Background: Thermus PilF is essential for pilus biogenesis and transformation.
Results: Zinc binding is essential for PilF complex stability, piliation, adhesion, and twitching motility.
Conclusion: PilF complex thermostability is essential for piliation but not for natural transformation.
Significance: This work highlights the role of zinc and ATP in AAA-ATPase stability and provides evidence that T4P and the DNA translocator are distinct systems.
Keywords: ATPases Associated with Diverse Cellular Activities (AAA), Cell Motility, DNA Transformation, Type IV Pili, Zinc
Abstract
The traffic AAA-ATPase PilF is essential for pilus biogenesis and natural transformation of Thermus thermophilus HB27. Recently, we showed that PilF forms hexameric complexes containing six zinc atoms coordinated by conserved tetracysteine motifs. Here we report that zinc binding is essential for complex stability. However, zinc binding is neither required for pilus biogenesis nor natural transformation. A number of the mutants did not exhibit any pili during growth at 64 °C but still were transformable. This leads to the conclusion that type 4 pili and the DNA translocator are distinct systems. At lower growth temperatures (55 °C) the zinc-depleted multiple cysteine mutants were hyperpiliated but defective in pilus-mediated twitching motility. This provides evidence that zinc binding is essential for the role of PilF in pilus dynamics. Moreover, we found that zinc binding is essential for complex stability but dispensable for ATPase activity. In contrast to many polymerization ATPases from mesophilic bacteria, ATP binding is not required for PilF complex formation; however, it significantly increases complex stability. These data suggest that zinc and ATP binding increase complex stability that is important for functionality of PilF under extreme environmental conditions.
Introduction
Uptake of naked DNA from the environment and incorporation of the DNA into the genome is a trait of many different bacteria. The cellular trait is also referred to as natural competence and was the driving force for the classical Griffith experiment demonstrating DNA as the hereditary material (1). Uptake of DNA from the environment into the bacterial cell requires the macromolecule DNA to traverse the cell envelope and the cytoplasmic membrane. Like the transport of other macromolecules, e.g. proteins, this process requires the concerted action of a number of proteins that in concert act to (i) bind the DNA at the outermost cell periphery, (ii) translocation into the periplasm of Gram-negative bacteria or the functional homolog in Gram-positives, and (iii) transport of the macromolecule across the cytoplasmic membrane.
The cellular machinery involved in uptake of DNA into the bacterial cell has been studied in Gram-positive model strains such as Bacillus subtilis or Streptococcus pneumoniae and in Gram-negative bacteria such as Neisseria gonorrhoeae and Acinetobacter baylyi (2, 3). These are all mesophiles thriving at ambient temperatures. In addition, genes are also shuffled around in hot environments, and this is regarded as a major driving force for microbial evolution (4). Moreover, DNA translocation machineries of thermophiles and hyperthermophiles are anticipated to have structural variations of the general theme to allow coping with high temperatures in which their hosts thrive. The Gram-negative bacterium Thermus thermophilus has become the model system to study natural competence in thermophiles (4). It has the outstanding feature that every cell in a culture is competent for natural transformation (5).
The DNA translocator of T. thermophilus has a tripartite structure: a secretin ring (PilQ) of unusual dimension that spans the entire cell periphery (6–8). It catalyzes the first step in DNA recognition (9). Second, it has a pilus-like structure (PilA1–4) mediating the transport of DNA into a DNase-resistant state and is suggested to be a dynamic structure (10). ComEA, ComEC, PilM, PilN, PilO, PilC, and PilD are proteins in the inner membrane of T. thermophilus and are required for the energy-dependent transport through the cytoplasmic membrane (11, 12). The mechanism of DNA transport through the cytoplasmic membrane is the least understood function in the process. In general, DNA translocator and type IV pili (T4P)2 systems of bacteria are rather similar. They are discussed as being identical or sometimes as having at least common components.
Translocation of DNA into the periplasm or to the membrane-integral DNA transporter is believed to be mediated by the dynamics of pilus-like substructures. Dynamics is given to the pilus-like structure by ATP hydrolysis, and retraction as well as detraction ATPases is suggested to catalyze retraction or expansion of the pilus-like structures. The AAA (ATPase associated with diverse cellular activities) ATPase PilF of T. thermophilus is essential for both natural transformation and pilus biogenesis and is suggested to catalyze extension of the pilus-like structures and T4P (13). The 98.2-kDa protein assembles into a hexameric ring (14). Recently, it was shown that the C-terminal region mediates ring formation, whereas the N-terminal region forms a disk-like structure. These distinct regions are separated by a stem-like structure (15). PilF has a tripartite structure with an N terminus encoding three general secretory domains (GSPII), a central region, and a C terminus that has the ATPase hydrolysis domains (Walker motifs A and B), and a conserved motif of four cysteine residues. Exchange of all four cysteine residues led to a complete loss of zinc binding (14). However, the role of this motif in cellular functions of PilF was not addressed. Here, we have followed up these observations and present a detailed study by subsequent removal of individual cysteine residues and the effect on piliation of T. thermophilus, pilus functions such as motility and adherence, natural competence, and thermostability of the PilF protein complex.
EXPERIMENTAL PROCEDURES
Organisms and Cultivation
All strains used in this study are listed in Table 1. The Escherichia coli strains DH5α and BL21 (DE3) were grown at 37 °C in LB medium (16). T. thermophilus HB27 was grown at 68 °C in TM+ complex medium (17). T. thermophilus mutants were grown on TM+ medium containing 5 or 15 μg/ml bleomycin (bleo) and 20 or 40 μg/ml kanamycin in liquid or solid medium, respectively.
TABLE 1.
Name and sources of the strains
Kan, kanamycin.
| Strain | Genotype | Source |
|---|---|---|
| T. thermophilus HB27 | Wild-type strain | (42) |
| ΔpilF::bleo | HB27, pilF (TTC1621) replaced by a bleomycin resistance marker, BleoR | (13) |
| wild-type | ΔpilF complemented with pDM12-pilFwt, BleoR, KanR | (13) |
| C781A (C1A) | ΔpilF complemented with pDM12-pilFC781A, BleoR, KanR | This study |
| C784A (C2A) | ΔpilF complemented with pDM12-pilFC784A, BleoR, KanR | This study |
| H783A/C784A (HC2A) | ΔpilF complemented with pDM12-pilFH783A/C784A, BleoR, KanR | This study |
| C816A (C3A) | ΔpilF complemented with pDM12-pilFC816A, BleoR, KanR | This study |
| C819A (C4A) | ΔpilF complemented with pDM12-pilFC819A, BleoR, KanR | This study |
| C781A/C784A (2CysA) | ΔpilF complemented with pDM12-pilFC781A/C784A, BleoR, KanR | This study |
| C781A/C784A/C816A (3CysA) | ΔpilF complemented with pDM12-pilFC781A/C784A/C816A, BleoR, KanR | This study |
| C781A/C784A/C816A/C819A (4CysA) | ΔpilF complemented with pDM12-pilFC781A/C784A/C816A/C819A, BleoR, KanR | This study |
Site-directed Mutagenesis of pilF and Purification of PilF Proteins
To analyze the individual role of the four cysteine residues of the tetracysteine motif (4Cys) in PilF, pET28a-pilF carrying the wild-type pilF gene was used to exchange the cysteine residues to alanine using the Phusion-site directed mutagenesis protocol from New England Biolabs (Frankfurt, Germany) (14). The primers used are listed in Table 2. The resulting plasmids pET28a-pilFC781A, pET28a-pilFC784A, pET28a-pilFH783A/C784A, pET28a-pilFC816A, pET28a-pilFC819A, pET28a-pilFC781A/H783A/C784A, pET28a-pilFC781A/H783A/C784A/C816A, and pET28a-pilFC781A/H783A/C784A/C816A/C819A were transferred into E. coli BL21 (DE3).
TABLE 2.
Primers and templates for site-directed mutagenesis of pilF
| Primer | Sequence 5′-3′ | Template plasmids for PCR | Resulting plasmids |
|---|---|---|---|
| pilF_pDM12_for | ATTACATATGCACCATCATCACCACCATCATAGCGTGCTCACCATAGGGGACAAAAGG | pET28a-pilF wild type and variants | pDM12-pilF wild type and variants |
| pilF_pDM12_rev | ACTTAGATGCGGCCGCTTACTCAATGGTACGCGCCAG | ||
| MutA781_for | GGTGGAGGTCAAGCCGGACCCC | pET28a-pilFwt | pDM12-pilFC781A |
| MutA781_rev | TTGCAGTGCTCGGCCACCCTGCGC | ||
| MutA784_for | AGCCGGACCCCGAGACCCTCAG | pET28a-pilFwt | pDM12-pilFC784A |
| MutA784_rev | TGACCTCCACCTTGGCGTGCTCGCACAC | ||
| MutA784_for | AGCCGGACCCCGAGACCCTCAG | pET28a-pilFC784A | pDM12-pilFH783A/C784A |
| MutA783/784_rev | TGACCTCCACCTTGGCGGCCTCGCACAC | ||
| MutA816_for | TGCGGCGGCACCGGGTACAAGGG | pET28a-pilFwt | pDM12-pilFC816A |
| MutA816_rev | CCGCTCGGCCCCCATGCCCTTGTAG | ||
| MutA819_for | GGGCATGGGGTGCGAGCGGGCCGGCGGCACC | pET28a-pilFwt | pDM12-pilFC819A |
| MutA819_rev | GGATGGCGTAGCGGCCCTTGTACCC | ||
| Mut2Cys_for | GCGCAGGGTGGCCGAGCACGCCAAGGTGGAG | pET28a-pilFwt | pDM12-pilFC781A/H783A/C784A |
| Mut2Cys_rev | CCTGAGGGTCTCGGGGTCCGGCTTGAC | ||
| Mut3Cys_for | GGCATGGGGGCCGAGCGGTG | pET28a-pilFC781A/H783AC784A | pDM12-pilFC781A/H783A/C784A/C816A |
| Mut3Cys_rev | CCTTGTACCCGGTGCCGCCG | ||
| Mut4Cys_for | CGAGCGGGCGGGCGGCACCG | pET28apilFC781A/H783A/C784A/C816A/C819A | pDM12pilFC781A/H783AC784A/C816A/C819A |
| Mut4Cys_rev | GGCGTAGCGGCCCTTGTACC |
Protein Purification
Overexpression was performed by cultivation of the recombinant E. coli BL21 (DE3) strains in a 10-liter fermenter (8 liters air/min, 600 rpm) at 37 °C, and pilF expression was induced by the addition of 1 mm isopropyl β-d-1-thiogalactopyranoside at A600 0.8–1. After induction, the temperature was reduced to 30 °C, and the cells were harvested 3.5 h after induction. Heat precipitation of E. coli proteins was carried out for 5–20 min at 68 °C depending on the PilF variant. PilF proteins were purified by nickel-nitrilotriacetic acid affinity chromatography and size exclusion chromatography as reported previously (14). The Gel Filtration HMW Calibration kit (GE Healthcare) was used as molecular weight standard. The protein concentration was analyzed according to Bradford (18).
Complementation of a ΔpilF::bleo Deletion Mutant with Wild-type pilF and Cysteine-depleted pilF Variants
The mutated pilF genes were amplified from the recombinant pET28a-pilF plasmids with the primers pilF_pDM12-for1 and pilF_NotI (Table 2). The amplified DNA fragments were digested with NdeI and NotI and ligated into the E. coli/Thermus shuttle vector pDM12. The resulting plasmids were designated pDM12-pilFC781A, pDM12-pilFC784A, pDM12-pilFH783A/C784A pDM12-pilFC816A, pDM12-pilF819A, pDM12-pilFC781A/H783A/C784A, pDM12-pilFC781A/H783A/784A/C816A, and pDM12-pilFC781A/H783A/784A/C816A/C819A (Table 2) and transformed into the T. thermophilus ΔpilF::bleo mutant via electroporation as described recently (13).
Analyses of PilF Complex Stability
The purified PilF complexes were diluted in Bicine buffer (50 mm Bicine, pH 8.5, 200 mm NaCl) to a concentration of 2 μg/μl, incubated for 0, 2.5, or 5 min at 70 °C, and subsequently analyzed by clear-native PAGE (19).
Analyses of the Zinc Content by Inductively-coupled Plasma Mass Spectrometry (ICP-MS)
The zinc content of the PilF complexes was analyzed by ICP-MS with 0.1 and 0.25 mg of protein as reported recently (20).
ATPase Assay
The determination of the ATPase activity was performed according to the method of Heinonen and Lahti (21). 200 μg of protein were diluted with 1 ml of TCM buffer (50 mm Tris, 50 mm MOPS, 50 mm CHES, 50 mm glycine, 150 mm KCl, 5 mm MgCl2, pH 9.5) to a final concentration of 0.2 μg of protein/μl and preincubated for 3 min at 70 °C before the reaction was started by the addition of ATP (final concentration 2.5 mm). The assay was performed at 70 °C, and 200-μl samples were taken at different time points. The reaction was stopped by the addition of 30 μl of 30% trichloroacetate. Precipitated protein was removed by centrifugation. 200 μl of the samples were mixed with 0.5 ml of acetone, 0.25 ml of 2.5 m sulfuric acid, and 0.25 ml of ammonium molybdate (10 mm)). The absorbance was measured after 10 min at 355 nm.
Thermofluor Assays
Heat stability of the wild-type PilF complex and the PilF mutant complexes was analyzed by thermofluor assays in a Rotor Gene Q5 Plex high resolution melt (HRM) thermocycler (Qiagen, Hilden, Germany). The fluorescent dye SYPRO® Orange was purchased from Invitrogen as a 5000× stock concentration in DMSO. 50 mm sodium acetate. pH 4, 4.5, 5, and 5.5, 50 mm MES, pH 6 and 6.5, 50 mm HEPES, pH 7 and 7.5, 50 mm Bicine, pH 8 and 8.5, and 50 mm CHES, pH 9 and 9.5, were used to analyze the effect of pH on thermostability of PilF. All buffer systems contained 200 mm NaCl.
10 μg of the PilF stock solution (5 μg of protein/μl buffer (Tris/HCl, 300 mm NaCl, pH 7.5)) were mixed with 10 μl of 2× buffer, 1 μl of 40× SYPRO® Orange, and 7 μl of H2Odest. The thermal protein denaturation was monitored by the increase in fluorescence intensity of the SYPRO® Orange dye as described by Niesen et al. (22). For fluorescence measurements the HRM channel was used with the excitation wavelength at 460 nm and the emission wavelength at 510 nm. All measurements were performed in triplicate.
The effect of AMP-PNP and ADP on the thermostability of the wild-type PilF complex and the mutated PilF complexes was analyzed by thermofluor assays in 50 mm Bicine buffer with 200 mm NaCl, pH 8.5, in the presence of different AMP-PNP and ADP concentrations ranging from 0 to 20 mm and from 0 to 10 mm, respectively.
Electron Microscopy
Electron microscopical analyses were performed according to Salzer et al. (13). 250 cells of the wild-type strain and the different mutants were investigated.
Twitching Motility Studies
Cells were grown on minimal medium (MM) plates containing 1% bovine serum albumin (BSA). The plates were incubated at 64 and 72 °C for 72 h and at 55 °C for 120 h, respectively. After staining with Coomassie Blue, cells were removed from the surface, and the twitching zone was visible as a clear zone, which appears as a dark zone after inversion of the pictures (13).
Microtiter Adhesion Assays
Microtiter adhesion assays were performed according to Salzer et al. (23). Cells were inoculated to an A600 of 0.05 and incubated for 3 days at 55 °C, 64 °C, or 72 °C. After incubation the optical density (600 nm) was measured. This step was followed by washing steps to remove non-sessile cells. Adhering cells were stained with 0.1% crystal violet solution. The remaining staining solutions were removed by washing three times with H2O. Crystal violet of the adhering cells was solved in ethanol, and the absorbance was measured at 570 nm. Adherence was calculated by the quotient of 570 nm to 600 nm.
Natural Transformation
Transformation studies were performed at 68 °C on TM+ agar medium using 5 μg of genomic DNA of a spontaneous streptomycin-resistant T. thermophilus HB27 mutant (24). The transformation frequency was calculated as transformants per living count. All transformation assays were performed in triplicate.
RESULTS
Generation of PilF Variants with Mutations in the Tetracysteine Motif
To elucidate the role of the individual cysteine residues in the various functions of PilF, single, double, triple, and quadruple cysteine mutants were generated by site-directed mutagenesis. Conserved motifs of PilF and the relative positions of the cysteine residues are indicated in Fig. 1A. Eight different variants were generated: C781A (C1A), C784A (C2A), H783A/C784A (HC2A), C816A (C3A), C819A (C4A), C781A/H783A/C784A (2CysA), C781A/H783A/C784A/C816A (3CysA) and C781A/H783A/C784A/C816A/C819A (4CysA). Histidine residues are also known to coordinate zinc (25); therefore, the variants exhibited also amino acid substitutions in the histidine residue next to one of the cysteine residues (His-783). The identity of the variants was confirmed by DNA sequencing. The variants were produced in E. coli BL21 (DE3) and purified by affinity and size exclusion chromatography.
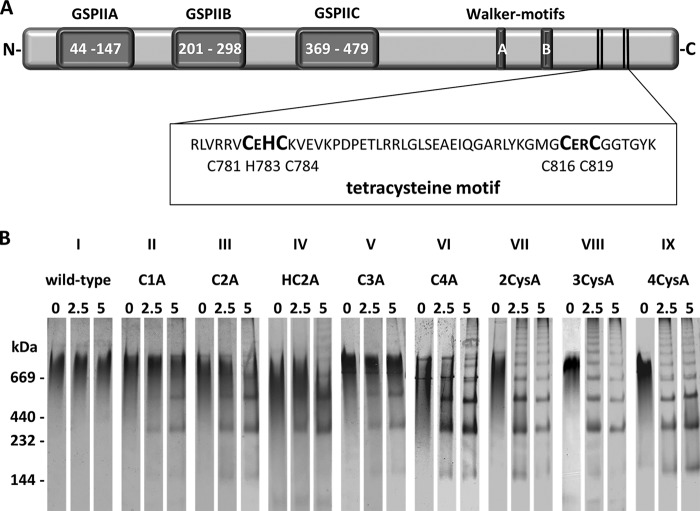
FIGURE 1.
Organization of the PilF domains (A) and heat stability of PilF variants (B). The general secretory pathway domains are abbreviated with GSPII. The Walker A and Walker B motifs are indicated. The conserved tetracysteine motif is enlarged. The purified complexes of the PilF variants were incubated for 0, 2.5, or 5 min at 70 °C, and then the samples were cooled on ice. 35 μg of the treated samples was separated by clear-native PAGE, and the proteins were stained with Coomassie Blue (B).
The Tetracysteine Motif Is Neither Essential for Oligomerization of PilF nor ATPase Activity
To examine the role of the cysteine residues in complex assembly the purified PilF wild-type protein and the mutant proteins were subjected to clear-native PAGE. The PilF wild-type protein was found to form a high molecular mass complex of >669 kDa (Fig. 1B, 0 min), which corresponds our recent findings that PilF forms hexameric complexes (14). The finding that all PilF variants had an apparent molecular mass comparable with the wild-type complex leads to the conclusion that the cysteine residues are not essential for complex assembly (Fig. 1B, II–IX, 0 min).
The close vicinity of the Walker A and the Walker B motifs to the tetracysteine motif at the C terminus of PilF (Fig. 1A) prompted us to analyze the effect of the cysteine mutations on ATPase activity. These studies revealed that ATPase activity was unaffected by the different substitutions (p > 0.05). The wild-type PilF complex exhibited an ATPase activity of 54 ± 1.6 nmol of Pi mg of protein−1 min−1, and all PilF complexes formed by the variants had comparable ATPase activities in the range from 46.32 to 53.16 nmol of Pi mg of protein−1 min−1 (see Table 4).
TABLE 4.
Features of the purified PilF complexes and phenotypes of the T. thermophilus pilF mutants
NA, not applicable.
| Strain/protein | Zinc per hexamera | ATPase activityb | Complex formationc | Thermostabilityd |
Piliatione |
Adherencef |
Twitching motilityf |
Transformation frequencyg | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Unfolding (t1) | Unfolding (t2) | 55 °C | 64 °C | 72 °C | 55 °C | 64 °C | 72 °C | 55 °C | 64 °C | 72 °C | |||||
| milliunits/mg | °C | ||||||||||||||
| HB27 | NA | NA | NA | NA | NA | ++ | + | + | ++ | + | + | + | + | + | + |
| ΔpilF::bleo | NA | NA | NA | NA | NA | − | − | − | − | − | − | − | − | − | − |
| Wild type | 6.03 | 54.00 | + | 80.8 | 90.5 | ++ | + | + | ++ | + | + | + | + | + | + |
| C1A | 5.65 | 46.32 | + | 78.6 | 91.0 | +++ | +/− | − | + | +/− | − | + | +/− | − | + |
| C2A | 5.16 | 49.86 | + | 71.6 | 91.2 | ++ | + | − | + | + | − | + | + | − | + |
| HC2A | 5.28 | 49.27 | + | 72.8 | 90.8 | ++ | + | − | + | + | − | + | + | − | + |
| C3A | 5.67 | 46.84 | + | 73.9 | 90.6 | + | +/− | − | + | +/− | − | + | +/− | − | + |
| C4A | 4.73 | 52.62 | + | 70.0 | 90.9 | − | − | − | + | − | − | − | − | − | +++ |
| 2CysA | 1.26 | 53.16 | + | 65.4 | 90.6 | +++ | − | − | + | + | − | − | − | − | + |
| 3CysA | 0.86 | 46.84 | + | 65.5 | 90.3 | +++ | − | − | + | + | − | − | − | − | + |
| 4CysA | 0.28 | 48.25 | + | 66.3 | 90.7 | +++ | − | − | + | + | − | − | − | − | + |
a Zinc content was analyzed by ICP-MS.
b ATPase activity of PilF variants was analyzed by the method described by Heinonen and Lahti (21).
c Complex formation of purified PilF proteins was analyzed by calibrated size exclusion chromatography.
d Thermostability was measured via thermofluor assays.
e Piliation was analyzed by electron microscopy (250 cells).
f Adherence was measured in microtiter adhesion assays. Twitching motility was analyzed on MM containing 1% BSA.
g Transformation frequencies were calculated as transformants per living count. +, wild-type phenotype; ++, increase compared to the wild-type; +++, strong increase compared to the wild-type; +/−, decrease compared to the wild-type; −, lack of the analyzed feature.
The Tetracysteine Motif Is Essential for Zinc Binding
Previous studies led to the detection of a six zinc atoms per hexameric PilF complex (20), and mutant studies indicated the conserved tetracysteine motif in zinc binding (14). To elucidate the role of the individual cysteine residues in zinc binding, the zinc content of the PilF complexes was analyzed by ICP-MS. The wild-type PilF protein had a zinc content of 6.03 ± 0.52 zinc atoms per hexamer, which was consistent with our recent finding (20). A similar zinc content was observed in variants in which only one cysteine was replaced (p > 0.05), except for the mutant protein C4A, where the cysteine 819 was replaced by alanine. In this mutant a slightly decreased zinc content of 4.73 ± 0.43 zinc atoms per hexamer was found (p < 0.05) (Fig. 2). PilF variants with a substitution in two or three cysteine residues had a dramatically reduced zinc content of 1.19 ± 0.13 and 0.99 ± 0.12 zinc atoms per PilF hexamer, respectively (p < 0.01) (Fig. 2, see Table 4). Deletion of all four cysteine residues abolished zinc binding completely, which is consistent with our recent findings (14, 20). Taken together, our results suggest that three cysteine residues still mediate zinc binding, whereas two cysteine residues cannot stably coordinate the zinc atom.
FIGURE 2.
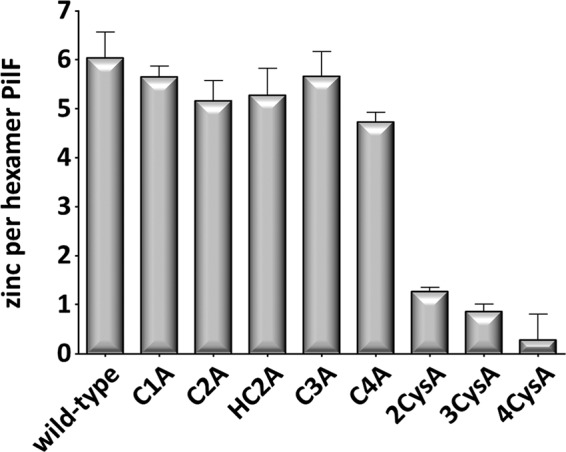
Determination of the zinc content of PilF variants via ICP-MS. 0.1 and 0.25 mg of purified PilF complexes were used for the determination of the zinc content by ICP-MS. Results were calculated from three biological independent experiments.
The Tetracysteine Motif and Zinc Binding Are Essential for Thermostability
Zinc binding of proteins is known to serve in both catalytic and structural capacities. However, most of the Zn(II)-4Cys are structurally relevant, important for the correct folding of proteins into their unique tertiary structure essential for biological function. To analyze the contribution of cysteine residues and zinc binding to the PilF complex stability, the PilF wild-type complex and the mutant complexes were subjected to heat treatment at 70 °C for 2.5 or 5 min. Separation of the PilF wild-type complex by clear-native PAGE revealed that heat treatment had no effect on oligomer stability (Fig. 1B). Preincubation of the variants C1A, C2A, HC2A, C3A, and C4A for 2.5 min at 70 °C still led to the detection of the >669-kDa complexes; however, in addition smaller oligomeric complexes of 200, 400, or 600 kDa (Fig. 1B, II–VI), corresponding to PilF dimers, tetramers, and hexamers, respectively, were found. Even higher amounts of these smaller oligomerization products were detected after 5 min of heat treatment (Fig. 1B, II–VI), whereas the amounts of the native PilF complex (>669 kDa) were significantly reduced (Fig. 1B, II–V) or even completely abolished (Fig. 1B, VI). The variant C4A (C819A) had the highest heat instability among the single mutants. After 5 min of heat treatment, the 669-kDa complex was completely abolished and in addition to the 200-, 400-. and 600-kDa complexes several high molecular mass complexes exceeding the apparent molecular mass of the wild-type complex were detected (Fig. 1B, VI). The finding that variants C1A, C2A, HC2A, and C3A were already significantly impaired in thermostability whereas the zinc content of these PilF complexes were nearly unaffected suggests that the individual cysteine residues contribute to complex stability.
Double, triple, and quadruple variants were even more unstable at 70 °C (Fig. 1B, VII–IX). The correlation of increasing instability and decreasing zinc content of the C4A mutant and the multiple cysteine mutants support our conclusion that zinc binding is essential for complex stability. Taken together, our results suggest that the cysteine residues and zinc binding are essential for thermostability of the PilF complex and that the cysteine residue 819 is most critical for thermostability. The detection of PilF complexes ranging from 200 kDa to high molecular mass complexes exceeding >669 kDa suggests the unstable mutant complexes dissociate during heat treatment and then associate again forming different multimeric PilF complexes.
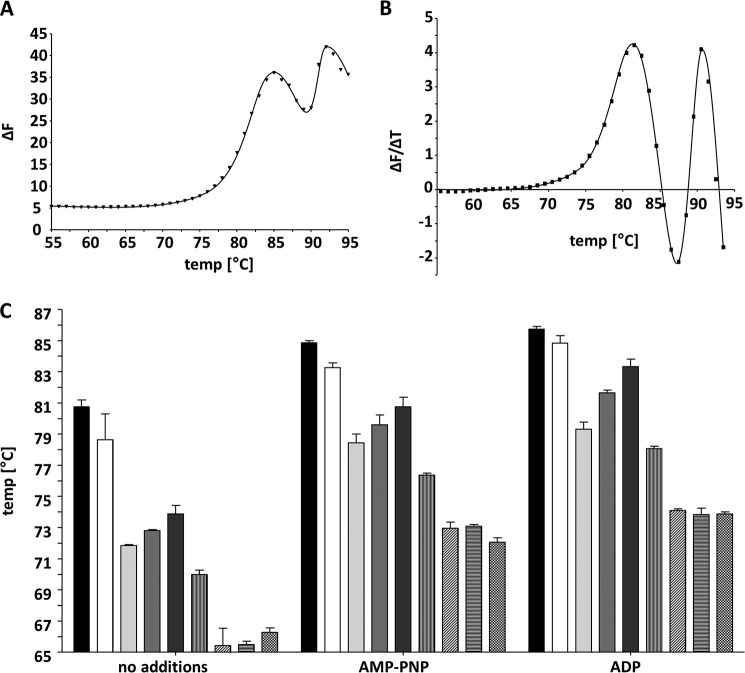
To analyze the effect of the tetracysteine motif on thermostability more quantitatively, we analyzed the effect of temperature on denaturation of PilF wild-type protein complex and the PilF complexes of the variants by thermofluor assays. A representative result of the melting curve at pH 8.5 and its first derivative are shown (Fig. 3, A and B). Interestingly, at each pH tested, a biphasic unfolding was found. In a buffer at pH 8.5 the first melting transition (t1) of the PilF complex was at 80.8 ± 0.8 °C, and the second (t2) was detected at 90.5° ± 0.0 °C (Fig. 3). Because the pH is known to have a significant effect on thermostability of protein complexes, the effect of the pH on thermostability of the PilF wild-type complex was analyzed. A decrease of the pH to 7 led to a decrease of t1 and t2 by 3.2 °C to 77.6 °C and by 7.2 °C to 83.3 °C, respectively (data not shown). An increase of the pH had an even more severe effect on the melting transitions, such as at pH 9.5 t1 decreased by 5.9 °C to 74.9 °C and t2 decreased by 9.2 °C to 81.3 °C (data not shown). Taken together, the highest thermostability was observed in 50 mm Bicine buffer, pH 8.5, containing 200 mm NaCl. Therefore, all further thermofluor assays with the PilF variants were performed in 50 mm Bicine buffer with 200 mm NaCl at pH 8.5.
FIGURE 3.
Thermofluor assays of the purified PilF wild-type complex. Thermofluor assays were performed at pH 8.5 (50 mm Bicine buffer with 200 mm NaCl). 20 μl of purified PilF complex (0.1 μg/μl) were used in thermofluor assays. The melting curve of PilF wild-type complex was measured in Rotor Gene Q5 Plex HRM thermo cycler (A). SYPRO® Orange was used to measure fluorescence in the HRM channel with the excitation wavelength at 460 nm and the emission wavelength at 510 nm. The first derivative of the melting curve is shown in B. Two transition temperatures were observed at 80.8 ± 0.8 °C and 90.5 ± 0.0 °C. The effect of cysteine mutations on AMP-PNP- and ADP-mediated stabilization of PilF complexes is shown in C. Unfolding of the PilF complexes was measured by thermofluor assays. The bars indicate the first melting transitions of different PilF complexes in the absence or in the presence of 5 mm AMP-PNP and 2.5 mm ADP, respectively. Black, wild type; white, C1A; light gray, C2A; middle gray, HC2A; dark gray, C3A; vertical stripes, C4A; diagonal stripes, 2CysA; horizontal stripes, 3CysA; black and white boxes, 4CysA.
All PilF variants had two temperature-induced unfolding events. The second (t2) unfolding event was unaffected by the mutations in the 4Cys motif, such as a second melting transition was observed at ∼90.5 °C with all PilF variants (see Table 4). In contrast, the first unfolding event differed significantly. A mutation of cysteine 781 resulted in a reduction of t1 by 2.2 °C (78.6 ± 1.7 °C) (Fig. 3C, see Table 4), and the mutation of C2A and HC2A reduced t1 by 9.2 °C to 71.6 ± 0.3 °C or by 8 °C to 72.8 ± 0.1 °C, respectively. The mutation of C3A reduced t1 by 6.9 °C (73.9 ± 0.6 °C) (Fig. 3C, see Table 4). Among the PilF variants with single cysteine substitutions, C4A had the most profound effect on t1. A decline of 10.8 °C down to 70 °C ± 0.3 °C was observed (Fig. 3C, see Table 4). This corresponds to our findings that cysteine 819 was most critical for heat stability and zinc binding of the PilF complexes. The effect of double, triple, and quadruple mutations within the 4Cys motif led to almost identical dramatic declines in t1 by ∼15 °C down to ∼66 °C (Fig. 3C).
AMP-PNP and ADP Increase Thermostability of PilF
Recently it has been shown that AMP-PNP binding results in structural changes of the PilF wild-type complex (15). To assess the effect of ATP binding on the thermostability of the wild-type PilF complex and the variants, we analyzed temperature-dependent denaturation using thermofluor assays in the presence of 5 mm AMP-PNP as the ATP analog or in the presence of 2.5 mm ADP in 50 mm Bicine buffer containing 200 mm NaCl, pH 8.5. Again we observed two transitions; both AMP-PNP and ADP increased t1 by 4 and 4.9 °C, respectively (Fig. 4, Table 3), whereas the second transition at ∼90.5 °C was unaffected by the addition of AMP-PNP or ADP. The finding, that two different concentrations of AMP-PNP (5 mm) or ADP (2.5 mm) resulted in the same change of Δt1, suggested different affinities of PilF to AMP-PNP and ADP. To test this hypothesis, varying concentrations of AMP-PNP (0–20 mm) and ADP (0 to 10 mm) were used in the thermofluor assays. In both cases a concentration-dependent increase of the first transition temperature (Fig. 4) was found. For AMP-PNP a dissociation constant at 83.2 °C of 1.67 ± 0.3 mm and for ADP at 83.3 °C of 0.38 ± 0.03 mm was calculated. Taken together, AMP-PNP and ADP stabilize the wild-type PilF complex, and the significantly higher affinity of PilF for ADP leads to an increased thermostability compared with AMP-PNP.
FIGURE 4.
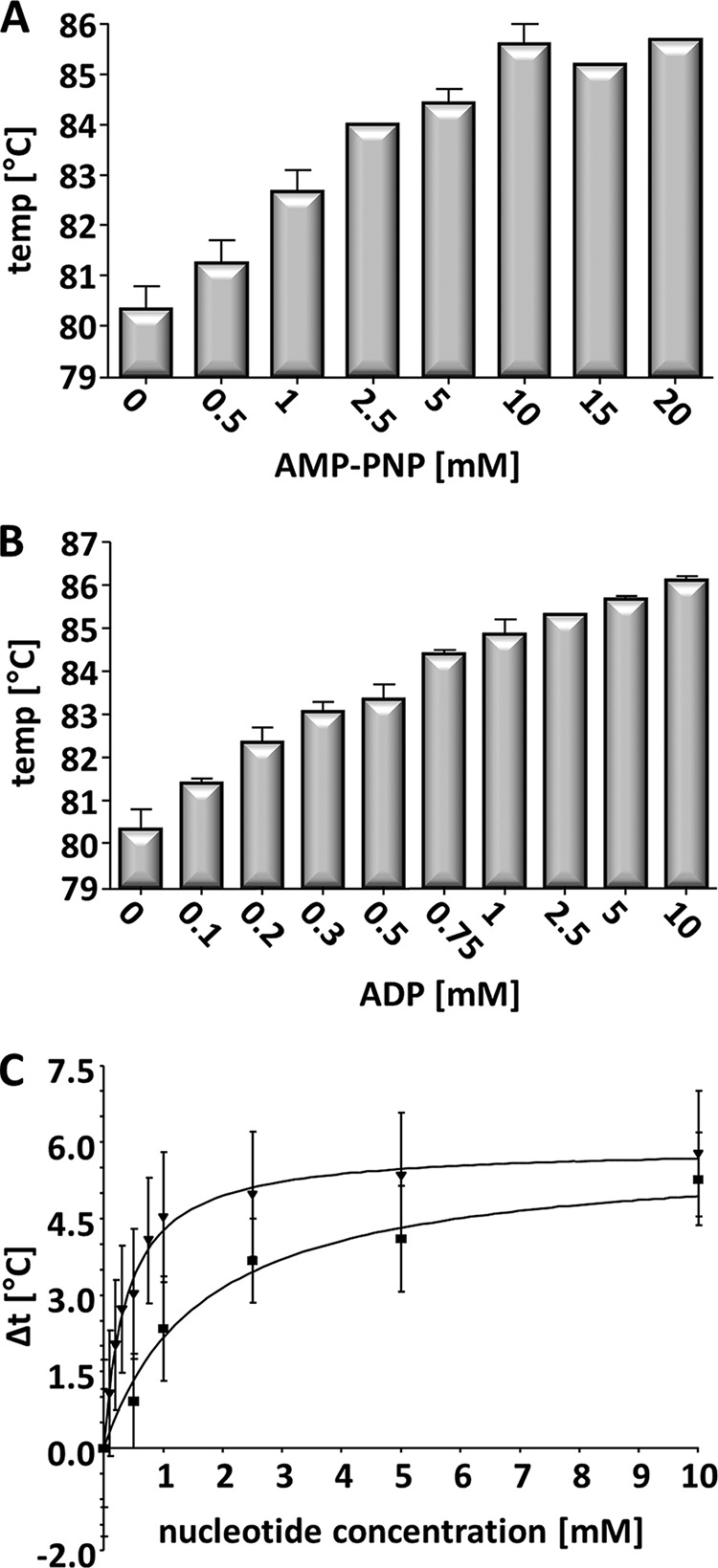
Dissociation temperature of PilF wild-type protein in presence of different concentrations of AMP-PNP (A) or ADP (B). Calculation of the dissociation constant by subtracting the melting transition without AMP-PNP or ADP from the melting transitions with different amounts of AMP-PNP (■) or ADP (▾) (C). The values are plotted against the nucleotide concentration. Dissociation constants are calculated by Graph Pad Prism 4.
TABLE 3.
Increase in melting transitions in the presence of AMP-PNP and ADP, respectively
| PilF variant | ΔtAMP-PNPa | ΔtADPb |
|---|---|---|
| °C | °C | |
| Wild type | +4.0 ± 0.6 | +4.9 ± 0.6 |
| C1A | +4.6 ± 1.7 | +6.2 ± 1.7 |
| C2A | +6.6 ± 0.6 | +7.5 ± 0.7 |
| HC2A | +6.8 ± 0.6 | +9.8 ± 0.2 |
| C3A | +6.9 ± 0.8 | +9.5 ± 0.7 |
| C4A | +6.4 ± 0.3 | +8.1 ± 0.3 |
| 2CysA | +7.5 ± 1.2 | +8.7 ± 1.1 |
| 3CysA | +7.6 ± 0.2 | +8.3 ± 0.5 |
| 4CysA | +5.8 ± 0.4 | +7.6 ± 0.3 |
a ΔtAMP-PNP was calculated by subtracting the melting transition without AMP-PNP from the melting transition with 5 mm AMP-PNP.
b ΔtADP was calculated by subtracting the melting transition without ADP from the melting transition with 2.5 mm ADP.
Effect of AMP-PNP and ADP on the Thermostability of the PilF Variants
As observed for the wild-type PilF, an increase in the first melting transition was detected in the presence of AMP-PNP or ADP for all PilF complexes formed by the PilF variants. However, AMP-PNP or ADP had different effects on the first transition temperature (Fig. 3C, Table 3). AMP-PNP led to an increase of the first melting peak temperature by 4–6.8 °C in wild-type PilF and the variants C1A, C2A, and HC2A (p < 0.05). An even higher stabilization effect was observed with the PilF variants C3A, C4A, 2CysA, 3CysA, and 4CysA. Here t1 was increased by 5.8 °C to 7.6 °C (p < 0.05). ADP had an even more severe effect with respect to stabilization of the PilF variants, such as t1 was elevated in the presence of ADP by 6.2 °C to 9.8 °C (Fig. 3C, Table 3), whereas t1 of the PilF wild-type complex was increased by ADP by 4.9 ± 0.6 °C (Fig. 3C, Table 3). Taken together, AMP-PNP and ADP mediate an elevated stabilization of the complexes formed by the cysteine-depleted PilF proteins in comparison to the PilF wild-type complex.
Role of the Cysteine Residues in Piliation
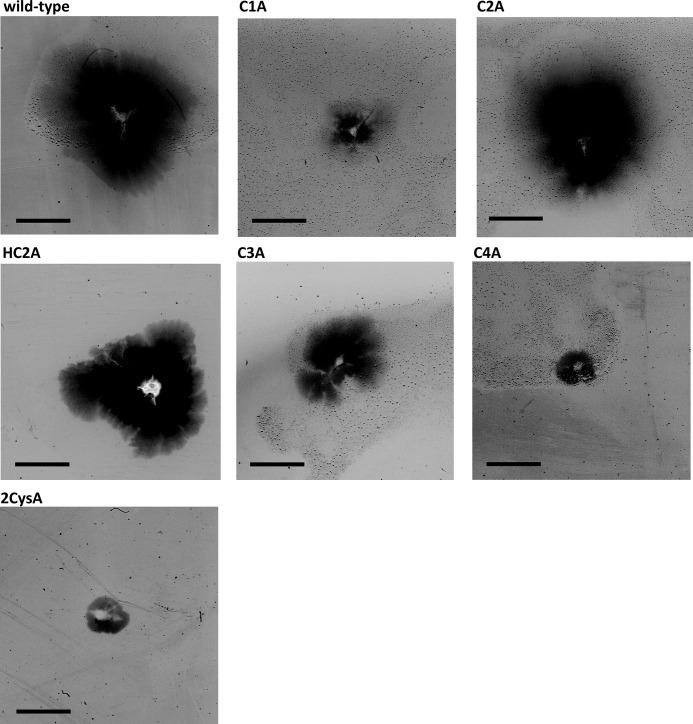
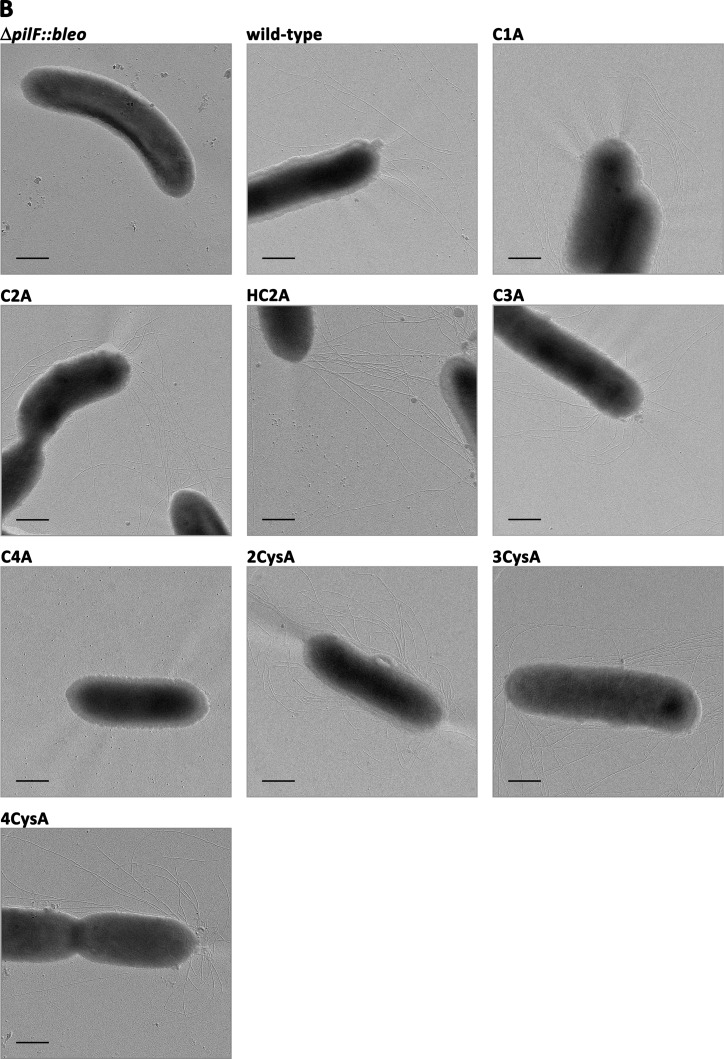
Previous studies showed that a ΔpilF::bleo mutant was completely impaired in piliation as observed by electron microscopy, which led to the conclusion that PilF is essential for pilus biogenesis (13). To analyze the role of the 4Cys motif in piliation the PilF variants were produced in trans in the ΔpilF::bleo T. thermophilus mutant strain. Therefore, the mutated pilF genes were cloned into the E. coli/Thermus shuttle vector pDM12. The recombinant plasmids were transformed into the T. thermophilus ΔpilF::bleo deletion mutant by electroporation, and the piliation phenotypes of the complemented ΔpilF::bleo mutants were analyzed by electron microscopy after growth on TM+ medium at 64 °C (Table 4). No pili were detected on the surface of the ΔpilF::bleo mutant, which corresponds to our previous results (13). Complementation of the ΔpilF::bleo mutant with the wild-type gene (pDM12-pilFwt) restored piliation. 80% of the cells were piliated with 7 ± 3 pilus structures per cell, which was comparable with the piliation phenotype of the HB27 wild-type strain (78% piliated cells, 6 ± 2 pili per cell). Similarly, complementation with pDM12-pilFC784A or pDM12-pilFH783A/C784A restored the piliation phenotype such that ∼77% or 83% of the cells were piliated, and 6 ± 1.5 or 8 ± 2.5 pili were detected per cell, respectively. In contrast, the construct pDM12-pilFC816A only partially restored the piliation phenotype (44% of the cells were piliated, and 1.5 ± 1 pili were detected per cell). Complementation with pDM12-pilFC781A resulted in a dramatically reduced piliation phenotype. Only 6.3% of the cells were piliated, and maximally 2 pili were detected per cell. The pilF mutant strains carrying pDM12-pilFC819A or multiple mutated pilF-genes were not piliated at all (Table 4). Taken together, all but cysteine residue 784 were implicated in or are even essential for the function of PilF in piliation.
The Tetracysteine Motif Is Essential for Twitching Motility and Adhesion
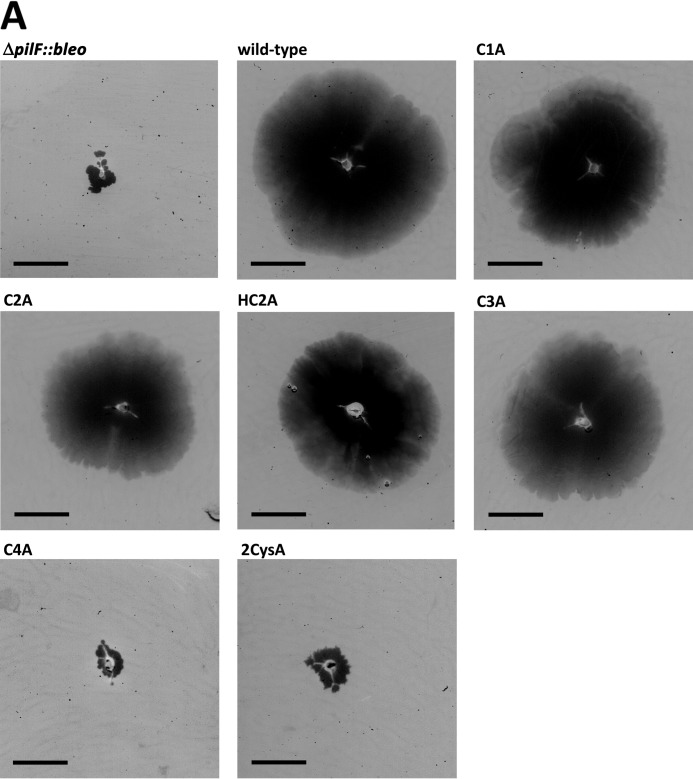
The pilus structures were found to be essential for twitching motility and adhesion of T. thermophilus (7, 13). To understand the role of the cysteine residues in pilus functions, twitching motility was analyzed. Therefore, the cells were grown on MM containing 1% BSA at 64 °C and stained with Coomassie Blue. Cells of the ΔpilF::bleo mutant carrying pDM12-pilFwt and the HB27 wild-type strain were used as controls and found to form large, uniform motility zones of 1 to 2 cm in diameter, whereas the ΔpilF::bleo mutant was completely defective in twitching motility (Fig. 5). These results correspond to our former findings (13). The variants C2A (C784A) and HC2A formed twitching zones comparable with those of the HB27 wild type (p > 0.05), but variants C1A (C781A), C3A (C816A), and C4A (C819A) were drastically impaired in twitching motility (p < 0.05) (Fig. 5, Table 4). These results are consistent with the significantly impaired piliation phenotypes of these three cysteine mutants.
FIGURE 5.
Effect of the cysteine mutations in PilF on twitching motility of T. thermophilus at 64 °C. Cells were grown for 3 days on MM containing 1% BSA under humid conditions. After growth plates were stained with Coomassie, cells were removed, and pictures were inverted. The scale bar corresponds to 0.5 cm.
To determine the role of the tetracysteine motif in T4P-mediated adhesion, microtiter adhesion assays were performed. Therefore, the complemented T. thermophilus ΔpilF::bleo mutants were grown in microtiter plates in 200 μl of medium at 64 °C. All PilF single cysteine mutants were significantly affected in adhesion (Fig. 6A, Table 4). Nearly no adhesion was even found with the double, triple, and quadruple cysteine mutants (Fig. 6A), which is consistent with the non-piliated phenotype of these mutants. Moreover, the finding that a single mutation in Cys-784 led to a 5-fold decrease in adhesion despite its nearly wild-type piliation phenotype suggests that the majority of the pili detected on the surface of this mutant are defective with respect to their adhesion function. This suggests that Cys-784 plays a major role in PilF-mediated assembly of adhesive pili.
FIGURE 6.
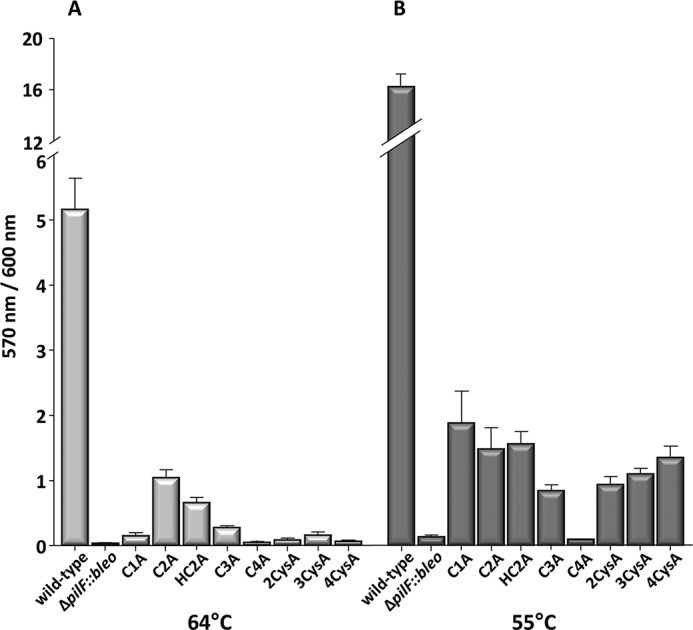
Effect of growth temperature on adhesion of pilF mutants. T. thermophilus cells were incubated in TM+ medium at 64 °C (A) or at 55 °C (B) in 96-well plates for 3 days. Adherence was calculated by the absorbance of crystal violet (570 nm) mediated by the adhered cells deviated by the amount of planktonic cells (600 nm). The mean was built from three independent cultures and in triplicate.
Growth Temperature Modulates the Motility, Adhesion, and Piliation Phenotype of the T. thermophilus PilF Mutants
Recently we reported that growth of T. thermophilus HB27 at low temperatures (55 °C) leads to hyperpiliation, thereby resulting in an increase in adhesion to plastic surfaces (23). This together with the finding that distinct cysteine residues of PilF play an essential role in motility, adhesion, and piliation raised the question of whether lowering the growth temperature would restore these phenotypes in the cysteine mutants. During growth at 55 °C the ΔpilF::bleo mutant carrying pDM12-pilFwt exhibited twitching zones with a diameter of 2–3 cm (Fig. 7A). This is comparable with those found with the T. thermophilus HB27 wild-type strain (13, 23). The piliation phenotype of the ΔpilF::bleo mutant carrying pDM12-pilFwt was also comparable with those of the HB27 wild type (13) such that 90.3% of the cells were piliated with 14 ± 3 pili per cell (Fig. 7B, Table 4). Interestingly, growth at 55 °C restored twitching motility in the ΔpilF::bleo mutant carrying pDM12-pilFC781A, pDM12-pilFC784A, pDM12-pilFH783A/C784A, or pDM12-pilFC816A (Fig. 7, C1A, C2A, HC2A, and C3A). The twitching zones of these mutants were comparable (p > 0.05) to those of the ΔpilF::bleo mutant carrying the wild-type pilF gene (Fig. 7A). Piliation analyses revealed that the mutant C1A had a strong increase in piliation with a non-countable number of pili. C2A and HC2A had 13 ± 2 and C3A had 6.5 ± 1.5 pili per cell (Fig. 7B, Table 4). The twitching phenotypes of the ΔpilF::bleo mutants carrying pDM12-pilFC819A (C4A) and of mutants with double (2CysA), triple (3CysA), and quadruple (4CysA) cysteine exchanges were not restored at 55 °C (p < 0.01) (Fig. 7A, Table 4).
FIGURE 7.
Effect of the growth temperature on twitching motility (A) and piliation (B) of T. thermophilus pilF mutants. Cells were grown at 55 °C for 5 days on MM containing 1% BSA. Plates were stained with Coomassie Blue, and the cells were removed after staining. Pictures were inverted. For piliation analyses the cells were transferred to copper grids and covered with 1.5-nm platinum/carbon in an angle of 25° and at minimum 250 cells per variant, and temperature was analyzed.
This raised the question of whether these mutants exhibited non-functional pili or were piliation-defective. To address this question the piliation phenotypes of these mutants were analyzed. Ninety-nine percent of the cells of the non-motile variants 2CysA, 3CysA, and 4CysA were piliated with a non-countable number of pili per cell (Fig. 7B, Table 4). The motility defect of the hyperpiliated 2CysA, 3CysA, and 4CysA mutants suggests that the pili on the surface were non-functional with respect to twitching motility. A cysteine mutant carrying a mutation in cysteine 819 (C4A) in PilF did not exhibit any pili on the surface at 55 °C (p < 0.01) (Fig. 7B). Therefore, we conclude that a single substitution of cysteine 819 has a dramatic effect on PilF function in pilus biogenesis.
To address the question of whether lowering the growth temperature to 55 °C restores the adhesion function of the pili formed by the cysteine mutants microtiter adhesion assays were performed. The ΔpilF::bleo mutant carrying the pDM12-pilFwt was used as the positive control and was found to exhibit a >3-fold increase in adhesion compared with adhesion at 64 °C (Fig. 6). These findings correspond to our recent observation with the HB27 wild type, which showed a significantly increased adhesion phenotype at low growth temperatures (23). After growth at 55 °C, the single mutants C1A, C2A, HC2A, and C3A exhibited an adhesion phenotype comparable with the adhesion phenotype of the wild-type cells grown at 64 °C (Fig. 6). The hyperpiliated, non-motile 2CysA, 3CysA, and 4CysA mutants also exhibited adhesion phenotypes comparable with those of the wild-type strain grown at 64 °C (Fig. 6B). Only the adhesion of the C4A cysteine mutant could not be restored at 55 °C, which is consistent with the absence of pili. Taken together the pili of the hyperpiliated double, triple, and quadruple mutants grown at 55 °C were non-functional with respect for twitching motility and impaired in adhesion. However, the restoration of the pilus biogenesis of the 2CysA, 3CysA, and 4CysA mutants at 55 °C supports our conclusion that the cysteine residues in PilF play an important role in PilF complex thermostability.
To verify the hypothesis that the cysteine residues are critical for thermostability of PilF, we enhanced the growth temperature to 72 °C and analyzed the piliation, twitching motility, and adherence of the cysteine mutants. As a control the ΔpilF::bleo mutant carrying pDM12-pilFwt was used. Analogously to recent findings for the HB27 wild-type strain, the piliation (5 ± 1.5 pili per cell, 77% piliated cells) and the adhesion (570 nm/600 nm = 0.5) were slightly reduced at elevated temperatures (23). Twitching motility at 72 °C was comparable (2.5–3 cm) to twitching at 64 °C. None of the PilF mutants exhibited twitching motility, adherence, or piliation at 72 °C (data not shown). These findings support our hypothesis that the tetracysteine motif is essential for thermostability of the PilF complex and, therefore, for a proper functioning of PilF in the pilus-mediated adhesion and twitching motility (Table 4).
Role of the Cysteine Residues in Transformation
PilF is essential for both piliation and natural transformation (13). Both systems are functionally linked, although there is substantial evidence that the pili themselves are not implicated in DNA uptake. This led to the question of whether the cysteine residues are important for the functionality of PilF in the DNA translocator. To analyze the role of the cysteine residues in natural transformation, the transformation frequencies of pilF mutants were analyzed. With the ΔpilF::bleo deletion mutant carrying the pilF wild-type gene, in trans transformation frequencies of 2.8 × 10−3 were obtained that were comparable with those of the HB27 wild-type strain (2.6 × 10−3) (Table 4). The mutants C1A, C2A, HC2A, C3A, 2CysA, 3CysA, and 4CysA were unaffected in natural transformations with 3.3 × 10−3, 2.6 × 10−3, 2.1 × 10−3, 2.2 × 10−3, 3.3 × 10−3, 2.7 × 10−3, and 3.6 × 10−3 transformants per living count, respectively (p > 0.05). These findings suggest that the cysteine residues are not essential for the function of PilF in natural transformation. Interestingly, the non-piliated C819A mutant exhibited a hypertransformability. Each cell was transformable (9.8 × 10−1 transformants per living count) (p < 0.01) (Table 4). This finding leads to the conclusion that the pilus structures are not required for natural transformation.
DISCUSSION
The AAA-ATPase PilF of T. thermophilus belongs to the PilF/PilB subfamily of traffic ATPases, which mediate the assembly of pilins into T4P and are also associated with natural competence and type II secretion systems (4, 26–29). These ATPases share highly conserved Walker A and Walker B boxes, histidine and aspartate boxes and a C-terminal tetracysteine motif (14, 30–32). The extended N terminus of PilF differs from all other known traffic ATPases and is predicted to contain three copies of a GSPII detected in proteins of type II secretion system (14, 33).
Recently we reported that PilF forms a homohexameric complex with six zinc atoms bound (20) Mutant studies suggested that zinc is coordinated by a conserved tetracysteine motif. The quadruple cysteine mutant proteins still assembled to hexameric PilF complexes, demonstrating that zinc binding is not essential for complex formation. In contrast PilF complexes comprising of PilF monomers with single cysteine substitutions still contained six zinc atoms. A coordination of zinc by just three cysteine residues or three histidine residues has also been found in alcohol dehydrogenases and carbonic anhydrases. Here the fourth residue is replaced by H2O (25). Analogously, we suggest that the fourth cysteine residue within the tetracysteine motif can be replaced by H2O.
However, one of the single cysteine substitutions, C819A, led to a statistically relevant decrease in zinc binding such as only ∼5 zinc atoms were detected in the hexameric complexes formed by the C819A mutant proteins (p < 0.05). The decrease in zinc binding of the C819A mutant protein complexes led to a decrease in thermostability. The correlation of reduced zinc binding and decreased thermostability of the C819A mutant protein complex indicates that zinc is of importance for complex stability. Analyses of PilF complexes formed by double and triple cysteine mutant proteins supported this conclusion as these PilF complexes showed a further decline in complex stability and were nearly completely devoid of zinc (maximal one zinc atom per complex). Moreover, the zinc-free PilF complex formed by the quadruple cysteine mutant protein exhibited the highest decline in complex stability. Taken together, these findings lead to the conclusion that zinc plays a major role in PilF complex stability. Moreover, these findings together with the broad distribution of tetracysteine motifs in polymerization AAA-ATPases lead to the conclusion that zinc binding is of great structural relevance. We suggest that zinc binding is important for correct folding and interactions of the AAA-ATPase subunits, thereby contributing to complex stability.
Analyses of the PilF complex stability by thermofluor assays unraveled two unfolding transitions of the PilF complexes, and only the first was affected by the cysteine mutations. The finding that even the quadruple mutant exhibited a wild-type ATPase activity at 70 °C together with the first unfolding event at 66.3 °C strongly suggests that the first unfolding event reflects the disassembly of the hexameric complexes, whereas the second transition represents unfolding of the monomers. This leads to the conclusion that the ATPase activity represents monomer-linked activity.
The assembly of some well investigated polymerization ATPases, such as GspE from Vibrio cholerae and TrbB from Helicobacter pylori requires ATP (34, 35). In contrast the assembly of the PilF complex in T. thermophilus does not require ATP. The ATP-independent assembly of PilF complex in T. thermophilus might be the result of an adaptation of T. thermophilus to its extreme environment.
This raises the question: Why does the assembly of the PilF complex of T. thermophilus not require ATP? An answer to this question might be derived from the unique triplicated GSPII domain at the N terminus of PilF. A triplicated GSPII domain has not been found in any other polymerization ATPase. It is tempting to speculate that these domains might function as an assistant platform for the ring formation of six C-terminal domains.
In many Gram-negative bacteria, pili and transformation were found to be functionally linked. However, despite this finding the relationship between these two features and the role of pilus structures in transformation is not understood. We found that multiple mutations in the tetracysteine motif resulted in a non-piliated phenotype but with normal transformation frequencies. Furthermore, one of the single cysteine mutants (C819A) did not exhibit any pilus structures but was hypertransformable. These findings provide clear evidence that in T. thermophilus pilus structures are not required for DNA uptake. This is different from the findings in other Gram-negative bacteria, such as N. gonorrhoeae and V. cholerae (36, 37). Here the pilus structures are suggested to be implicated in DNA uptake.
How could the hypertransformability of mutant C819A be explained? The C819A mutation had the most profound effect on PilF complex stability, zinc binding, and piliation, which suggests that Cys-819 plays a major role in the structural organization of the PilF monomers. A mutation in Cys-819 might abolish interactions of the PilF complex with components of the T4P assembly platform, leading to the non-piliated phenotype of this mutant. As a consequence, all of the PilF complexes might be available for interactions with components of the DNA translocator, such as the T4P-unrelated pilins PilA1–3 (10) leading to hypertransformability.
The absence of pili on the multiple cysteine mutants correlated with the significant decrease of zinc binding to the mutated PilF complexes, indicating that zinc binding is important for pilus biogenesis. However, a decrease in growth temperature to 55 °C restores the piliation phenotype of all, but one (Cys-819), cysteine mutants. This provides clear evidence that the tetracysteine motif and zinc binding are not essential for the PilF-mediated pilus assembly. Taken together, the finding that the piliation defects of cysteine mutants can be overcome by lowering the growth temperature strongly suggests that the increased instabilities of the PilF mutant complexes are responsible for the piliation defects of the mutants grown at 64 °C. The increased piliation of all PilF mutants and the wild type after growth at lower growth temperatures is consistent with our recent finding that the growth temperature has a significant effect of expression of T4P genes (23). We found that a decrease in growth temperature significantly increases the expression of the structural subunit of the pili, pilA4, leading to an increase in piliation (23). However, it has to be noted that despite their hyperpiliation, the multiple cysteine mutants were non-motile even at 55 °C and reduced in adhesion. Lowering the temperature to 55 °C is sufficient to increase the amount of stable PilF complexes thereby leading to pilus assembly. The adhesion and motility defects of the piliated mutants lead to the conclusion that cysteine residues and/or zinc binding of the PilF complex are essential for pilus functions.
The cysteine mutants with multiple mutations were non-motile but hyperpiliated. How can this be explained? One possibility might be that the mutations significantly affect the interactions of PilF with the antagonistic ATPases PilT1 and PilT2, which recently have been found to be essential for T4P-mediated twitching motility and adhesion of T. thermophilus (13). This would abolish depolymerization of pilus structures, thereby leading to the hyperpiliated phenotype. Another clue to answer this question might be derived from the finding that the pilus-mediated surface motility of Myxococcus xanthus and V. cholerae requires the translocation of polymerization ATPases such as PilB from the leading to the lagging pole and vice versa (38–40). For M. xanthus it was shown that this relocation of the ATPase requires parts of the cytoskeleton and a Ras-like G protein, MglA, and its cognate GTPase-activating protein (GAP), MglB (38). Proteins that show homologies to these proteins were also found in T. thermophilus and might be also required for the relocation of PilF from T. thermophilus (41). The multiple cysteine mutations in the tetracysteine motif and the absence of zinc in the mutant PilF complexes might have a significant effect on the structure of the PilF complexes and/or the transfer of structural changes through the PilF complex, thereby affecting the interaction of PilF complexes with cytoskeletal components. This might inhibit the movement of PilF from one pole to the other and thereby diminish or even abolish twitching motility.
Acknowledgment
We are grateful to Bernd Ludwig (Goethe University, Frankfurt, Germany) for the kind gift of plasmid pDM12.
This work was supported by a grant from the Deutsche Forschungsgemeinschaft (AV 9/6-1).
- T4P
- type IV pili
- AAA-ATPase
- ATPase associated with diverse cellular activities
- bleo
- bleomycin
- GSPII
- general secretory domain
- ICP-MS
- inductively coupled plasma-mass spectrometry
- AMP-PNP
- adenylylimidodiphosphate
- Bicine
- N,N-bis(2-hydroxyethyl)glycine
- CHES
- 2-(cyclohexylamino)ethanesulfonic acid
- HRM
- high resolution melt
- (C1A)
- C781A
- C2A
- C784A
- HC2A
- H783A/C784A
- C3A
- C816A
- C4A
- C819A
- 2CysA
- C781A/H783A/C784A
- 3CysA
- C781A/H783A/C784A/C816A
- 4CysA
- C781A/H783A/C784A/C816A/C819A
- MM
- minimal medium.
REFERENCES
- 1. Lorenz M. G., Wackernagel W. (1994) Bacterial gene transfer by natural genetic transformation in the environment. Microbiol. Rev. 58, 563–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Averhoff B., Graf I. (2008) The natural transformation system of Acinetobacter baylyi ADP1: a unique DNA transport machinery. In Acinetobacter Molecular Biology (Gerischer U., ed.) pp. 119–140, Caister Academic Press, Norfolk, UK [Google Scholar]
- 3. Dubnau D. (1999) DNA uptake in bacteria. Annu. Rev. Microbiol. 53, 217–244 [DOI] [PubMed] [Google Scholar]
- 4. Averhoff B. (2009) Shuffling genes around in hot environments: the unique DNA transporter of Thermus thermophilus. FEMS Microbiol. Rev. 33, 611–626 [DOI] [PubMed] [Google Scholar]
- 5. Koyama Y., Hoshino T., Tomizuka N., Furukawa K. (1986) Genetic transformation of the extreme thermophile Thermus thermophilus and of other Thermus spp. J. Bacteriol. 166, 338–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Burkhardt J., Vonck J., Averhoff B. (2011) Structure and function of PilQ, a secretin of the DNA transporter from the thermophilic bacterium Thermus thermophilus HB27. J. Biol. Chem. 286, 9977–9984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Burkhardt J., Vonck J., Langer J. D., Salzer R., Averhoff B. (2012) Unusual N-terminal ααβαββα fold of PilQ from Thermus thermophilus mediates ring formation and is essential for piliation. J. Biol. Chem. 287, 8484–8494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Friedrich A., Prust C., Hartsch T., Henne A., Averhoff B. (2002) Molecular analyses of the natural transformation machinery and identification of pilus structures in the extremely thermophilic bacterium Thermus thermophilus strain HB27. Appl. Environ. Microbiol. 68, 745–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schwarzenlander C., Averhoff B. (2006) Characterization of DNA transport in the thermophilic bacterium Thermus thermophilus HB27. FEBS J. 273, 4210–4218 [DOI] [PubMed] [Google Scholar]
- 10. Friedrich A., Rumszauer J., Henne A., Averhoff B. (2003) Pilin-like proteins in the extremely thermophilic bacterium Thermus thermophilus HB27: implication in competence for natural transformation and links to type IV pilus biogenesis. Appl. Environ. Microbiol. 69, 3695–3700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schwarzenlander C., Haase W., Averhoff B. (2009) The role of single subunits of the DNA transport machinery of Thermus thermophilus HB27 in DNA binding and transport. Environ. Microbiol. 11, 801–808 [DOI] [PubMed] [Google Scholar]
- 12. Rumszauer J., Schwarzenlander C., Averhoff B. (2006) Identification, subcellular localization, and functional interactions of PilMNOWQ and PilA4 involved in transformation competency and pilus biogenesis in the thermophilic bacterium Thermus thermophilus HB27. FEBS J. 273, 3261–3272 [DOI] [PubMed] [Google Scholar]
- 13. Salzer R., Joos F., Averhoff B. (2014) Type IV pilus biogenesis, twitching motility, and DNA uptake in Thermus thermophilus: discrete roles of antagonistic ATPases PilF, PilT1, and PilT2. Appl. Environ. Microbiol. 80, 644–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rose I., Biuković G., Aderhold P., Müller V., Grüber G., Averhoff B. (2011) Identification and characterization of a unique, zinc-containing transport ATPase essential for natural transformation in Thermus thermophilus HB27. Extremophiles 15, 191–202 [DOI] [PubMed] [Google Scholar]
- 15. Collins R. F., Hassan D., Karuppiah V., Thistlethwaite A., Derrick J. P. (2013) Structure and mechanism of the PilF DNA transformation ATPase from Thermus thermophilus. Biochem. J. 450, 417–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bertani G. (1951) Studies on lysogenesis. J. Bacteriol. 62, 293–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Oshima T., Imahori K. (1971) Isolation of an extreme thermophile and thermostability of its transfer ribonucleic acid and ribosomes. J. Gen. Appl. Microbiol. 17, 513–517 [Google Scholar]
- 18. Bradford M. M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 19. Wittig I., Schägger H. (2005) Advantages and limitations of clear-native PAGE. Proteomics 5, 4338–4346 [DOI] [PubMed] [Google Scholar]
- 20. Salzer R., Herzberg M., Nies D. H., Biuković G., Grüber G., Müller V., Averhoff B. (2013) The DNA uptake ATPase PilF of Thermus thermophilus: a reexamination of the zinc content. Extremophiles 17, 697–698 [DOI] [PubMed] [Google Scholar]
- 21. Heinonen J. K., Lahti R. J. (1981) A new and convenient colorimetric determination of inorganic orthophosphate and its application to the assay of inorganic pyrophosphatase. Anal. Biochem. 113, 313–317 [DOI] [PubMed] [Google Scholar]
- 22. Niesen F. H., Berglund H., Vedadi M. (2007) The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat. Protoc. 2, 2212–2221 [DOI] [PubMed] [Google Scholar]
- 23. Salzer R., Kern T., Joos F., Averhoff B. (2014) Environmental factors affecting the expression of type IV pilus genes as well as piliation of Thermus thermophilus. FEMS Microbiol. Lett. 357, 56–62 [DOI] [PubMed] [Google Scholar]
- 24. Friedrich A., Hartsch T., Averhoff B. (2001) Natural transformation in mesophilic and thermophilic bacteria: identification and characterization of novel, closely related competence genes in Acinetobacter sp. strain BD413 and Thermus thermophilus HB27. Appl. Environ. Microbiol. 67, 3140–3148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Vallee B. L., Auld D. S. (1990) Zinc coordination, function, and structure of zinc enzymes and other proteins. Biochemistry 29, 5647–5659 [DOI] [PubMed] [Google Scholar]
- 26. Chen I., Dubnau D. (2003) DNA transport during transformation. Front. Biosci. 8, 544–556 [DOI] [PubMed] [Google Scholar]
- 27. Hobbs M., Mattick J. S. (1993) Common components in the assembly of type 4 fimbriae, DNA transfer systems, filamentous phage and protein-secretion apparatus: a general system for the formation of surface-associated protein complexes. Mol. Microbiol. 10, 233–243 [DOI] [PubMed] [Google Scholar]
- 28. Nunn D. (1999) Bacterial type II protein export and pilus biogenesis: more than just homologies? Trends Cell Biol. 9, 402–408 [DOI] [PubMed] [Google Scholar]
- 29. Peabody C. R., Chung Y. J., Yen M. R., Vidal-Ingigliardi D., Pugsley A. P., Saier M. H., Jr. (2003) Type II protein secretion and its relationship to bacterial type IV pili and archaeal flagella. Microbiology 149, 3051–3072 [DOI] [PubMed] [Google Scholar]
- 30. Robien M. A., Krumm B. E., Sandkvist M., Hol W. G. (2003) Crystal structure of the extracellular protein secretion NTPase EpsE of Vibrio cholerae. J. Mol. Biol. 333, 657–674 [DOI] [PubMed] [Google Scholar]
- 31. Jakovljevic V., Leonardy S., Hoppert M., Søgaard-Andersen L. (2008) PilB and PilT are ATPases acting antagonistically in type IV pilus function in Myxococcus xanthus. J. Bacteriol. 190, 2411–2421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Camberg J. L., Sandkvist M. (2005) Molecular analysis of the Vibrio cholerae type II secretion ATPase EpsE. J. Bacteriol. 187, 249–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lu C., Turley S., Marionni S. T., Park Y. J., Lee K. K., Patrick M., Shah R., Sandkvist M., Bush M. F., Hol W. G. (2013) Hexamers of the type II secretion ATPase GspE from Vibrio cholerae with increased ATPase activity. Structure 21, 1707–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Krause S., Barcena M., Pansegrau W., Lurz R., Carazo J. M., Lanka E. (2000) Sequence-related protein export NTPases encoded by the conjugative transfer region of RP4 and by the cag pathogenicity island of Helicobacter pylori share similar hexameric ring structures. Proc. Natl. Acad. Sci. U.S.A. 97, 3067–3072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shiue S. J., Kao K. M., Leu W. M., Chen L. Y., Chan N. L., Hu N. T. (2006) XpsE oligomerization triggered by ATP binding, not hydrolysis, leads to its association with XpsL. EMBO J. 25, 1426–1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lång E., Haugen K., Fleckenstein B., Homberset H., Frye S. A., Ambur O. H., Tønjum T. (2009) Identification of neisserial DNA binding components. Microbiology 155, 852–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Seitz P., Pezeshgi Modarres H., Borgeaud S., Bulushev R. D., Steinbock L. J., Radenovic A., Dal Peraro M., Blokesch M. (2014) ComEA is essential for the transfer of external DNA into the periplasm in naturally transformable Vibrio cholerae cells. PLoS Genet. 10, e1004066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bulyha I., Lindow S., Lin L., Bolte K., Wuichet K., Kahnt J., van der Does C., Thanbichler M., Søgaard-Andersen L. (2013) Two small GTPases act in concert with the bactofilin cytoskeleton to regulate dynamic bacterial cell polarity. Dev. Cell 25, 119–131 [DOI] [PubMed] [Google Scholar]
- 39. Keilberg D., Søgaard-Andersen L. (2014) Regulation of bacterial cell polarity by small GTPases. Biochemistry 53, 1899–1907 [DOI] [PubMed] [Google Scholar]
- 40. Seitz P., Blokesch M. (2013) DNA-uptake machinery of naturally competent Vibrio cholerae. Proc. Natl. Acad. Sci. U.S.A. 110, 17987–17992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Miertzschke M., Koerner C., Vetter I. R., Keilberg D., Hot E., Leonardy S., Søgaard-Andersen L., Wittinghofer A. (2011) Structural analysis of the Ras-like G protein MglA and its cognate GAP MglB and implications for bacterial polarity. EMBO J. 30, 4185–4197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Oshima T., Imahori K. (1974) Description of Thermus thermophilus (Yoshida and Oshima) comb. nov., a nonsporulating thermophilic bacterium from a Japanese thermal spa. Int. J. Syst. Bacteriol. 24, 102–112 [Google Scholar]