Background: Post-translational modification is involved in regulating IRE1 signaling.
Results: CHIP ubiquitinated IRE1 and regulated the IRE1/TRAF2/JNK pathway.
Conclusion: Ubiquitination is a post-translational modification that is required for IRE1 signal transduction.
Significance: This study may provide the first evidence that the IRE1/TRAF2/JNK pathway is regulated by ubiquitination.
Keywords: ER Stress, Senescence, Signal Transduction, Ubiquitin Ligase, Unfolded Protein Response (UPR), CHIP, IRE1
Abstract
Deciphering the inositol-requiring enzyme 1 (IRE1) signaling pathway is fundamentally important for understanding the unfolded protein response (UPR). The ubiquitination of proteins residing on the endoplasmic reticulum (ER) membrane has been reported to be involved in the UPR, although the mechanism has yet to be fully elucidated. Using immunoprecipitation and mass spectrometry, IRE1 was identified as a substrate of the E3 ligase CHIP (carboxyl terminus of HSC70-interacting protein) in HEK293 cells under geldanamycin-induced ER stress. Two residues of IRE1, Lys545 and Lys828, were targeted for Lys63-linked ubiquitination. Moreover, in CHIP knockdown cells, IRE1 phosphorylation and the IRE1-TRAF2 interaction were nearly abolished under ER stress, which may be due to lacking ubiquitination of IRE1 on Lys545 and Lys828, respectively. The cellular responses were evaluated, and the data indicated that CHIP-regulated IRE1/TRAF2/JNK signaling antagonized the senescence process. Therefore, our findings suggest that CHIP-mediated ubiquitination of IRE1 contributes to the dynamic regulation of the UPR.
Introduction
In the endoplasmic reticulum (ER),3 synthesized proteins mature and are assembled for delivery to target organelles. The homeostasis of the folded proteins in the ER is monitored and maintained via a coordinated adaptive pathway termed the unfolded protein response (UPR) (1). Pathophysiological insults lead to the accumulation of unfolded proteins in the ER and cause ER stress. In response to the accumulation of unfolded proteins, cells adapt to this stressful condition via the UPR, initially to restore normal cellular function by halting protein translation and activating signaling pathways that increase the expression of molecular chaperones involved in protein folding (2, 3). In the UPR, inositol-requiring enzyme 1 (IRE1) is one of three key sensors that reside on the ER membrane. It is generally accepted that IRE1 has dual functions to cope with ER stress. On the one hand, in response to ER stress, IRE1 dimerizes to activate its cytosolic RNase domain and to cleave a 26-nucleotide intronic sequence from XBP1 (X-box DNA binding protein 1) mRNA (4). The resulting XBP1, an activator of UPR genes, activates the expression of numerous chaperone genes to improve the protein folding capacity in the ER (5, 6). On the other hand, activated IRE1 recruits TNF receptor-associated factor 2 (TRAF2) to the ER membrane and activates the pro-apoptotic ASK1 (apoptosis signal-regulating kinase)-JNK pathway (7).
The mechanisms by which IRE1 is regulated in the UPR have yet to be explored. Recently, the functions of IRE1 have been reported to be regulated via post-translational modifications such as ADP-ribosylation by poly(ADP-ribose) polymerase 16 (8), dephosphorylation by protein phosphatase 2C on endoplasmic reticulum (9), and ubiquitination by the E3 ligase HRD1 (10). These studies really have initiated greater understanding about the working mechanism of IRE1, but they have also introduced further questions. Except the proteins anchored on ER membrane, can the proteins not located on ER perform modifications to IRE1 under ER stress? Are there additional types of post-translational modifications added to IRE1 under ER stress? As IRE1 responds to ER stress through two pathways, XBP-1 splicing and IRE1/TRAF2/JNK, how does an IRE1 modification regulate them, one or both pathways? These questions prompted us to investigate IRE1 modifications more thoroughly.
CHIP (carboxyl terminus of HSC70-interacting protein) has been identified as a regulator that maintains protein homeostasis, particularly by targeting various pathological unfolded proteins in numerous tissues and cells (11). It is generally accepted that CHIP is not localized to a specific organelle, whereas it is found on the ER outer membrane (12). Many studies have shown that CHIP is an E3 ubiquitin ligase for endoplasmic reticulum-associated degradation (ERAD) (13, 14), a process involved in cell adaptation to ER stress. Because ERAD is often detected around the ER membrane, a logical deduction is that CHIP may exert some functions on the ER membrane under ER stress. However, whether CHIP participates in the UPR, the specific branch of the UPR that CHIP participates in and how the information of CHIP ubiquitination is delivered in response to ER stress are still largely unknown. In this study, we performed IP assays followed by a proteomic screen and identified one E3 ligase, CHIP, in the IRE1 immunoprecipitates. CHIP interacted with and ubiquitinated IRE1 in a Lys63-linked manner. The ubiquitination event was likely critical for regulating the IRE1/TRAF2/JNK pathway and affecting cellular senescence. Our findings suggest that the post-translational modification of IRE1 by the E3 ligase CHIP contributes to the dynamic control of IRE1 signaling in response to ER stress.
EXPERIMENTAL PROCEDURES
Vector Construction for Recombinant Expression, Site-directed Mutagenesis, and RNA Interference
For recombinant expression in HEK293 cells, full-length human IRE1, CHIP, TRAF2, OTUB1 (OTU domain-containing ubiquitin aldehyde-binding protein 1), and ubiquitin (Ub) cDNAs were amplified from HEK293 cDNA by RT-PCR and cloned into pCMV-HA and pCMV-MYC. For pulldown experiments with GST-fused recombinant protein, the CHIP gene was inserted into pGEX-4T-1 to generate the pGEX-4T-1-CHIP fusion. For in vitro ubiquitination assay with His tag-fused recombinant protein, the CHIP gene was inserted into pET-30a(+) to generate the pET-30a-CHIP fusion.
For the site-directed mutagenesis, plasmids containing the CHIP, IRE1, and Ub genes were mutated via single site-directed mutagenesis (Agilent, Santa Clara, CA) using the corresponding mutated primers that contained the indicated mutated amino acid. The mutation sites were predicted using the BDM-PUB (Computational Prediction of Protein Ubiquitination Sites with a Bayesian Discriminant Method) algorithm.
For silencing gene expression in HEK293 cells, specific synthetic oligonucleotides were designed: IRE1, 5′-GATCCGCCTGACGAAACTTCCTTTTTCAAGAGAAAAGGAAGTTTCGTCAGGCTTTTTGAAA-3′ and 5′-AGCTTTTCCAAAAAGCCTGACGAAACTTCCTTTTCTCTTGAAAAAGGAAGTTTCGTCAGGCG-3′; and CHIP, 5′-GATCCGAAGCGCTGGAACAGCATTCTCAAGAGAAATGCTGTTCCAGCGCTTCTTTTTTGGAAA-3′ and 5′-AGCTTTTCCAAAAAAGAAGCGCTGGAACAGCATTTCTCTTGAGAATGCTGTTCCAGCGCTTCG-3′. These oligonucleotides were annealed and ligated into pSilencer 3.0-H1 using the BamHI and HindIII restriction sites. All the constructed vectors were verified by sequencing.
Reagents and Cell Lines
Antibodies against IRE1 (H-190), CHIP (G-2), protein disulfide isomerase (H-160), BIP (H-128), p-JNK (G-7), actin (C4), and XBP1 (M-186) were purchased from Santa Cruz Biotechnology (Dallas, TX). Other antibodies were purchased from several commercial sources: p-IRE1 (NB100–2323) was purchased from Novus Biologicals (Littleton, CO), LC-3 (L7543) was purchased from Sigma-Aldrich, p-p38 (4511) was purchased from Cell Signaling Technology (Danvers, MA), and MYC was purchased from Beijing Protein Innovation (Beijing, China).
Geldanamycin (G3381) and cycloheximide (C4859) were purchased from Sigma-Aldrich, Bafilomycin A1 (88899-55-2) was obtained from Fermentek (Jerusalem, Israel), and SB203580 (S1863) and SP600125 (S1876) were purchased from Beyotime (Haimen, China). HEK293 cells were obtained from American Type Culture Collection and were cultured in RPMI 1640 (Invitrogen) supplemented with 10% inactivated FBS, 100 units/ml penicillin, and 100 μg/ml streptomycin at 37 °C and 5% CO2.
Immunofluorescent Confocal Microscopy
HEK293 cells were placed on glass cover slips, and subconfluent cells were treated. The treated and untreated cells were fixed in 3.7% paraformaldehyde and permeabilized with 0.3% Triton X-100 in PBS at room temperature. The indicated primary antibodies diluted in PBS containing 1% bovine serum albumin and 0.3% Triton X-100 were incubated with the cells at 4 °C overnight. The secondary antibodies, goat anti-rabbit IgG-TRITC, or goat anti-mouse IgG-FITC (Santa Cruz Biotechnology), were incubated with the cells for 1 h at room temperature. All of the labeled cells were examined using a Leica TCSSP5 confocal fluorescence imaging microscope (Leica Microsystems, Wetzler, Germany) equipped with a 63× oil immersion objective with a numerical aperture of 1.4 at room temperature. The confocal images were acquired using LAS AF software, version 2.0 (Leica Microsystems). DAPI was used to stain the nuclei.
Western Blot
The lysates or affinity beads were boiled in 20 μl of Laemmli buffer containing 50 mm Tris-HCl, pH 6.8, 30% glycerol, 4% SDS, and 1% β-mercaptoethanol. The samples were subjected to 8% or 10% SDS-PAGE, and the electrophoresed gels were electrotransferred onto polyvinylidene difluoride membranes (Millipore, Billerica, MA). The blotted membranes were probed with the indicated primary antibodies at various dilutions and then incubated with HRP-conjugated secondary antibodies. The immunorecognition signals were detected using ECL and were acquired using ImageQuant ECL (GE Healthcare).
Immunoprecipitation and Pulldown to Capture Associated Proteins
Transiently or stably transfected HEK293 cells were collected from culture dishes and lysed with radioimmune precipitation assay buffer, pH 7.5, containing 50 mm Tris-HCl, 150 mm NaCl, 1% Triton X-100, and a protease inhibitor mixture. For co-immunoprecipitation, the cell lysates were precleared with 25 μl of protein A/G PLUS-agarose beads (Santa Cruz Biotechnology) for 4 h and then incubated with antibody (2 μg) overnight at 4 °C, after which 25 μl of protein A/G PLUS-agarose beads were added. After washing with ice-cold radioimmune precipitation assay buffer, the proteins bound to the beads were analyzed by Western blot.
The GST-CHIP fusion protein was expressed in Escherichia coli BL21 cells, and the overexpressed recombinant fusion protein was purified by affinity chromatography with glutathione-Sepharose beads (GE Healthcare). The GST or GST-CHIP recombinant protein was incubated with the precleared HEK293 cell lysates harvested from transiently IRE1-expressing cells. Glutathione-Sepharose beads were added to the mixture, which was incubated overnight at 4 °C. After washing with ice-cold radioimmune precipitation assay buffer, the proteins bound to the beads were analyzed by Western blot.
Transient and Stable Transfection
The constructed vectors were transfected into HEK293 cells using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Six hours after transfection, the medium was refreshed, and the cells were maintained in an incubator at 37 °C in 5% CO2. Genes expression or knockdown was examined by Western blot 24–48 h after transient gene transfection in HEK293 cells. For stable gene knockdowns, the transfected cells were cultured and selected in medium containing 600 μg/ml G418. G418-resistant cell colonies were isolated for further culture, and the expression of the target gene was assessed by Western blot.
In Vitro Ubiquitination Assay
The His-CHIP fusion protein as the E3 ligase was expressed in E. coli BL21 cells, and the recombinant fusion protein was purified by affinity chromatography with nickel-nitrilotriacetic acid beads. The recombinant protein GST-IRE1 (amino acids 465–977) as the ubiquitination substrate was purchased from Sino Biological Inc. (Beijing, China). The in vitro ubiquitination assay kit was purchased from Enzo Life Sciences (Plymouth Meeting, PA). Ubiquitination was analyzed following protocols recommended by the manufacturer.
SA-β-gal Assay to Evaluate Cellular Senescence
HEK293 cells were seeded into six-well plates, and after 24 h, the cells were treated as indicated. SA-β-gal staining was performed according to the manufacturer's protocol (Cell Signaling Technology). To estimate the amount of senescence, the percentage of cells positive for SA-β-gal staining was calculated in several randomly selected representative fields (n = 10).
Identification of IRE1-interacting Proteins
The immunocomplexes precipitated with protein A/G beads bound to an IRE1 antibody were treated with Laemmli buffer and analyzed by 12% SDS-PAGE. The electrophoresed gels were visualized using silver nitrate staining and then cut into several pieces. The gel slices were dried using acetonitrile followed by alkylation with iodoacetamide and reduction with DTT. The treated gel pieces were incubated with trypsin (Promega, Madison, WI) overnight at 4 °C. The peptides generated from tryptic digestion were injected into a Dionex LC-Packings Ultimate3000 HPLC (Amsterdam, the Netherlands) mounted with a C18 column (75 μm × 150 mm) and run with a linear acetonitrile gradient (0–60%) at a flow rate of 400 nl/min. The eluted peptides were injected directly into a Q Exactive hybrid quadrupole-Orbitrap MS (Thermo Fisher, Bremen, Germany) using nanoESI spray at an ion spray of 1800 V. Intact masses were measured at a resolution of 70,000 (at m/z 200) in the Orbitrap using an AGC target value of 3 × 106 charges. The top 20 peptide signals (charge states 2+ and higher) were submitted to MS/MS in the HCD cell (m/z 2 isolation width, 27% normalized collision energy). The output files from the LC-MS/MS analyses were converted to mgf files using Protein Discovery 1.3 (Thermo Fisher, Bremen, Germany) and delivered to the Mascot 2.3 search engine (Matrix Science, Boston, MA), in which the data were queried against the Swiss-Prot database 57.15 for human proteins with the following search parameters: peptide mass tolerance, 15 ppm; fragment mass tolerance, 20 mmu; methionine oxidation and the amino terminus of peptides changed from Gln to pyroGlu as variable modifications; and carbamidomethylation of cysteine as a static modification.
Statistical Analysis
All experimental data were generated from experiments that were performed at least in triplicate, and the corresponding data are presented as the means ± S.D. The significant differences between two groups were determined using Student's t test, and the null hypothesis was rejected in cases of p < 0.05. The quantitative evaluation of the Western blot images was performed using ImageQuant TL software (Piscataway, NJ).
RESULTS
CHIP Interacted with IRE1
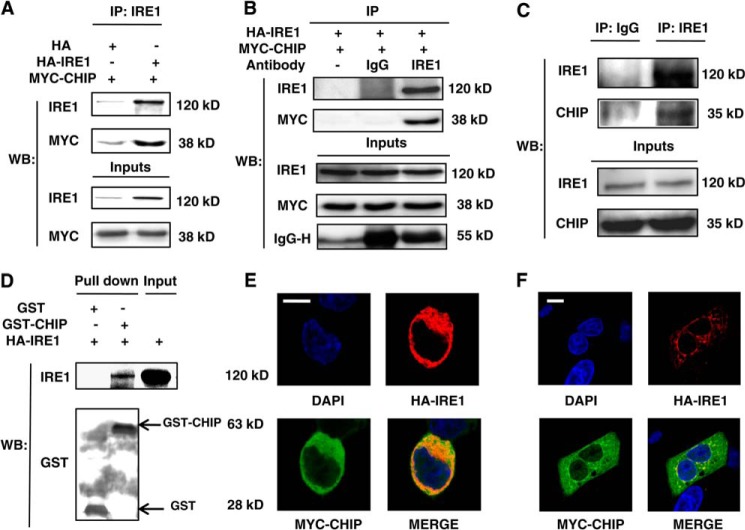
In HEK293 cells with IRE1 overexpression, the IRE1 antibody was used to immunoprecipitate protein complexes associated with IRE1. The immunoprecipitated proteins were separated by SDS-PAGE and stained with silver nitrate. All the stained bands were excised, digested with trypsin, and delivered to LC-MS/MS for protein identification. As shown in Table 1, five ubiquitination-related proteins were solely identified in the immunoprecipitates with the IRE1 antibody: three E3 ligase proteins (CHIP and two SCF (Skp1-Cullin-F-box) subunits (FBXL21 and FBXO11)) and two deubiquitinases (OTUB1 and UBHL1). This finding indicates that ubiquitination may be a critical modification on IRE1. The detection of CHIP is intriguing based on its function in ERAD. We conducted co-immunoprecipitation assays to confirm the interaction of CHIP and IRE1 found by mass spectrometry. CHIP was found in the IRE1 immunoprecipitates from the cells co-transfected with HA-IRE1 but was limitedly detected in the cells transfected with CHIP alone (Fig. 1A). When IgG was used as an immunoprecipitation reagent in the same experiment, no CHIP was detected by the MYC antibody (Fig. 1B). Furthermore, endogenous IRE1 and CHIP were co-immunoprecipitated using an IRE1 antibody and normal IgG as the control (Fig. 1C). Next, GST and recombinant GST-CHIP were utilized to pull down overexpressed IRE1 in HEK293 cells. As depicted in Fig. 1D, the immunosignal against IRE1 was present only in the pulldown products of GST-CHIP. Finally, the co-localization of CHIP and IRE1 in cells was examined by confocal microscopy, which implicates that CHIP partially co-localizes with IRE1 (Fig. 1E). Because the nucleus occupies two-thirds of the volume of the entire HEK293 cell, we chose another cell line, A549, to repeat the co-localization experiment. The confocal images depicted in Fig. 1F demonstrated that IRE1 was present in the region around the nucleus where the ER was located, whereas CHIP was found throughout the cytoplasm with certain distribution patterns. Moreover, CHIP was enriched in some areas, and these areas co-localized with IRE1 as well. These results indicate that CHIP interacts with IRE1.
TABLE 1.
Ubiquitination-related proteins in IRE1 immunoprecipitates
The IRE1 immunoprecipitates were analyzed by LC/MS-MS. Five ubiquitination-related proteins were identified and are listed according to their scores.
| Protein name | Accession number | Unique peptide | Peptide sequence | Spectrum | Theoretical molecular mass |
|---|---|---|---|---|---|
| kDa | |||||
| CHIP | Q9UNE7 | 2 | LNFGDDIPSALR | 5 | 35 |
| SPLTQEQLIPNLAMK | |||||
| OTUB1 | Q96FW1 | 1 | IQQEIAVQNPLVSER | 1 | 29 |
| UCHL1 | P09936 | 2 | QFLSETEK | 3 | 25 |
| QIEELKGQEVSPK | |||||
| FBXL21 | Q9UKT6 | 1 | FVRLCER | 3 | 48 |
| FBXO11 | Q86XK2 | 2 | LPDEVVLK | 6 | 102 |
| DNKIMNNQDAIEK |
FIGURE 1.
CHIP interacted with IRE1. A, the lysates of HEK293 cells co-transfected with HA-IRE1/MYC-CHIP or HA-EMPTY/MYC-CHIP were immunoprecipitated with an IRE1 antibody and analyzed by Western blot with antibodies against IRE1 and MYC. B, the lysates of HEK293 cells co-transfected with HA-IRE1/MYC-CHIP were immunoprecipitated with IgG, IRE1 antibody, or without antibody, respectively. The immunoprecipitated proteins were analyzed by Western blot with antibodies against IRE1 and MYC. C, HEK293 cell lysates were immunoprecipitated with an IRE1 antibody or normal IgG (control), followed by Western blot with antibodies against IRE1 and CHIP. D, HA-IRE1-transfected HEK293 cell lysates were pulled down using GST-CHIP or GST (25 μg), followed by Western blot with antibodies against IRE1 and GST. E, confocal microscopy images were obtained from HEK293 cells co-transfected with HA-IRE1 (red) and MYC-CHIP (green); the nuclei were imaged by DAPI staining (blue). Bar, 10 μm. F, confocal microscopy images were obtained from A549 cells co-transfected with HA-IRE1 (red) and MYC-CHIP (green); the nuclei were imaged by DAPI staining (blue). Bar, 10 μm. IP, immunoprecipitation; WB, Western blot.
CHIP Ubiquitinated IRE1
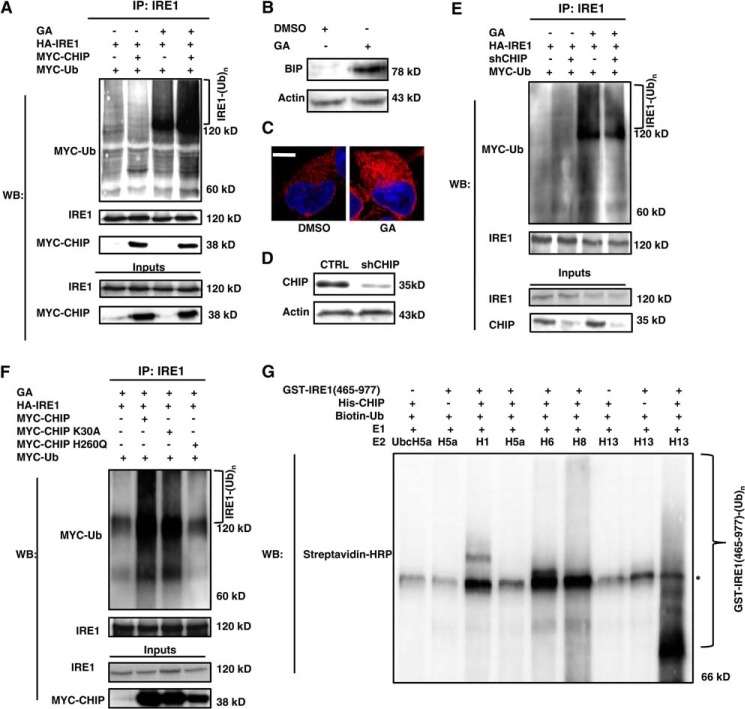
IRE1 is a substrate of an E3 ligase, and the ubiquitination of IRE1 could increase its degradation rate (10). Because CHIP is a typical E3 ligase that ubiquitinates its substrates and regulates their turnover rates, we inquired whether CHIP ubiquitinated IRE1. In transfected cells under normal conditions, IRE1 was not obviously ubiquitinated, regardless of whether the cells were overexpressing CHIP (Fig. 2A). Because IRE1 is a transducer of and is activated in response to ER stress, we utilized geldanamycin (GA) (15) to induce ER stress; the effects of GA were confirmed using the ER stress indicators BIP (binding immunoglobulin protein) and protein disulfide isomerase (Fig. 2, B and C). In response to ER stress, IRE1 was significantly ubiquitinated, and this ubiquitination was augmented by CHIP overexpression (Fig. 2A). Considering the possibility of CHIP autoubiquitination as reported by Woo et al. (16), we performed experiments to test whether ER stress could induce CHIP autoubiquitination to rule out the possibility that the ubiquitination bands found in Fig. 2A were the ubiquitinated forms of CHIP. The results indicate that GA could not induce CHIP autoubiquitination (data not shown). Next, we transfected the CHIP shRNA vector into HEK293 cells and obtained CHIP knockdown stable cell lines (Fig. 2D). The ubiquitination of IRE1 was markedly attenuated by endogenous CHIP knockdown, further indicating that endogenous CHIP ubiquitinates IRE1 (Fig. 2E). The two functional sites in CHIP, Lys30 (interacts with HSC70) and His260 (catalytic activity) (17), were mutated and used to evaluate which site was necessary for CHIP to ubiquitinate IRE1. As presented in Fig. 2F, K30A CHIP slightly weakened IRE1 ubiquitination compared with wild-type CHIP; however, the ubiquitination was nearly abolished by H260Q CHIP. The evidences above indicate that CHIP ubiquitinates IRE1 in vivo. We next investigated whether CHIP directly ubiquitinated IRE1. We obtained His-CHIP and GST-fused IRE1 cytoplasmic tail (amino acids 465–977) recombinants and measured the IRE1 ubiquitination using an in vitro ubiquitination kit. As shown in Fig. 2G, CHIP could directly ubiquitinate GST-IRE1 (amino acids 465–977) in vitro, whereas the ubiquitination only occurred with UbcH13/Uev2. These results demonstrate that CHIP acts as an E3 ligase to ubiquitinate IRE1.
FIGURE 2.
CHIP ubiquitinated IRE1. A, HEK293 cells were transfected with HA-IRE1, MYC-CHIP, MYC-Ub, or MYC-empty as indicated. After 24 h, the cells were treated with or without 1 μm GA for 12 h, followed by IP with an IRE1 antibody and Western blot with IRE1 and MYC antibodies. B, HEK293 cell lysates treated with GA or DMSO were analyzed by Western blot with antibodies against BIP and actin. C, confocal microscopy images were obtained from HEK293 cells treated with GA or DMSO; protein disulfide isomerase staining (red) indicates ER stress, and the nuclei were visualized by DAPI staining (blue). Bar, 10 μm. D, HEK293 cells were transfected with an shRNA vector targeting CHIP or an empty vector; the stably transfected cell lines were selected using 600 μg/ml G418. CHIP expression in these cell lines was estimated by Western blot using antibodies against CHIP and actin. CTRL: control. E, CHIP RNAi and control stable cells were transfected with HA-IRE1 and MYC-Ub. F, HEK293 cells were transfected with HA-IRE1, MYC-CHIP, MYC-CHIP mutants, MYC-Ub, or MYC-empty as indicated. All the cells in E and F were treated and assayed as in A. G, recombinant protein GST-fused IRE1 cytoplasmic tail (amino acids 465–977) and His-tagged CHIP were added in combination with Ub machinery components (Biotin-Ub, E1, UbcH5a, Ubc13, and several other E2s) as indicated. After a 1-h incubation in 37 °C, samples were boiled in SDS sample buffer and subjected to Western blot with streptavidin-HRP to recognize ubiquitinated IRE1. *, nonspecific bands. CTRL, control; IP, immunoprecipitation; WB, Western blot.
CHIP Ubiquitinated IRE1 on Lys545 and Lys828 in a Lys63-linked Manner
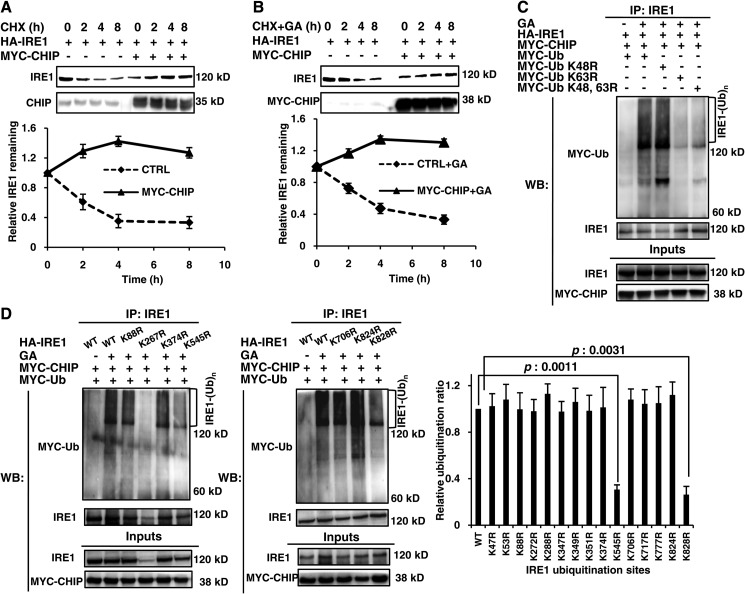
Generally, CHIP brings the ubiquitin chain to its substrates and stimulates their degradation (18, 19). Therefore, we investigated whether CHIP-mediated IRE1 ubiquitination increased the turnover of IRE1. IRE1 abundance was consistent in both CHIP knockdown cells (Fig. 2E) and CHIP overexpressing cells (Fig. 2A). Cycloheximide, an inhibitor of protein synthesis, was used to further evaluate the impact of CHIP on IRE1. As illustrated in Fig. 3A, exogenous IRE1 expression gradually decreased after cycloheximide treatment in a time-dependent manner, with a nearly 70% decrease in IRE1 by 8 h. When CHIP was overexpressed in these cells, the degradation of IRE1 was not enhanced but blocked in some level. A similar phenomenon occurred in cells with increased ER stress (Fig. 3B).
FIGURE 3.
CHIP ubiquitinated IRE1 on Lys545 and Lys828 in a Lys63-linked manner. A, HEK293 cells were transfected with HA-IRE1, MYC-CHIP, or MYC-empty as indicated for 24 h. The cells were treated with 50 μg/ml cycloheximide for various time points between 0 and 8 h and analyzed by Western blot with an IRE1 antibody. B, the same IRE1 degradation assay in A was performed with 1 μm GA treatment. C, HEK293 cells were transfected with HA-IRE1, MYC-CHIP, MYC-Ub, or MYC-Ub mutants as indicated. D, HEK293 cells were transfected with MYC-CHIP, MYC-Ub, HA-IRE1, or HA-IRE1 mutants as indicated in Table 2. The cells in C and D were treated with or without 1 μm GA for 12 h, followed by IP with an IRE1 antibody and Western blot with IRE1 and MYC antibodies. All data are from at least three independent experiments. The p values were calculated using Student's t test. CHX, cycloheximide; CTRL, control; IP, immunoprecipitation; WB, Western blot.
For further evaluation of the effect of CHIP abundance on the degradation of endogenous IRE1, we performed the degradation assay in HEK293 stable cell lines with/without CHIP RNAi. The Western blot images showed similar patterns in these two stable cell lines, and the same phenomenon was found under ER stress condition (data not shown). These results indicate that CHIP does not induce endogenous IRE1 degradation. Combining the evidence from these figures, we conclude that CHIP does not induce IRE1 degradation.
Substrate ubiquitination by CHIP does not always result in enhanced degradation. Ubiquitination at Lys48 favors protein degradation, whereas ubiquitination at Lys63 likely participates in regulating signal transduction (20). Therefore, we identified the ubiquitin ligation site(s) on IRE1 by co-transfecting wild-type or mutant ubiquitin with CHIP and IRE1. Compared with wild-type ubiquitin in cells under ER stress, K48R ubiquitin did not significantly change IRE1 ubiquitination; however, K63R and double-mutated ubiquitin nearly depleted IRE1 ubiquitination (Fig. 3C). These results suggest that CHIP ubiquitinates IRE1 in a Lys63-linked manner. Furthermore, it is believed that the manner of ubiquitination depends primarily on the ubiquitin-conjugating enzyme (E2) that is involved in the ubiquitination process (20). As shown in Fig. 2G, we found that CHIP could only ubiquitinate the IRE1 cytoplasmic tail (amino acids 465–977) with UbcH13. This observation is consistent with the report by Zhang et al. (21) that the E2 UbcH13 collaborated with CHIP to specifically forms a Lys63-linked ubiquitin chain, which assisted CHIP to ubiquitinate its substrates. Taking all the evidence above into account, we come to a conclusion that CHIP ubiquitinates IRE1 in a Lys63-linked manner.
Next, we examined the ubiquitin-conjugating sites on IRE1. Utilizing the Bayesian discriminant method for ubiquitination sites, the 16 top ranked sites were individually mutated from lysine to arginine (Table 2) to determine their ubiquitination potential. Except for K267R, the other 15 IRE1 mutants could be overexpressed in cells and recognized by the IRE1 antibody. Compared with wild-type IRE1, the K545R and K828R IRE1 mutants exhibited a significant change in ubiquitination in response to ER stress (Fig. 3D), demonstrating that these two lysine residues are likely to be CHIP modification sites.
TABLE 2.
Predicted IRE1 ubiquitination sites using the Bayesian discriminant method
The 16 top ranking predicted ubiquitination sites are listed in the table.
| Peptide | Position | Score |
|---|---|---|
| CSGSSASKAGSSPSL | 545 | 3.53 |
| GSLHAVSKRTGSIKW | 47 | 2.27 |
| EGSHPFGKSLQRQAN | 777 | 1.95 |
| SKRTGSIKWTLKEDP | 53 | 1.88 |
| SDFGLCKKLAVGRHS | 717 | 1.84 |
| PQKRPSAKHVLKHPF | 824 | 1.67 |
| VGRITKWKYPFPKET | 267 | 1.59 |
| SLYTLGSKNNEGLTK | 88 | 1.57 |
| TPTLYVGKYSTSLYA | 288 | 1.45 |
| FDPGLKSKNKLNYLR | 349 | 1.44 |
| PSAKHVLKHPFFWSL | 828 | 1.31 |
| KWKYPFPKETEAKSK | 272 | 1.29 |
| PGLKSKNKLNYLRNY | 351 | 1.29 |
| PNAHGKIKAMISDFG | 706 | 1.29 |
| VKFDPGLKSKNKLNY | 347 | 1.21 |
| TPLSASTKMLERFPN | 374 | 1.13 |
CHIP Participated in the IRE1/TRAF2/JNK Pathway
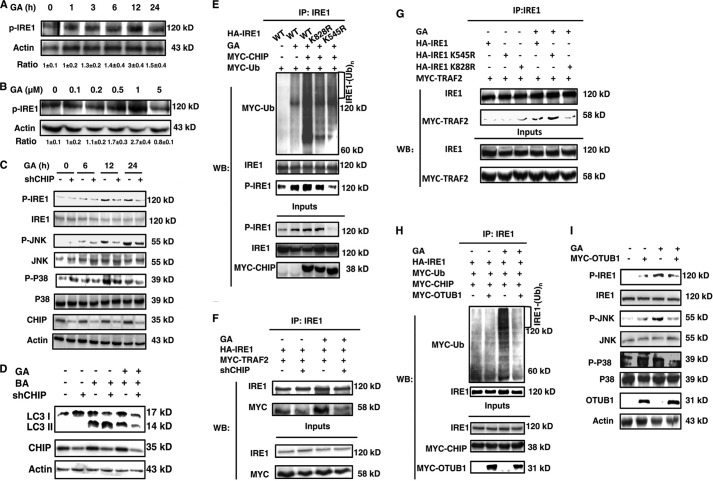
Because Lys63 ubiquitination by CHIP has been reported to be involved in signal transduction (22), we determined whether CHIP played a functional role in IRE1 signaling pathways in response to ER stress. We first verified that GA activated IRE1-related pathways. We found that GA induced IRE1 phosphorylation in a dose-dependent manner within the GA concentration from 0 to 1 μm and in a time-dependent manner within 12 h. However, the phosphorylation of IRE1 was attenuated at 24 h or with 5 μm GA (Fig. 4, A and B). The influence of CHIP on IRE1/XBP1 function was then examined. There was no difference in XBP1 activation between CHIP knockdown and control cell lines, implying that the CHIP-IRE1 interaction does not regulate the IRE1-XBP1 pathway (data not shown). Next, the effect of CHIP on the IRE1/TRAF2/JNK function was evaluated. In response to ER stress, the IRE1-TRAF2 interaction activates ASK1, a downstream protein that relays the UPR signal to the stress-activated protein kinase JNK (7). Therefore, the phosphorylation status of the related proteins was examined. The phosphorylation of IRE1, JNK and p38 decreased in response to CHIP knockdown under ER stress (Fig. 4C). The macroautophagy phenotype, which is regulated by JNK, downstream of IRE1 (23), was then assessed. The result corresponded with the IRE1/TRAF2/JNK activation status (Fig. 4D). Then we wondered whether the phosphorylation of IRE1 correlated with its ubiquitination. We found that the phosphorylation status of IRE1 was increased when it was ubiquitinated by CHIP. However, the K545R but not K828R mutant form of IRE1 abolished its phosphorylation under ER stress (Fig. 4E). This finding strengthens the deduction that the phosphorylation of IRE1 depends on its ubiquitination. In addition, we found that the interaction between exogenous IRE1 and TRAF2 was enhanced under ER stress in control cells, which agrees with the previous observation that the IRE1-TRAF2 complex only forms in response to ER stress (7); however, in CHIP knockdown cells, this interaction was completely inhibited (Fig. 4F). To examine which residue, Lys545 or Lys828, ubiquitination of IRE1 is critical for the IRE1/TRAF2 interaction, the two vectors with IRE1 mutated as K545R or K828R were transfected into HEK293 cells, followed by the same experiment as above to assess the IRE1/TRAF2 interaction. As illustrated in Fig. 4G, the K828R mutant nearly abolished this interaction even under the same ER stress; however, the K545R mutant did not significantly affect the interaction. This result suggests that the ubiquitination of IRE1 at Lys828 is critically important for IRE1/TRAF2 interaction, which is possibly involved in the regulation of the IRE1/TRAF2/JNK branch. Therefore, the evidence presented above demonstrates that CHIP likely regulates the IRE1/TRAF2/JNK pathway.
FIGURE 4.
CHIP participated in IRE1/TRAF2/JNK signal transduction. A and B, the lysates of HEK293 cells treated with 1 μm GA for different time points (A) or various concentrations of GA for 12 h (B) were immunoblotted with antibodies against phospho-IRE1 and actin. C, CHIP RNAi and control stable cell lysates were immunoblotted using antibodies against total and phosphorylated IRE1, JNK, or p38 after various treatment periods with 1 μm GA. D, CHIP RNAi and control stable cells were treated with or without DMSO, 1 μm GA, and 200 nm bafilomycin A1 as indicated. The lysates were immunoblotted with antibodies against LC3 and actin. E, HEK293 cells were transfected with HA-IRE1, HA-IRE1 K545R, HA-IRE1 K828R, MYC-CHIP, and MYC-Ub as indicated. F, CHIP RNAi and control stable cells were transfected with HA-IRE1 and MYC-TRAF2 as indicated. G, HEK293 cells were transfected with HA-IRE1, HA-IRE1 K545R, HA-IRE1 K828R, and MYC-TRAF2 as indicated. H, HEK293 cells were transfected with HA-IRE1, MYC-CHIP, MYC-Ub, MYC-OTUB1, or MYC-empty as indicated. All the transfected cells described in E–H were treated with or without 1 μm GA for 12 h, followed by IP with an IRE1 antibody and Western blot with IRE1 (E), phosphorylated IRE1 and MYC antibodies, or IRE1 and MYC antibodies (F–H). I, HEK293 cells were transfected with MYC-OTUB1 or MYC-empty as indicated. The transfected cells were treated with or without 1 μm GA for 12 h followed by Western blot with total and phosphorylated (where appropriate) antibodies against MYC, actin, IRE1, JNK, and p38. BA, bafilomycin A1; IP, immunoprecipitation; WB, Western blot.
Moreover, as shown in Table 1, the deubiquitinase OTUB1 was also present in the IRE1 immunoprecipitates. Therefore, we hypothesized that OTUB1 might deubiquitinate IRE1 and thereby regulate the IRE1/TRAF2/JNK pathway. In HEK293 cells, overexpression of OTUB1 abolished CHIP-mediated IRE1 ubiquitination under ER stress (Fig. 4H). Moreover, the activation of IRE1, JNK, and p38 was also remarkably inhibited in cells overexpressing OTUB1 (Fig. 4I). Based on these observations, it is reasonable to deduce that OTUB1 removes the ligated ubiquitin on IRE1 under ER stress, thereby regulating the IRE1/TRAF2/JNK pathway. Thus, ubiquitination is indeed a modification involved in the activation of the IRE1/TRAF2/JNK pathway.
The IRE1/TRAF2/JNK Pathway Antagonized Cellular Senescence under ER Stress
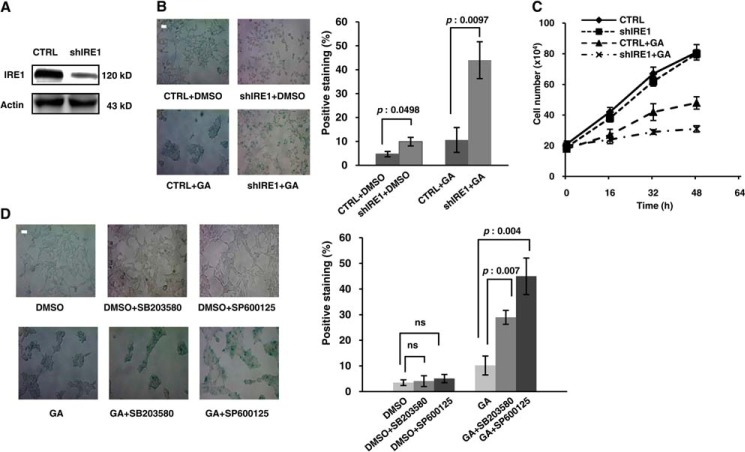
The involvement of IRE1 in multiple cellular processes, such as proliferation, apoptosis, and autophagy, has been well established (23–25). A recent report showed that IRE1 was involved in senescence. Denoyelle et al. (26) determined that IRE1 and XBP1 played a pro-senescence role in mutant Ras-driven senescence. In this study, we examined whether the IRE1/TRAF2/JNK pathway played a role in senescence. A stable cell line was selected after transfection of an shRNA vector targeting IRE1 (Fig. 5A). There was no difference in the ratio of senescent cells between IRE1 knockdown cells and control cells; however, senescence markedly increased in IRE1 knockdown cells under ER stress (Fig. 5B). This result indicates that IRE1 potentially regulates cellular senescence in response to ER stress. Additionally, by counting cell numbers, we generated the growth curves of IRE1 knockdown stable cell lines and normal stable cell lines under the condition with/without GA treatment. The result shown in Fig. 5C was consistent with the observation obtained from SA-β-gal staining (Fig. 5B). Furthermore, to clarify whether GA performs a specific effect to induce senescence process, we tested GA and two other ER stress inducers, DTT and tunicamycin, in a parallel experiment. All three reagents were able to cause senescence augmentation in IRE1 knockdown stable cell lines, implying that GA does have a function on cell behavior comparable with those of other ER stress inducers (data not shown). We then studied whether this senescence event correlated with the IRE1/TRAF2/JNK pathway. A JNK inhibitor (SP600125) and a p38 inhibitor (SB203580) were used to inhibit the activation of this pathway. As depicted in Fig. 5D, the senescence ratio after GA treatment was significantly increased regardless of which inhibitor, SP600125 or SB203580, was added, suggesting that activation of the IRE1/TRAF2/JNK pathway antagonizes cellular senescence.
FIGURE 5.
The IRE1/TRAF2/JNK pathway antagonized cellular senescence under ER stress. A, HEK293 cells were transfected with a shRNA vector against IRE1 or an empty vector, and the stably transfected cells were selected using 600 μg/ml G418. IRE1 expression in these cell lines was estimated by Western blot using antibodies against IRE1 and actin. B, the IRE1 RNAi and control stable cell lines were treated with or without 1 μm GA for 24 h and assayed for β-gal staining. At least 500 cells were counted, and the data are presented as the means ± S.D. from at least three independent experiments. C, the same number of IRE1 RNAi and control stable cells were treated with or without 1 μm GA and were quantified using a cell counter at different time points. D, HEK293 cells were co-treated with or without 1 μm GA and 10 μm SB203580 or SP600125 for 24 h and assayed for β-gal staining. The positive cells were quantified as in B. The p values were calculated using Student's t test. Bar, 50 μm. CTRL, control; ns, not significant (p > 0.05).
CHIP Antagonized the Senescence of HEK293 Cells under ER Stress
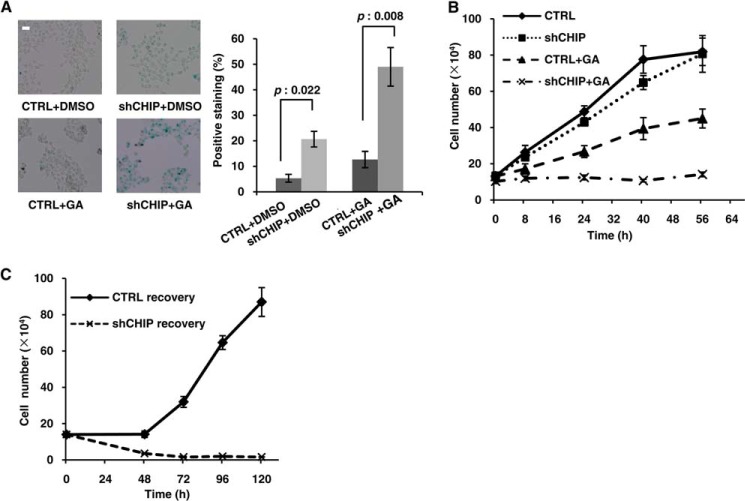
The involvement of CHIP in cellular senescence is well documented (27). As described above, CHIP facilitated IRE1 ubiquitination, and IRE1 participated in the senescence pathway in response to ER stress. Therefore, we ascertained the role of CHIP in IRE1-related senescence. First, senescence and cell growth were examined in cells with or without CHIP knockdown. The results showed that ∼20% of the cells were positive for β-gal (Fig. 6A), and the cell growth rate was slightly decreased in CHIP knockdown cells (Fig. 6B). Next, we evaluated cellular behavior under ER stress. In CHIP normal cells, the β-gal staining was almost unchanged, but the cell proliferation rate decreased by nearly 50% in response to ER stress. In CHIP knockdown cells, ∼60% of the cells were strongly positive for β-gal, and cell proliferation nearly ceased in response to ER stress (Fig. 6, A and B). Finally, after GA treatment, the recovery ability of these two cell lines was determined. The stressed and reseeded CHIP knockdown cells continued without proliferating until cell death occurred; however, the cells with normal CHIP expression almost completely recovered their proliferation ability within 48 h (Fig. 6C). As irreversibility is a common feature of senescent cells, the data suggests that CHIP is indeed involved in the cellular senescence process under ER stress.
FIGURE 6.
CHIP antagonized cellular senescence under ER stress. A, CHIP RNAi and control stable cell lines were treated with or without 1 μm GA for 24 h and assayed for β-gal staining. At least 500 cells were counted, and the data are presented as the means ± S.D. from at least three independent experiments. B, the same number of CHIP RNAi and control stable cells were treated with or without 1 μm GA and were quantified using a cell counter at different time points. C, CHIP RNAi and control stable cell lines were treated with 1 μm GA for 48 h. The cells were trypsinized, and equal numbers of cells were reseeded onto dishes; the subsequent cell count was determined at different time points. The data were quantified as in B. The p values were calculated using Student's t test. Bar, 50 μm. CTRL, control; ns, not significant (p > 0.05).
DISCUSSION
We have systematically investigated one of the post-translational modifications of IRE1 in HEK293 cells under ER stress. Our data revealed that CHIP interacted with IRE1, leading to its ubiquitination, and that this ubiquitination event regulated the IRE1/TRAF2/JNK pathway, which possibly affected cellular senescence. These discoveries expand our understanding of the UPR, especially regarding the functions of CHIP under ER stress. Although some results described above are consistent with previous observations or conclusions, some remain controversial with others.
CHIP has been demonstrated to participate in the ERAD pathway. Cytoplasmic CHIP is believed to enhance the ubiquitination of retrotranslocated proteins and then to expedite the degradation of unfolded proteins (28). In contrast to the traditional view, the discoveries described in this study suggest that the ubiquitination of IRE1 by CHIP is likely to mediate signaling rather than protein degradation under ER stress. Similar to our results, Yang et al. (22) claimed that the CHIP ubiquitination could affect signal transduction but not protein degradation in RAW264.7 cells, whereas Gao et al. (10) perceived that an ER resident E3-ligase did ubiquitinate IRE1. We therefore postulate that CHIP performs as a regulator with dual functions. The ERAD machinery is abundantly activated once ER stress is induced. We believe that because the proteasome-dependent degradation of unfolded proteins is generally a time-consuming process, whereas the UPR is a rapid response to ER stress, the degradation path is not always efficient enough to remove unfolded proteins while coping with greater ER stress. According to our model of dual functions, we assume that CHIP-mediated ubiquitination is an economical response to ER stress, which functions both by facilitating the degradation of unfolded proteins and by modifying IRE1 to activate the UPR pathway.
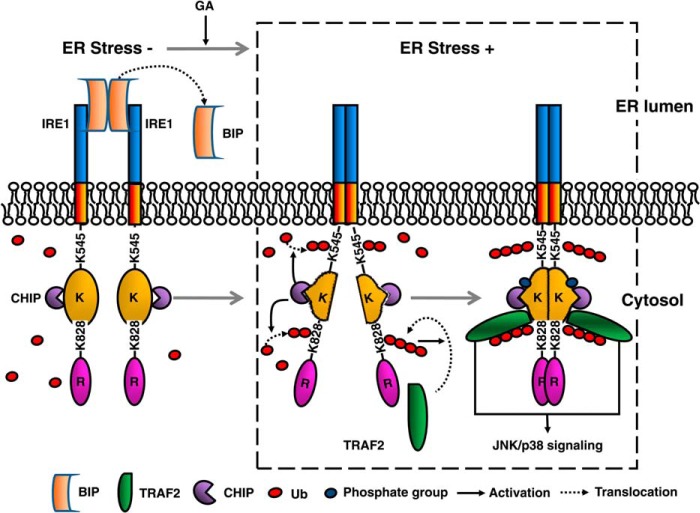
We identified one ubiquitin-conjugating site, Lys545, on IRE1 that was likely to be associated with IRE1 phosphorylation. It is well accepted that phosphorylation of IRE1 is required for the formation of IRE1 dimers, whereas IRE1 dimerization leads to its conformational changes and further stimulates IRE1 kinase and RNase activity (29–31). Based on topological analysis, Lys545 is located at the connecting domain that links the transmembrane and cytosolic domains. Therefore, we assume that the K545R mutation abolishes the juxtaposition of the cytosolic domains of two IRE1 monomers so as to impair the IRE1 phosphorylation. In addition, we observed that the IRE1-TRAF2 interaction was weakened in CHIP knockdown cells and was dependent on IRE1 ubiquitination at Lys828 under ER stress. OTUB1, a deubiquitinase, could almost completely abolish CHIP-mediated IRE1 ubiquitination and thus inactivated IRE1/JNK and p38 signaling. An analysis of IRE1 topology suggests that this ubiquitination site is not located in the RNase domain (mapped to 837–977) but likely resides in the kinase domain, where it is essential for TRAF2 binding (7). A logical deduction is that the ubiquitination at Lys828 acts as a platform for TRAF2 anchoring. Our observation is in partial agreement with the findings from Arshad et al. (32). Under ER stress induced by staurosporine, the authors observed that the knockdown of RNF13, an E3 ligase, impaired the IRE1/TRAF2 signaling axis and caused JNK inactivation. Therefore, we propose IRE1 ubiquitination as a key modification for regulation of IRE1/TRAF2/JNK signaling under ER stress. Summarizing all the analysis above, a mechanism model for the hypothesis is illustrated in Fig. 7.
FIGURE 7.
Schematic diagram to illustrate the ubiquitination of IRE1 by CHIP and the effect to the relevant signaling.
Senescence has been reported to be regulated by the UPR. As mentioned above, the ectopic expression of H-RAS G12V in primary human melanocytes regulated the UPR and induced senescence (26); on the other hand, aging was accelerated in CHIP knock-out mice (27). CHIP was gradually activated during senescence and regulated the abundance of p53 and oxidized proteins (33). The question remains as to what senescence mechanisms are associated with the UPR and CHIP. Our results summarized above seem to offer a proper answer to this question, which is basically in agreement with the observation that the activity of JNK is tightly correlated with senescence development (34). Furthermore, we determined that CHIP knockdown impaired the IRE1/TRAF2/JNK pathway, and this impairment was accompanied by decreased autophagy in HEK293 cells. Abundant evidence has demonstrated that this pathway provides important stimulation for initiating autophagy (23, 35), which may antagonize senescence (36, 37). Our findings are consistent with these reports. It is well known that CHIP could ubiquitinate lots of substrates that may regulate senescence. The directly experimental evidence that supports senescence regulated by IRE1 ubiquitination related with CHIP is not gained yet. However, the correlation evidence described in this study has set a solid foundation for the hypothesis that CHIP participates in the UPR, at least through IRE1-related signaling transduction, and partially regulates senescence.
This work was supported by National High Technology Research and Development Program Grant 2012AA020206 and National Basic Research Program of China Grant 2011CB910704.
- ER
- endoplasmic reticulum
- GA
- geldanamycin
- IRE1
- inositol-requiring enzyme 1
- TRAF2
- TNF receptor-associated factor 2
- UPR
- unfolded protein response
- ERAD
- ER-associated degradation
- Ub
- ubiquitin
- SA-β-gal
- senescence-associated-β-galactosidase staining.
REFERENCES
- 1. Bernales S., Papa F. R., Walter P. (2006) Intracellular signaling by the unfolded protein response. Annu. Rev. Cell Dev. Biol. 22, 487–508 [DOI] [PubMed] [Google Scholar]
- 2. Walter P., Ron D. (2011) The unfolded protein response: from stress pathway to homeostatic regulation. Science 334, 1081–1086 [DOI] [PubMed] [Google Scholar]
- 3. Merksamer P. I., Papa F. R. (2010) The UPR and cell fate at a glance. J. Cell Sci. 123, 1003–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Calfon M., Zeng H., Urano F., Till J. H., Hubbard S. R., Harding H. P., Clark S. G., Ron D. (2002) IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature 415, 92–96 [DOI] [PubMed] [Google Scholar]
- 5. Lee A. H., Iwakoshi N. N., Glimcher L. H. (2003) XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol. Cell. Biol. 23, 7448–7459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shaffer A. L., Shapiro-Shelef M., Iwakoshi N. N., Lee A. H., Qian S. B., Zhao H., Yu X., Yang L., Tan B. K., Rosenwald A., Hurt E. M., Petroulakis E., Sonenberg N., Yewdell J. W., Calame K., Glimcher L. H., Staudt L. M. (2004) XBP1, downstream of Blimp-1, expands the secretory apparatus and other organelles, and increases protein synthesis in plasma cell differentiation. Immunity 21, 81–93 [DOI] [PubMed] [Google Scholar]
- 7. Urano F., Wang X., Bertolotti A., Zhang Y., Chung P., Harding H. P., Ron D. (2000) Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science 287, 664–666 [DOI] [PubMed] [Google Scholar]
- 8. Jwa M., Chang P. (2012) PARP16 is a tail-anchored endoplasmic reticulum protein required for the PERK- and IRE1α-mediated unfolded protein response. Nat. Cell Biol. 14, 1223–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lu G., Ota A., Ren S., Franklin S., Rau C. D., Ping P., Lane T. F., Zhou Z. H., Reue K., Lusis A. J., Vondriska T., Wang Y. (2013) PPM1l encodes an inositol requiring-protein 1 (IRE1) specific phosphatase that regulates the functional outcome of the ER stress response. Mol. Metab. 2, 405–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gao B., Lee S. M., Chen A., Zhang J., Zhang D. D., Kannan K., Ortmann R. A., Fang D. (2008) Synoviolin promotes IRE1 ubiquitination and degradation in synovial fibroblasts from mice with collagen-induced arthritis. EMBO Reports 9, 480–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Connell P., Ballinger C. A., Jiang J., Wu Y., Thompson L. J., Höhfeld J., Patterson C. (2001) The co-chaperone CHIP regulates protein triage decisions mediated by heat-shock proteins. Nat. Cell Biol. 3, 93–96 [DOI] [PubMed] [Google Scholar]
- 12. Schmidt B. Z., Watts R. J., Aridor M., Frizzell R. A. (2009) Cysteine string protein promotes proteasomal degradation of the cystic fibrosis transmembrane conductance regulator (CFTR) by increasing its interaction with the C terminus of Hsp70-interacting protein and promoting CFTR ubiquitylation. J. Biol. Chem. 284, 4168–4178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. El Khouri E., Le Pavec G., Toledano M. B., Delaunay-Moisan A. (2013) RNF185 is a novel E3 ligase of endoplasmic reticulum-associated degradation (ERAD) that targets cystic fibrosis transmembrane conductance regulator (CFTR). J. Biol. Chem. 288, 31177–31191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pabarcus M. K., Hoe N., Sadeghi S., Patterson C., Wiertz E., Correia M. A. (2009) CYP3A4 ubiquitination by gp78 (the tumor autocrine motility factor receptor, AMFR) and CHIP E3 ligases. Arch. Biochem. Biophys. 483, 66–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Davenport E. L., Moore H. E., Dunlop A. S., Sharp S. Y., Workman P., Morgan G. J., Davies F. E. (2007) Heat shock protein inhibition is associated with activation of the unfolded protein response pathway in myeloma plasma cells. Blood 110, 2641–2649 [DOI] [PubMed] [Google Scholar]
- 16. Woo C. H., Le N. T., Shishido T., Chang E., Lee H., Heo K. S., Mickelsen D. M., Lu Y., McClain C., Spangenberg T., Yan C., Molina C. A., Yang J., Patterson C., Abe J. (2010) Novel role of C terminus of Hsc70-interacting protein (CHIP) ubiquitin ligase on inhibiting cardiac apoptosis and dysfunction via regulating ERK5-mediated degradation of inducible cAMP early repressor. FASEB J. 24, 4917–4928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Xu W., Marcu M., Yuan X., Mimnaugh E., Patterson C., Neckers L. (2002) Chaperone-dependent E3 ubiquitin ligase CHIP mediates a degradative pathway for c-ErbB2/Neu. Proc. Natl. Acad. Sci. U.S.A. 99, 12847–12852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ahmed S. F., Deb S., Paul I., Chatterjee A., Mandal T., Chatterjee U., Ghosh M. K. (2012) The chaperone-assisted E3 ligase C terminus of Hsc70-interacting protein (CHIP) targets PTEN for proteasomal degradation. J. Biol. Chem. 287, 15996–16006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li X., Huang M., Zheng H., Wang Y., Ren F., Shang Y., Zhai Y., Irwin D. M., Shi Y., Chen D., Chang Z. (2008) CHIP promotes Runx2 degradation and negatively regulates osteoblast differentiation. J. Cell Biol. 181, 959–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lim K. L., Lim G. G. (2011) K63-linked ubiquitination and neurodegeneration. Neurobiol. Dis. 43, 9–16 [DOI] [PubMed] [Google Scholar]
- 21. Zhang M., Windheim M., Roe S. M., Peggie M., Cohen P., Prodromou C., Pearl L. H. (2005) Chaperoned ubiquitylation: crystal structures of the CHIP U box E3 ubiquitin ligase and a CHIP-Ubc13-Uev1a complex. Mol. Cell 20, 525–538 [DOI] [PubMed] [Google Scholar]
- 22. Yang M., Wang C., Zhu X., Tang S., Shi L., Cao X., Chen T. (2011) E3 ubiquitin ligase CHIP facilitates Toll-like receptor signaling by recruiting and polyubiquitinating Src and atypical PKC{zeta}. J. Exp. Med. 208, 2099–2112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ogata M., Hino S., Saito A., Morikawa K., Kondo S., Kanemoto S., Murakami T., Taniguchi M., Tanii I., Yoshinaga K., Shiosaka S., Hammarback J. A., Urano F., Imaizumi K. (2006) Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol. Cell. Biol. 26, 9220–9231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tabas I., Ron D. (2011) Integrating the mechanisms of apoptosis induced by endoplasmic reticulum stress. Nat Cell Biol. 13, 184–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Thorpe J. A., Schwarze S. R. (2010) IRE1α controls cyclin A1 expression and promotes cell proliferation through XBP-1. Cell Stress Chaperones 15, 497–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Denoyelle C., Abou-Rjaily G., Bezrookove V., Verhaegen M., Johnson T. M., Fullen D. R., Pointer J. N., Gruber S. B., Su L. D., Nikiforov M. A., Kaufman R. J., Bastian B. C., Soengas M. S. (2006) Anti-oncogenic role of the endoplasmic reticulum differentially activated by mutations in the MAPK pathway. Nat. Cell Biol. 8, 1053–1063 [DOI] [PubMed] [Google Scholar]
- 27. Min J. N., Whaley R. A., Sharpless N. E., Lockyer P., Portbury A. L., Patterson C. (2008) CHIP deficiency decreases longevity, with accelerated aging phenotypes accompanied by altered protein quality control. Mol. Cell. Biol. 28, 4018–4025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Younger J. M., Chen L., Ren H. Y., Rosser M. F., Turnbull E. L., Fan C. Y., Patterson C., Cyr D. M. (2006) Sequential quality-control checkpoints triage misfolded cystic fibrosis transmembrane conductance regulator. Cell 126, 571–582 [DOI] [PubMed] [Google Scholar]
- 29. Ali M. M., Bagratuni T., Davenport E. L., Nowak P. R., Silva-Santisteban M. C., Hardcastle A., McAndrews C., Rowlands M. G., Morgan G. J., Aherne W., Collins I., Davies F. E., Pearl L. H. (2011) Structure of the Ire1 autophosphorylation complex and implications for the unfolded protein response. EMBO J. 30, 894–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhou J., Liu C. Y., Back S. H., Clark R. L., Peisach D., Xu Z., Kaufman R. J. (2006) The crystal structure of human IRE1 luminal domain reveals a conserved dimerization interface required for activation of the unfolded protein response. Proc. Natl. Acad. Sci. U.S.A. 103, 14343–14348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kimata Y., Ishiwata-Kimata Y., Ito T., Hirata A., Suzuki T., Oikawa D., Takeuchi M., Kohno K. (2007) Two regulatory steps of ER-stress sensor Ire1 involving its cluster formation and interaction with unfolded proteins. J. Cell Biol. 179, 75–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Arshad M., Ye Z., Gu X., Wong C. K., Liu Y., Li D., Zhou L., Zhang Y., Bay W. P., Yu V. C., Li P. (2013) RNF13, a RING finger protein, mediates endoplasmic reticulum stress-induced apoptosis through the inositol-requiring enzyme (IRE1α)/c-Jun NH2-terminal kinase pathway. J. Biol. Chem. 288, 8726–8736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sisoula C., Trachana V., Patterson C., Gonos E. S. (2011) CHIP-dependent p53 regulation occurs specifically during cellular senescence. Free Radic. Biol. Med. 50, 157–165 [DOI] [PubMed] [Google Scholar]
- 34. Lee J. J., Lee J. H., Ko Y. G., Hong S. I., Lee J. S. (2010) Prevention of premature senescence requires JNK regulation of Bcl-2 and reactive oxygen species. Oncogene 29, 561–575 [DOI] [PubMed] [Google Scholar]
- 35. Raciti M., Lotti L. V., Valia S., Pulcinelli F. M., Di Renzo L. (2012) JNK2 is activated during ER stress and promotes cell survival. Cell Death Dis. 3, e429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Demidenko Z. N., Zubova S. G., Bukreeva E. I., Pospelov V. A., Pospelova T. V., Blagosklonny M. V. (2009) Rapamycin decelerates cellular senescence. Cell Cycle 8, 1888–1895 [DOI] [PubMed] [Google Scholar]
- 37. Demidenko Z. N., Blagosklonny M. V. (2008) Growth stimulation leads to cellular senescence when the cell cycle is blocked. Cell Cycle 7, 3355–3361 [DOI] [PubMed] [Google Scholar]