Background: The function of many proteins is regulated through post-translational methylation.
Results: The previously uncharacterized human methyltransferase FAM86A and its yeast homologue Yjr129c methylate eukaryotic translation elongation factor 2 (eEF2), altering translational frameshifting.
Conclusion: Evolutionarily conserved FAM86A methyltransferase modulates the function of eEF2.
Significance: The activity of a novel protein-modifying enzyme is discovered and is shown to have functional consequences.
Keywords: Enzyme, Enzyme Catalysis, Post-translational Modification (PTM), Protein Methylation, Translation Elongation Factor
Abstract
The components of the cellular protein translation machinery, such as ribosomal proteins and translation factors, are subject to numerous post-translational modifications. In particular, this group of proteins is frequently methylated. However, for the majority of these methylations, the responsible methyltransferases (MTases) remain unknown. The human FAM86A (family with sequence similarity 86) protein belongs to a recently identified family of protein MTases, and we here show that FAM86A catalyzes the trimethylation of eukaryotic elongation factor 2 (eEF2) on Lys-525. Moreover, we demonstrate that the Saccharomyces cerevisiae MTase Yjr129c, which displays sequence homology to FAM86A, is a functional FAM86A orthologue, modifying the corresponding residue (Lys-509) in yeast eEF2, both in vitro and in vivo. Finally, Yjr129c-deficient yeast cells displayed phenotypes related to eEF2 function (i.e. increased frameshifting during protein translation and hypersensitivity toward the eEF2-specific drug sordarin). In summary, the present study establishes the function of the previously uncharacterized MTases FAM86A and Yjr129c, demonstrating that these enzymes introduce a functionally important lysine methylation in eEF2. Based on the previous naming of similar enzymes, we have redubbed FAM86A and Yjr129c as eEF2-KMT and Efm3, respectively.
Introduction
Methylation represents one of the most common biochemical reactions occurring in the cell, catalyzed by methyltransferases (MTases)3 and involving the transfer of methyl groups from donor to substrate molecules. The most common methyl donor is S-adenosyl-l-methionine (AdoMet), and the human genome encodes over 200 putative AdoMet-dependent MTases, the function of most of which is still unknown (1). Collectively, the MTases can methylate a wide range of substrates, from simple oxidized arsenic or chloride ions to complex macromolecules, such as nucleic acids and proteins (reviewed in Ref. 2). Methylation of proteins typically occurs on lysine or arginine residues; however, several other amino acids, such as histidine, aspartate, glutamate, or glutamine, can also be methylated (3–5). Lysine can be modified on its ϵ-amino group with either one, two, or three methyl groups, the addition of which decreases the number of potential hydrogen bonds and increases the bulk of the residue yet leaves the overall positive charge unaffected. Lysine methylation can thus influence several aspects of protein function, including stability, enzymatic activity, subcellular localization, interaction with other molecules, and the introduction of additional post-translational modifications (for a review, see Ref. 6).
Lysine methylation has been extensively studied in the context of histone proteins, where it is usually localized to the flexible N-terminal tail and is read by effector proteins, which regulate chromatin structure and gene expression (for a review, see Ref. 7). All of the known human MTases involved in histone methylation belong to the SET domain superfamily, with the exception of DOT1L, which contains a seven-stranded β-sheet (7BS) core fold characteristic for the largest class of human AdoMet-dependent MTases, the 7BS-MTases (8). In addition to DOT1L, only four other human 7BS-MTases have been shown to specifically methylate lysine residues: CaM-KMT methylates calmodulin (9), VCP-KMT methylates p97/VCP (10, 11), HSPA-KMT methylates HSP70 proteins (11, 12), and METTL22 methylates Kin17 (11). These four enzymes are closely related and, together with six other putative human MTases, including the one described in this work, constitute the human members of the so-called MTase family 16 (MTF16) (10).
The components of the protein translation machinery have long been observed to be modified with methyl groups on lysine residues, from ribosomal proteins to translation factors, both in fungal and mammalian cells (13–16) (for a review, see Ref. 17). However, little progress has been made in understanding the functional significance of these modifications or in identifying the responsible MTases. For example, in the span of 3 decades since the first reports of lysine methylation of eukaryotic elongation factor 1A (eEF1A), only two MTases acting on eEF1A lysines have been recently identified in yeast, still leaving several methylated lysine residues without a known MTase (15, 18).
Eukaryotic elongation factor 2 (eEF2) is a GTPase that catalyzes the translocation of the nascent protein chain from the A-site to the P-site of the ribosome, an essential function of every cell. It is highly conserved throughout Eukarya and also has a functional homologue, elongation factor G (EF-G) in prokaryotes. From yeast to humans, eEF2 is subject to several known post-translational modifications that regulate or adjust its activity, such as the inactivating threonine phosphorylation by an eEF2-specific kinase (19) or the constitutive formation of a unique diphthamide residue through sequential modifications of a histidine residue (for a review, see Ref. 20). Diphthamide is the target of several bacterial toxins that inactivate eEF2 via ADP-ribosylation, halting protein translation and resulting in cell death (21, 22). Protein synthesis is a highly energy-consuming process, and the post-translational modification of eEF2 serves as one of the main mechanisms for regulating protein synthesis at the level of elongation, which is crucial for cell survival during stress (23, 24).
Here we describe a novel modification of eEF2, the trimethylation of a conserved lysine residue (Lys-525 in human/Lys-509 in yeast sequence), which appears to occur throughout the evolutionary tree in humans, rabbits, and rats and in the yeast Saccharomyces cerevisiae. We show the previously uncharacterized human methyltransferase FAM86A and its yeast homologue Yjr129c to be the enzymes catalyzing the corresponding methylation reaction. In addition, knockout of the YJR129C gene in S. cerevisiae cells abolishes eEF2 methylation, proving that Yjr129c is the sole enzyme responsible for this methylation in yeast. Interestingly, lack of eEF2 methylation increases the sensitivity of the yeast toward sordarin, a translation inhibitor that directly targets fungal eEF2; it also increases the occurrence of −1 ribosomal frameshifting events, indicating that lysine methylation is required for optimal eEF2 function.
EXPERIMENTAL PROCEDURES
Cloning and Mutagenesis
ORFs were amplified with Phusion DNA polymerase (Thermo Scientific). The primers were designed to generate PCR products flanked by restriction sites appropriate for T4 DNA ligase-mediated insertion into the vectors. The vectors used were pET28a (Novagen), pYES260 (obtained from EUROSCARF), pNTAPa (Agilent Technologies), and pcDNA5/FRT/TO (Invitrogen). In addition, to create the pcDNA5/FRT/TO_TAP-FAM86A constructs, two rounds of PCRs were performed, first amplifying the calmodulin binding peptide (CBP) and streptavidin binding peptide (SBP) containing tag (tandem affinity purification (TAP) tag), containing the first 15 nucleotides of FAM86A at the 3′-end, and FAM86A, containing the last 15 nucleotides of the SBP tag at the 5′-end. These products were then mixed and used as template to amplify TAP-FAM86A with the flanking primers with appropriate restriction sites. Mutagenesis was performed using the PrimerX program to design the mutagenic primers and the QuikChange II site-directed mutagenesis kit (Stratagene). The identity of all constructs was verified by DNA sequencing.
Bioinformatic Analyses
The National Center for Biotechnology Information (NCBI) protein-protein Basic Local Alignment Tool (BLAST) was used for identification of proteins homologous to human FAM86 and eEF2 proteins (25). For the genomic distribution of human FAM86 genes, data were extracted from Ensembl (26) (release 71, April 2013, human genome assembly GRCh37). Multiple protein sequence alignments were performed using the Muscle algorithm embedded in Jalview (version 2.8) (27). Secondary structure of proteins was predicted from primary sequence using the PSIPRED algorithm (28).
Generation and Cultivation of Human Cell Lines
Stable cell lines for inducible overexpression of CBP-SBP-tagged FAM86A (TAP-FAM86A) or of the CBP-SBP tag alone (TAP) were generated using the Flp-InTM T-RExTM-293 system (Invitrogen) according to the manufacturer's instructions. Briefly, Flp-InTM T-RExTM-293 cells were cotransfected with pcDNA5/FRT/TO containing TAP-FAM86A or TAP and pOG44 (Invitrogen) using FuGENE® 6 transfection reagent (Roche Applied Science), and colonies were selected using 200 μg/ml hygromycin B (Invitrogen). Cells were thereafter maintained in Dulbecco's modified Eagle's medium (DMEM) (Lonza) supplemented with 10% (v/v) tetracycline-free fetal bovine serum (Clontech), 1× GlutaMAXTM (Invitrogen), and 100 units/ml penicillin and streptomycin (Lonza).
Tandem Affinity Purification
TAP was performed using the InterPlay mammalian TAP system (Agilent) according to the manufacturer's instructions with a few exceptions. Briefly, cells containing either TAP-FAM86A or TAP were induced using 1 μg/ml doxycycline (Clontech) for 24 h. The purification of TAP-FAM86A was performed additionally with the inclusion of either 100 μm AdoMet or S-adenosyl-l-homocysteine (AdoHcy) in all buffers to examine whether binding of putative substrates may be influenced by the presence of co-substrate or inhibitor, as reported previously (29). Proteins were finally eluted from calmodulin resin using NuPAGE gel loading buffer (Invitrogen).
S. cerevisiae Strains, Media, and Growth Assays
Wild-type (WT) BY4742 (MATα; his3Δ1; leu2Δ0; lys2Δ0; ura3Δ0) and yjr129cΔ mutant JEY9581 (MAT α; his3Δ1; leu2Δ0; lys2Δ0; ura3Δ0; yjr129c::kanMX4) strains were acquired from Euroscarf. The isogenic strain JEY10800 (Mat α; his3Δ1; leu2Δ0; lys2Δ0; ura3Δ0; yjr129c::URA3), which was used to test for paromomycin sensitivity and to confirm sordarin sensitivity, was made by transforming BY4742 with a PCR product consisting of the URA3 cassette flanked by regions of homology toward YJR129C (primer sequences: GAATGAAGATCTATTCTACGATAGATTACATCAGCGGTGCCCTGGAAAATACGAGCAGATTGTACTGAGAGTGC and GTTTCGTGATCTTGTAGATCCGTATGGGTGCAATCAAAGGTTTGAATTGCAGGGCCTGGAACAACACTCAACCCTATC). Deletion of YJR129C was confirmed by PCR. Yeast cells were grown in either YPD (1% Bacto yeast extract, 2% peptone, 2% glucose) or synthetic defined medium containing yeast nitrogen base with ammonium sulfate (Sigma-Aldrich), appropriate amino acid dropout mix (Clontech), and 2% glucose. Temperature sensitivity and sensitivity to drugs were assayed by growing the yeast cultures to the same A600 in YPD and then spotting 200 μl of 10-fold serial dilutions onto YPD plates, containing either no drug or one of the following: anisomycin (MP Biomedicals), cycloheximide (Biovision), hygromycin (Invitrogen), nourseothricin (Werner Bioagents), paromomycin sulfate salt (Sigma-Aldrich), rapamycin (Cayman Chemical), or sordarin sodium salt (Santa Cruz Biotechnology, Inc.). The plates were incubated at 25, 30, and/or 37 °C for 2–4 days. To assay the sensitivity to sordarin in liquid culture, the yeast strains were grown to an A600 of 0.22 in YPD at 30 °C with shaking at 250 rpm, at which point sordarin was added, and the cultures were allowed to grow further at either 30 or 37 °C; A600 of the cultures was measured at the indicated time points.
In Vitro Methyltransferase Assays
2 μg of recombinant His6-eEF2 proteins or 50 μg of yeast or HEK293 protein extracts were diluted in MTase buffer (50 mm Tris, pH 7.5, 5 mm MgCl2, 50 mm NaCl) and treated with 10–100 pmol of His6-FAM86A or His6-Yjr129c enzymes in the presence of either 0.5–1 μCi of [3H]AdoMet (for fluorography) or 1 mm cold AdoMet (for mass spectrometry (MS) analysis) for 1 h at 37 °C. The reaction was stopped by transfer to ice and the addition of NuPAGE gel loading buffer (Invitrogen). Products of the reaction were resolved on SDS-PAGE and either 1) stained with Coomassie Blue and the band of interest excised for MS analysis or 2) transferred to PVDF membrane (Immobilon), which was subsequently dried, sprayed with En3Hance spray (PerkinElmer Life Sciences), and exposed to Eastman Kodak Co. BioMax MS film (Sigma-Aldrich) at −80 °C.
Cell Treatment with AdOx
Flp-InTM T-RExTM-293 cells were grown to ∼80% confluence, and then 20 μm AdOx was added, and cells were grown for an additional 18–24 h. Cells were trypsinized, harvested by centrifugation, and frozen at −20 °C until further use.
Lysis and Fractionation of Lysate
To lyse the Flp-InTM T-RExTM-293 cells, 50 mg of wet cell pellet were resuspended in 200 μl of lysis buffer (50 mm Tris-HCl, pH 7.6, 100 mm NaCl, 1% Triton X-100, 5% glycerol, 1 mm DTT, and 1× Complete (EDTA-free) protease inhibitor mixture (Roche Applied Science)) for 10 min at 4 °C and then sonicated. To lyse rat tissues, the tissues were frozen in liquid nitrogen and crushed with a mortar and pestle in lysis buffer. To lyse yeast cells, the cultures were harvested at an A600 of 1.0 and lysed in lysis buffer, with 150 mm NaCl and without Triton X-100, by vortexing with glass beads. Lysates were cleared by centrifugation at 4 °C. Relative protein concentrations of the lysates were determined with the NanoDrop 2000 spectrophotometer (Thermo Scientific). For methylation site identification, eEF2 from yeast lysates was partially purified first on cation exchange (S-column) and then on anion exchange (Q-column) as described (30). To partially purify eEF2 from mammalian lysates, an aliquot of cell lysate (1–2 mg of total protein content) was diluted using dilution buffer (50 mm Tris-HCl, pH 7.6, 1% Triton X-100, 5% glycerol, and 1 mm DTT) to adjust NaCl concentration to 33 mm. Diluted cell lysate was loaded onto a Q-column equilibrated in dilution buffer. Material bound to the column was eluted using dilution buffer with a step gradient of increasing NaCl concentrations. Typically, four 100-μl fractions were collected: 0.15Q (between 0.033 and 0.15 m NaCl), eEF2-containing fraction 0.3Q (0.15–0.3 m NaCl), 0.5Q (0.3–0.5 m NaCl), and 0.75Q (0.5–0.75 m NaCl).
MS Analysis
Reverse phase (C18) nano-online liquid chromatographic MS/MS analysis of proteolytic peptides was performed using a system consisting of two Agilent 1200 HPLC binary pumps (nano and capillary) with autosampler, column heater, and integrated switching valve. This LC system was coupled via a nano-electrospray ion source to a LTQ-Orbitrap mass spectrometer (Thermo Fisher Scientific). In-gel proteolytic digestion was performed with trypsin (Sigma-Aldrich) or AspN (Roche Applied Science). For the analyses, 5 μl of peptide solution was injected into the 5 × 0.3-mm extraction column filled with Zorbax 300 SB-C18 of 5-μm (diameter) particle size (Agilent). Samples were washed with mobile phase (0.1% formic acid and 3% acetonitrile in water). The flow rate was 10 μl/min provided by the capillary pump. After 7 min, the integrated switching valve was activated, and peptides were eluted in the back-flush mode from the extraction column onto a 150 × 0.075-mm C18, 3-μm resin column (GlycproSIL C18–80Å, Glycpromass, Stove, Germany). The mobile phase consisted of acetonitrile and MS grade water, both containing 0.1% formic acid. Chromatographic separation was achieved using a binary gradient from 5 to 55% of acetonitrile in water in 60 min. The flow rate was 0.2 μl/min provided by the nanoflow pump. Mass spectra were acquired in the positive ion mode, applying a data-dependent automatic switch between survey scan and tandem mass spectra (MS/MS) acquisition. Peptide samples were analyzed with a collision-induced dissociation fragmentation method, acquiring one Orbitrap survey scan in the mass range of m/z 200–2000 followed by MS/MS of the six most intense ions in the Orbitrap. The target value in the LTQ-Orbitrap was 1,000,000 for survey scan at a resolution of 30,000 at m/z 400, using lock masses for recalibration to improve the mass accuracy of precursor ions.
MS data were analyzed with the in-house maintained human, rat, and yeast protein sequence database and a database containing eEF2 protein sequence using SEQUESTTM. The mass tolerances of a fragment ion and a parent ion were set as 0.5 Da and 10 ppm, respectively. Methionine oxidation and cysteine carbamidomethylation was selected as a variable modification. A false discovery rate of 0.01 was required for proteins and peptides with a minimum length of 6 amino acids. eEF2 un-, mono-, di-, and trimethylated peptide MS spectra were manually searched by Qual Browser (version 2.0.7).
Recombinant Protein Expression and Purification
For the expression of His6-tagged methyltransferases, pET28a_FAM86A and pET28a_YJR129C constructs were transfected into the Escherichia coli BL21-CodonPlus (DE3)-RIPL expression strain (Agilent). Cells were cultured with appropriate antibiotics at 37 °C and shaking at 250 rpm until they reached A600 of 0.6. The temperature was then reduced to 16 °C, and the expression of recombinant proteins was induced by the addition of 100 μm isopropyl 1-thio-β-d-galactopyranoside and allowed to proceed for 16 h. Cells were then harvested by centrifugation. Cell pellets were lysed in lysis buffer (50 mm Tris, pH 7.6, 300 mm NaCl, 10% (w/v) glycerol, 30 mm imidazole, 3 mm β-mercaptoethanol, 1× Complete protease inhibitor mixture (Roche Applied Science), 0.5 mg/ml lysozyme (Sigma-Aldrich), and 10 units/ml Benzonase (Sigma-Aldrich)) for 30 min on ice and sonicated. The resultant lysates were cleared by centrifugation, and recombinant proteins were allowed to bind to nickel-nitrilotriacetic acid-agarose (Qiagen) at 4 °C for 16 h. The resin was washed with wash buffer (50 mm Tris, pH 7.6, 300 mm NaCl, 10% (w/v) glycerol, 30 mm imidazole, 3 mm β-mercaptoethanol), and His6-tagged proteins were eluted in wash buffer supplemented with 300 mm imidazole. Eluates were buffer-exchanged to storage buffer (100 mm NaCl, 20 mm Tris, pH 6.8, and 1 mm DTT) by sequential concentration and dilution using Vivaspin 20 ultrafiltration columns with a molecular mass cut-off of 10 kDa (Sartorius AG). Proteins were aliquoted and stored at −80 °C. For the expression of His6-tagged S. cerevisiae eEF2 proteins, pYES260 constructs were transformed into competent yjr129cΔ BY4742 yeast using the LiAc/SS carrier method (31), and transformants were selected on SD −Ura plates. The cells were cultured at 30 °C and shaking at 250 rpm; recombinant protein expression was induced at A600 of 0.6 by changing from YPD medium to YPG (1% Bacto yeast extract, 2% peptone, 2% galactose); and the cells were further cultured overnight. The cultures were then pelleted; lysed in lysis buffer (50 mm potassium phosphate, pH 7.6, 300 mm KCl, 10 mm imidazole, 1× Complete protease inhibitor mixture (Roche Applied Science), 1 mm DTT) by vortexing with acid-washed glass beads (Sigma); and cleared by centrifugation. The resulting lysates were allowed to bind to nickel-nitrilotriacetic acid-agarose (Qiagen) at 4 °C for 2 h. The resin was washed with wash buffer (50 mm potassium phosphate, pH 7.6, 300 mm KCl, 20 mm imidazole, 1 mm DTT), and the His6-tagged proteins were eluted with wash buffer containing 250 mm imidazole. The eluates were then buffer-exchanged to storage buffer as described above. Protein concentrations were determined using the BCA protein assay kit (Pierce).
Protein Synthesis Rate
To measure in vivo [35S]methionine incorporation, yeast strains were grown in 20 ml of liquid YPD medium at 30 °C to an A600 of 0.3 and then switched to SD −Met/−Cys medium and grown for an additional 1 h, at which point 600 μCi of Met-35S label (Hartmann Analytic) was added. 100 μl of the cell suspension was harvested at the indicated time points, pelleted, precipitated in 1 ml of 10% TCA, and filtered through GF/C glass microfiber filters (Whatman). Acid-insoluble radioactivity was measured by liquid scintillation counting using Ultima Gold XR (PerkinElmer Life Sciences).
Luciferase Assay Measurements
Cells from liquid cultures of S. cerevisiae strains grown to A600 0.4–0.6 were lysed for 25 s using passive lysis buffer (all reagents from the Promega Dual-Luciferase reporter assay system). Firefly luciferase activity was measured (10-s integration time) using reconstituted luciferase assay buffer in a GloMax 20/20 luminometer (Promega). Renilla luciferase activity (10-s integration time) was determined subsequent to quenching of firefly luciferase activity using Stop & Glo Buffer. Signals were >100-fold above background. The ratio of firefly luciferase to Renilla luciferase activity is reported. To account for potential differences in translational processivity, the percentage of ribosomal frameshifting of a strain was determined as the ratio of the firefly luciferase/Renilla luciferase signals of a ribosomal frameshifting site-bearing insert to that of a control sequence. A variant of the previously reported pRS425-iD2000 vector was used, which contains the HIV-1 programmed ribosomal frameshifting site (32) between the Renilla and firefly luciferase ORFs (33). pRS425-iD2000 contains a high copy number 2 μ origin, a LEU2 selection marker, a strong glycolytic PGK1 promoter, the dual luciferase reporter cassette, and a downstream CYC1 transcription terminator for mRNA stabilization. The probability (p values) that two populations are the same was tested using Student's t test (two-tailed; assuming groups have unequal variance).
RESULTS
Bioinformatics Analysis of Human FAM86 and Its Homologues
We have previously established several of the human members of MTF16 as lysine-specific protein MTases (KMTs), and in the present work, we set out to elucidate the function of a previously uncharacterized member, FAM86. Whereas the genomes of most mammals have a single FAM86 gene, FAM86 in higher primates (Homininae) is contained within a tumor break-prone segmental duplication region, which has spread to multiple genomic locations (34). Thus, human FAM86 was reported to display substantial copy number variations, ranging from 17 to 21 copies between the three investigated genomes. In the human reference genome (GRCh37), 15 FAM86 sequences are annotated, distributed between seven chromosomes (Table 1). Of these, the majority (11) represent non-transcribed pseudogenes, whereas the remaining four, FAM86A, FAM86B1, FAM86B2, and FAM86C1, are annotated as protein coding.
TABLE 1.
Genomic distribution of human FAM86 genes
| Gene | Gene ID | Location |
|---|---|---|
| FAM86A | ENSG00000118894 | 16:5134305–5147809:−1 |
| FAM86B1 | ENSG00000186523 | 8:12039605–12051642:−1 |
| FAM86B2 | ENSG00000145002 | 8:12282913–12293915:−1 |
| FAM86C1 | ENSG00000158483 | 11:71498556–71512282:1 |
| FAM86B3Pa | ENSG00000173295 | 8:8086117–8102387:1 |
| FAM86C2Pa | ENSG00000160172 | 11:67559119–67572807:−1 |
| FAM86DPa | ENSG00000244026 | 3:75470703–75484261:−1 |
| FAM86EPa | ENSG00000251669 | 4:3943487–3957146:−1 |
| FAM86FPa | ENSG00000164845 | 12:8385108–8395544:−1 |
| FAM86GPa | ENSG00000166492 | 11:3431582–3443726:−1 |
| FAM86HPa | ENSG00000253540 | 3:129817935–129830315:−1 |
| FAM86JPa | ENSG00000171084 | 3:125635463–125648867:1 |
| FAM86KPa | ENSG00000163612 | 4:9155022–9167177:1 |
| FAM86LPa | ENSG00000242731 | 7:6971315–6979234:-1 |
| FAM86MPa | ENSG00000186234 | 4:9694119–9704578:1 |
a FAM86 pseudogenes.
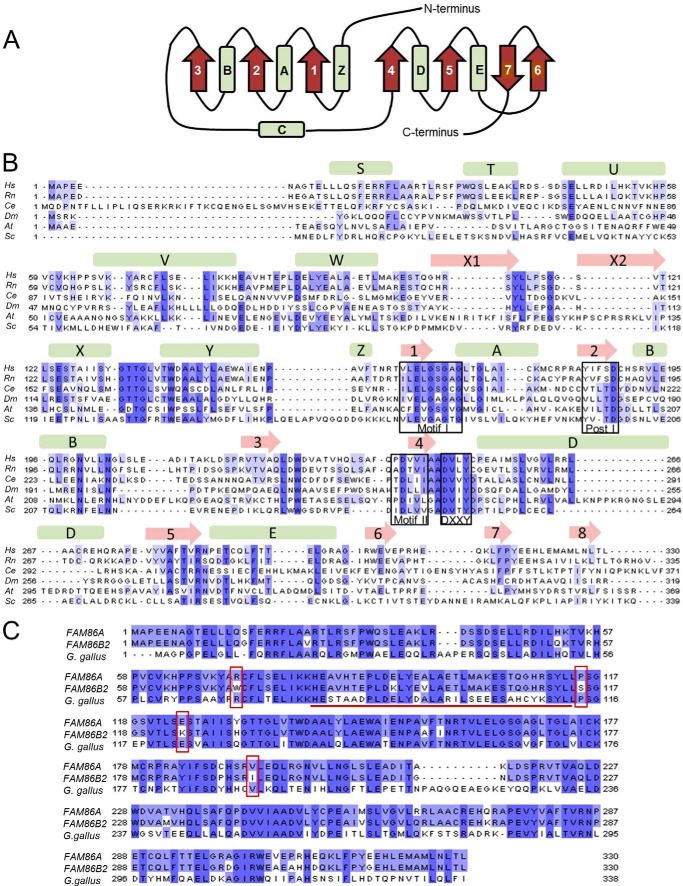
Given the recent appearance of the extra copies of FAM86 genes in humans, it is likely that some of the four annotated FAM86 proteins are non-functional. To investigate which of the four human FAM86 genes encode biologically relevant, functional proteins, we compared the protein sequences with those of predicted proteins from organisms that have a single FAM86 gene. Putative FAM86 homologues appear to be present in all eukaryotes, and an alignment of FAM86 sequences from various organisms revealed the motifs characteristic of 7BS AdoMet-dependent MTases (denoted Motif I, Post I, and Motif II) (35). Also, secondary structure prediction indicated the overall conserved 7BS core fold. In addition, the alignment showed a (D/E)XX(Y/F) motif downstream of Motif II typical of human MTF16 (Fig. 1, A and B) (36, 10). Two of the four annotated human FAM86 proteins, namely FAM86C1 and FAM86B1, are truncated relative to the vertebrate “consensus” protein of ∼330–340 amino acids. FAM86C1 (165 amino acids) is devoid of a substantial part of the 7BS fold, whereas FAM86B1 (296 amino acids) lacks a segment of 34 amino acids in the N-terminal region. In contrast, FAM86A and FAM86B2 are both 330 amino acids long and only differ by 26 point substitutions.
FIGURE 1.
FAM86 is a 7BS-MTase conserved across Eukarya. A, the canonical core folds of the 7BS-MTases, α-helices, and β-strands are represented as barrels and arrows, respectively. B, alignment of FAM86 protein sequences from Homo sapiens (Hs; NP_958802.1), Rattus norvegicus (Rn; NP_001100445.1), Caenorhabditis elegans (Ce; NP_498985.3), Drosophila melanogaster (Dm; NP_573368.2), Arabidopsis thaliana (At; NP_198092.1), and S. cerevisiae (Sc; NP_012663.2). Boxes indicate motifs characteristic for 7BS-MTases (Motif I, Post I, and Motif II) as well as the (D/E)XX(Y/F) motif typical of MTase family 16. The predicted secondary structure for human FAM86A is shown above the alignment. C, alignment of human FAM86A and FAM86B2 protein sequences (NP_958802.1 and NP_001131082.1) with the Gallus gallus FAM86 homologue (NP_001025798.1). The G. gallus sequence is included as a representative of a typical vertebrate FAM86 protein. The underlined segment indicates residues missing in the FAM86B1 protein. Residues that are conserved in vertebrates but differ between FAM86A and FAM86B2 are boxed.
To investigate which of the two proteins, FAM86A or FAM86B2, is likely to represent the ancestral functional sequence, we analyzed the degree of purifying selection. To this end, we generated an alignment of vertebrate FAM86 proteins and assessed which of the 26 substituted residues corresponded to conserved positions (data not shown). Interestingly, 4 of the 26 amino acid substitutions were found at positions conserved in vertebrates, and in all of these cases, FAM86A, but not FAM86B2, adhered to the vertebrate consensus (Fig. 1C). In conclusion, our initial bioinformatics analysis indicated human FAM86A to be a functional protein with probable orthologues in a wide range of other eukaryotes. Therefore, and also because we were unable to amplify FAM86B2 cDNA from human cells, we chose to focus the experimental part of our study on FAM86A.
Identification of Lys-509 in Yeast eEF2 as a Substrate for FAM86 Proteins
Because other MTF16 MTases have been shown to interact avidly with their substrates, we set out to search for FAM86A substrates among its interaction partners. To this end, we performed tandem affinity purification of TAP-tagged FAM86A after inducing its overexpression in a human embryonic kidney cell line (Flp-In 293 T-REx). We reasoned that the binding of FAM86 to a putative substrate may be influenced by the presence of the methyl donor AdoMet or its demethylated counterpart AdoHcy, and the purification buffer was supplemented with either AdoMet, AdoHcy, or no co-substrate, and in all three cases, a protein of ∼100 kDa was pulled down by the FAM86A bait (Fig. 2A). Subsequent MS analysis of the corresponding bands identified the protein as eEF2.
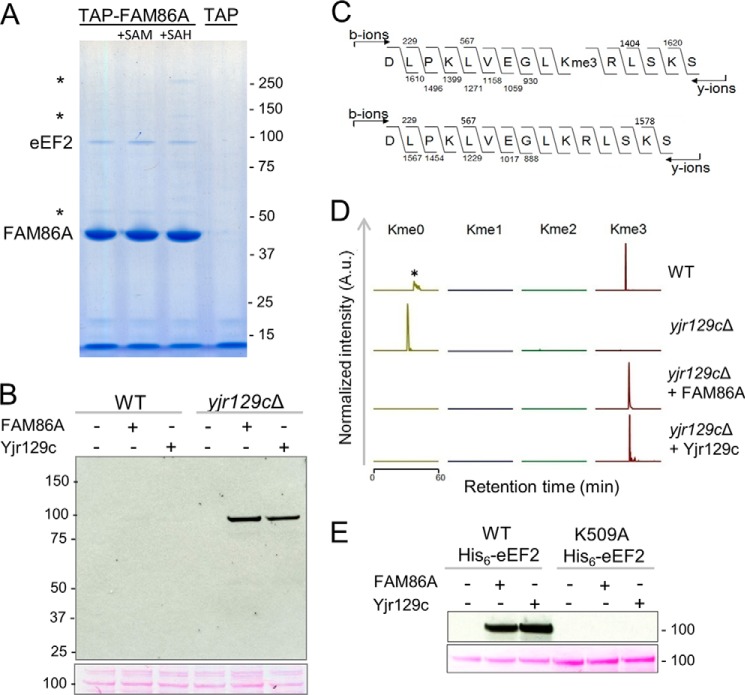
FIGURE 2.
FAM86A and its yeast homologue Yjr129c are eEF2 methyltransferases. A, eEF2 co-purifies with FAM86A in TAP. Shown is a Coomassie Blue-stained SDS-polyacrylamide gel of TAP eluates from a Flp-In T-REx HEK293-derived cell line treated with doxycycline to induce expression of SBP- and CBP-tagged FAM86A (TAP-FAM86A) or the CBP-SBP tag alone (TAP). Purification buffers were supplemented with AdoMet (SAM) or AdoHcy (SAH) where indicated. The identity of the bait and co-purifying proteins was determined by peptide mass fingerprinting; asterisks indicate β-tubulin (50 kDa), pyruvate carboxylase (125 kDa), and acetyl-CoA carboxylase 1 (265 kDa). Molecular weight markers (in kilodaltons) are shown on the right. B, eEF2 from yjr129cΔ yeast extracts is efficiently methylated by FAM86A and Yjr129c. Top, MTase reactions with recombinant FAM86A or Yjr129c, [3H]AdoMet, and WT or yjr129cΔ yeast extracts separated by SDS-PAGE and subjected to fluorography; bottom, Ponceau S stain of the 100-kDa region of the same membrane. C, trimethylation of Lys-509 in eEF2. Shown are MS/MS fragmentation patterns of representative AspN-generated trimethylated peptides from the WT strain (top) and unmethylated peptides from the yjr129cΔ strain (bottom). D, Lys-509 in eEF2 is the target of methylation by the MTases Yjr129c and FAM86A. Shown are MS chromatograms of AspN-generated Lys-509-containing peptides from partially purified eEF2 from WT or yjr129cΔ yeast extracts. When indicated, the extracts had been incubated with recombinant Yjr129c or FAM86A in the presence of AdoMet. *, an unrelated peptide. A.u., arbitrary units. E, mutagenesis of Lys-509 in eEF2 abolishes FAM86A and Yjr129c-mediated methylation. Recombinant FAM86A or Yjr129c was incubated in the presence of [3H]AdoMet with WT or K509A His6-eEF2, expressed, and purified from yjr129cΔ mutant yeast. The MTase reactions were separated by SDS-PAGE and subjected to fluorography (top) and Ponceau S staining (bottom).
In order to examine whether eEF2 is a substrate for FAM86A-mediated methylation in vitro, we required a source of unmethylated eEF2. In agreement with the published literature, our attempts to produce the WT and several truncations of His-tagged human eEF2 in E. coli were unsuccessful, and therefore, we turned to a eukaryotic system for production of unmethylated eEF2. We chose the yeast S. cerevisiae, because eEF2 is highly conserved between humans and yeast, and it has previously been expressed and purified from this organism.
Importantly, the S. cerevisiae genome contains a putative FAM86A homologue, Yjr129c, albeit of rather low sequence similarity (27% identity to human FAM86A), and we reasoned that it may be necessary to use a yjr129cΔ knock-out strain to obtain unmethylated eEF2. First, we investigated whether FAM86A or its putative yeast homologue could methylate endogenous yeast eEF2. To this end, we prepared total cell extracts from the WT and yjr129cΔ mutant S. cerevisiae strains and incubated them with [3H]AdoMet and the E. coli-expressed recombinant MTase FAM86A or Yjr129c. The products of the reactions were separated by SDS-PAGE and transferred to a PVDF membrane, which was analyzed for incorporated radioactivity by fluorography (Fig. 2B). A single radioactive band of ∼100 kDa, corresponding to the molecular weight of eEF2, appeared in yjr129cΔ mutant samples treated with either of the MTases but not in the untreated yjr129cΔ extract. Interestingly, no band appeared in the WT samples, suggesting that in WT S. cerevisiae, the ∼100-kDa substrate, which is probably eEF2, may already be highly methylated and thus refractory to further methylation. Taken together, these results show that Yjr129c is the functional yeast homologue of FAM86A and suggest that both of these MTases can methylate yeast eEF2.
In order to identify the site(s) of methylation, we partially purified endogenous eEF2 from the WT and yjr129cΔ mutant yeast strains and subjected it to MS analysis, with or without prior treatment by purified FAM86A or Yjr129c proteins (Fig. 2, C and D). Importantly, in eEF2 from WT cells, Lys-509 was found exclusively in the trimethylated state, as already suggested by results from Fig. 2B, whereas the same residue in eEF2 from yjr129cΔ mutant cells was unmodified. In addition, after treatment with either MTase, eEF2 from yjr129cΔ yeast became fully trimethylated. To investigate whether Lys-509 is the only residue targeted by the MTases, we expressed and purified His6-tagged WT or K509A mutant yeast eEF2 from yjr129cΔ cells. Indeed, WT His6-eEF2 was efficiently methylated by both MTases, whereas no methylation of the K509A mutant could be detected (Fig. 2E), indicating that Lys-509 is the only possible acceptor site for these enzymes. In summary, the above results show that both Yjr129c and FAM86 are able to trimethylate a single residue in yeast eEF2, Lys-509, and implicate Yjr129c as the yeast MTase responsible for introducing this modification.
Methylation of Human eEF2 by FAM86 Proteins
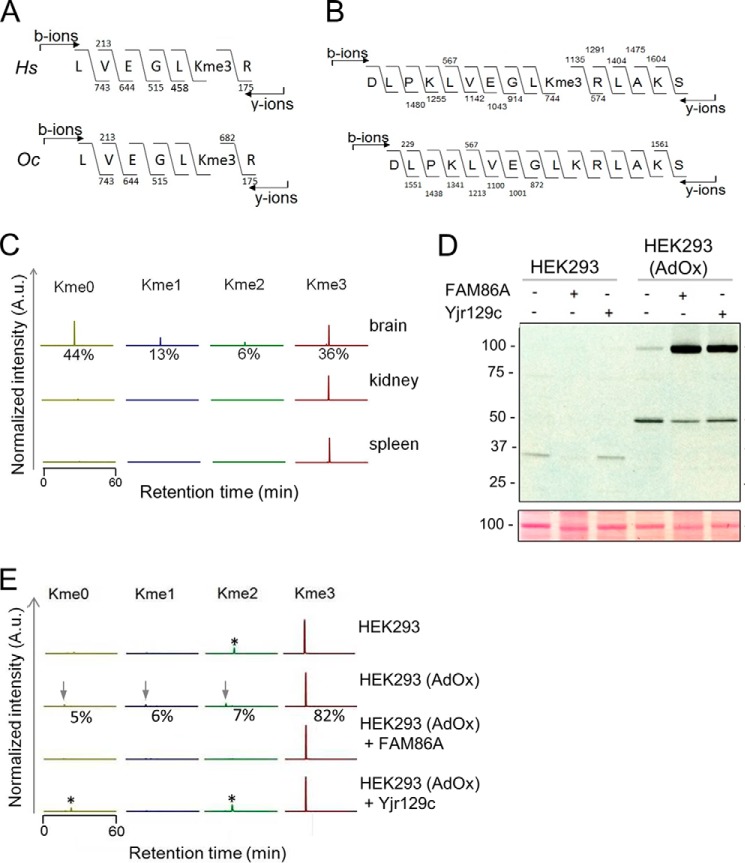
Next, we wanted to test, in a similar manner, whether the human and yeast MTases were able to methylate human eEF2 because the lysine residue analogous to Lys-509 in yeast eEF2 is present in the human protein (Lys-525). In fact, this lysine appears to be conserved in all eukaryotic eEF2 homologues analyzed, implicating this residue as evolutionarily important. However, we were unable to successfully express human His6-eEF2 in yjr129cΔ yeast cells and therefore chose to use cultured human cells to investigate the methylation of human eEF2. To address whether Lys-525 methylation of eEF2 also occurs in vivo in mammalian cells, we analyzed its methylation status in human HEK293 cells, in rabbit reticulocyte lysate, and in several rat tissues. The MS analysis showed that this residue was present exclusively in the trimethylated state in both human and rabbit cells (Fig. 3A). The analogous eEF2 lysine from rat kidney and spleen was also fully trimethylated. However, in the brain of the rat, it was substantially hypomethylated; about half was completely unmethylated, and only a third was trimethylated, with small amounts of mono- and dimethylated Lys-525 also present (Fig. 3, B and C).
FIGURE 3.
Methylation of mammalian eEF2. A, human and rabbit eEF2 is trimethylated on Lys-525. Shown is MS/MS fragmentation of a trypsin-generated human (HEK293) (Hs, top) and rabbit (Oc, bottom) eEF2-derived peptide, supporting Lys-525 as the target of methylation. B, eEF2 is trimethylated on Lys-525 in the rat. Shown is MS/MS fragmentation of AspN-generated trimethylated (top) and unmethylated (bottom) eEF2-derived peptides from the rat brain. C, eEF2 methylation status in a panel of rat tissues. Chromatograms were gated for different methylation states of AspN-generated Lys-525-containing peptides (as in B) from partially purified eEF2 from rat brain, kidney, and spleen. Relative intensities of signals for the different methylation states are indicated in percentages. A.u., arbitrary units. D, eEF2 from a hypomethylated human embryonic kidney cell line is efficiently methylated by both FAM86A and Yjr129c. A protein extract from HEK293 cells that had either been left untreated (HEK293) or treated with 20 μm AdOx to induce hypomethylation (HEK293(AdOx)) was incubated with recombinant FAM86A or Yjr129c in the presence of [3H]AdoMet. The MTase reactions were separated by SDS-PAGE and subjected to fluorography (top) and Ponceau S staining (bottom). E, Lys-525 in human eEF2 is subject to FAM86A- and Yjr129c-mediated trimethylation in vitro. eEF2 was partially purified from extracts from untreated and AdOx-treated cells and then subjected to LC-MS/MS analysis. When indicated, eEF2 had been incubated with recombinant Yjr129c or FAM86A in the presence of AdoMet. Chromatograms were gated for different methylation states of trypsin-generated Lys-525-containing eEF2 peptides (as shown in A). Arrows, peptides of interest; *, unrelated peptides. Relative intensities of signals for the different methylation states are indicated in percentages for the hypomethylated cell line.
In order to investigate whether FAM86A may be responsible for the Lys-525 methylation that is present in human cells, we needed to acquire human eEF2 that was not fully trimethylated. Knockout or knockdown of FAM86A could prove difficult due to the presence of a large number of similar genes and transcripts. To overcome this problem, we treated HEK293 cells with adenosine dialdehyde (AdOx), an agent that indirectly inhibits AdoMet-dependent methyl transfer by blocking the hydrolysis of AdoHcy, the by-product of methylation and competitive inhibitor of AdoMet, creating a hypomethylated environment in the cell (37). We then methylated the extracts from these hypomethylated cells with either FAM86A or Yjr129c. Interestingly, both MTases could methylate a ∼100-kDa protein in the lysate from AdOx-treated cells (Fig. 3D). MS analysis of the protein confirmed its identity as eEF2 and showed that in the AdOx-treated cells, Lys-525 is found in all states of methylation: unmodified (5%) and mono- (6%), di- (7%), and trimethylated (82%) (Fig. 3E). Importantly, after incubation of the lysate from AdOx-treated cells with FAM86A, Lys-525 became 100% trimethylated, demonstrating that FAM86A is capable of methylating this site in human eEF2. Recombinant yeast Yjr129c could also methylate human eEF2 to full trimethylation. Interestingly, no radiolabeled eEF2 band was visible in the AdOx-free samples after treatment with either MTase, confirming that eEF2 in these cells is indeed fully methylated at Lys-525, as suggested by the MS analysis (Fig. 3, D and E). In summary, these results identify Lys-525 in human eEF2 as target of trimethylation and strongly indicate that human FAM86A catalyzes the corresponding methylation reaction in vivo.
Biological Significance of eEF2 Methylation
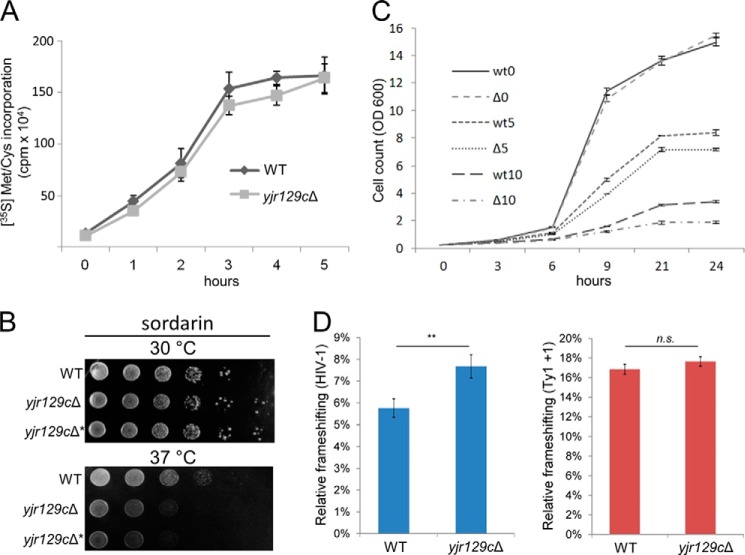
In order to evaluate the biological role of eEF2 methylation, we searched for possible phenotypes of the yjr129cΔ mutant yeast. The translation machinery seemed to be largely unaffected because the growth rate of the yjr129cΔ strain was comparable with that of WT yeast on YPD both at the optimal growth temperature of 30 °C and at 25 and 37 °C (data not shown). We have also checked whether total protein synthesis was affected in the yjr129cΔ yeast by measuring the rate of incorporation of 35S-labeled methionine and cysteine amino acids into polypeptides in the WT versus knock-out strain (Fig. 4A). However, no significant difference in translation rate was observed, indicating that methylation of Lys-509 is not critical for efficient protein translation, at least under the growth conditions investigated. When comparing growth on YPD plates containing drugs that inhibit protein translation, the yjr129cΔ mutant did not exhibit altered sensitivity to rapamycin, cycloheximide, nourseothricin, and hygromycin at 30 °C or to paromomycin and anisomycin at either 30 or 37 °C (data not shown).
FIGURE 4.
Phenotypic analysis of the yjr129cΔ yeast strain. A, comparison of overall protein synthesis between WT and yjr129cΔ yeast strains. Cultures were grown to the same A600 in SD −Met/−Cys medium; a mix of 35S-labeled methionine and cysteine was added; and total protein synthesis was measured at each time point by TCA precipitation of total protein, followed by scintillation counting. Error bars, S.D. (n = 4). B, hypersensitivity of the yjr129cΔ yeast strain to sordarin on solid medium at 37 °C. Dilutions of WT yeast cells and two independently derived yjr129cΔ mutants (JEY9581 (yjr129cΔ) and JEY10800 (yjr129cΔ*)) were grown on YPD plates containing 5 μg/ml sordarin at 30 or 37 °C for 3 days. C, hypersensitivity of the yjr129cΔ yeast strain to sordarin in liquid medium. The graph shows the growth of the WT and yjr129cΔ (JEY9581) strains in liquid YPD medium containing 0 μg/ml (wt0 and Δ0), 5 μg/ml (wt5 and Δ5), or 10 μg/ml (wt10 and Δ10) sordarin over a period of 24 h at 37 °C. Error bars, range of duplicates. D, dual luciferase assays investigating the effects of YJR129C deletion on ribosomal frameshifting. YJR129C deletion significantly alters programmed ribosomal frameshifting of the HIV-1 gag-pol but not the Ty1 +1 site, relative to the luciferase activity of control sequences, which was unaffected. n = 3; mean ± S.D.; **, p < 0.01; n.s., not significant.
However, at 37 °C, but not at 30 °C, the mutant strain was more sensitive than the WT to the antifungal eEF2-specific translation inhibitor sordarin (Fig. 4B). Sordarin is a naturally occurring antifungal compound that binds to eEF2 in a cleft in close proximity to Lys-509 and negatively impacts translation by locking eEF2 onto the ribosome (38, 39). Altered sordarin sensitivity was confirmed by monitoring the rate of growth of the yjr129cΔ mutant in liquid YPD medium containing 0, 5, or 10 μg/ml sordarin. At 37 °C, the mutant strain exhibited a considerably slower growth rate than the WT in the presence of sordarin but not in YPD alone (Fig. 4C).
To investigate whether methylation may have an effect on the accuracy of translation in yeast, we used a dual luciferase assay measuring programmed −1 ribosomal frameshifting of the human immunodeficiency virus (HIV-1) gag-pol site, or programmed +1 ribosomal frameshifting of the yeast Ty1 retrotransposable element (32). In this assay, Renilla and firefly luciferase cDNAs are separated by a frameshifting site or a control sequence (33); their relative ratio allows quantification of ribosomal frameshifting. The control sequence was used to account for effects of the insert sequence on luciferase stability/activity or potential differences in ribosomal processivity. Fig. 4D shows the relative percentage in frameshift as compared with controls. The accuracy of translation was unaltered in the Ty1 +1 context, yet the yjr129cΔ mutant displayed increased ribosomal frameshifting in the HIV-1 context, suggesting that Lys-509 methylation may maintain translational accuracy by preventing −1 ribosomal frameshift.
DISCUSSION
In this work, we have established that the previously uncharacterized human MTase FAM86A specifically trimethylates Lys-525 in eEF2 and that the yeast enzyme Yjr129c targets the corresponding residue in yeast eEF2 (Lys-509). Moreover, Yjr129c-deficient yeast displayed increased ribosomal frameshifting and sensitivity toward the eEF2-specific drug sordarin, indicating that this methylation is functionally important. In yeast, two MTases acting on translation elongation factors have been identified previously (Efm1 and Efm2) (18, 40), and accordingly, we rename Yjr129c as Efm3 (elongation factor methyltransferase 3); based on the human lysine MTase nomenclature, we redub FAM86A as eEF2-KMT.
The modified lysine in eEF2 lies in domain III of the protein, on the outer surface of a highly conserved α-helix (39), and appears to be present in eEF2 homologues from all eukaryotic organisms. eEF2-KMT homologues are also predicted to exist across the eukaryotic branch of the evolutionary tree, and the conservation of both the methylated residue and the methyltransferase suggests an important role for the modification. Interestingly, although the translational machinery is highly conserved between the prokaryotes and eukaryotes, the modified lysine is absent in the prokaryotic eEF2 homologue, EF-G, and prokaryotes also do not appear to have an MTase with homology to eEF2-KMT.
Previously, we and others have unraveled the function of the four human MTF16 members C2orf34 (CaM-KMT), METTL21D (VCP-KMT), METTL21A (HSPA-KMT), and METTL22 (KIN-KMT), showing that these enzymes target specific Lys residues in calmodulin, valosin-containing protein (VCP), 70-kDa heat-shock proteins (HSPA), and the DNA repair protein Kin17 (KIN), respectively (9–12). Thus, the five human MTF16 members characterized thus far (including eEF2-KMT) are lysine-specific, and it is likely that this activity is shared by some of the five remaining uncharacterized human proteins in this family. However, S. cerevisiae Yil110w, which is the putative yeast orthologue of one such protein, METTL18, was recently shown to be a histidine-specific MTase targeting the ribosomal protein Rpl3, indicating that the human MTF16 may not exclusively comprise KMTs (41).
The human MTF16 enzymes characterized thus far share some interesting features; they appear to target a single lysine residue in a single protein (or in a group of highly similar paralogues), and they have a strong affinity for their targets, which are highly abundant cellular proteins. Regarding the latter, the methods used are biased toward the identification of abundant substrates, so one should not exclude the possibility that additional substrates can be found among other, less abundant proteins. On a different note, it is interesting that Efm3 and eEF2-KMT proved to be functional orthologues despite their limited sequence similarity (27% identity). For example, human HSPA-KMT and VCP-KMT, which are considerably more similar (37% identity), have different functions, underscoring that prediction of functional orthology from sequence similarity is non-trivial.
Yeast carrying inactivating mutations in genes in the evolutionarily conserved diphthamide biosynthesis pathway (DPH1-DPH7), are devoid of the diphthamide modification in eEF2. Most of these mutants lack an obvious growth or protein synthesis phenotype, but, like the efm3Δ mutant, they exhibit increased ribosomal frameshifting and display altered sensitivity to certain translation elongation indicator drugs, especially under thermal stress (42). In mice, however, the loss of single DPH genes leads to abnormal embryonic development and even lethality (43, 44). No studies have reported any phenotypes or genetic disorders associated with eEF2-KMT inactivation, but it was recently reported that inactivation of another, yet uncharacterized, human MTF16 member, METTL23, caused intellectual disability (45, 46).
Although we did not directly investigate whether the GTPase activity of eEF2 was influenced by lysine methylation, the overall protein synthesis rate was unaffected in the efm3Δ mutant yeast, suggesting that GTPase activity is unlikely to be significantly affected. Similarly, previous studies have found that methylation of two chaperone ATPases, Hsp70 and VCP, by their respective MTF16 KMTs did not greatly affect their ATPase activity (10, 12).
The efm3Δ mutant yeast displayed hypersensitivity toward the naturally occurring antifungal translation inhibitor sordarin, which specifically binds to eEF2 from certain species of fungi and locks it on the ribosome, stalling protein synthesis and leading to cell death. This indicates that the methylation state of Lys-509 affects the interaction of S. cerevisiae eEF2 with sordarin, and, accordingly, sensitivity toward this drug has been found to be determined by the sequence of amino acids in the “sordarin specificity region” (residues 517–524 in S. cerevisiae eEF2) at the entrance to the sordarin binding cleft (47), which is in close proximity to the Lys-509 in the three-dimensional structure of eEF2 (39).
When examining the positioning of the methylated lysine in various structures of eEF2 in complex with the ribosome, it becomes clear that this residue is facing the ribosomal protein RPS23, where it may contact a conserved asparagine residue (Asn-99 in S. cerevisiae). Interestingly, RPS23 homologues from E. coli and S. cerevisiae have long been established as “accuracy centers” of the ribosome, with various mutations contributing to either increased or decreased accuracy of translation (48, 49), and, recently, the hydroxylation of RPS23 was found to be critical for translational fidelity (33). Together with our observation that the lack of Lys-509 methylation in yeast eEF2 promotes −1 ribosomal frameshifting, this suggests that the eEF2-RPS23 interaction may indeed be affected in the efm3Δ mutant yeast and that eEF2-KMT/Efm3-mediated methylation of eEF2 may play an important role in optimizing its interaction with RPS23.
Methylation of ribosomal proteins and other proteins involved in translation is common in both eukaryotes and prokaryotes, and some of these methylations appear to be dynamic and may be of regulatory importance. For example, the extent of ribosomal protein methylation has been shown to vary with the cell cycle in synchronized HeLa cells (50), and the degree of monomethylation of ribosomal protein RPL29 at a lysine residue has been shown to vary in different rat tissues, from almost complete monomethylation in the brain to only 27% methylation in the liver (51). Also, eEF1A, which is responsible for delivering tRNAs to the ribosome, displays altered lysine methylation status during germination in Mucor racemosus (52) and in 3T3B mouse cells upon transformation with the simian virus SV40 (16), suggesting that methylation of this elongation factor may be of regulatory importance. Although we detected eEF2 exclusively in the trimethylated state in most of the tested mammalian cells and tissues, we found eEF2 to be largely hypomethylated in the rat brain. Clearly, future research should address whether this variability in eEF2 methylation reflects a regulatory mechanism. Moreover, numerous lysine-specific demethylases have been discovered during the last decade, targeting primarily histones but also other proteins, and it will be of great interest to study if any of these can demethylate eEF2.
While the present work was in the process of being submitted for publication, an article appeared in the literature reporting findings similar to part of the current work, showing that Yjr129c from S. cerevisiae is responsible for methylating Lys-509 in yeast eEF2 (53).
Research in the laboratory of P. Ø. F. is supported by the University of Oslo and by grants from the Norwegian Cancer Society and the Research Council of Norway.
- MTase
- methyltransferase
- eEF2
- eukaryotic translation elongation factor 2
- eEF1A
- eukaryotic translation elongation factor 1A
- 7BS
- seven-stranded β-sheet
- MTF16
- methyltransferase family 16
- KMT
- lysine methyltransferase
- AdoMet
- S-adenosyl-l-methionine
- CBP
- calmodulin binding peptide
- SBP
- streptavidin binding peptide
- TAP
- tandem affinity purification
- AdoHcy
- S-adenosyl-l-homocysteine
- AdOx
- adenosine dialdehyde
- VCP
- valosin-containing protein.
REFERENCES
- 1. Petrossian T. C., Clarke S. G. (2011) Uncovering the human methyltransferasome. Mol. Cell. Proteomics 10, 10.1074/mcp.M110.000976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Thayer J. S. (2002) Review: biological methylation of less-studied elements. Appl. Organomet. Chem. 16, 677–691 [Google Scholar]
- 3. Nyman T., Schüler H., Korenbaum E., Schutt C. E., Karlsson R., Lindberg U. (2002) The role of MeH73 in actin polymerization and ATP hydrolysis. J. Mol. Biol. 317, 577–589 [DOI] [PubMed] [Google Scholar]
- 4. Figaro S., Scrima N., Buckingham R. H., Heurgué-Hamard V. (2008) HemK2 protein, encoded on human chromosome 21, methylates translation termination factor eRF1. FEBS Lett. 582, 2352–2356 [DOI] [PubMed] [Google Scholar]
- 5. Sprung R., Chen Y., Zhang K., Cheng D., Zhang T., Peng J., Zhao Y. (2008) Identification and validation of eukaryotic aspartate and glutamate methylation in proteins. J. Proteome Res. 7, 1001–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Huang J., Berger S. L. (2008) The emerging field of dynamic lysine methylation of non-histone proteins. Curr. Opin. Genet. Dev. 18, 152–158 [DOI] [PubMed] [Google Scholar]
- 7. Greer E. L., Shi Y. (2012) Histone methylation: a dynamic mark in health, disease and inheritance. Nat. Rev. Genet. 13, 343–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fauman E. B., Blumenthal R. M., Cheng X. (1999) Structure and evolution of AdoMet-dependent methyltransferases. in S-Adenosylmethionine-dependent Methyltransferases: Structures and Functions (Cheng X., Blumenthal R. M., ed) pp. 1–38, World Scientific Publishing, Singapore [Google Scholar]
- 9. Magnani R., Dirk L. M. A., Trievel R. C., Houtz R. L. (2010) Calmodulin methyltransferase is an evolutionarily conserved enzyme that trimethylates Lys-115 in calmodulin. Nat. Commun. 1, 43. [DOI] [PubMed] [Google Scholar]
- 10. Kernstock S., Davydova E., Jakobsson M., Moen A., Pettersen S., Mælandsmo G. M., Egge-Jacobsen W., Falnes P. Ø. (2012) Lysine methylation of VCP by a member of a novel human protein methyltransferase family. Nat. Commun. 3, 1038. [DOI] [PubMed] [Google Scholar]
- 11. Cloutier P., Lavallée-Adam M., Faubert D., Blanchette M., Coulombe B. (2013) A newly uncovered group of distantly related lysine methyltransferases preferentially interact with molecular chaperones to regulate their activity. PLoS Genet. 9, e1003210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jakobsson M. E., Moen A., Bousset L., Egge-Jacobsen W., Kernstock S., Melki R., Falnes P. Ø. (2013) Identification and characterization of a novel human methyltransferase modulating Hsp70 protein function through lysine methylation. J. Biol. Chem. 288, 27752–27763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chang F. N., Navickas I. J., Chang C. N., Dancis B. M. (1976) Methylation of ribosomal proteins in HeLa cells. Arch. Biochem. Biophys. 172, 627–633 [DOI] [PubMed] [Google Scholar]
- 14. Kruiswijk T., Kunst A., Planta R. J., Mager W. H. (1978) Modification of yeast ribosomal proteins methylation. Biochem. J. 175, 221–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hiatt W. R., Garcia R., Merrick W. C., Sypherd P. S. (1982) Methylation of elongation factor 1 α from the fungus Mucor. Proc. Natl. Acad. Sci. U.S.A. 79, 3433–3437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Coppard N. J., Clark B. F. C., Cramer F. (1983) Methylation of elongation factor 1α in mouse 3T3B and 3T3BSV40 cells. FEBS Lett. 164, 330–334 [DOI] [PubMed] [Google Scholar]
- 17. Polevoda B., Sherman F. (2007) Methylation of proteins involved in translation. Mol. Microbiol. 65, 590–606 [DOI] [PubMed] [Google Scholar]
- 18. Lipson R. S., Webb K. J., Clarke S. G. (2010) Two novel methyltransferases acting upon eukaryotic elongation factor 1A in Saccharomyces cerevisiae. Arch. Biochem. Biophys. 500, 137–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ryazanov A. G., Davydova E. K. (1989) Mechanism of elongation factor 2 (EF-2) inactivation upon phosphorylation. Phosphorylated EF-2 is unable to catalyze translocation. FEBS Lett. 251, 187–190 [DOI] [PubMed] [Google Scholar]
- 20. Su X., Lin Z., Lin H. (2013) The biosynthesis and biological function of diphthamide. Crit. Rev. Biochem. Mol. Biol. 48, 515–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Oppenheimer N. J., Bodley J. W. (1981) Diphtheria toxin: site and configuration of ADP-ribosylation of diphthamide in elongation factor 2. J. Biol. Chem. 256, 8579–8581 [PubMed] [Google Scholar]
- 22. Zhang Y., Liu S., Lajoie G., Merrill A. R. (2008) The role of the diphthamide-containing loop within eukaryotic elongation factor 2 in ADP-ribosylation by Pseudomonas aeruginosa exotoxin A. Biochem. J. 413, 163–174 [DOI] [PubMed] [Google Scholar]
- 23. Kaul G., Pattan G., Rafeequi T. (2011) Eukaryotic elongation factor-2 (eEF2): its regulation and peptide chain elongation. Cell Biochem. Funct. 29, 227–234 [DOI] [PubMed] [Google Scholar]
- 24. Leprivier G., Remke M., Rotblat B., Dubuc A., Mateo A. R., Kool M., Agnihotri S., El-Naggar A., Yu B., Somasekharan S. P., Faubert B., Bridon G., Tognon C. E., Mathers J., Thomas R., Li A., Barokas A., Kwok B., Bowden M., Smith S., Wu X., Korshunov A., Hielscher T., Northcott P. A., Galpin J. D., Ahern C. A., Wang Y., McCabe M. G., Collins V. P., Jones R. G., Pollak M., Delattre O., Gleave M. E., Jan E., Pfister S. M., Proud C. G., Derry W. B., Taylor M. D., Sorensen P. H. (2013) The eEF2 kinase confers resistance to nutrient deprivation by blocking translation elongation. Cell 153, 1064–1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. McGinnis S., Madden T. L. (2004) BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res. 32, W20–W25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Flicek P., Amode M. R., Barrell D., Beal K., Billis K., Brent S., Carvalho-Silva D., Clapham P., Coates G., Fitzgerald S., Gil L., Girón C. G., Gordon L., Hourlier T., Hubbard T. J., Hunt S., Johnson N., Juettemann T., Kähäri A. K., Keenan S., Kulesha E., Martin F. J., Maurel T., McLaren W. M., Murphy D. N., Nag R., Overduin B., Pignatelli M., Pritchard B., Pritchard E., Riat H. S., Ruffier M., Sheppard D., Taylor K., Thormann A., Trevanion S. J., Vullo A., Wilder S. P., Wilson M., Zadissa A., Aken B. L., Birney E., Cunningham F., Harrow J., Herrero J., Hubbard T. J. P., Kinsella R., Muffato M., Parker A., Spudich G., Yates A., Zerbino D. R. (2014) Ensembl 2014. Nucleic Acids Res. 42, D749–D755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Waterhouse A. M., Procter J. B., Martin D. M., Clamp M., Barton G. J. (2009) Jalview Version 2: a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. McGuffin L. J., Bryson K., Jones D. T. (2000) The PSIPRED protein structure prediction server. Bioinformatics 16, 404–405 [DOI] [PubMed] [Google Scholar]
- 29. Chen T., Nayak N., Majee S. M., Lowenson J., Schäfermeyer K. R., Eliopoulos A. C., Lloyd T. D., Dinkins R., Perry S. E., Forsthoefel N. R., Clarke S. G., Vernon D. M., Zhou Z. S., Rejtar T., Downie A. B. (2010) Substrates of the Arabidopsis thaliana protein isoaspartyl methyltransferase 1 identified using phage display and biopanning. J. Biol. Chem. 285, 37281–37292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jørgensen R., Carr-Schmid A., Ortiz P. A., Kinzy T. G., Andersen G. R. (2002) Purification and crystallization of the yeast elongation factor eEF2. Acta Crystallogr. D Biol. Crystallogr. 58, 712–715 [DOI] [PubMed] [Google Scholar]
- 31. Gietz R. D., Schiestl R. H. (2007) Frozen competent yeast cells that can be transformed with high efficiency using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2, 1–4 [DOI] [PubMed] [Google Scholar]
- 32. Jacks T., Power M. D., Masiarz F. R., Luciw P. A., Barr P. J., Varmus H. E. (1988) Characterization of ribosomal frameshifting in HIV-1 gag-pol expression. Nature 331, 280–283 [DOI] [PubMed] [Google Scholar]
- 33. Loenarz C., Sekirnik R., Thalhammer A., Ge W., Spivakovsky E., Mackeen M. M., McDonough M. A., Cockman M. E., Kessler B. M., Ratcliffe P. J., Wolf A., Schofield C. J. (2014) Hydroxylation of the eukaryotic ribosomal decoding center affects translational accuracy. Proc. Natl. Acad. Sci. U.S.A. 111, 4019–4024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Darai-Ramqvist E., Sandlund A., Müller S., Klein G., Imreh S., Kost-Alimova M. (2008) Segmental duplications and evolutionary plasticity at tumor chromosome break-prone regions. Genome Res. 18, 370–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Petrossian T. C., Clarke S. G. (2009) Multiple motif scanning to identify methyltransferases from the yeast proteome. Mol. Cell Proteomics 8, 1516–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Katz J. E., Dlakić M., Clarke S. (2003) Automated identification of putative methyltransferases from genomic open reading frames. Mol. Cell Proteomics 2, 525–540 [DOI] [PubMed] [Google Scholar]
- 37. Bartel R. L., Borchardt R. T. (1984) Effects of adenosine dialdehyde on S-adenosylhomoysteine hydrolase and S-adenosylmethionine-dependent transmethylations in mouse L929 cells. Mol. Pharmacol. 25, 418–424 [PubMed] [Google Scholar]
- 38. Justice M. C., Hsu M. J., Tse B., Ku T., Balkovec J., Schmatz D., Nielsen J. (1998) Elongation factor 2 as a novel target for selective inhibition of fungal protein synthesis. J. Biol. Chem. 273, 3148–3151 [DOI] [PubMed] [Google Scholar]
- 39. Jørgensen R., Ortiz P. A., Carr-Schmid A., Nissen P., Kinzy T. G., Andersen G. R. (2003) Two crystal structures demonstrate large conformational changes in the eukaryotic ribosomal translocase. Nat. Struct. Biol. 10, 379–385 [DOI] [PubMed] [Google Scholar]
- 40. Couttas T. A., Raftery M. J., Padula M. P., Herbert B. R., Wilkins M. R. (2012) Methylation of translation-associated proteins in Saccharomyces cerevisiae: identification of methylated lysines and their methyltransferases. Proteomics 12, 960–972 [DOI] [PubMed] [Google Scholar]
- 41. Webb K. J., Zurita-Lopez C. I., Al-Hadid Q., Laganowsky A., Young B. D., Lipson R. S., Souda P., Faull K. F., Whitelegge J. P., Clarke S. G. (2010) A novel 3-methylhistidine modification of a yeast ribosomal protein Rpl3 is dependednt upon the YIL110W methyltransferase. J. Biol. Chem. 285, 37598–37606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Uthman S., Bär C., Scheidt V., Liu S., ten Have S., Giorgini F., Stark M. J., Schaffrath R. (2013) The amidation step of diphthamide biosynthesis in yeast requires DPH6, a gene identified through mining the DPH1-DPH5 interaction network. PLOS Genet. 9, e1003334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Liu S., Wiggins J. F., Sreenath T., Kulkarni A. B., Ward J. M., Leppla S. H. (2006) Dph3, a small protein required for diphthamide biosynthesis, is essential in mouse development. Mol. Cell Biol. 26, 3835–3841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Webb T. R., Cross S. H., McKie L., Edgar R., Vizor L., Harrison J., Peters J., Jackson I. J. (2008) Diphthamide modification of eEF2 requires a J-domain protein and is essential for normal development. J. Cell Sci. 121, 3140–3145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Reiff R. E., Ali B. R., Baron B., Yu T. W., Ben-Salem S., Coulter M. E., Schubert C. R., Hill R. S., Akawi N. A., Al-Younes B., Kaya N., Evrony G. D., Al-Saffar M., Felie J. M., Partlow J. N., Sunu C. M., Schembri-Wismayer P., Alkuraya F. S., Meyer B. F., Walsh C. A., Al-Gazali L., Mochida G. H. (2014) METTL23, a transcriptional partner of GABPA, is essential for human cognition. Hum. Mol. Genet. 23, 3456–3466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bernkopf M., Webersinke G., Tongsook C., Koyani C. N., Rafiq M. A., Ayaz M., Müller D., Enzinger C., Aslam M., Naeem F., Schmidt K., Gruber K., Speicher M. R., Malle E., Macheroux P., Ayub M., Vincent J. B., Windpassinger C., Duba H. C. (2014) Disruption of the methyltransferase-like 23 gene METTL23 causes mild autosomal recessive intellectual disability. Hum. Mol. Gen. 23, 4015–4023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Shastry M., Nielsen J., Ku T., Hsu M. J., Liberator P., Anderson J., Schmatz D., Justice M. C. (2001) Species-specific inhibition of fungal protein synthesis by sordarin: identification of a sordarin-specificity region in eukaryotic elongation factor 2. Microbiology 147, 383–390 [DOI] [PubMed] [Google Scholar]
- 48. Alksne L. E., Anthony R. A., Liebman S. W., Warner J. R. (1993) An accuracy center in the ribosome conserved over 2 billion years. Proc. Natl. Acad. Sci. U.S.A. 90, 9538–9541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Synetos D., Frantziou C. P., Alksne L. E. (1996) Mutations in yeast ribosomal proteins S28 and S4 affect the accuracy of translation and alter the sensitivity of the ribosomes to paromomycin. Biochim. Biophys. Acta 1309, 156–166 [DOI] [PubMed] [Google Scholar]
- 50. Chang F. N., Navickas I. J., Au C., Budzilowicz C. (1978) Identification of the methylated ribosomal proteins in HeLa cells and the fluctuation of methylation during the cell cycle. Biochim. Biophys. Acta 518, 89–94 [DOI] [PubMed] [Google Scholar]
- 51. Williamson N. A., Raliegh J., Morrice N. A., Wettenhall R. E. (1997) Post-translational processing of rat ribosomal proteins. Ubiquitous methylation of Lys22 within the zinc-finger motif of RL40 (carboxy-terminal extension protein 52) and tissue-specific methylation of Lys4 in RL29. Eur. J. Biochem. 246, 786–793 [DOI] [PubMed] [Google Scholar]
- 52. Fonzi W. A., Katayama C., Leathers T., Sypherd P. S. (1985) Regulation of protein synthesis factor EF-1 α in Mucor racemosus. Mol. Cell Biol. 5, 1100–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhang L., Hamey J. J., Hart-Smith G., Erce M. A., Wilkins M. R. (2014) Elongation factor methyltransferase 3: a novel eukaryotic lysine methyltransferase. Biochem. Biophys. Res. Commun. 451, 229–234 [DOI] [PubMed] [Google Scholar]