Background: Protein factors function in both FeS cluster assembly on and transfer from the Isu scaffold protein.
Results: Common Isu residues are critical for binding both frataxin homologue (Yfh1) assembly factor and heat shock protein 70 (Hsp70) transfer factor.
Conclusion: Yfh1 and Hsp70 binding is mutually exclusive.
Significance: Mutual exclusive binding of assembly/transfer factors may be important in transitioning between cluster assembly and transfer.
Keywords: Frataxin, Iron-Sulfur Protein, Mitochondria, Molecular Chaperone, Protein-Protein Interaction, Saccharomyces cerevisiae
Abstract
In mitochondria FeS clusters, prosthetic groups critical for the activity of many proteins, are first assembled on Isu, a 14-kDa scaffold protein, and then transferred to recipient apoproteins. The assembly process involves interaction of Isu with both Nfs1, the cysteine desulfurase serving as a sulfur donor, and the yeast frataxin homolog (Yfh1) serving as a regulator of desulfurase activity and/or iron donor. Here, based on the results of biochemical experiments with purified wild-type and variant proteins, we report that interaction of Yfh1 with both Nfs1 and Isu are required for formation of a stable tripartite assembly complex. Disruption of either Yfh1-Isu or Nfs1-Isu interactions destabilizes the complex. Cluster transfer to recipient apoprotein is known to require the interaction of Isu with the J-protein/Hsp70 molecular chaperone pair, Jac1 and Ssq1. Here we show that the Yfh1 interaction with Isu involves the PVK sequence motif, which is also the site key for the interaction of Isu with Hsp70 Ssq1. Coupled with our previous observation that Nfs1 and Jac1 binding to Isu is mutually exclusive due to partially overlapping binding sites, we propose that such mutual exclusivity of cluster assembly factor (Nfs1/Yfh1) and cluster transfer factor (Jac1/Ssq1) binding to Isu has functional consequences for the transition from the assembly process to the transfer process, and thus regulation of the biogenesis of FeS cluster proteins.
Introduction
In all three domains of life proteins having iron-sulfur (FeS) cluster prosthetic groups play important roles in critical cellular processes such as redox reactions, catalysis, and sensing of stress (1, 2). In eukaryotic cells, mitochondria, which inherited their iron-sulfur cluster (ISC)4 biogenesis system from bacterial ancestors, play an essential role in maturation of FeS cluster containing proteins functioning in all subcellular compartments (3). The proteins involved in FeS clusters biogenesis are highly conserved. In eukaryotic cells, the majority of these proteins are essential for cell viability, underscoring the biological importance of FeS cluster proteins. In both mitochondrial and bacterial ISC systems, a highly conserved 14-kDa protein (called Isu in the yeast Saccharomyces cerevisiae) plays a central role in FeS cluster biogenesis, serving both as a scaffold for de novo cluster assembly and as a platform for cluster transfer onto recipient apoproteins (3). Both the assembly and transfer steps of FeS cluster biogenesis involve the interaction of Isu with protein factors (4). Because the ISC system of S. cerevisiae is the focus of this report, we use the gene/protein nomenclature for this organism throughout. In S. cerevisiae, Isu is encoded by the closely related and functionally exchangeable paralogous genes, ISU1 and ISU2 (5, 6). Isu1 plays the major functional role due to its higher level of expression.
De novo assembly of FeS clusters requires Isu interaction with two proteins, the cysteine desulfurase Nfs1, which releases sulfur from cysteine (7), and the yeast frataxin ortholog, Yfh1, which is a putative iron donor and/or regulator of desulfurase enzymatic activity (8–11). Yfh1 and Nfs1 interact both with each other and with Isu, thereby forming the FeS cluster assembly complex, which constitutes the structural and functional unit responsible for cluster synthesis within the Isu scaffold (9, 10, 12–16). In bacteria the cysteine desulfurase (IscS) functions as a single polypeptide. However, in eukaryotes, Nfs1 functions as a stable heterodimer in complex with a small accessory protein Isd11 (referred to as Nfs1(Isd11) throughout) (17–21). Isd11 is proposed to both stabilize Nfs1 and regulate its catalytic activity (22). Although the role of Nfs1(Isd11) as a sulfur donor for FeS cluster synthesis is indisputable, the role of frataxin, which is associated with the neurodegenerative disease Fredreich ataxia (23) in this process is hotly debated (24, 25). Recent experimental data strongly support the view that the major biological function of frataxin is directly related to its role as a component of the FeS cluster assembly complex, but whether it serves as an iron donor, regulator of desulfurase activity, or both is unresolved (26–28).
In both mitochondria and bacteria, FeS cluster transfer from Isu onto recipient apoproteins requires a dedicated J-protein/Hsp70 molecular chaperone pair (29). Similar to other members of the J-protein class (30), Jac1 binding to its client protein (i.e. Isu) targets it for binding to its Hsp70 partner (i.e. Ssq1) (31, 32). It is thought that conformational changes of the cluster-loaded holo-Isu induced upon Hsp70 binding triggers the release of the FeS cluster and its transfer onto a recipient apoprotein (33–35). In both bacteria and mitochondria, the C-terminal domain of Jac1 binds to a specific region on the surface of Isu (36–38), with the most important players of a large interaction interface being three hydrophobic residues of Jac1 (32) interacting with three hydrophobic residues of Isu1 (39). The Hsp70-Isu interaction is also highly sequence specific, interacting via a highly conserved Pro, Val, Lys (PVK) motif (40, 41), which is part of a flexible loop localized in proximity to the FeS binding pocket (35).
Although the role of the assembly complex for de novo FeS cluster synthesis is well established (3, 26) and the importance of the Hsp70/Jac1 chaperones for FeS cluster transfer to recipient apoproteins has solid experimental support (29), relatively little is known about the specific interactions required for assembly complex formation and how the molecular architecture and protein-protein interactions involved in its formation affect transition to the chaperone-mediated transfer reaction. To address these issues, we have pursued structure/function analyses, using S. cerevisiae as a model system. Previously we showed that Nfs1 and Jac1 utilize overlapping binding sites on Isu and that their interactions with Isu are mutually exclusive (39). Here, we show that the stable association of Yfh1 within the tripartite FeS cluster assembly complex is dependent on both its Nfs1(Isd11)- and Isu1-binding interfaces. Furthermore, we demonstrate that Yfh1 binding to Isu1 involves the PVK sequence motif, which is also the binding site for Ssq1 (41). Thus binding of assembly complex factors (Nfs1/Yfh1) to form the assembly complex and action of the transfer factors (Jac1/Ssq1) are mutually exclusive. We hypothesize that this exclusivity likely has functional consequences for the transition from the cluster assembly step to the cluster transfer step in the biogenesis of FeS cluster proteins.
EXPERIMENTAL PROCEDURES
Yeast Strains, Plasmids, Media, and Chemicals
All strains were of the S. cerevisiae W303 background. Cells expressing Yfh1W131A were created by sporulation and dissection of a yfh1-Δ/YFH1 strain (42) harboring a centromeric plasmid carrying the yfh1W131A gene under control of the YFH1 promoter and LEU2 marker. To assess the level of Nfs1 variants, strains having the WT gene at the normal chromosomal location under the control of the glucose-repressible GAL1–10 promoter were used (43). To construct an expression vector for the GST-Yfh1 fusion protein, the mature sequence of YFH1 (codons 52–174) was cloned into the glutathione S-transferase (GST)-expressing vector pGEX-KG (44) using BamHI and XhoI restriction sites. YFH1, ISU1, and NFS1 mutants were generated in pRS315-YFH1, pRS314-ISU1, and pRS316-NFS1 using the Stratagene QuikChange protocol (41), as were all mutants in Escherichia coli expression vectors. yfh-Δ (42) and nfs1-Δ (43) strains were described previously. Yeast were grown on YPD (1% yeast extract, 2% peptone, and 2% glucose) or synthetic media as described (45). All chemicals, unless stated otherwise, were purchased from Sigma.
Protein Purification
The sequences encoding the mature form of Yfh1 (residues 52–174) with a C-terminal polyhistidine tag was cloned into pET-3a (Novagen, Madison, WI). Protein overexpression was induced for 4 h at 30 °C in E. coli C41(DE3) by adding isopropyl 1-thio-d-galactopyranoside at a final concentration of 1 mm. Cells were harvested and lysed using a French press in buffer Y1 (50 mm Tris-HCl, pH 8.0, 50 mm NaCl, 1 mm phenylmethanesulfonyl fluoride (PMSF), 10% glycerol). The cell lysate was subjected to His-Bind Resin (Novagen) chromatography. After washing steps with buffer Y1, Y2 (Y1 + 0.5 m NaCl), and Y3 (Y1 + 20 mm imidazole), proteins were eluted with a linear 20–200 mm imidazole gradient in buffer Y1. Fractions containing pure Yfh1 protein, were collected, pooled, concentrated on Centriprep 10K (Millipore), dialyzed against buffer F (50 mm Tris-HCl, pH 8.0, 10% glycerol, and 50 mm NaCl), and stored at −70 °C.
Expression of the mature form of GST-Yfh1 and GST protein alone were induced in the E. coli strain C41(DE3) carrying the pGEX-KG-YFH1 or pGEX-KG plasmid, respectively, by addition of 1 mm isopropyl 1-thio-d-galactopyranoside at A600 = 0.6. After 4 h growth at 30 °C, cells were harvested and lysed using a French press in buffer L (25 mm Tris-HCl, pH 8.0, 200 mm NaCl, 1 mm PMSF, 1 mm dithiothreitol (DTT), 10% glycerol, and 0.05% Triton X-100). After a clarifying spin, the supernatant was loaded onto a 1-ml glutathione-agarose column (Fluka) equilibrated with 10 volumes of buffer L. Next, the column was washed with 100 ml of buffer L without PMSF, 10 volumes of buffer L with 0.5 m NaCl, and 10 volumes of buffer L with 10 mm MgCl2 and 1 mm ATP. After a final wash with 10 volumes of buffer L, proteins were eluted with buffer E (25 mm Tris-HCl, pH 8.0, 200 mm NaCl, 1 mm DTT, 10% glycerol, 0.05% Triton X-100, and 50 mm reduced glutathione). Fractions containing GST-Yfh1 (or GST) were pooled, dialyzed against buffer F (50 mm Tris-HCl, pH 8.0, 10% glycerol and 50 mm NaCl), and stored at −70 °C.
The mature form of Ssq1 (residues 19–657) with a C-terminal polyhistidine tag was purified from E. coli strain BL21(DE3) that co-expressed SSQ1 and HEP1, which encodes a small accessory protein, from the plasmid pRSFDuet1-SSQ1His/HEP1 (46). Hep1 stabilizes Ssq1 when expressed in the bacterial cells but, as Ssq1 does not form a stable complex with Hep1, it is not present in the Ssq1 final preparation. Expression was induced by addition of 1 mm isopropyl 1-thio-d-galactopyranoside at A600 = 0.6 and cells were grown at 25 °C for 4 h. Cells were harvested, resuspended in C1 buffer (20 mm HEPES-KOH, pH 7.5, 150 mm KCl, 20 mm imidazole, 10% glycerol, 1 mm PMSF, 2 mm magnesium acetate), and lysed using a French press. After a clarifying spin, the supernatant was loaded onto His-Bind Resin (Novagen) equilibrated in buffer C1 and after washing with buffer C2 (C1 + 0.05% Triton X-100), C3 (C1 + 10 mm MgCl2, 1 mm ATP), and C4 (C1 + 1 m KCl), proteins were eluted with a 40–250 mm imidazole gradient in buffer C1. Fractions containing pure Ssq1 were collected, pooled, and dialyzed to buffer C5 (20 mm HEPES-KOH, pH 8.0, 100 mm KCl, 10% glycerol, 5 mm β-mercaptoethanol). Ssq1 was aliquoted and stored at −70 °C.
Wild-type (WT) and mutant Nfs1(Isd11), possessing a polyhistidine tag at the N terminus, were purified as described previously (39). WT and mutant Jac1, with a polyhistidine tag at the C terminus, were purified as described previously (31), except E. coli strain C41(DE3) was used for expression. Recombinant Isu1 with a polyhistidine tag at the C terminus, and WT and mutant Isu1-GST fusion proteins were purified as described previously (Refs. 31 and 32, respectively). In all cases, the purity of WT and variant protein preparations, as judged by using SDS-PAGE separation and Coomassie Blue staining, was 90% or greater (data not shown). In all cases, protein concentrations, determined using the Bradford (Bio-Rad) assay with bovine serum albumin as a standard, are expressed as the concentration of monomers.
Pulldown Assay
Pulldown experiments were performed as described in Ref. 39. In short, 2.5 μm GST-Yfh1 WT or mutant proteins was incubated with the indicated concentrations of Nfs1(Isd11) and/or Isu1 in PD buffer (40 mm Hepes-KOH, pH 7.5, 5% (v/v) glycerol, 100 mm KCl, 1 mm DTT, 10 mm MgCl2) for 20 min at 25 °C to allow complex formation. Reduced glutathione-immobilized agarose beads were pre-equilibrated with 0.1% bovine serum albumin, 0.1% Triton X-100, and 10% (v/v) glycerol in PD buffer. 40 μl of beads (∼20 μl bead volume) were added to each reaction and incubated at 4 °C for 1 h with rotation. The beads were washed three times with 500 μl of PD buffer. Proteins bound to the beads were incubated with 20 μl of 2-fold concentrated Laemmli sample buffer (125 mm Tris-HCl, pH 6.8, 5% sodium dodecyl sulfate, 10% 2-mercaptoethanol, 20% (v/v) glycerol) for 10 min at 100 °C and aliquots were loaded on SDS-PAGE and visualized by Coomassie Blue staining. After quantitation by densitometry values were plotted in GraphPad Prism using 1:1 binding hyperbola to fit data and plotted as relative units with maximum binding given a value of 1. A similar procedure was used for Isu1-GST pulldown assays. In the case of interaction of Isu1-GST with Ssq1, 2.5 μm Isu1-GST, WT or mutant proteins was incubated with 4 μm Ssq1 and the indicated concentrations of Jac1 in PD buffer with 2 mm ATP for 30 min at 25 °C to allow complex formation. Samples were then treated as described above.
Cysteine Desulfurase Enzymatic Activity
The enzymatic activity of Nfs1(Isd11) was measured as sulfide production using cysteine as the substrate as previously described (47). In the standard assay, 0.5 μm Nfs1(Isd11) was incubated alone or with other proteins, as indicated, in 220 μl of CD buffer (20 mm Hepes-KOH, pH 7.5, 5% glycerol, 100 mm KCl, 6 mm DTT, 10 μm pyridoxal phosphate) for 15 min at 30 °C. The reaction was initiated by the addition of 1.0 mm l-cysteine. Following an incubation of 20 min at 30 °C, the reaction was terminated by the addition of 0.1 ml of 20 mm N,N-dimethyl-p-phenylenediamine sulfate in 7.2 n HCl and 0.1 ml of 30 mm FeCl3 in 1.2 n HCl. After further incubation in the dark for 20 min, the absorption of methylene blue was measured at 667 nm.
Cellular Levels of Proteins
To compare levels of Yfh1 or Nfs1 variants in whole cell lysates a culture volume equivalent to 1 ml of A600 = 1.0 was harvested. The pellet was resuspended in 100 μl of 2-fold concentrated Laemmli sample buffer followed by addition of 15 μl of glass beads (0.5 mm). Cell lysates were prepared by bead-beating at 4 °C for 3 min in a cell disruptor. Proteins were solubilized by incubation at 100 °C for 5 min. Insoluble material was removed by centrifugation, and proteins in the supernatant were separated using SDS-PAGE gels. The resolved proteins were transferred electrophoretically to nitrocellulose. Yfh1 or Nfs1 variants were detected by the enhanced chemiluminescence method (48) using anti-Yfh1 or anti-Nfs1 polyclonal antiserum. When analyzing mitochondrial protein, mitochondria (containing 2.5 μg of protein) were solubilized in 2-fold concentrated Laemmli buffer, and analyzed as described above.
Evolutionary Analysis
Amino acid sequences of orthologs of the scaffold protein (ISU1), cysteine desulfurase (NFS1), and frataxin (YFH1) were obtained by retrieving sequences from InterPro (49) database entries: IPR011339 for ISU1, IPR010240 for NFS1, and IPR002908 for YFH1. The Proteobacterial dataset included all sequences listed under the taxon Proteobacteria in InterPro except sequences listed under Alphaproteobacteria, which constituted their own dataset. The Eukaryotic dataset included sequences listed under Eukaryota. The retrieved sequences were filtered to include only sequences from species for which orthologs of all three genes were present in each dataset. Sequences were aligned using MAFFT version 7.123b with default options (50) and amino acid frequencies were determined at positions homologous to Isu1, Nfs1, and Yfh1 from S. cerevisiae (complete data available on request from J. M.).
RESULTS AND DISCUSSION
Stable Yfh1 Binding Requires the Presence of both Nfs1(Isd11) and Isu1
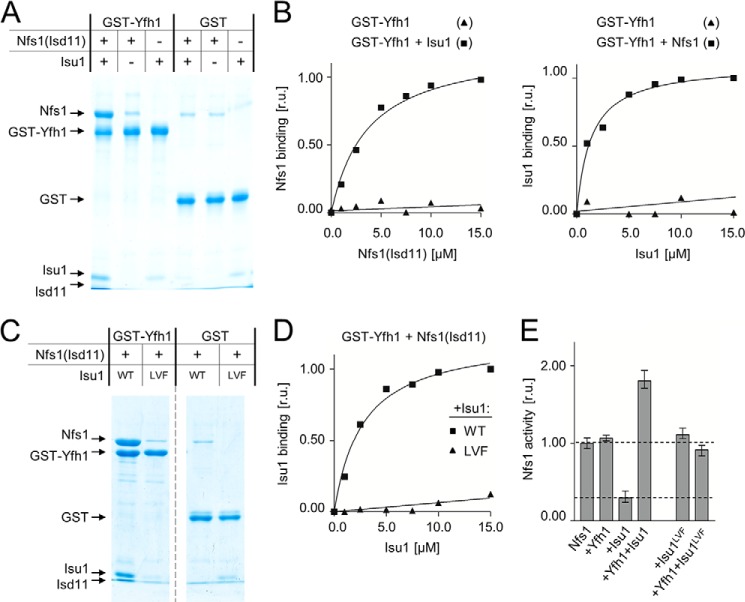
As a first step in defining the molecular mechanism of the interaction of Yfh1 with its binding partners Nfs1(Isd11) and Isu, we developed a semiquantitative pulldown assay that allowed us to measure protein-protein interactions in vitro. Encouraged by the earlier finding that a GST-Yfh1 fusion protein is fully functional in vivo (8), we developed a pulldown assay. GST-Yfh1 (2.5 μm) was preincubated with an excess of both Nfs1(Isd11) and Isu1 (15 μm each). Both Nfs1(Isd11) and Isu1 were efficiently pulled down along with GST-Yfh1 (Fig. 1A). In contrast, if Isu1 was left out of the reaction, no complex between GST-Yfh1 and Nfs1(Isd11) was detected above background levels. Similarly, we did not observe GST-Yfh1 interaction with Isu1 if Nfs1(Isd11) was left out of the reaction. Next, we varied the concentration of Nfs1(Isd11) or Isu1 independently, keeping the other in excess (15 μm compared with 2.5 μm GST-Yfh1). When GST-Yfh1 and Isu1 were incubated with increasing concentrations of wt Nfs1(Isd11) from 1 to 15 μm (Fig. 1B), Nfs1(Isd11) bound to GST-Yfh1 in a concentration-dependent manner. The opposite was also the case when the concentration of Isu1 was varied. These results indicate that Yfh1 formed a stable complex with Isu1 and Nfs1(Isd11), but only when both binding partners were present in the reaction mixture.
FIGURE 1.
Stable Yfh1 binding requires the presence of both Nfs1(Isd11) and Isu1. GST-Yfh1 pulldown (A–D). Glutathione resin was used to pull down GST-Yfh1 (or GST) and any associated proteins. Pelleted proteins were separated by SDS-PAGE and stained. A, 2.5 μm GST-Yfh1 or GST was mixed with 15 μm Nfs1(Isd11) and/or Isu1 as indicated (±). B, 2.5 μm GST-Yfh1 was mixed with 15 μm Isu1 and the indicated concentrations of Nfs1(Isd11) (left panel) or with 15 μm Nfs1(Isd11) and the indicated concentrations of Isu1 (right panel). Densitometry values plotted as relative units (r.u.) with maximum binding Nfs1 (left) and Isu1 (right) were set at 1. C, 2.5 μm GST-Yfh1 or GST was mixed with 15 μm Nfs1(Isd11) and WT Isu1 or Isu1L63A/V72A/F94A variant (LVF). Broken line indicates where the gel was cropped. D, 2.5 μm GST-Yfh1 was mixed with 15 μm Nfs1(Isd11) and indicated concentrations of Isu1 (WT) or Isu1L63A/V72A/F94A variant (LVF). Samples were quantified as described in B. E, cysteine desulfurase activity of Nfs1(Isd11) measured in the absence and presence of the indicated proteins. Activity of Nfs1(Isd11) alone was set to 1. Bars represent average value for three independent measurements, with error bars indicating the range of the measurements.
Disruption of Isu1-Nfs1 Interaction Prevents Stable Binding of Yfh1 to Either Nfs1(Isd11) or Isu1
The results described above are consistent with the idea that the physical interaction between Isu1 and Nfs1(Isd11) is required for stable interaction of Yfh1. To test this idea directly, we took advantage of an Isu1 variant, Isu1L63A/V72A/F94A, which we previously demonstrated to be defective in interacting with Nfs1(Isd11) (39). Indeed, in the presence of Isu1L63A/V72A/F94A, neither Nfs1(Isd11) nor Isu1L63A/V72A/F94A were pulled down with GST-Yfh1 (Fig. 1, C and D), indicating that formation of a stable Isu1-Nfs1(Isd11) complex is required for Yfh1 binding to form the tripartite assembly complex.
It had been previously demonstrated with mammalian orthologs that Yfh1 stimulates the enzymatic activity of cysteine desulfurase, but only in the presence of Isu, whereas Isu, in the absence of Yfh1, inhibits desulfurase activity (2, 9, 10, 51). We decided to take advantage of this functional test of the interaction between Yfh1 and its binding partners. First, we carried out controls to determine whether the yeast proteins behaved similarly to their mammalian orthologs. We observed results similar to those reported for the mammalian proteins. When Yfh1 was added to Nfs1(Isd11) no effect on desulfurase activity was observed. However, when Isu1 was added to Nfs1(Isd11) cysteine desulfurase activity was reduced by ∼70% (Fig. 1E). Upon addition of Yfh1 and Isu1 together desulfurase activity was stimulated ∼1.7-fold. In contrast, when Isu1 WT was replaced by Isu1L63A/V72A/F94A, the variant defective in interaction with Nfs1, no inhibitory effect on Nfs1 desulfurase activity was observed. Moreover, addition of Yfh1 in the presence of Isu1L63A/V72A/F94AA had no stimulatory effect on the enzymatic activity of Nfs1.
Together, the results described above strongly support the idea that Yfh1 only stably interacts with Isu and/or Nfs1(Isd11) when they are in complex. Formation of stable Nfs1(Isd11)-Isu or orthologous complexes was demonstrated previously by biochemical experiments utilizing purified proteins (9, 14, 16, 39, 52) or mitochondrial and cellular extracts (10, 15). It was also postulated (15) that Yfh1 binds the pre-formed Nfs1(Isd11)-Isu complex. Here we demonstrate that the simultaneous presence of Yfh1 binding partners Isu1 and Nfs1(Isd11) is not sufficient for the tripartite assembly complex formation, as when they are unable to interact with each other due to substitutions that prevent Isu1 interaction with Nfs1(Isd11), stable Yfh1 binding does not occur.
Prediction of Amino Acid Residues Involved in Yfh1 Interactions with Isu1 and Nfs1(Isd11)
The biochemical analysis described above suggests that Yfh1 stably interacts with a pre-formed Isu1-Nfs1(Isd11) complex. Mechanistically this requirement could be interpreted in two different ways. (i) Simultaneous interactions of Yfh1 with both Isu1 and Nfs1(Isd11) are required for tripartite complex stability, due to the additive effect of two “low affinity” interactions. (ii) Yfh1 interacts with a single component (i.e. either Nfs1 or Isu1), but the “high affinity” interaction site is only present in the Isu1-Nfs1(Isd11) complex, not the individual proteins, due to a conformational change caused upon interaction of Isu1 and Nfs1(Isd11).
To understand the critical interactions needed for formation of the tripartite assembly complex, we decided to dissect the Yfh1-Isu1 and Yfh1-Nfs1(Isd11) interactions, with a goal of isolating variants that severely affected each individual interaction. As no structural data are available for the mitochondrial FeS cluster assembly complex, we took advantage of a structural model of the homologous bacterial complex (13). In this model, cysteine desulfurase (IscS) and Isu scaffold (IscU) interact with each other and with a monomer of bacterial frataxin (CyaY), which inserts deeply into the cleft formed between IscU and IscS (Fig. 2). Thus, CyaY could form binding interfaces with both IscU and IscS. Using this model (13), together with published experimental data from a variety of systems (8–12, 15, 53–55), we predicted amino acid residues critical for the Yfh1 interaction with each binding partner. Our predictions are depicted in Fig. 2. Below, we describe in more detail our rationale behind each prediction and present its experimental validation.
FIGURE 2.
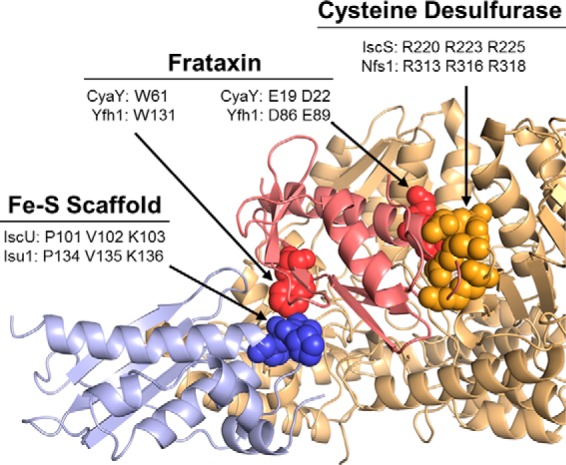
Structural model of bacterial FeS cluster assembly complex. PDB file of bacterial FeS assembly complex (13) was kindly provided by Annalisa Pastore (King's College, London). Residues predicted to be critical for frataxin interaction with the FeS cluster scaffold and cysteine desulferase are listed for bacterial proteins (CyaY, IscU, and IscS) and their yeast orthologs (Yfh1, Isu1, and Nfs1, respectively), as indicated.
Trp131 Is Critical for Yfh1 Interaction with Isu1
Several lines of evidence indicate that the evolutionarily conserved Trp131 is critical for interaction of Yfh1 with Isu1 (2, 15, 20, 55, 56). Using our in vitro assay, we tested whether GST-Yfh1 with Trp131 replaced with alanine was able to pull down Isu1 and Nfs1(Isd11) (Fig. 3A). The ability of GST-Yfh1W131A to form the assembly complex, as monitored by Isu1 binding, was strongly reduced. Even at the highest Isu1 concentration (15 μm) binding was reduced by ∼90%. Moreover, consistent with the reduced binding ability, Yfh1W131A did not stimulate desulfurase activity when mixed with Nfs1(Isd11) and Isu1 (Fig. 3B). To assess the in vivo effects of the Trp131 → Ala alteration, we constructed a yfh1W131A strain. yfh1W131A cells grew slowly (Fig. 3C), even though the level of expression of Yfh1W131A was similar to that of WT as determined using immunoblot analysis (Fig. 3D). Together, these results suggest that Trp131 plays a critical role in Yfh1 binding to Isu1 to form the tripartite assembly complex.
FIGURE 3.
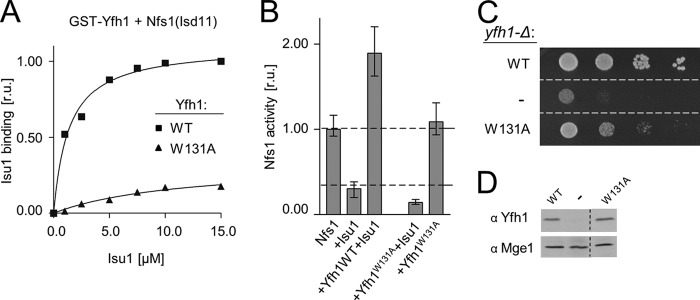
Replacement of Yfh1 Trp131 by alanine results in defective interaction with Isu1 of the Isu1-Nfs1(Isd11) complex. A, 2.5 μm GST-Yfh1 (WT) or GST-Yfh1W131A (W131A) was mixed with 15 μm Nfs1(Isd11) and at the indicated concentrations of Isu1. Glutathione resin was added to pull down the GST-Yfh1 and associated proteins, which were separated by SDS-PAGE and stained. Densitometry values plotted as relative units (r.u.) with maximum binding set at 1. B, cysteine desulfurase enzymatic activity of Nfs1(Isd11) was measured in the absence and presence of the indicated proteins. Desulfurase activity of Nfs1(Isd11) alone was set to 1. Bars represent the average value for three independent measurements, with error bars indicating the range of measurements. C, 10-fold serial dilutions of yfh1-Δ cells harboring plasmid-borne copy of YFH1 (WT), yfh1W131A (W131A), or plasmid lacking an insert (−), as indicated, were plated on glucose-rich medium and incubated at 30 °C for 3 days. D, equivalent amounts of cells described in C were solubilized as described under “Experimental Procedures” and subjected to immunoblot analysis using antibodies specific for Yfh1 and, as a control, Mge1. Strains (C) and samples (D) irrelevant to this study were removed from the image as indicated by a dotted line.
Isu1 Residues Pro134, Val135, Lys136, Which Are Critical for Ssq1 Binding, Are Also Important for Yfh1 Interaction with Isu1-Nfs1(Isd11) Complex
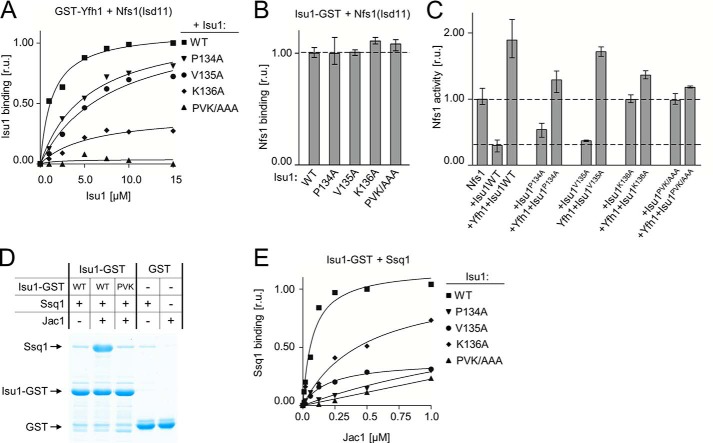
In analyzing the bacterial model (Fig. 2) to predict residues of Isu1 potentially involved in interaction with Yfh1 we noticed that a flexible loop of IscU having the evolutionary invariant Pro, Val, Lys (PVK) motif (29, 35) is in close proximity to bacterial frataxin Trp61, a site homologous to Trp131 of Yfh1. This juxtaposition was very intriguing, as this PVK motif is the only known binding site of the Hsp70s necessary for cluster transfer in the bacterial and mitochondrial systems (29, 41). To test the idea that the PVK motif might be critical for Yfh1, as well as Hsp70, binding, we tested an Isu1 variant having alanines substituted for Pro134, Val135, and Lys136 (Isu1PVK/AAA) in our GST-Yfh1 pulldown assay. The amount of Isu1PVK/AAA pulled down by GST-Yfh1 in the presence of excess Nfs1(Isd11) was at the background level (Fig. 4A), indicating the importance of the PVK motif.
FIGURE 4.
PVK motif of Isu1 is critical for stable Yfh1 interaction with the Isu1-Nfs1(Isd11) complex. A, 2.5 μm GST-Yfh1 and 15 μm Nfs1(Isd11) was mixed with the indicated concentrations of Isu1 (WT) or indicated variant having alanine substitutions in place of Pro134, Val135, and/or Lys136, as indicated. Glutathione resin was added to pull down the GST-Yfh1 and associated proteins, which were separated by SDS-PAGE and stained. Densitometry values plotted as relative units (r.u.) with maximum binding of WT Isu1 protein set at 1. B, 2.5 μm Isu1-GST (WT) or the indicated replacement variant was mixed with 15 μm Nfs1(Isd11). Glutathione resin was added to pull down the GST-Yfh1 and associated proteins, which were separated by SDS-PAGE and stained. Binding of WT was set at 1. Bars represent average value for three independent measurements, with error bars indicating the range of the measurements. C, cysteine desulfurase activity of Nfs1(Isd11) was measured in the absence and presence of the indicated proteins. Activity of Nfs1(Isd11) alone was set to 1. Bars represent average value for three independent measurements, with error bars indicating the range of the measurements. D, 2.5 μm Isu1-GST (WT) or Isu1P134A/V135A/K136A-GST variant (PVK) or GST were incubated in the presence (+) or absence (−) of 4 μm Ssq1 and/or 0.25 μm Jac1. Glutathione resin was added to pull down GST and associated proteins, which were separated by SDS-PAGE and stained. E, 2.5 μm Isu1-GST, WT, or the indicated variants, were mixed with 4 μm Ssq1 and varying concentrations of Jac1. Glutathione resin was added to pull down GST and associated proteins, which were separated by SDS-PAGE and stained. Densitometry values plotted as relative units (r.u.) with maximum binding of Ssq1 set at 1.
Next we tested single alanine substitutions of the PVK motif (Fig. 4A). Altering Lys136 had the most severe effect on binding, with alteration of Pro134 or Val135 only moderately reducing binding. At the highest Isu1K136A concentration used (15 μm) the amount of Isu1K136A pulled down by GST-Yfh1 was reduced by ∼80% (Fig. 4A). As a control, we also tested whether the PVK alteration affected interaction with Nfs1(Isd11), as stable Yfh1 binding to Isu1 requires the Isu1-Nfs1(Isd11) interaction (see Fig. 1). We used our previously published Isu1-GST pulldown assay (39). Each of the Isu1PVK/AAA variants pulled down Nfs1(Isd11) as efficiently as the WT control (Fig. 4B), indicating that substitutions within the PVK motif did not affect the ability of Isu1 to form a stable complex with Nfs1(Isd11). We conclude that in the context of assembly complex formation, the PVK motif of Isu1 is directly involved in Yfh1 binding.
We also compared the relative importance of the three residues of the PVK motif for interaction with Yfh1 and Hsp70 Ssq1. We incubated Isu1PVK/AAA-GST variants (2.5 μm) with excess purified Ssq1 protein (4 μm). We also included substoichiometric amounts of Jac1 co-chaperone, as it serves to target Isu1-GST for Ssq1 binding, thus promoting formation of a stable Ssq1-Isu1-GST complex (31, 41) (Fig. 4, D and E). When the Isu1PVK/AAA-GST variant was used, the amount of Ssq1 pulled down was reduced to background levels, even at the highest Jac1 concentration tested (1 μm) (Fig. 4, D and E). However, individual residues of the PVK motif had different contributions to Ssq1 binding compared with Yfh1 binding. For example, whereas the Lys136 → Ala substitution had the strongest effect on Yfh1 binding (Fig. 4A) its effect on interaction of Ssq1 was the weakest (Fig. 4E). In contrast, the strongest defect for Ssq1 binding was observed with the Pro134 → Ala substitution, which had only a moderate effect on Yfh1 binding. We conclude that the PVK motif plays a critical role for Isu1 interaction with both Yfh1 and Ssq1, but individual residues contribute differently to the two interactions. However, the dependence of both on the PVK motif for binding, suggests that Yfh1 and Ssq1 interactions with Isu1 are mutually exclusive.
Because of the potential importance of such mutual exclusivity, we also tested the effect of the alanine replacements within the PVK motif on the cysteine desulfurase activity of Nfs1(Isd11) (Fig. 4C). In the presence of the Isu1PVK/AAA variants, Yfh1 displayed a reduced ability to stimulate desulfurase activity. These defects are consistent with the reduced ability of Yfh1 to bind Nfs1(Isd11)-Isu1 complexes having substitutions within the PVK motif.
Interestingly, addition of either Isu1PVK/AAA or Isu1K136A to Nfs1(Isd11) revealed an unexpected behavior. These two Isu1 variants did not inhibit desulfurase activity, despite the fact that both bind Nfs1(Isd11) as efficiently as WT protein (Fig. 4B). In contrast, the Isu1P134A and Isu1V135A variants significantly inhibited desulfurase activity as well as WT Isu1. These results point to the interpretation that the PVK motif, and more specifically, the Lys136 residue may play a significant role in the inhibition of the desulfurase enzymatic activity. However, the mechanistic basis of this inhibition and its biological importance are not well understood.
Charged Residues of Yfh1 and Nfs1 Are Critical for Their Interaction in the Context of the Assembly Complex Formation
Structural and experimental data available for bacterial homologs (Fig. 2) (13) and experimental results published for mitochondrial proteins (24, 53–55, 57) suggested negatively charged Asp86 and Glu89 of Yfh1 as potential interacting partners with positively charged Arg313, Arg316, and Arg318 of Nfs1. Reasoning that replacement by alanines may have a lesser effect compared with charge reversal, we purified variants of Yfh1 and Nfs1 having substitutions of the residues to either alanine (Yfh1DE/AA and Nfs1RRR/AAA) or to a residue of the opposite charge (Yfh1DE/KK and Nfs1RRR/EEE). Indeed, although Yfh1DE/AA had a markedly reduced ability to pull down Nfs1(Isd11) and Isu1, no Yfh1DE/KK binding above background levels was observed (Fig. 5A). The Yfh1DE/KK variant was also unable to stimulate Nfs1(Isd11) desulfurase activity in the presence of Isu1 (Fig. 5B). Moreover, cells expressing the DE/KK variant as the only copy of Yfh1, displayed a slow growth phenotype at all temperatures tested (Fig. 5C and data not shown), despite the fact that the Yfh1DE/KK was expressed at levels comparable with WT (Fig. 5D). These results are consistent with Asp86 and Glu89 playing a significant role in Yfh1 interaction with Nfs1(Isd11) to form the assembly complex, as well as verifying the observation that a stable association of Yfh1 with both Nfs1(Isd11) and Isu1 requires simultaneous interactions with both binding partners.
FIGURE 5.
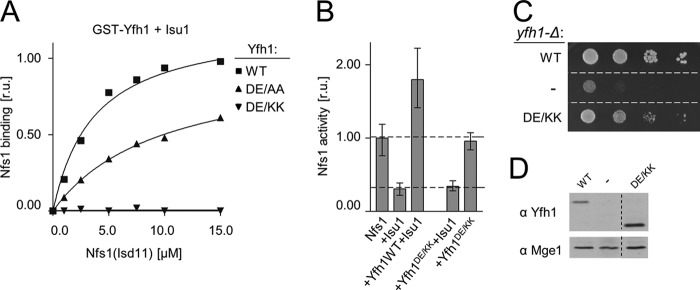
Replacement of residues Asp86 and Glu89 of Yfh1 results in reduced interaction with Nfs1 of the Isu1-Nfs1(Isd11) complex. A, 2.5 μm GST-Yfh1, WT, or variants having Asp86 and Glu89 replaced by Ala or Lys (DE/AA and DE/KK, respectively) was mixed with 15 μm Isu1 and the indicated concentrations of Nfs1(Isd11). Glutathione resin was added to pull down the GST-Yfh1 and associated proteins, which were then separated by SDS-PAGE and stained. Densitometry values plotted as relative units (r.u.) with maximum binding of Nfs1 set at 1. Bars represent average values for three independent measurements, with error bars indicating the range of the measurements. B, cysteine desulfurase activity of Nfs1(Isd11) was measured in the absence and presence of the indicated proteins. Desulfurase activity of Nfs1(Isd11) alone was set to 1. Bars represent the average value for three independent measurements, with error bars indicating the range of measurements. C, 10-fold serial dilution of yfh1-Δ cells harboring plasmid-borne copy of YFH1 (WT), yfh1D86K/E89K (DE/KK) or plasmid lacking an insert (−) were plated on glucose-rich medium and incubated at 30 °C for 3 days. D, equivalent amounts of cells described in C were solubilized and subjected to immunoblot analysis using antibodies specific for Yfh1 and, as a control, Mge1. Strains (C) and lanes (D) irrelevant to this study were removed from the images as indicated by dotted line.
Analysis of the Nfs1RRR variants revealed that either substitution to alanine or to glutamic acid caused a severe reduction in binding. No detectable pulldown by GST-Yfh1 was observed in either case, indicating that Arg313, Arg316, and Arg318 are critical for Yfh1 binding. To ensure the validity of our interpretation, we tested whether Nfs1RRR/AAA and Nfs1RRR/EEE were able to form a stable complex with Isu1, using the Isu1-GST pulldown assay as described above. We found that both Nfs1 variants formed the Nfs1(Isd11)-Isu1-GST complex as efficiently as WT protein (Fig. 6B). We also found that Yfh1 was unable to stimulate the desulfurase activity of either Nfs1RRR/AAA(Isd11) or Nfs1RRR/EEE(Isd11), despite the fact that the basal enzymatic activity of these variants was similar to the WT control (Fig. 6C).
FIGURE 6.
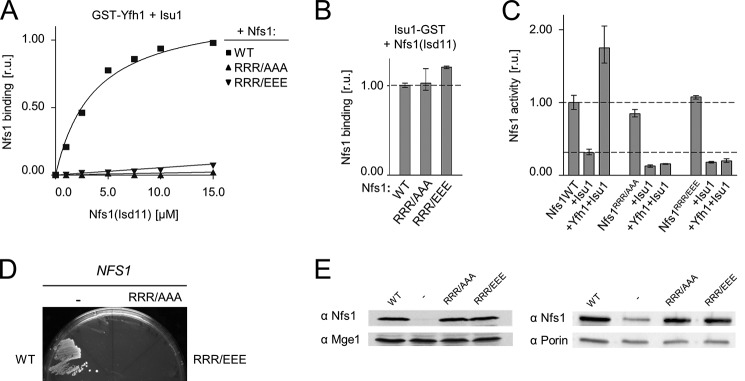
Replacement of residues Arg313, Arg316, and Arg318 of Nfs1 results in reduced binding of Yfh1 to the Isu1-Nfs1(Isd11) complex. A, 2.5 μm GST-Yfh1 was mixed with 15 μm Isu1 and the indicated concentrations of Nfs1(Isd11); wild-type (WT), Nfs1R313A/R316A/R318A (RRR/AAA), or Nfs1R313E/R316E/R318E (RRR/EEE). Glutathione resin was added to pull down GST-Yfh1 and associated proteins, which were separated by SDS-PAGE and stained. Densitometry values plotted as relative units (r.u.) with maximum binding of WT Nfs1 protein set at 1. B, 2.5 μm Isu1-GST was mixed with 15 μm Nfs1(Isd11) (WT) or the indicated replacement variants. Samples were treated and quantitated as in A. C, enzymatic activity of cysteine desulfurase Nfs1(Isd11) (WT) and the indicated replacement variants were measured in the absence and presence of other proteins, as indicated. Desulfurase activity of WT Nfs1(Isd11) alone was set to 1. B and C, bars represent average values for three independent measurements, with presented error bars indicating the range of the measurements. D, nfs1-Δ cells harboring an URA3-marked plasmid containing the WT copy of NFS1 and a second plasmid harboring either NFS1 (WT) or nfs1RRR/AAA or nfs1RRR/EEE or plasmid lacking an insert (−) were plated on glucose-minimal medium containing 5-fluoroorotic acid, which selects for cells having lost the plasmid containing the URA3 marker. The plate was incubated at 30 °C for 3 days. E, lysates of GAL-NFS1 cells transformed with plasmids having no insert (−) or harboring either a WT copy of NFS1 (WT), nfs1RRR/AAA, or nfs1RRR/EEE, as indicated, under the control of the native NFS1 promoter were prepared 24 h after transfer from galactose- to glucose-based medium and separated by SDS-PAGE. Immunoblots were probed with antibodies specific to Nfs1 and Mge1 (left) or porin (right), as a loading control. Left, whole cell lysates. Right, mitochondrial lysates.
We also tested the effect of these alterations on in vivo function. Cells expressing either the RRR/AAA or RRR/EEE variant as the only copy of Nfs1 were inviable (Fig. 6D). To ensure that the null phenotypes were due to altered protein function, not low expression, we used a yeast strain in which Nfs1 WT expression was driven by the GAL-10 promoter and, thus, repressed upon glucose addition. After transformation of these cells with a plasmid harboring a mutant NFS1 allele under control of the native NFS1 promoter, cultures were shifted from galactose- to glucose-based medium. WT Nfs1 was depleted below the level of immunodetection, whereas the levels of both Nfs1RRR/AAA and Nfs1RRR/EEE were similar to that of Nfs1 in a WT strain (Fig. 6E). We conclude that, despite normal expression levels, neither the RRR/AAA nor the RRR/EEE Nfs1 variants are able to support cell growth.
These in vivo results were unexpected, as replacement of the residues forming the apparently complementary binding interface of Yfh1 resulted in a slow growth, not a null, phenotype (see Fig. 5C). One possible explanation is that our model-based prediction of residues forming the Yfh1-Nfs1 interface is not precise. The three residues substituted on the Nfs1 side could be contributing more to the interaction than the two residues substituted on the Yfh1 side. This interpretation is consistent with our observation that Yfh1DE/AA had markedly reduced, but measurable, binding capacities in the pulldown assay (Fig. 5A), whereas no Nfs1RRR/AAA binding was detected (Fig. 6A). Alternatively, this site of Nfs1 may be involved in more than one protein-protein interaction. Data published recently for the bacterial system (58, 59), indicates that the homologous RRR residues of the Nfs1 ortholog are not only involved in binding of bacterial frataxin, but are also critical for interaction with ferrodoxin, a protein whose activity is needed for the reduction of sulfur during the process of FeS cluster assembly (3, 59). As ferrodoxin is an essential component of the FeS cluster biogenesis system, reduced binding by the Nfs1RRR/AAA variant could well explain the null phenotype. In addition, these RRR residues of Nfs1 were also identified as part of a highly conserved RRRPR motif of Nucleotide Localization Sequences (NLS) (60, 61). Substitutions within the NLS may potentially prevent targeting of Nfs1 to the nucleus and thus affect its nuclear function(s). Additional studies are needed to test these possibilities.
Residues Involved in Yfh1 Interaction with Isu1 and Nfs1(Isd11) in the Context of the FeS Cluster Assembly Complex Formation Are Evolutionary Conserved
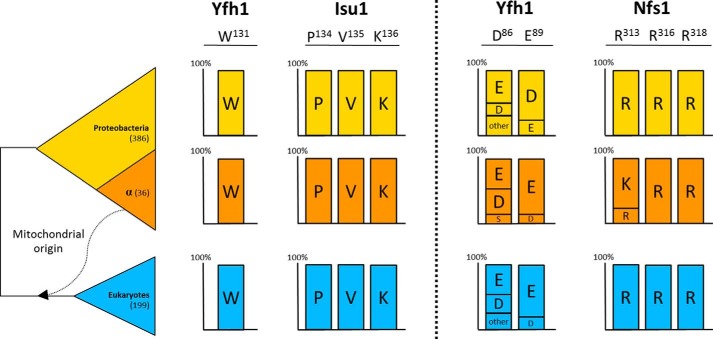
To place our experimental findings in a wider evolutionary context, we identified residues homologous to those involved in the Yfh1-Isu1 and Yfh1-Nfs1 interactions across phylogeny. First, we identified orthologs of Yfh1, Isu1, and Nfs1 in fully sequenced genomes of 422 bacterial and 199 eukaryotic species. Our analyses revealed that residues involved in Yfh1 interaction with Isu1 are extremely evolutionary conserved, as both Trp131 of Yfh1 and Pro134, Val135, and Lys136 of Isu1 are invariant across all analyzed orthologous sequences (Fig. 7). In the case of residues involved in Yfh1 interaction with Nfs1, we observed that sites occupied by Asp86 and Glu89 of Yfh1 are occupied mostly by acidic residues in both bacterial and eukaryotic orthologs, indicating that the chemical nature of the Yfh1 interaction with Nfs1 is evolutionary conserved. An even higher level of conservation was observed on the Nfs1 side of the interaction. In most orthologs, sites occupied by Arg313, Arg316, and Arg318 of Nfs1 were invariant across phylogeny. In α-proteobacteria, Arg313 was replaced by Lys in most orthologs, which maintains the chemical nature of the site involved in Yfh1 interaction. Overall, the phylogenetic analyses revealed a high conservation of the residues involved in interaction of Yfh1 with Isu1 and Nfs1, supporting the idea that that the molecular mechanism of the interaction of these proteins in the context of the FeS assembly complex formation, is also highly evolutionarily conserved.
FIGURE 7.
Residues involved in Yfh1 interaction with the Isu1-Nfs1(Isd11) complex are evolutionary conserved. Residues occupying positions homologous to those involved in Yfh1-Isu1 and Yfh1-Nfs1 interactions with Isu1-Nfs1(Isd11) complex are indicated for orthologous proteins from proteobacteria (386 species), α-proteobacteria (36 species), and eukaryotes (199 species). The percentage of species having given residues is indicated.
Conclusions
Our results indicate that formation of a stable tripartite assembly complex requires the interaction of Yfh1 with both its binding partners, Nfs1(Isd11) and Isu1. In the alternative hypothesis, that the interaction between Isu1 and Nfs1(Isd11) caused a conformational change in one of the partners resulting formation of a high affinity binding site were true, we would have expected to find one of the interactions to be of predominant importance. This was not the case, as similar effects of individual binding site replacements were observed, regardless of whether formation of the tripartite assembly complex was detected by the GST based pulldown technique or by measuring Nfs1(Isd11) desulfurase activity. Overall, an ordered maturation of the FeS cluster assembly complex, with Isu1 and Nfs1(Isd11) interacting first and Yfh1 binding to the preformed Nfs1(Isd11)-Isu1 heterodimer via interactions with both proteins, is the most likely scenario in vivo. The Isu-Nfs1(Isd11) heterodimer is readily detectable under a variety of conditions both in vivo and in vitro (9, 10, 14–16, 39, 52). Although interaction between Yfh1 (or its orthologs) and Isu and Nfs1 individually have been detected under some conditions (12, 13, 62), individual interactions among pairs of components of the tripartite complex are possible (Nfs1(Isd11)-Yfh1 or Yfh1-Isu1), yet, an ordered mechanism is likely dominant, due to the high stability of the Nfs1(Isd11)-Isu1 heterodimer.
We also conclude from our results that the PVK motif of Isu is involved in both critical stages of FeS cluster biogenesis, cluster assembly, and cluster transfer, as it is necessary for binding of Yfh1 as well as Hsp70 Ssq1. We previously demonstrated that two other proteins involved in cluster assembly (Nfs1) and cluster transfer (Jac1) also share overlapping, mutually exclusive binding sites on the surface of Isu1 (Fig. 8A) (39). Together these results suggest that exclusivity of binding could have important functional consequences, initiating and directing the ordered transition from the FeS assembly complex to the FeS transfer reaction catalyzed by the Hsp70 chaperone system. How might such exclusivity help orchestrate such a progression? We proposed previously (39) that Jac1 co-chaperone facilitates displacement of holo-Isu1 from the FeS assembly complex by interfering with the Nfs1-Isu1 interaction. Results presented here indicate that disruption of the Isu1-Nfs1(Isd11) interaction also destabilizes the Isu1-Yfh1 interaction, as the latter depends on Nfs1(Isd11)-Isu1 binding. Thus, holo-Isu1 released from Nfs1(Isd11) by the action of Jac1 will also result in Yfh1 release, thereby exposing the PVK site for Hsp70 interaction (Fig. 8B). Once Ssq1 binds the PVK motif, Isu1 is “protected” against Yfh1 rebinding, even if Nfs1(Isd11) rebinds to Isu1 after dissociation of Jac1. Thus the chaperones may play a regulatory role by controlling the flow of FeS clusters from the assembly complex to the recipient proteins. Further work comparing cluster-bound and cluster-free forms of the Isu scaffold and the interacting proteins is needed to verify the models discussed above.
FIGURE 8.
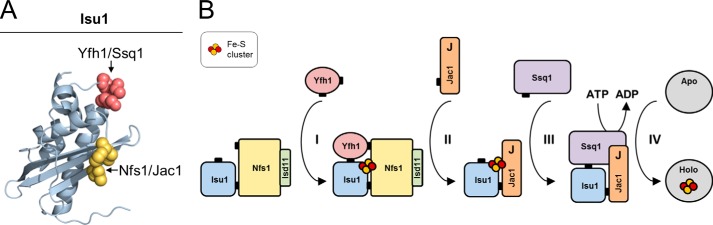
Ordered transition from FeS cluster assembly to transfer. A, homology model of Isu1 (39) with highlighted residues involved in Yfh1-Ssq1 (red) and Nfs1-Jac1 (yellow) interaction with Isu1, as part of the FeS cluster assembly complex. B, Yfh1 binds the pre-formed Isu1-Nfs1(Isd11) complex (I) facilitating FeS cluster synthesis. Jac1 displaces holo-Isu1 from the assembly complex (II) to form the holo-Isu1-Jac1 complex. Ssq1 binds the PVK motif of holo-Isu1 in complex with Jac1 (III). J-domain (J) of Jac1 stimulates the ATPase activity of Ssq1 facilitating FeS cluster transfer (IV) to the recipient apoprotein.
Acknowledgments
We thank Annalisa Pastore (King's College London) for providing PDB files of the bacterial FeS assembly complex and Roland Lill (Philipps-Universität Marburg, Marburg, Germany) and Patrick D'Silva (Indian Institute of Science, Bangalore, India) for sharing plasmids.
This work was supported, in whole or in part, by National Institutes of Health Grant GM27870 (to E. A. C), Polish National Science Center Grant DEC-2012/06/A/NZ1/00002 (to J. M.), and Foundation for Polish Science International Ph.D. Projects Grant MPD/2010/5 co-financed by the European Union European Regional Development Fund, Operational Program Innovative Economy 2007–2013 (to J. M.).
- ISC
- iron-sulfur cluster
- Nfs1(Isd11)
- Nfs1 in complex with accessory protein Isd11.
REFERENCES
- 1. Johnson D. C., Dean D. R., Smith A. D., Johnson M. K. (2005) Structure, function, and formation of biological iron-sulfur clusters. Annu. Rev. Biochem. 74, 247–281 [DOI] [PubMed] [Google Scholar]
- 2. Li H., Gakh O., Smith D. Y., 4th, Ranatunga W. K., Isaya G. (2013) Missense mutations linked to friedreich ataxia have different but synergistic effects on mitochondrial frataxin isoforms. J. Biol. Chem. 288, 4116–4127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lill R., Hoffmann B., Molik S., Pierik A. J., Rietzschel N., Stehling O., Uzarska M. A., Webert H., Wilbrecht C., Mühlenhoff U. (2012) The role of mitochondria in cellular iron-sulfur protein biogenesis and iron metabolism. Biochim. Biophys. Acta 1823, 1491–1508 [DOI] [PubMed] [Google Scholar]
- 4. Stehling O., Lill R. (2013) The role of mitochondria in cellular iron-sulfur protein biogenesis: mechanisms, connected processes, and diseases. Cold Spring Harb. Perspect. Biol. 5, a011312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Garland S. A., Hoff K., Vickery L. E., Culotta V. C. (1999) Saccharomyces cerevisiae ISU1 and ISU2: members of a well-conserved gene family for iron-sulfur cluster assembly. J. Mol. Biol. 294, 897–907 [DOI] [PubMed] [Google Scholar]
- 6. Gerber J., Neumann K., Prohl C., Mühlenhoff U., Lill R. (2004) The yeast scaffold proteins Isu1p and Isu2p are required inside mitochondria for maturation of cytosolic Fe/S proteins. Mol. Cell Biol. 24, 4848–4857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Smith A. D., Agar J. N., Johnson K. A., Frazzon J., Amster I. J., Dean D. R., Johnson M. K. (2001) Sulfur transfer from IscS to IscU: the first step in iron-sulfur cluster biosynthesis. J. Am. Chem. Soc. 123, 11103–11104 [DOI] [PubMed] [Google Scholar]
- 8. Gerber J., Mühlenhoff U., Lill R. (2003) An interaction between frataxin and Isu1/Nfs1 that is crucial for Fe/S cluster synthesis on Isu1. EMBO Rep. 4, 906–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tsai C. L., Barondeau D. P. (2010) Human frataxin is an allosteric switch that activates the Fe-S cluster biosynthetic complex. Biochemistry 49, 9132–9139 [DOI] [PubMed] [Google Scholar]
- 10. Colin F., Martelli A., Clémancey M., Latour J. M., Gambarelli S., Zeppieri L., Birck C., Page A., Puccio H., Ollagnier de Choudens S. (2013) Mammalian frataxin controls sulfur production and iron entry during de novo Fe4S4 cluster assembly. J. Am. Chem. Soc. 135, 733–740 [DOI] [PubMed] [Google Scholar]
- 11. Pandey A., Gordon D. M., Pain J., Stemmler T. L., Dancis A., Pain D. (2013) Frataxin directly stimulates mitochondrial cysteine desulfurase by exposing substrate-binding sites, and a mutant FeS cluster scaffold protein with frataxin-bypassing ability acts similarly. J. Biol. Chem. 288, 36773–36786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cook J. D., Kondapalli K. C., Rawat S., Childs W. C., Murugesan Y., Dancis A., Stemmler T. L. (2010) Molecular details of the yeast frataxin-Isu1 interaction during mitochondrial Fe-S cluster assembly. Biochemistry 49, 8756–8765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Prischi F., Konarev P. V., Iannuzzi C., Pastore C., Adinolfi S., Martin S. R., Svergun D. I., Pastore A. (2010) Structural bases for the interaction of frataxin with the central components of iron-sulphur cluster assembly. Nat. Commun. 1, 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shi R., Proteau A., Villarroya M., Moukadiri I., Zhang L., Trempe J. F., Matte A., Armengod M. E., Cygler M. (2010) Structural basis for Fe-S cluster assembly and tRNA thiolation mediated by IscS protein-protein interactions. PLoS Biol. 8, e1000354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schmucker S., Martelli A., Colin F., Page A., Wattenhofer-Donzé M., Reutenauer L., Puccio H. (2011) Mammalian frataxin: an essential function for cellular viability through an interaction with a preformed ISCU/NFS1/ISD11 iron-sulfur assembly complex. PLoS One 6, e16199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Marinoni E. N., de Oliveira J. S., Nicolet Y., Raulfs E. C., Amara P., Dean D. R., Fontecilla-Camps J. C. (2012) (IscS-IscU)2 complex structures provide insights into Fe2S2 biogenesis and transfer. Angew. Chem. Int. Ed. Engl. 51, 5439–5442 [DOI] [PubMed] [Google Scholar]
- 17. Wiedemann N., Urzica E., Guiard B., Müller H., Lohaus C., Meyer H. E., Ryan M. T., Meisinger C., Mühlenhoff U., Lill R., Pfanner N. (2006) Essential role of Isd11 in mitochondrial iron-sulfur cluster synthesis on Isu scaffold proteins. EMBO J. 25, 184–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Adam A. C., Bornhövd C., Prokisch H., Neupert W., Hell K. (2006) The Nfs1 interacting protein Isd11 has an essential role in Fe/S cluster biogenesis in mitochondria. EMBO J. 25, 174–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Richards T. A., van der Giezen M. (2006) Evolution of the Isd11-IscS complex reveals a single α-proteobacterial endosymbiosis for all eukaryotes. Mol. Biol. Evol. 23, 1341–1344 [DOI] [PubMed] [Google Scholar]
- 20. Shan Y., Napoli E., Cortopassi G. (2007) Mitochondrial frataxin interacts with ISD11 of the NFS1/ISCU complex and multiple mitochondrial chaperones. Hum. Mol. Genet. 16, 929–941 [DOI] [PubMed] [Google Scholar]
- 21. Shi Y., Ghosh M. C., Tong W. H., Rouault T. A. (2009) Human ISD11 is essential for both iron-sulfur cluster assembly and maintenance of normal cellular iron homeostasis. Hum. Mol. Genet. 18, 3014–3025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pandey A., Golla R., Yoon H., Dancis A., Pain D. (2012) Persulfide formation on mitochondrial cysteine desulfurase: enzyme activation by a eukaryote-specific interacting protein and Fe-S cluster synthesis. Biochem. J. 448, 171–187 [DOI] [PubMed] [Google Scholar]
- 23. Campuzano V., Montermini L., Moltò M. D., Pianese L., Cossée M., Cavalcanti F., Monros E., Rodius F., Duclos F., Monticelli A., Zara F., Cañizares J., Koutnikova H., Bidichandani S. I., Gellera C., Brice A., Trouillas P., De Michele G., Filla A., De Frutos R., Palau F., Patel P. I., Di Donato S., Mandel J. L., Cocozza S., Koenig M., Pandolfo M. (1996) Friedreich's ataxia: autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science 271, 1423–1427 [DOI] [PubMed] [Google Scholar]
- 24. Stemmler T. L., Lesuisse E., Pain D., Dancis A. (2010) Frataxin and mitochondrial FeS cluster biogenesis. J. Biol. Chem. 285, 26737–26743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Vaubel R. A., Isaya G. (2013) Iron-sulfur cluster synthesis, iron homeostasis and oxidative stress in Friedreich ataxia. Mol. Cell. Neurosci. 55, 50–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pastore A., Adinolfi S. (2014) Chronochemistry in neurodegeneration. Front. Mol. Neurosci. 7, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Martelli A., Puccio H. (2014) Dysregulation of cellular iron metabolism in Friedreich ataxia: from primary iron-sulfur cluster deficit to mitochondrial iron accumulation. Front. Pharmacol. 5, 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bridwell-Rabb J., Fox N. G., Tsai C. L., Winn A. M., Barondeau D. P. (2014) Human frataxin activates Fe-S cluster biosynthesis by facilitating sulfur transfer chemistry. Biochemistry 53, 4904–4913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Vickery L. E., Cupp-Vickery J. R. (2007) Molecular chaperones HscA/Ssq1 and HscB/Jac1 and their roles in iron-sulfur protein maturation. Crit. Rev. Biochem. Mol. Biol. 42, 95–111 [DOI] [PubMed] [Google Scholar]
- 30. Kampinga H. H., Craig E. A. (2010) The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat. Rev. Mol. Cell Biol. 11, 579–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dutkiewicz R., Schilke B., Knieszner H., Walter W., Craig E. A., Marszalek J. (2003) Ssq1, a mitochondrial Hsp70 involved in iron-sulfur (Fe/S) center biogenesis. Similarities to and differences from its bacterial counterpart. J. Biol. Chem. 278, 29719–29727 [DOI] [PubMed] [Google Scholar]
- 32. Ciesielski S. J., Schilke B. A., Osipiuk J., Bigelow L., Mulligan R., Majewska J., Joachimiak A., Marszalek J., Craig E. A., Dutkiewicz R. (2012) Interaction of J-protein co-chaperone Jac1 with Fe-S scaffold Isu is indispensable in vivo and conserved in evolution. J. Mol. Biol. 417, 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chandramouli K., Johnson M. K. (2006) HscA and HscB stimulate [2Fe-2S] cluster transfer from IscU to apoferredoxin in an ATP-dependent reaction. Biochemistry 45, 11087–11095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bonomi F., Iametti S., Morleo A., Ta D., Vickery L. E. (2008) Studies on the mechanism of catalysis of iron-sulfur cluster transfer from IscU[2Fe2S] by HscA/HscB chaperones. Biochemistry 47, 12795–12801 [DOI] [PubMed] [Google Scholar]
- 35. Bonomi F., Iametti S., Morleo A., Ta D., Vickery L. E. (2011) Facilitated transfer of IscU-[2Fe2S] clusters by chaperone-mediated ligand exchange. Biochemistry 50, 9641–9650 [DOI] [PubMed] [Google Scholar]
- 36. Andrew A. J., Dutkiewicz R., Knieszner H., Craig E. A., Marszalek J. (2006) Characterization of the interaction between the J-protein Jac1p and the scaffold for Fe-S cluster biogenesis, Isu1p. J. Biol. Chem. 281, 14580–14587 [DOI] [PubMed] [Google Scholar]
- 37. Füzèry A. K., Tonelli M., Ta D. T., Cornilescu G., Vickery L. E., Markley J. L. (2008) Solution structure of the iron-sulfur cluster cochaperone HscB and its binding surface for the iron-sulfur assembly scaffold protein IscU. Biochemistry 47, 9394–9404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kim J. H., Füzéry A. K., Tonelli M., Ta D. T., Westler W. M., Vickery L. E., Markley J. L. (2009) Structure and dynamics of the iron-sulfur cluster assembly scaffold protein IscU and its interaction with the cochaperone HscB. Biochemistry 48, 6062–6071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Majewska J., Ciesielski S. J., Schilke B., Kominek J., Blenska A., Delewski W., Song J. Y., Marszalek J., Craig E. A., Dutkiewicz R. (2013) Binding of the chaperone Jac1 protein and cysteine desulfurase Nfs1 to the iron-sulfur cluster scaffold Isu protein is mutually exclusive. J. Biol. Chem. 288, 29134–29142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hoff K. G., Cupp-Vickery J. R., Vickery L. E. (2003) Contributions of the LPPVK motif of the iron-sulfur template protein IscU to interactions with the Hsc66-Hsc20 chaperone system. J. Biol. Chem. 278, 37582–37589 [DOI] [PubMed] [Google Scholar]
- 41. Dutkiewicz R., Schilke B., Cheng S., Knieszner H., Craig E. A., Marszalek J. (2004) Sequence-specific interaction between mitochondrial Fe-S scaffold protein Isu and Hsp70 Ssq1 is essential for their in vivo function. J. Biol. Chem. 279, 29167–29174 [DOI] [PubMed] [Google Scholar]
- 42. Voisine C., Schilke B., Ohlson M., Beinert H., Marszalek J., Craig E. A. (2000) Role of the mitochondrial Hsp70s, Ssc1 and Ssq1, in the maturation of Yfh1. Mol. Cell Biol. 20, 3677–3684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mühlenhoff U., Gerber J., Richhardt N., Lill R. (2003) Components involved in assembly and dislocation of iron-sulfur clusters on the scaffold protein Isu1p. EMBO J. 22, 4815–4825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Guan K. L., Dixon J. E. (1991) Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal. Biochem. 192, 262–267 [DOI] [PubMed] [Google Scholar]
- 45. Sherman F. (1991) Getting started with yeast. Methods Enzymol. 194, 3–21 [DOI] [PubMed] [Google Scholar]
- 46. Pareek G., Samaddar M., D'Silva P. (2011) Primary sequence that determines the functional overlap between mitochondrial heat shock protein 70 Ssc1 and Ssc3 of Saccharomyces cerevisiae. J. Biol. Chem. 286, 19001–19013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jaschkowitz K., Seidler A. (2000) Role of a NifS-like protein from the cyanobacterium Synechocystis PCC 6803 in the maturation of FeS proteins. Biochemistry 39, 3416–3423 [DOI] [PubMed] [Google Scholar]
- 48. Thorpe G. H., Kricka L. J., Moseley S. B., Whitehead T. P. (1985) Phenols as enhancers of the chemiluminescent horseradish peroxidase-luminol-hydrogen peroxide reaction: application in luminescence-monitored enzyme immunoassays. Clin. Chem. 31, 1335–1341 [PubMed] [Google Scholar]
- 49. Hunter S., Jones P., Mitchell A., Apweiler R., Attwood T. K., Bateman A., Bernard T., Binns D., Bork P., Burge S., de Castro E., Coggill P., Corbett M., Das U., Daugherty L., Duquenne L., Finn R. D., Fraser M., Gough J., Haft D., Hulo N., Kahn D., Kelly E., Letunic I., Lonsdale D., Lopez R., Madera M., Maslen J., McAnulla C., McDowall J., McMenamin C., Mi H., Mutowo-Muellenet P., Mulder N., Natale D., Orengo C., Pesseat S., Punta M., Quinn A. F., Rivoire C., Sangrador-Vegas A., Selengut J. D., Sigrist C. J., Scheremetjew M., Tate J., Thimmajanarthanan M., Thomas P. D., Wu C. H., Yeats C., Yong S. Y. (2012) InterPro in 2011: new developments in the family and domain prediction database. Nucleic Acids Res. 40, D306–D312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Katoh K., Standley D. M. (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bridwell-Rabb J., Iannuzzi C., Pastore A., Barondeau D. P. (2012) Effector role reversal during evolution: the case of frataxin in Fe-S cluster biosynthesis. Biochemistry 51, 2506–2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Urbina H. D., Silberg J. J., Hoff K. G., Vickery L. E. (2001) Transfer of sulfur from IscS to IscU during Fe/S cluster assembly. J. Biol. Chem. 276, 44521–44526 [DOI] [PubMed] [Google Scholar]
- 53. Foury F., Pastore A., Trincal M. (2007) Acidic residues of yeast frataxin have an essential role in Fe-S cluster assembly. EMBO Rep. 8, 194–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Li H., Gakh O., Smith D. Y., 4th, Isaya G. (2009) Oligomeric yeast frataxin drives assembly of core machinery for mitochondrial iron-sulfur cluster synthesis. J. Biol. Chem. 284, 21971–21980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Leidgens S., De Smet S., Foury F. (2010) Frataxin interacts with Isu1 through a conserved tryptophan in its β-sheet. Hum. Mol. Genet. 19, 276–286 [DOI] [PubMed] [Google Scholar]
- 56. Tsai C. L., Bridwell-Rabb J., Barondeau D. P. (2011) Friedreich's ataxia variants I154F and W155R diminish frataxin-based activation of the iron-sulfur cluster assembly complex. Biochemistry 50, 6478–6487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Bridwell-Rabb J., Winn A. M., Barondeau D. P. (2011) Structure-function analysis of Friedreich's ataxia mutants reveals determinants of frataxin binding and activation of the Fe-S assembly complex. Biochemistry 50, 7265–7274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kim J. H., Frederick R. O., Reinen N. M., Troupis A. T., Markley J. L. (2013) [2Fe-2S]-ferredoxin binds directly to cysteine desulfurase and supplies an electron for iron-sulfur cluster assembly but is displaced by the scaffold protein or bacterial frataxin. J. Am. Chem. Soc. 135, 8117–8120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yan R., Konarev P. V., Iannuzzi C., Adinolfi S., Roche B., Kelly G., Simon L., Martin S. R., Py B., Barras F., Svergun D. I., Pastore A. (2013) Ferredoxin competes with bacterial frataxin in binding to the desulfurase IscS. J. Biol. Chem. 288, 24777–24787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Nakai Y., Nakai M., Hayashi H., Kagamiyama H. (2001) Nuclear localization of yeast Nfs1p is required for cell survival. J. Biol. Chem. 276, 8314–8320 [DOI] [PubMed] [Google Scholar]
- 61. Naamati A., Regev-Rudzki N., Galperin S., Lill R., Pines O. (2009) Dual targeting of Nfs1 and discovery of its novel processing enzyme, Icp55. J. Biol. Chem. 284, 30200–30208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wang T., Craig E. A. (2008) Binding of yeast frataxin to the scaffold for Fe-S cluster biogenesis, Isu. J. Biol. Chem. 283, 12674–12679 [DOI] [PMC free article] [PubMed] [Google Scholar]