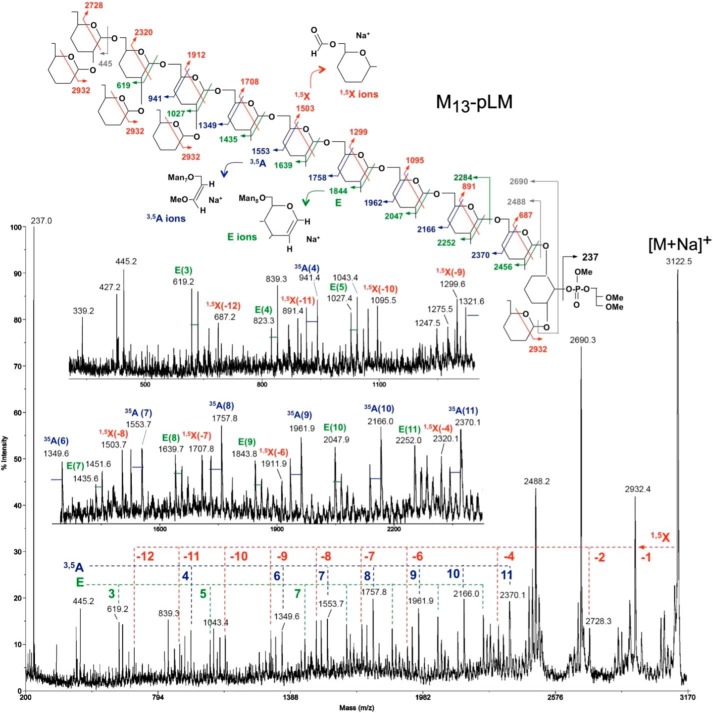
Glycosyl linkage analysis (18, 19) revealed t-Manp, 2-Manp, 6-Manp, and 2,6-Manp in the approximate ratios of 6:1:9:4. For structural details, the quantitatively major components containing 12, 13, 14, and 15 mannosyl units were analyzed by high energy CID MS/MS on the parent [M+Na]+ ion. An exemplary MALDI-MS/MS spectrum for M13-pLM (sodiated parent ion at m/z 3122.5) is shown in Fig. 1 along with a schematic drawing illustrating the deduced fragmentation pattern and the structures of the major product ion series. Interpretation is based primarily on the experience derived from similar high energy CID MS/MS analysis of permethylated N- and O-glycans as reported previously (20) and the known structural features of LM. We have further limited the possible range of structural isomers to an α-6-mannan core, with single-α2-Man branching along the backbone attached to O-6 of inositol, which is further substituted by another single Man at O-2 and a phosphoglycerol unit at O-1.
FIGURE 1.
The mass spectrum, formation of selected ions, and cleavage rational of the CID MS/MS analysis of m/z 3122.5, the M+Na+ ion of methylated M13-pLM produced by M. smegmatis ΔMSMEG_4241. A simplified structure is used where the OMe groups are left off from carbons 3 and 4, and the OMe group on carbon 2 is indicated by a single line. All fragments illustrated are complexed with a Na+ ion. The key 1,5X and 3,5A ions are indicated in red and blue, respectively; the less well understood E ions are shown in green. The formation of 0,4A ions is not illustrated for simplicity but identified on the spectra itself with the blue bars emanating for the 3,5A ions (28 atomic mass units apart). The formation of the major ion at m/z 2690 is not understood. One possibility is that it is formed via concerted loss of the phosphoglycerol moiety together with the single Man on Inositol at O-2 as indicated. The ion at m/z 2488 corresponds to a sodiated mannan chain released from the inositol with a hydrogen transfer yielding at 1-anhydromannose at the reducing end. Another major ion found in all spectra is that at m/z 237, which corresponds to sodiated phosphoglycerol.
The 1,5X series was particularly informative in determining the positions of branching along the six-linked mannan backbone. The 1,5X cleavage ions carry the reducing end of the molecule, and the branching residues can be located by “missing” ions in the series. Thus the largest of the series, i.e. the sodiated ion at m/z 2932, can be derived from cleavage at any of the terminal Manp residues, whereas the next cleavage, where 2 Manp residues are lost, forms the ion at m/z 2728 (Fig. 1). The 1,5X cleavage that would loose 3 Manp residues to yield an ion at m/z 2524 is missing, but the next in series, namely loss of four Manp residues, does occur to yield an ion at m/z 2320. This cleavage pattern is indicative of a branched non-reducing terminus as depicted in Fig. 1. In accordance to this picture, the ion at m/z 2116 is missing because of the next branch point, but the presence of the remainder of the ion series at m/z 1912, 1708, 1503, 1299, 1095, 891, and 687 suggests that the rest of the mannan chain is mostly unbranched up to the inositol residue.
The 3,5A ions, which contain the reducing end of the LM molecule, confirm the same branching pattern. Together with the 0,4A ions at 28 atomic mass units lower (not annotated for simplicity), they also directly demonstrate that the backbone is 6-linked. This 3,5A ion series starts at m/z 2370, which contains 11 complete Manp residues and C-6, -5, and -4 from an additional Manp at the reducing end. It continues with ions at m/z 2166, 1962, 1758, 1553, and 1349 showing the same 6 unbranched Manp residues as the 1,5X ion series. The ion at m/z 1145 is missing due to the first branch at the non-reducing end, but the series continues again with an ion at m/z 941, and then no further ions in this series are seen. m/z 737 should be missing, but we cannot explain the lack of an ion at m/z 533. It is possible that the ion at m/z 533 is not formed due to some structural constraints at the non-reducing end of the molecule.
The same linear structure at the reducing end and branched structure at the non-reducing end is also supported by the series of E ions beginning at m/z 2456, proceeding downward to m/z 1435 with each intervening mass present (m/z 2456, 2252, 2048, 1843, 1639, 1435), and then m/z 1027 where the Manp at O-2 is eliminated, with the ion at m/z 1231 conspicuously absent. Both m/z 823 and m/z 619 are present; m/z 619 is formed as illustrated in Fig. 1 but m/z 823 is not expected. It could possibly be formed from the ion at m/z 1027 by the expulsion of a non-branched internal Manp, as documented previously for chemical ionization MS of per-O-methylated oligosaccharide alditols (21). In conclusion, the E, 3,5A, and 1,5X series all demonstrate the presence of 6 unbranched 6-linked Manp residues attached to the inositol ring. Nevertheless, we cannot absolutely rule out the possibility of other slightly different arrangements of branching occurring at the reducing end of some or all of the molecules affording the same parent ion at m/z 3122.5
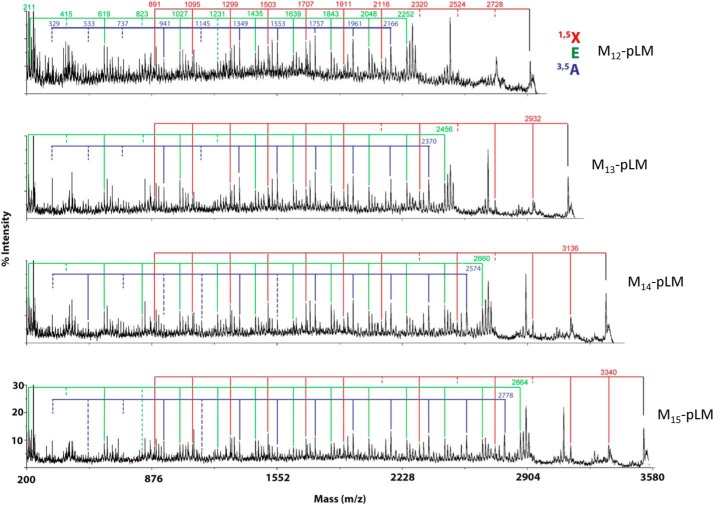
The MS/MS data of other size pLMs also supports related structures to that of Man13-pLM. Analysis of the CID MS/MS of the M+Na+ ions at m/z 3530 of M15-plM suggests that the additional two mannosyl residues are present as an additional branched unit at the non-reducing end and retain the six linear 6-linked mannosyl residues at the reducing end. In contrast, M12-pLM and M14-pLM (Fig. 2) appear to be mixtures with five or seven internal linear 1,6-mannosyl units and have two, three or four terminal mannosyl units attached to the O-2 of mannosyl units in the main chain. These major structures are shown in Fig. 3). It should be noted that although the data support these structures as the quantitatively major arrangements, we cannot rule out small amounts of additional isomers.
FIGURE 2.
The MS/MS spectra of M12-pLM M13-pLM, M14-pLM, and M15-pLM produced by M. smegmatis ΔMSMEG_4241. The key 1,5X and 3,5A are indicated as well as the less well understood E ions (see Fig. 1 for their formation). The presence and absence of the various ions in these three series were used to deduce the structure presented in Fig. 4 as was discussed in detail for M13-pLM in the first section of “Results.”
FIGURE 3.
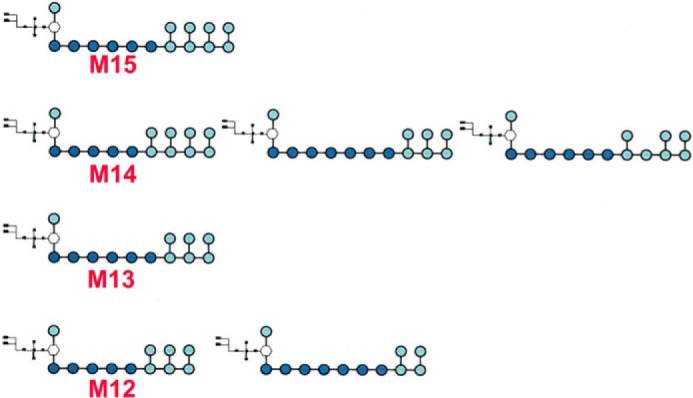
The major species of pLM produced by M. smegmatis ΔMSMEG_4241.
Analysis of the LM-like Product Synthesized by M. smegmatis ΔMSMEG_6387
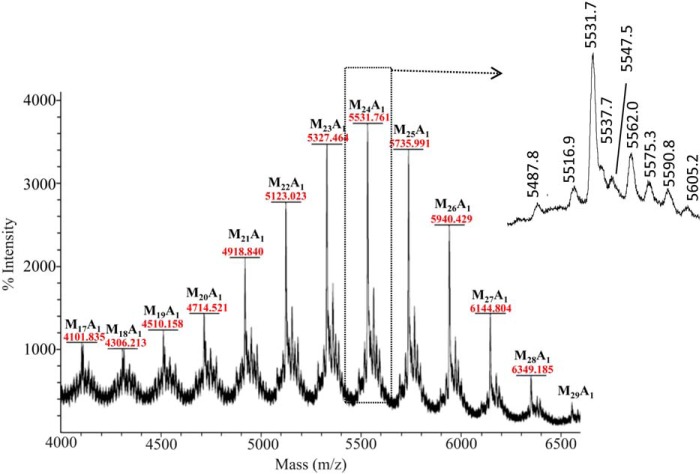
MSMEG_6387 encodes the α-1,5-AraT (EmbC), which is expected to be the second enzyme involved in the arabinosylation of LAM after the “priming enzyme,” which puts the first arabinosyl residue on the mannan chain (this enzyme is not yet identified). In the hope that the embC knock-out mutant would accumulate the LAM mannan substituted with one or more arabinosyl residues (depending on how many arabinan chains are present), we analyzed the LM produced by M. smegmatis ΔMSMEG_6387 by MALDI-TOF MS, which by yielding the molecular weight of the components allows for determination of exactly how many, if any, arabinosyl residues are present. We showed previously that LM from WT M. smegmatis contains no arabinosyl residues (14). In dramatic contrast, the LM-like material produced by M. smegmatis ΔMSMEG_6387, regardless of the number of mannosyl residues, was arabinosylated with a single arabinosyl residue (Fig. 4). These LAM biosynthetic precursors containing a single arabinosyl residue ranged from 17 to 29 mannosyl residues with 21–26 mannosyl residues being quantitatively major. Importantly, the results show that only a single arabinan is present on each mannan core of LAM. Very small amounts of ions corresponding to two arabinosyl residues and no arabinosyl residues were also detected as shown in the insert to Fig. 4, but these ions were in the midst of inexplicable ions, and we hesitate to assign them. Previous work has shown that the arabinan is attached to the mannan core at O-2 of a 2,6-Manp residue in M. tuberculosis LAM (8).
FIGURE 4.
The positive ion MALDI-TOF MS methylated “LM-like” polymer produced by M. smegmatis ΔMSMEG_6387. The phosphatidylinositol is de-esterified and methylated as shown for LM13 in Fig. 1. The M+Na+ ions of species, which contain a single arabinosyl residue and 17–29 mannosyl residues attached to a methylated inositol-phosphoryl-glycerol (Inos-P-Gro), dominate the spectrum. At lower m/z values (data not shown), methyl glycosides of mannosyl oligomers plus and minus a single arabinosyl are found. The methyl glycoside series with the single arabinosyl unit persist up to Ara1Man26 (see the ion at m/z 5537.7 in the inset (mass off by 1 atomic mass units). The formation of these ions is not understood; they might be fragment ions from the higher mass ions or actual oligomers that formed before or during the methylation procedure; in either case it seems likely that somehow they were derived from the parent LM-like molecule. Each major Ara-Mann-Inos-P-Gro is surrounded by ions of 44 and 14 atomic mass units lower in mass and 16, 44, and 60 atomic mass units higher in mass as shown for the sodiated ion resulting from Ara1-Man24-Inos-P-Gro at m/z 5531.3. We cannot explain these ions with any degree of certainty. The ion at m/z 5516.9 is consistent with sodiated Ara1-Man24-Inos-P-Gro with a single OH group from under-methylation. The ion at m/z 5487.6 is consistent with sodiated Ara2-Man23-Inos-P-Gro and the ion at m/z 5575.3 with sodiated Man25-Inos-P-Gro. If these assignments are correct, very occasionally two arabinan chains could be present on LAM, and in this mutant some non-arabinosylated LM might be present. However, given their very weak intensity and the inexplicable ions at m/z 5547.058, m/z 5562.023, and m/z 5590.8, these assignments must be considered tentative.
Analysis of the Mannan Core of the LAM-like Molecule Synthesized by M. smegmatis ΔMSMEG_4247
M. smegmatis ΔMSMEG_4247, a knock-out mutant of the α-1,2-ManT, makes a LAM of a smaller size than the WT parent strain with no detectable LM-like lipoglycan due to the lack of branching (and possible extension) of the mannan core (16, 22). We, therefore, analyzed the mannan core present in this LAM-like molecule upon treatment with Cellulomonas arabinases (23) to remove most of the arabinan. The material retained after passage through a 10,000 molecular mass cutoff membrane was analyzed directly (without methylation) by negative ion MALDI-TOF MS as shown in Fig. 5 (analysis after methylation gave similar results, but the spectrum was stronger in the negative ion non-methylated sample). The predominant acylation pattern was C-16 and C-19 (diacylated). LM precursors with and without a single arabinosyl residue were found from Man10 through Man18 confirming the results above that only a single arabinan chain is attached to the mannan. Presumably, all these mannans were linear due to the lack of the α-1,2-ManT.
FIGURE 5.
The negative ion MALDI-TOF MS of the native lipomannan core of the LAM-like molecule produced by M. smegmatis ΔMSMEG_4247. The predominant species are acylated with two C-16 fatty acyl groups and one C-19 fatty acyl group. They differ in the number of mannosyl and arabinosyl residues present.
DISCUSSION
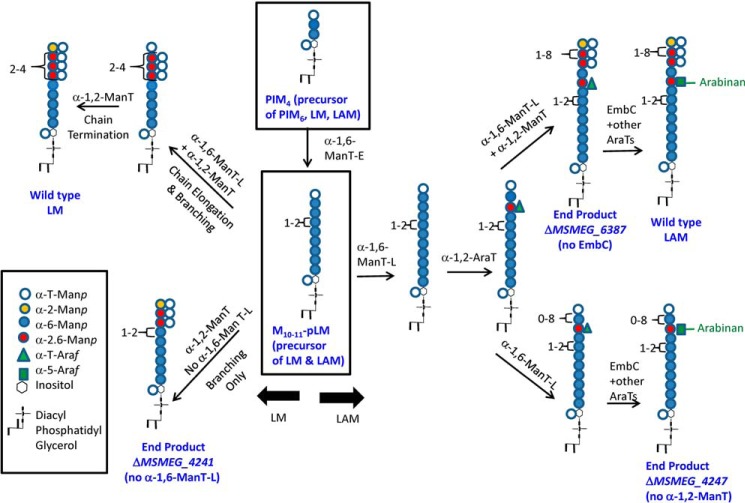
The biosynthesis of LM, LAM, and arabinogalactan is complex and carefully controlled. One of the key biosynthetic distinctions is the different arabinosylation pattern on LAM in contrast to arabinogalactan. More germane to this study is the destination of PIM4 to its three possible end products, PIM6, or as shown in Fig. 6, LM or LAM. The branch point in the pathway to PIM6 occurs by the action of PimE, an α-1,2-ManT that adds a mannosyl residue to O-2 of the third α-(1,6)-mannosyl unit attached at O-6 of the inositol (24). However, in the pathway to LM and LAM, the PIM4 is elongated by as yet to be identified α-1,6-ManT(s), designated as α-1,6-ManT-E. The key branch point between LAM and LM is postulated to be the final linear product of ManT-E, which contains a linear α-1,6-mannan of around 9 residues (M10–11-pLM). In the absence of the second α-1,6-ManT, α-1,6-ManT-L, this product can be acted on by the branching ManT (α-1,2-ManT, the gene product of MSMEG_4247) to form the branched end product produced by M. smegmatis ΔMSMEG_4241 and analyzed in detail in this report (see the lower left of Fig. 6). In contrast, the priming α-1,2-AraT cannot act on M10–11-pLM in the absence of α-1,6-ManT-L, and no LAM is formed in this mutant. When α-1,6-ManT-L and α-1,2-AraT are present, M10–11-pLM can be arabinosylated and elongated, and in the absence of the α-1,2-ManT, all of M10–11-pLM is converted to LAM (with a linear mannan) as found in M. smegmatis ΔMSMEG_4247. In the presence of all three enzymes, both LAM and LM are formed, and the distribution of the two macromolecules is presumably controlled by the ratios of the three proteins. The mannan with (LAM) or without (LM) the priming arabinosyl residue is further elongated and branched. As shown in elegant experiments by the Morita and co-workers (22), the degree of elongation of the mannan is dependent on the ratio of the α-1,2-ManT to the α-1,6-ManT-L. Presumably, the α-1,2-ManT prefers to add an α-Manp to a 6-linked Manp but can add a mannosyl residue to the terminal α-Manp unit, and when it does so, it terminates the growth of the mannan as α-1,6-ManT-L cannot act on this substrate. Thus, depending on the relative abundance of linear α-1,6-Manp residues to α-t-Manp residue (which itself is dependent on the activity of α-1,6-ManT-L), the mannan chain is further elongated or terminated.
FIGURE 6.
A working model of the biosynthetic pathway of LM and LAM as deduced from these and earlier studies. The key intermediate, M10–11-pLM, is shown in the center and lead to wild-type LM and LAM and to the polymers produced by the different constructs studied herein. The non-reducing terminal is shown as fully branched, but it is most likely that some of the branches at the non-reducing termini are occasionally missing.
The arabinan is elongated in the presence of the α-1,5-AraT gene product of embC (and other AraT(s)) as shown in the top right of Fig. 6 to result in WT LAM. Alternatively, as shown in the bottom right of Fig. 6, LAM with an unbranched linear mannan is formed in the absence of the α-1,2-ManT. In the absence of EmbC, the penultimate product shown in the top right of Fig. 6 is formed as analyzed in M. smegmatis ΔMSMEG_6387. Presumably, mannan elongation and branching and arabinan synthesis can go on simultaneously.
The biosynthetic model of Fig. 6 is based on the present experimental evidence but also has testable assumptions and leads to further questions. Some of these include the assumption that the α-1,2-AraT adds an arabinosyl residue to a linear α-1,6-Manp as shown in Fig. 6 and not to the t-Manp at the end of the mannan chain. Presumably, if the arabinosyl residue was added to the end of the chain this would inhibit its further mannosylation. The model further predicts that the arabinosyl residue is generally in the middle of the mannan chain; detailed structure analysis of the end product produced by M. smegmatis ΔMSMEG_6387 and ΔMSMEG_4247 should confirm this. An unanswered question is whether the inability of the α-1,2-AraT to act when α-1,6-ManT-L is missing is due to the size of the substrate (as shown in Fig. 6) or due to lack of protein/protein interactions assuming that LM and LAM are assembled by a multienzyme complex; the presence of Ara-Man10-inositol-phosphoryl-glycerol in the lipoglycan products of M. smegmatis ΔMSMEG_4247 might suggest (in contrast to the pathway presented in Fig. 6) that the size of the substrate is not the cause of the lack of priming Araf transfer in the ΔMSMEG_4241 (α-1,6-ManT-L) knock-out mutant. And finally, one of the most basic questions remains, which is how the arabinan synthesis is directed to LAM rather than arabinogalactan type arabinan. These questions can be addressed at least in part by further detailed analysis of the polymers of the mutants described herein, by the generation of additional mutants, and finally by the expression and purification of active AraTs and ManTs followed by the study of their catalytic activity on specifically prepared substrates.
Acknowledgments
The MALDI-TOF/TOF MS/MS data were acquired at the previous National Research Program for Genomic Medicine Core Facilities for Proteomics and Glycomics (NSC 99-3112-B-001-025) and current Core Facilities for Protein Structural Analysis at Academia Sinica, supported under the Taiwan National Core Facility Program for Biotechnology, NSC 100-2325-B-001-029.
This work was supported, in whole or in part, by National Institutes of Health Grant AI064798 (NIAID).
- LAM
- lipoarabinomannan
- Manp
- mannopyranosyl
- PIM
- phosphatidylinositol mannoside
- Araf
- arabinofuranosyl
- ManT
- mannosyltransferase
- LM
- lipomannan
- pLM
- precursor LM
- AraT
- arabinosyltransferase
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- CID
- collision-induced dissociation.
REFERENCES
- 1. Angala S. K., Belardinelli J. M., Huc-Claustre E., Wheat W. H., Jackson M. (2014) The cell envelope glycoconjugates of Mycobacterium tuberculosis. Crit. Rev. Biochem. Mol. Biol. 10.3109/10409238.2014.925420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gilleron M., Jackson M., Nigou J., Puzo G. (2008) Structure, activities and biosynthesis of the phosphatidyl-myo-inositol-based lipoglycans. In The Mycobacterial Cell Envelope (Daffé M., Reyrat J.-M., eds.) pp. 75–105, American Society for Microbiology, Washington, D. C [Google Scholar]
- 3. Torrelles J. B., Schlesinger L. S. (2010) Diversity in Mycobacterium tuberculosis mannosylated cell wall determinants impacts adaptation to the host. Tuberculosis 90, 84–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hunter S. W., Gaylord H., Brennan P. J. (1986) Structure and antigenicity of the phosphorylated lipopolysaccharide antigens from the leprosy and tubercle bacilli. J. Biol. Chem. 261, 12345–12351 [PubMed] [Google Scholar]
- 5. Hunter S. W., Brennan P. J. (1990) Evidence for the presence of a phosphatidylinositol anchor on the lipoarabinomannan and lipomannan of Mycobacterium tuberculosis. J. Biol. Chem. 265, 9272–9279 [PubMed] [Google Scholar]
- 6. Chatterjee D., Hunter S. W., McNeil M., Brennan P. J. (1992) Lipoarabinomannan: multiglycosylated form of the mycobacterial mannosylphosphatidylinositols. J. Biol. Chem. 267, 6228–6233 [PubMed] [Google Scholar]
- 7. Khoo K.-H., Dell A., Morris H. R., Brennan P. J., Chatterjee D. (1995) Structural definition of acylated phosphatidylinositol mannosides from Mycobacterium tuberculosis: definition of a common anchor for lipomannan and lipoarabinomannan. Glycobiology 5, 117–127 [DOI] [PubMed] [Google Scholar]
- 8. Chatterjee D., Bozic C. M., McNeil M., Brennan P. J. (1991) Structural features of the arabinan component of the lipoarabinomannan of Mycobacterium tuberculosis. J. Biol. Chem. 266, 9652–9660 [PubMed] [Google Scholar]
- 9. Shi L., Berg S., Lee A., Spencer J. S., Zhang J., Vissa V., McNeil M. R., Khoo K.-H., Chatterjee D. (2006) The carboxy terminus of EmbC from Mycobacterium smegmatis mediates chain length extension of the arabinan in lipoarabinomannan. J. Biol. Chem. 281, 19512–19526 [DOI] [PubMed] [Google Scholar]
- 10. Chatterjee D., Lowell K., Rivoire B., McNeil M. R., Brennan P. J. (1992) Lipoarabinomannan of Mycobacterium tuberculosis: capping with mannosyl residues in some strains. J. Biol. Chem. 267, 6234–6239 [PubMed] [Google Scholar]
- 11. Treumann A., Xidong F., McDonnell L., Derrick P. J., Ashcroft A. E., Chatterjee D., Homans S. W. (2002) 5-Methylthiopentose: a new substituent on liporabinomannan in Mycobacterium tuberculosis. J. Mol. Biol. 316, 89–100 [DOI] [PubMed] [Google Scholar]
- 12. Turnbull W. B., Shimizu K. H., Chatterjee D., Homans S. W., Treumann A. (2004) Identification of the 5-methylthiopentose substituent in Mycobacterium tuberculosis lipoarabinomannan. Angew. Chem. Int. Ed. Engl. 43, 3918–3922 [DOI] [PubMed] [Google Scholar]
- 13. Joe M., Sun D., Taha H., Completo G. C., Croudace J. E., Lammas D. A., Besra G. S., Lowary T. L. (2006) The 5-deoxy-5-methylthio-xylofuranose residue in mycobacterial lipoarabinomannan: absolute stereochemistry, linkage position, conformation, and immunomodulatory activity. J. Am. Chem. Soc. 128, 5059–5072 [DOI] [PubMed] [Google Scholar]
- 14. Kaur D., McNeil M. R., Khoo K.-H., Chatterjee D., Crick D. C., Jackson M., Brennan P. J. (2007) New insights into the biosynthesis of mycobacterial lipomannan arising from deletion of a conserved gene. J. Biol. Chem. 282, 27133–27140 [DOI] [PubMed] [Google Scholar]
- 15. Mishra A. K., Alderwick L. J., Rittmann D., Tatituri R. V., Nigou J., Gilleron M., Eggeling L., Besra G. S. (2007) Identification of an α(1→6) mannopyranosyltransferase (MptA) involved in Corynebacterium glutamicum lipomannan biosynthesis, and identification of its orthologue in Mycobacterium tuberculosis. Mol. Microbiol. 65, 1503–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kaur D., Berg S., Dinadayala P., Gicquel B., Chatterjee D., McNeil M. R., Vissa V. D., Crick D. C., Jackson M., Brennan P. J. (2006) Biosynthesis of mycobacterial lipoarabinomannan: role of a branching mannosyltransferase. Proc. Natl. Acad. Sci. U.S.A. 103, 13664–13669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhang N., Torrelles J. B., McNeil M. R., Escuyer V. E., Khoo K.-H., Brennan P. J., Chatterjee D. (2003) The Emb proteins of mycobacteria direct arabinosylation of lipoarabinomannan and arabinogalactan via an N-terminal recognition region and a C-terminal synthetic region. Mol. Microbiol. 50, 69–76 [DOI] [PubMed] [Google Scholar]
- 18. Ciucanu I., Kerek F. (1984) A simple and rapid method for the permethylation of carbohydrates. Carbohydr. Res. 131, 209–217 [Google Scholar]
- 19. Daffe M., Brennan P. J., McNeil M. (1990) Predominant structural features of the cell wall arabinogalactan of Mycobacterium tuberculosis as revealed through characterization of oligoglycosyl alditol fragments by gas chromatography/mass spectrometry and by 1H and 13C NMR analyses. J. Biol. Chem. 265, 6734–6743 [PubMed] [Google Scholar]
- 20. Yu S. Y., Wu S. W., Khoo K. H. (2006) Distinctive characteristics of MALDI-Q/TOF and TOF/TOF tandem mass spectrometry for sequencing of permethylated complex type N-glycans. Glycoconj. J. 23, 355–369 [DOI] [PubMed] [Google Scholar]
- 21. McNeil M. (1983) Elimination of internal glycosyl residues during chemical ionization-mass spectrometry of per-O-alkylated oligosaccharide-alditols. Carbohydr. Res. 123, 31–40 [Google Scholar]
- 22. Sena C. B., Fukuda T., Miyanagi K., Matsumoto S., Kobayashi K., Murakami Y., Maeda Y., Kinoshita T., Morita Y. S. (2010) Controlled expression of branch-forming mannosyltransferase is critical for mycobacterial lipoarabinomannan biosynthesis. J. Biol. Chem. 285, 13326–13336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. McNeil M. R., Robuck K. G., Harter M., Brennan P. J. (1994) Enzymatic evidence for the presence of a critical terminal hexa-arabinoside in the cell walls of Mycobacterium tuberculosis. Glycobiology 4, 165–173 [DOI] [PubMed] [Google Scholar]
- 24. Morita Y. S., Sena C. B., Waller R. F., Kurokawa K., Sernee M. F., Nakatani F., Haites R. E., Billman-Jacobe H., McConville M. J., Maeda Y., Kinoshita T. (2006) PimE is a polyprenol phosphate-mannose-dependent mannosyltransferase that transfers the fifth mannose of phosphatidylinositol mannoside in mycobacteria. J. Biol. Chem. 281, 25143–25155 [DOI] [PubMed] [Google Scholar]