Background: Formins build essential actin-based structures.
Results: The tails of Cappuccino and other formins contribute to both nucleation and processive filament elongation by binding actin monomers and filaments, respectively.
Conclusion: Formin tails tune actin assembly. Their role in processivity was not previously recognized.
Significance: Identifying the functions of the tail domain will lead to an understanding of how Capu and other formins function and are regulated.
Keywords: Actin, Cytoskeleton, Drosophila, Formin, Oocyte, Capu, TIRF
Abstract
Formins are multidomain proteins that assemble actin in a wide variety of biological processes. They both nucleate and remain processively associated with growing filaments, in some cases accelerating filament growth. The well conserved formin homology 1 and 2 domains were originally thought to be solely responsible for these activities. Recently a role in nucleation was identified for the Diaphanous autoinhibitory domain (DAD), which is C-terminal to the formin homology 2 domain. The C-terminal tail of the Drosophila formin Cappuccino (Capu) is conserved among FMN formins but distinct from other formins. It does not have a DAD domain. Nevertheless, we find that Capu-tail plays a role in filament nucleation similar to that described for mDia1 and other formins. Building on this, replacement of Capu-tail with DADs from other formins tunes nucleation activity. Capu-tail has low-affinity interactions with both actin monomers and filaments. Removal of the tail reduces actin filament binding and bundling. Furthermore, when the tail is removed, we find that processivity is compromised. Despite decreased processivity, the elongation rate of filaments is unchanged. Again, replacement of Capu-tail with DADs from other formins tunes the processive association with the barbed end, indicating that this is a general role for formin tails. Our data show a role for the Capu-tail domain in assembling the actin cytoskeleton, largely mediated by electrostatic interactions. Because of its multifunctionality, the formin tail is a candidate for regulation by other proteins during cytoskeletal rearrangements.
Introduction
Formin proteins construct actin filament networks that function in cell division, polarization, and motility (1, 2). They are multifunctional proteins that nucleate new filaments (3, 4), remain processively associated with growing filament ends (5), and cross-link actin filaments and microtubules (6, 7). The Drosophila formin Cappuccino (Capu)2 is essential for polarity establishment during oocyte development (8). Capu is associated with a diffuse mesh of actin filaments (F-actin) that spans the oocyte cytoplasm (9), and loss of Capu results in female sterility (10). Capu is a member of the FMN group of formins (11), and its mammalian homolog FMN2 is associated with a similar F-actin mesh structure in the mouse oocyte (12). Aside from the well conserved formin homology (FH) 1 and 2 domains and an N-terminal regulatory domain (13), Capu contains a short C-terminal tail (Fig. 1A) (14, 15). The tail is conserved among FMN family formins and has a consensus motif distinct from that of the Diaphanous autoinhibitory domain (DAD) found in Diaphanous-related formins (Fig. 1B).
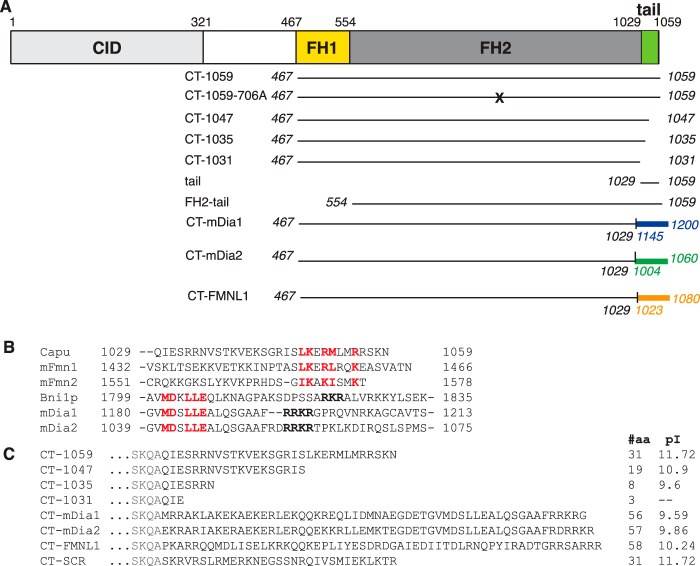
FIGURE 1.
Domain structure of Capu. A, diagram of full-length Capu (isoform A). Capu contains FH domains, a Capu inhibitory domain (CID) (13), and a tail domain. Fragments used in this work are shown below the full-length construct. The mutation I706A in the CT-1059-706A construct is analogous to the mutation I1431A in Bni1p (38). B, a multiple sequence alignment of tails from Capu and mouse FMN formins and DAD domains from other formins. The core tail and DAD motifs are boldface in red and show little sequence homology between DAD and tail. C, sequences of the tail region for constructs used here, including truncations, chimeras, and the scrambled tail. For each construct, the length (#aa) and pI are shown to the right of the sequence. aa, amino acids.
Across formin families, “tail” is a loosely defined term, referring to the region between the FH2 domain and the C terminus. The tail length varies greatly from 25–30 amino acids in Capu and other FMN formins (Fig. 1B) to 308 amino acids in INF2, 106 amino acids in mDia1, 162 amino acids in mDia2 (7), and 82 amino acids in FMNL3 (16). Some of these longer tails include DAD domains and partial Wiskott-Aldrich syndrome protein homology 2 (WH2) domains (16, 17). Aside from their roles in intramolecular regulation, tails are sites for interaction with the actin and microtubule cytoskeletons. This has been demonstrated for the mammalian formins FMNL3 (16), mDia1 (18), mDia2, and INF2 (7) and also for the microtubule binding activity of Capu (19).
Not surprisingly, because of their functional importance, the formin tails are also binding sites for other accessory actin assembly factors. The WH2-based nucleator Spire binds to Capu-tail (14, 15), and this interaction is essential for Drosophila development (20). The tumor suppressor adenomatous polyposis coli binds to mDia1-tail to form a nucleation “rocket launcher” complex (21). Similarly, in budding yeast, the activator Bud6 binds to the C-terminal tail of the formin Bni1p to deliver actin monomers to Bni1p and stimulate its actin assembly activity (22).
To better understand how C-terminal tails function in formin-mediated actin assembly, we studied the role of Capu-tail in filament nucleation and processive elongation in vitro. Our approach was to biochemically characterize a series of Capu truncations and chimeras, adding tails from other formins to Capu-FH1FH2. This is the first study of how actin assembly is influenced by the tail of an FMN family formin, and we find some similarities and differences with other formin tails characterized to date. Similar to other formins, Capu-tail plays roles in actin filament nucleation and binds directly to actin monomers, albeit with a lower affinity than what has been reported for other tails. We also identify a role for Capu-tail that has not been described previously for other formin tails: association with the filament end, which results in increased processivity. Consistent with these being conserved roles among formin tails, replacement of Capu-tail with DAD domains tunes both the nucleation and processivity of Capu. Finally, we find that the tail is necessary but not sufficient for the F-actin bundling activity of Capu. Formin tails in general tend to be highly positively charged, and we found that for Capu, electrostatic interactions are necessary for its actin assembly activity. Some of these interactions involve specific residues within the tail, whereas others are nonspecific, only requiring charge. We propose a simple structural model for how Capu functions during processive elongation.
EXPERIMENTAL PROCEDURES
DNA Constructs
Isoform A of Drosophila Capu (Flybase CG no. CG3399) was used as a template to make the fragments of Capu described below. Chimeric proteins CT-mDia1 corresponding to Capu(467–1028)-mDia1(1145–1200), CT-mDia2 corresponding to Capu(467–1028)-mDia2(1004–1060), and CT-FMNL1 corresponding to Capu(467–1028)-FMNL1(1023–1080) were constructed using the splicing by overlap extension PCR method (23). The DAD domain boundaries were chosen on the basis of the well defined DAD domain of mDia1 (24). A Perl random number generator was used to create a scrambled tail sequence. The coding sequence was constructed by DNA assembly (25) from oligonucleotides and spliced onto Capu to create CT-SCR or Capu(467–1028)-scrambled(1029–1059). All chimeric and truncated versions of Capu were cloned into a modified version of the pET15b plasmid with an N-terminal His tag. The RPEL1 gene was created by DNA assembly (25) to match the coding sequence for amino acids 60–97 of the murine MAL/MRTF-A (myocardin-related transcription factor A) gene (26) and cloned into the pGEX6P2 vector.
Protein Expression, Purification, and Labeling
All Capu fragments that contained the FH2 domain were expressed and purified as described previously (15, 19). Concentrations of constructs containing the FH1 and FH2 domains were quantified using the molar extinction coefficient 75,188 M−1 cm−1 for the CT-1059 dimer (27). Concentrations for proteins containing the FH2 domain as well as for GST-tail are given in dimer for all data presented below. The tail and RPEL1 peptides were expressed as GST fusions. They were purified and cleaved from GST as described previously for Capu-tail (15). The GST-tail fusion was purified as described previously (19). The concentrations of Capu-FH2, tail, GST-tail, and RPEL1 were obtained using SDS-PAGE and quantitative Sypro-Red staining (Invitrogen). Acanthamoeba castellanii actin and Drosophila profilin (Chic) were purified as described previously (13, 28). Actin was labeled with pyrene-iodoacetamide (28), Oregon Green 488-iodoacetamide (Invitrogen) (13), or EZ-link maleimide-PEG2-biotin (Thermo Scientific) (19).
Circular Dichroism Spectroscopy
Purified, frozen glycerol protein stocks were dialyzed overnight into 50 mm potassium phosphate (pH 7.5), 1 mm DTT and then precleared by centrifugation at 100,000 × g for 15 min at 4 °C. We monitored thermal denaturation of 2 μm Capu at 222 nm using a JASCO J-715 circular dichroism spectrophotometer with Peltier temperature control. The temperature range was scanned at 1 °C/min, with a 1-nm bandwidth and 8-s response time.
Fluorescence Spectroscopy
Pyrene-actin polymerization assays were carried out essentially as described (15). Briefly, 4 μm magnesium-G-actin (5% pyrene-labeled) was polymerized in 1× KMEH (50 mm KCl, 1 mm MgCl2, 1 mm EGTA, 10 mm HEPES (pH 7)). For steady-state G-actin sequestering assays, 2 μm F-actin (2% pyrene-labeled) was incubated overnight at 4 °C with varying concentrations of tail, RPEL1, or Latrunculin-B. Pyrene-actin fluorescence was measured using a Tecan F200 plate reader with filters for λexcitation = 360 ± 17 nm and λemission = 400 ± 10 nm. The concentration of barbed ends in Fig. 2, C and F, was calculated using the slope of the polymerization trace and the kinetic rate constants for addition of ATP-actin monomers to the barbed ends of filaments (29).
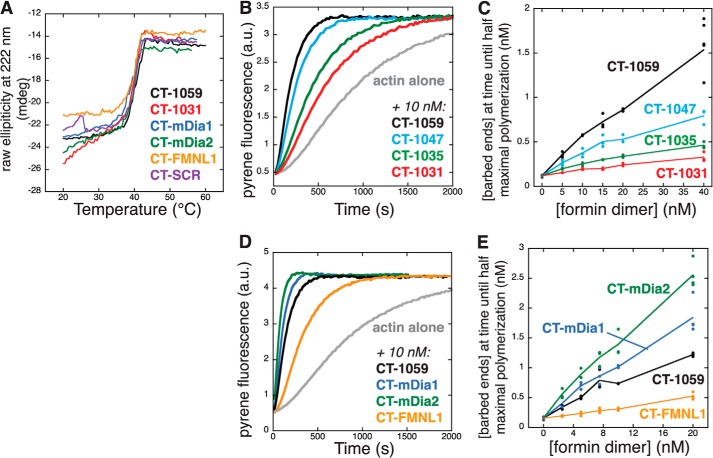
FIGURE 2.
Formin tails play a role in actin filament nucleation. A, thermal stability of full-length, truncated, and chimeric Capu. Secondary structure content was observed by circular dichroism at 222 nm over a range of temperatures (mdeg, millidegrees). The apparent Tm values were ≈40 °C for all constructs (2 μm dimer in 50 mm potassium phosphate (pH 7.5), 1 mm DTT). B and C, truncation of the tail (CT-1047, CT-1035, and CT-1031) compromises Capu-induced filament assembly. D and E, replacing the tail with DAD domains from other formins tunes the activity of Capu. B–E, 4 μm actin (5% pyrene-labeled) was polymerized in the presence of a range of formin dimer concentrations. Raw data for 10 nm formin dimer are shown in B and D (a.u., arbitrary units). C and E, barbed end concentrations were calculated assuming the same on and off rate as actin alone (see “Experimental Procedures” and Fig. 4). The line connects the mean barbed end concentrations at each formin concentration.
For tail/actin binding assays, calcium-G-actin was incubated for at least 15 min on ice with a 2-fold molar excess of Latrunculin-B. Samples with 40 nm Alexa Fluor 488-labeled Capu-tail plus varying concentrations of tail, RPEL1, or actin were diluted to final buffer conditions (1 mm MgCl2, 1 mm EGTA, 10 mm HEPES (pH 7), 0.5 mm thesit) with NaCl added as indicated. Anisotropy was measured on a Tecan M1000 using G-factor correction (λexcitation = 470 nm, λemission = 535 nm). The competition anisotropy data in Fig. 3A was fit using EQTK, a forthcoming analysis tool for coupled equilibria using algorithms from Refs. 30, 31. Briefly, we performed a nonlinear regression using a coupled equilibrium competitive binding model to find an equilibrium dissociation constant (Kd), given a Kd for actin and the labeled tail peptide (630 nm).
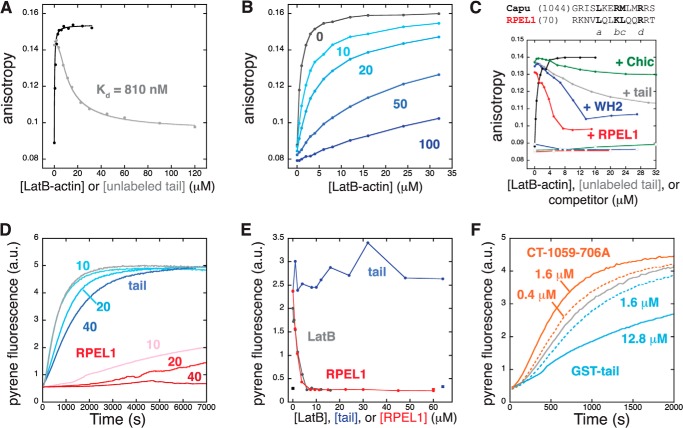
FIGURE 3.
The Capu-tail binds to monomeric actin. A, monomeric Latrunculin-B-magnesium-G-actin increases the anisotropy of 40 nm Alexa Fluor 488-labeled Capu-tail in a dose-dependent manner (black curve). Competition with unlabeled tail in the presence of 6 μm Latrunculin-B-magnesium-G-actin gives a Kd of 810 nm (gray curve). B, the interaction between Alexa Fluor 488-tail and G-actin is highly sensitive to salt. For each titration of 40 nm Alexa Fluor 488-labeled Capu-tail with Latruculin-B-magnesium-G-actin, the concentration of NaCl is indicated (Kd values: 10 mm NaCl, 2.4 μm; 20 mm, 5.0 μm; 50 mm, 20 μm; 100 mm, 71 μm). C, the RPEL1 domain of MRTF (red) and the WH2-B domain of Spire (blue) affect the tail/actin interaction. A sequence alignment of the core Capu tail and core RPEL1 domain with the essential actin-binding residues is shown in boldface (26). Profilin competes less effectively. Open symbols are for titration of competitor in the absence of actin. D, the tail slows filament growth (blue curves) but does not sequester actin monomers like RPEL (red curves). The concentration of tail or RPEL1 is given in micromolar. E, At steady state, the tail does not sequester actin monomers. The indicated amounts of tail, RPEL1, or Latruculin-B were incubated overnight with 2 μm F-actin (10% pyrene-labeled). The fluorescence of 2 μm G-actin (10% pyrene-labeled) is shown in the presence or absence of tail or RPEL1. F, a dimeric tail does not nucleate filaments and has weak sequestering activity at the concentrations tested (light blue: 1.6 μm monomer, dashed line; 12.8 μm monomer, solid line). CT-1059 containing a mutation of the conserved isoleucine 706 (CT-1059-706A) shows weak nucleation activity (orange: 0.4 μm monomer, dashed line; 1.6 μm monomer, solid line). D and F, polymerization assays contained 4 μm magnesium-G-actin (5% pyrene-labeled).
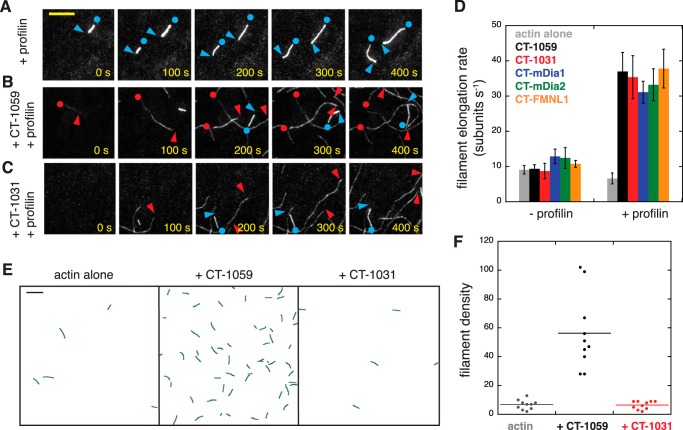
Total Internal Reflection Fluorescence Microscopy
PEGylated glass coverslips were prepared using amino-propyl-triethoxy-silane and a mix of N-hydrosuccinimide-PEG-methoxy and N-hydrosuccinimide-PEG-biotin as described previously (13). The final conditions in the flow chambers were 0.6–1 μm magnesium-G-actin (25–30% Oregon Green -labeled) in 1× KMEH, 0.2% methylcellulose, 50 mm DTT, 0.2 mm ATP, 50 μg/ml κ-casein (Sigma), 20 mm glucose, 250 μg/ml glucose oxidase, and 50 μg/ml catalase. Prior to imaging, flow cells of ≈15-μl volume were assembled using double sticky tape and prepared as follows: 1) 2 min with 50 μl of 1% Pluronic F-127 (Sigma), 50 μg/ml κ-casein, 1× PBS; 2) wash with 1× TIRF buffer (1× KMEH, 0.2% methylcellulose, 50 mm DTT, 0.2 mm ATP, 20 mm glucose); 3) 30–60 s with 25 μl of 40 nm streptavidin; 4) wash with 25 μl of 1× TIRF buffer; 5) 30–60 s with 25 μl of 80 nm biotinylated heavy meromyosin or 200 nm biotin-phalloidin (no difference in rate was observed between the two immobilization methods); 6) wash with 50 μl of 1× TIRF buffer; and 7) addition of 50 μl of magnesium-G-actin with additional proteins.
For the processivity measurements in Fig. 5, following blocking with streptavidin and the subsequent wash (step 4 above), Alexa Fluor 647-phalloidin-F-actin seeds (1–10 nm, 0.5–1% biotin-labeled) were immobilized on the coverslip and washed with 1× TIRF buffer, and then 50–200 nm Capu was added to the slide. The actin polymerization mix was added directly to those flow chambers. Images were collected every 10 s on a DMI6000 TIRF microscope (Leica). Data were analyzed using the JFilament plugin (32) to Fiji (33). Linear regressions to obtain filament growth rates were done using NumPy (34).
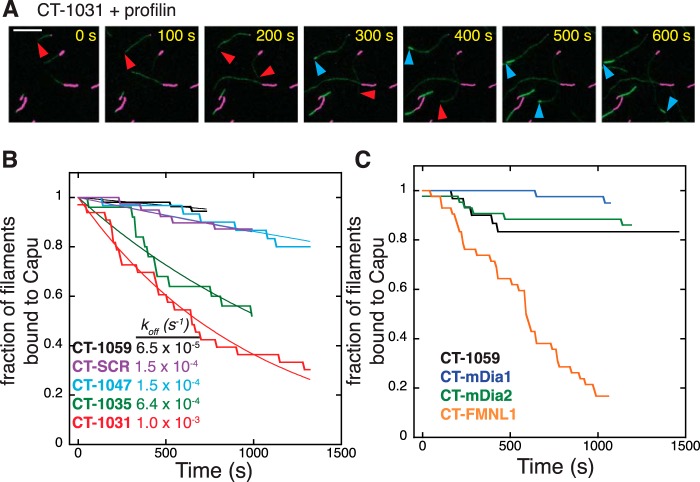
FIGURE 5.
Formin tails are a determinant of processivity. A, TIRF images showing CT-1031 dissociating from the barbed end. F-actin seeds (0.5% biotin-labeled) were stabilized with Alexa Fluor 647-phalloidin. Those seeds were incubated with 220 nm CT-1031. Finally, 0.6 μm actin (30% Oregon Green-labeled) and 3 μm profilin (without formin) were added to the chamber. Magenta, seeds; green, polymerizing actin. Red arrowheads indicate the formin-bound barbed end. Blue arrowheads point to the unbound barbed end following dissociation. Scale bar = 10 μm. B, truncation of the tail reduces the processivity of Capu, whereas scrambling the tail sequence does not compromise this activity. Seeds were incubated with 200 nm CT-1059, 65 nm CT-SCR, 250 nm CT-1047, 65 nm CT-1035, or 220 nm CT-1031 prior to adding actin. Each curve represents data from ≥25 filaments from at least two slides. Curves were fit to an exponential decay function to obtain off rates (koff). Sample imaging for CT-1059 and CT-1031 is shown in supplemental Movies 4 and 5. C, chimeric Capu constructs have varied processivity. Experiments were similar to those in B, except that the seeds were preincubated with 40 nm CT-1059, CT-mDia1, CT-mDia2, or CT-FMNL1.
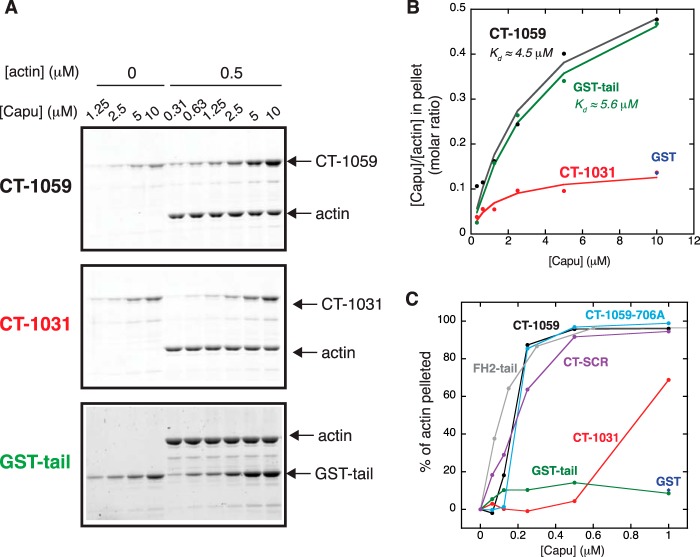
Actin Filament Cosedimentation Assays
Actin (10 μm) was polymerized in 1× KMEH for 1 h at room temperature before adding a 1:1 molar ratio of phalloidin. Capu was precleared by centrifugation at 117,000 × g for 20 min at 4 °C. After polymerization, filaments were diluted to a final concentration of 0.5 μm in the presence of 0, 0.625, 1.25, 2.5, 5, and 10 μm CT-1059, CT-1031, or GST-tail. The polymerized filaments were transferred using cut pipette tips to avoid shearing. The mixtures were incubated for 20 min at room temperature, followed by centrifugation at 89,000 × g for 10 min in 4 °C. The pellets were resolved by SDS-PAGE. The gels were stained with SyproRed and visualized using a Pharos FX Plus molecular imager with Quantity One software (Bio-Rad). Experimental data were fit using the McGhee von Hippel model (35) for a one-dimensional lattice as described previously for Capu/microtubule interactions (19).
Actin bundling experiments were similar to the actin binding experiments. Actin was diluted to a final concentration of 4 μm in the presence of varying concentrations of Capu. The filaments and protein solutions were incubated for 20 min at room temperature, followed by centrifugation at 14,000 × g for 5 min at 4 °C. The concentration of filaments in the supernatant were analyzed to create the plot in Fig. 6B.
FIGURE 6.
The Capu-tail is necessary but not sufficient for F-actin bundling. A and B, F-actin binding by CT-1059, CT-1031, GST-tail, and GST was measured by high-speed cosedimentation with 0.5 μm phalloidin-stabilized F-actin. Sypro Red-stained gels were analyzed to generate the titration curves in B. Only pellet fractions are shown. Filled circles are experimental data, and lines show a fit to the McGhee von Hippel model. C, the F-actin bundling activity of Capu was measured by low speed cosedimentation. Each construct was incubated with 4 μm phalloidin-stabilized F-actin.
RESULTS
Formin Tails Play a Role in Actin Filament Nucleation
In a previous study, we made constructs that lacked all or part of the Capu-tail and found that they had compromised filament assembly activities (Figs. 1A and 2B) (15). To better understand this observation, we measured the stability and filament assembly activity of a series of Capu truncations and chimeras. To assess whether removal or replacement of the C-terminal 28 amino acids compromised the structure of the FH2 domain, we carried out thermal denaturation experiments using circular dichroism spectroscopy. All Capu constructs underwent irreversible unfolding transitions at ≈40 °C (Fig. 2A), indicating that the truncated or chimeric versions of Capu were not significantly destabilized compared with the WT. The apparent melting temperature of Capu is similar to the thermal stability measured by differential scanning calorimetry for the mDia1 core FH2 domain (36). On the basis of the similar stabilities of WT, truncated, and chimeric Capu, we attribute the variation in filament assembly activity in Fig. 2, B—E, to the presence and identity of the tail domain as opposed to gross structural defects in the FH2 domain.
We compared actin assembly by CT-1059, CT-1047, CT-1035, and CT-1031 in bulk pyrene actin polymerization assays (Fig. 2, B and C). Bulk polymerization assays are sensitive to both the filament nucleation and elongation functions of formins. We observed large differences in the earliest time points, which suggested that the loss of filament assembly activity was due to a loss of nucleation activity. This is consistent with the observation that the mDia1-DAD domain enhances nucleation without affecting the rate of filament elongation (18). We later used TIRF microscopy to confirm that the Capu-mediated barbed end growth rate is unaffected by the presence or absence of the tail (Fig. 4D) and that CT-1059 produces more filaments than CT-1031 (Fig. 4, E and F). On the basis of a model in which the barbed end growth rate is unaffected by truncation of the tail, we calculated the concentration of barbed ends present at the time until half-maximal polymerization (t½, a regime in which we expect barbed end growth to account for ≈95% of filament polymerization) over a range of Capu concentrations (Fig. 2C). For the concentrations tested, CT-1031, the fragment of Capu that is missing the tail, produced fewer filament barbed ends, and the activities of CT-1035 and CT-1047 were intermediate to CT-1059 and CT-1031. Therefore, progressive truncation of the tail decreases the nucleation rate of Capu.
FIGURE 4.
The FH1 and FH2 domains determine the filament elongation rate. A–C, single actin filaments were observed by TIRF microscopy in the presence of profilin (A) plus 2 nm CT-1059 (B) or 66 nm CT-1031 (C). Conditions were 1 μm magnesium-G-actin (20% Oregon Green-labeled) and 5 μm profilin. Blue arrowheads show the barbed ends not bound to formin. Blue dots show pointed ends. Red arrowheads indicate formin-bound barbed ends. D, quantification of filament growth rates for truncated and 2 nm chimeric Capu in the absence and presence of profilin. For each condition, bar graphs show the mean ± S.D. (n ≥ 20 filaments from two to four slides). For the profilin alone rate, more than 20 filaments were analyzed from 12 different slides, including slides with and without formin present. Supplemental Movies 1–3 correspond to the images in A–C. E, skeletonized images showing the entire field of view for 1 μm magnesium-G-actin ± 4 nm CT-1059 or CT-1031. F, for each condition, the number of filaments for 10 different fields of view (five each from two different flow chambers). The field size was 82 × 82 μm. Scale bars = 10 μm.
Across different families, formins have varying nucleation activity (1, 6, 37). Because both the Capu-tail and mDia1-DAD enhance nucleation, we asked whether the tails play an active role in determining the nucleation rate. Specifically, we asked whether a DAD domain from another formin might increase or decrease the nucleation activity of the FH2 domain of Capu. We constructed chimeric formins with the FH1 and FH2 domains of Capu and the DAD domains of mDia1, mDia2, or FMNL1 (Fig. 1). These DAD domains were chosen because their parent formins display a range of nucleation activities (6). The observed changes in activity for the chimeras compared with CT-1059 correlated with the identity of their DAD domain. The mDia1 and mDia2 DAD domains had an activating effect, whereas the FMNL1 DAD reduced the nucleation activity of Capu (Fig. 2, D and E). Chimeras with the full-length tails of mDia1 and mDia2 were insoluble or unstable (data not shown). However, at least for mDia1, it has been shown that the DAD domain is the crucial domain for actin nucleation, whereas the rest of the tail is dispensable for this activity (18). Taken together, the data in Fig. 2 show that Capu-tail has an important function in the filament nucleation activity of Capu and that this property is conserved among formins. Specifically, formin tails are necessary for efficient nucleation, and they contribute to the rate of filament formation.
The Capu-tail Binds to Monomeric Actin
The importance of the tail domain in filament nucleation activity led us to ask whether the Capu-tail peptide alone bound monomeric actin. A similar interaction was reported for the DAD domain of mDia1 (18) and the WH2/DAD-containing tails of FMNL3 and INF2 (16, 17). We used a fluorescence anisotropy assay to test for binding between an Alexa Fluor 488-labeled Capu-tail and Latrunculin-B-bound magnesium-G-actin. We observed a saturable increase in fluorescence anisotropy when G-actin was added to the labeled Capu-tail, indicating a specific interaction (black curve, Fig. 3A). Addition of an excess of unlabeled Capu-tail reduced this effect (gray curve, Fig. 3A). We fit the competition binding data to obtain a Kd of 810 nm for the Capu-tail·G-actin complex. The data in Fig. 3A were collected under conditions with no added salt. As increasing amounts of NaCl were added to the binding buffer, the affinity of actin for Alexa Fluor 488-tail was reduced (Fig. 3B). Therefore, Capu-tail·actin binding is mediated by electrostatic interactions that are weakened in standard polymerization buffer (50 mm KCl).
To learn how the Capu-tail interacts with monomeric actin, we compared its sequence and binding to other known actin monomer binding motifs. The tail peptide does not have significant sequence homology to WH2 domains or to DAD domains except for the presence of several basic residues. The Capu-tail sequence is similar to the amphipathic helix of RPEL domains (Fig. 3C), which binds between subdomains 1 and 3 of actin in the hydrophobic cleft (26). Similar to mDia1, but unlike FMNL3, we observed minimal competition in the anisotropy assay when Drosophila profilin was added (16, 18). In contrast, both Spire-WH2-B and the RPEL1 peptide from murine MAL/MRTF-A reduce the anisotropy of Alexa Fluor 488-labeled Capu-tail in the presence of actin (Fig. 3C). However, the reproducibly sharply bent competition curve cannot be fit with a standard 1:1 competitive binding model. Therefore, we speculate that WH2 and RPEL1 affect the ability of Capu-tail to bind actin in a mechanism that is not fully described by a competitive binding model.
The interaction with monomeric actin also led us to ask how the isolated Capu-tail peptide affects actin polymerization, particularly in comparison with the RPEL1 peptide. At a 10-fold molar excess under polymerizing conditions (Fig. 3D), the Capu-tail peptide slows the rate of actin polymerization but does not change the steady-state plateau signal, whereas the RPEL1 peptide strongly sequesters magnesium-G-actin from polymerizing (Kd ≈ 1 μm in 100 mm NaCl) (26). Consistent with this finding, the Capu-tail does not alter the steady-state concentration of F-actin at a 32-fold molar excess following overnight incubation (Fig. 3E). Because Capu-tail cannot sequester like RPEL, we conclude that Capu-tail has a weak and transient interaction with monomers.
Next we asked whether dimerizing the tail by adding a GST tag would create a nucleation-competent construct as observed for other formins (16, 18). Similarly, a dimeric tail could be built using the FH2 mutant I706A (CT-1059-706A), a point mutation that disrupts a key actin binding site in the “knob” region of the FH2 domain (38). We observed some filament nucleation with CT-1059-706A (Fig. 3F, orange curves) and no filament nucleation with GST-tail (Fig. 3F, blue curves), indicating that dimerization of the tail is not sufficient to create an actin nucleator. This is in contrast with GST-mDia1-DAD, which can accelerate filament nucleation on its own but fits with the finding that the FH2 domain enhances actin monomer binding (18). GST-tail has a more pronounced effect on actin polymerization than monomeric tail does (Fig. 3, D and F), indicating that the dimerized tail may bind cooperatively to actin monomers. Taken together, our actin monomer binding data indicate that the tail has a weak interaction with monomers, most likely by binding to the hydrophobic cleft between subdomains 1 and 3 of actin.
Formin Tails Do Not Determine the Rate of Filament Growth
Formins bind to filament barbed ends, modulating their growth rate (5, 37). We asked whether formin tails affect barbed end association. First we measured filament growth rates in the presence of 1 μm magnesium-G-actin (25% Oregon Green 488-labeled). CT-1059 does not slow the growth of filament barbed ends in the absence of profilin (Fig. 4 and supplemental Movies 1 and 2) (13). In the presence of a 5-fold molar excess of Drosophila profilin (Chic), CT-1059 accelerates filament growth ∼6-fold over profilin-actin alone to a growth rate of ≈35 subunits s−1 (Fig. 4D and supplemental Movie S3). The fast-growing filaments are dim, indicating that labeling actin at Cys-374 reduces its binding affinity for Chic, as reported for other types of profilin (37).
When the tail is truncated from Capu (CT-1031) or replaced by a DAD domain, the filament elongation rate in the presence or absence of profilin is unaffected (Fig. 4D). Similarly, the presence or absence of the DAD domain of mDia1 did not affect the acceleration of barbed end growth from profilin-actin (18). These data and the direct observation of nucleation by CT-1031 versus CT-1059 (Fig. 4, E and F) support our conclusion that the slower rate of actin assembly in Fig. 2 is a result of fewer actin filament nuclei instead of reduced barbed end growth. We also conclude that the formin-mediated barbed end growth rate is determined by the FH1 and FH2 domains of formins and not by their tails.
Formin Tails Are a Determinant of Processivity
All fungal and metazoan formins characterized to date remain processively associated with the filament barbed end for 103-105 rounds of subunit addition (39). It is believed that this is an important physiological function because formins prevent capping protein from blocking barbed end growth (5). When measuring filament growth rates in the presence of profilin, we observed variability in the number of dissociation events for the constructs tested in Fig. 4. Dissociation events were counted as cases where a fast-growing, dim filament switched to slow-growing and bright (Fig. 5A and supplemental Movies 4 and 5). Dissociation events were not measurable in the absence of profilin because Capu-bound filaments grow at rates similar to unbound filaments. To quantify differences in processivity, we used a seeded growth assay where F-actin seeds containing biotin-labeled actin were immobilized on a glass coverslip and incubated with Capu before adding 0.6 μm magnesium-G-actin (30% Oregon Green-labeled, not biotin-labeled). The purpose of this setup was to minimize interactions between the formin-bound growing end of the filament and the coverslip surface.
After monomers were added and unbound formin was washed out, the fraction of fast-growing, dim filaments was tracked over time. All constructs used in Fig. 5B had similar filament growth rates (data not shown). We observed a faster decay in the fraction of filaments that were bound to Capu for CT-1031 and CT-1035 compared with CT-1059 and CT-1047, with the lowest processivity for CT-1031 (Fig. 5B). These curves were fit to a single exponential decay to obtain an off rate (koff, Fig. 5B). The characteristic run length (barbed end growth rate/koff) for CT-1059 is ≈310,000 subunits (20 subunits s−1/0.000065 s−1) and only 20,000 subunits for CT-1031. The two shorter truncations, CT-1047 and CT-1035, had intermediate off rates (Fig. 5B). We also tested CT-SCR, a construct in which the tail residues were arranged randomly (scrambled) to give a new sequence with the same total charge and theoretical pI as the original (Fig. 1C) (19). CT-SCR had a similar off rate as CT-1059 and CT-1047 (Fig. 5B). Because the processivity decreases monotonically as the C terminus is truncated and is similar for CT-1059 and CT-SCR, we conclude that nonspecific electrostatic interactions between the tail and filament contribute to processive barbed end attachment.
The chimeric Capu constructs also showed variability in their processivities (Fig. 5C). CT-mDia1 and CT-mDia2 had run lengths similar to or longer than CT-1059. CT-FMNL1 dissociated more readily from filament ends. Dissociation curves for chimeras were not readily fit by a single exponential decay. Our TIRF data indicate that although the growth rate is independent of the tail sequence, formin tails enhance processive association with the barbed end.
The Capu-tail Is Necessary but Not Sufficient for F-actin Bundling
We reasoned that enhanced processivity could be due to Capu-tail binding actin filaments. Capu has been shown to bind to F-actin and microtubules (19, 27, 40). Binding to microtubules was recently mapped to the Capu-tail and FH2 domain (19), but binding to F-actin has not been studied as carefully. Because binding of the Spire-kinase non-catalytic C-lobe domain competes with F-actin binding by CT-1059 (27), we asked whether the tail was the F-actin binding site on Capu-1059. In a high-speed cosedimentation assay (Fig. 6, A and B), removal of the tail (CT-1031) reduced F-actin binding compared with CT-1059. This is consistent with data for Capu/microtubule binding (19) and also FMNL3/F-actin binding, as assayed by filament severing (16). GST-tail bound F-actin with an affinity (Kd ≈ 5.6 μm) similar to CT-1059 (Kd ≈ 4.5 μm), whereas GST alone had no or minimal binding to F-actin. The affinity of Capu for F-actin is weaker than measured previously using a competition experiment with Spire-KIND (27). In our earlier report, filament binding was measured using protein with a C-terminal polyhistidine tag, which probably strengthened the interaction with the negatively charged filament because it was near the C-terminal F-actin interaction site. On the basis of these data, we conclude that the tail interacts with F-actin and that this interaction is a major contributor to the affinity of CT-1059 for F-actin. As discussed below, we believe that filament binding by Capu-tail and other formin tails results in enhanced processivity.
In addition to F-actin binding, CT-1059 also bundles filaments (27). We used a low-speed cosedimentation assay and TIRF microscopy to examine the role of the tail in bundling. When F-actin is titrated with increasing CT-1059 (Fig. 6C), there is a sharp transition between bundled and unbundled F-actin at a 1:16 molar ratio of CT-1059 dimer to F-actin, corresponding to ≈1% decoration of F-actin with CT-1059 (on the basis of the affinity for F-actin measured in Fig. 6B). In TIRF microscopy (supplemental Movie S6), we observed the formation of both parallel and antiparallel F-actin bundles in the presence of CT-1059-706A. Consistent with the F-actin binding data, CT-1031 had reduced or no bundling activity compared with CT-1059 (Fig. 6B and supplemental Movie S7). CT-1059-706A, FH2-tail, and CT-SCR all have bundling activities similar to CT-1059. Surprisingly, GST-tail on its own does not bundle F-actin despite the fact that it binds F-actin (Fig. 6B) and is a dimer. This suggests that undefined FH2 contacts, separate from the actin-knob binding site, along with the tail contacts, are necessary for bundling. Perhaps the binding orientation of the FH2-containing construct (CT-1059) is different from that of GST-tail so that, in the CT-1059·F-actin complex, the tails on a single dimer are forced to contact different filaments and, therefore, bundle. On the basis of these data and the lack of bundling by GST-tail, we conclude that nonspecific, electrostatic interactions with the tail are important but not sufficient for F-actin bundling.
DISCUSSION
The Role of Formin Tails in Filament Nucleation and G-actin Binding
The role of Capu-tail in nucleation is consistent with recent reports about Diaphanous-related formins (16, 18) and INF2 (41) in that the formin tail is a site for monomer binding and, thus, aids the FH2 domain in filament nucleation. The low affinity of the Capu-tail/G-actin interaction is notable when comparing it to other formin tails. The WH2-like tails of INF2 and FMNL3 bind G-actin with submicromolar or low micromolar affinity, respectively, in buffers containing 50 mm salt (16, 17). The mDia1 DAD domain has a high micromolar affinity in a buffer containing 200 mm salt (18). The Capu-tail has a low micromolar affinity in the absence of salt, and the affinity drops drastically as salt is added (Fig. 3B). On the basis of competition anisotropy measurements, it is likely that Capu-tail binds in or near the hydrophobic cleft of G-actin, similar to the RPEL and WH2 domains (26, 42).
Capu-tail also interacts with the WH2-containing actin nucleator Spire through the Spire-KIND domain (14, 15). Because of the extensive interaction interface between Spire-KIND and tail (15, 43) and the inhibitory effect of the KIND domain (27), we speculate that KIND binding and G-actin binding are mutually exclusive. Sequestering this actin binding region is at least part of the mechanism by which KIND inhibits the nucleation activity of Capu (15). Because tail/actin interactions are ≈1000-fold weaker than the tail/KIND interaction, we do not expect that actin monomers would effectively compete with this interaction to activate Capu, as reported recently for INF2 (41). Instead, we speculate that Spire binding may be a mode of enhancing the weak monomer binding of the tail, given that binding of each Spire molecule places four WH2 domains in proximity to the Capu-FH2 domain. Likewise, mDia1 binds adenomatous polyposis coli through its tail, and Bni1 binds Bud6 through its tail. In both of these cases, the formin tails have been shown to enhance nucleation (18). Perhaps the binding partner amplifies this property.
Finally, we note that chimeric Capu has nucleation rates similar to or faster than WT CT-1059 (Fig. 2E and Ref. 19). This may reflect both a conserved mechanism and the promiscuity of the hydrophobic cleft as an interaction site for G-actin binding proteins. In the Bni1p/actin cocrystal structure, this cleft interacts with a highly conserved helix in the FH2 knob (38). An attractive model for Capu-mediated filament nucleation is the handoff mechanism between the tail and FH2-knob that has been proposed for FMNL3 (44).
The Role of Formin Tails in Processive Elongation and Filament Binding
We describe a novel role for the formin tail, namely that it enhances processivity. We observed this activity directly in single filament elongation assays with our various Capu truncations and chimeras. We also see evidence for such activity for the influence of the DAD domain on processivity in bulk assays presented by Goode and coworkers (supplemental Fig. S1 in Ref. 18). They observed differences in actin assembly that could be due to a reduction in protection from capping protein when the DAD domain was truncated off of mDia1. The FH2 domain also contributes to processivity. The chimeras CT-mDia1 and CT-mDia2 have processivities similar to CT-1059 (≈310,000 subunits), and mDia1 and mDia2 FH1-FH2-tail constructs have lower processivities (≈37,500 and 92,000 subunits, respectively) (37).
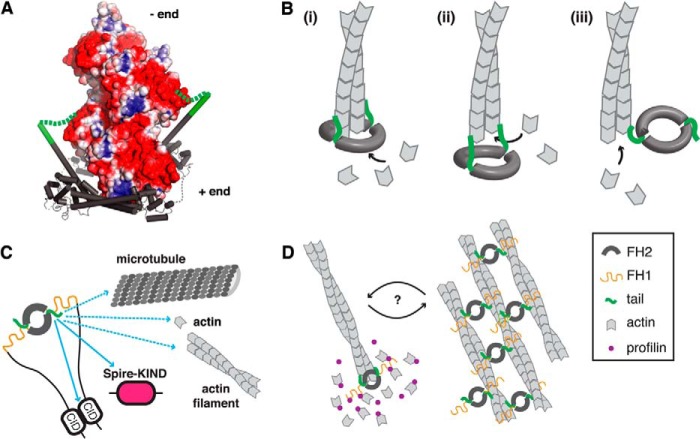
We propose a structural model for the tail as a nonspecific electrostatic tether to the filament. Shown in Fig. 7A is the DAAM1 crystal structure (PDB code 2J1D (45)) overlaid on the Bni1-actin cocrystal structure (PDB code 1Y64 (38)) and an x-ray fiber diffraction structure of F-actin (PDB code 2ZWH (46)). The surface of the actin filament has large swaths with negative electrostatic potential. The region that would most likely correspond to the Capu-tail sequence (shown in green) is oriented toward the pointed end and well positioned to make favorable electrostatic interactions with the growing filament. This interaction could help the formin stay bound to the barbed end in the putative open state when FH2 domain contacts are not fully satisfied (39). The structural model in Fig. 7A depicts the open conformation proposed by Rosen and coworkers (38). The same tail/filament interaction could be present in the open state proposed by Paul and Pollard (47, 48). It is possible that the tail could interact with the filament in an even more open FH2 configuration (Fig. 7B, ii) or in an orientation that leaves the barbed end completely free but the tail bound near the barbed end (Fig. 7B, iii).
FIGURE 7.
Model for interactions between the Capu tail and actin. A, structural model of the interaction between the tail and elongating actin filament on the basis of the structures of yeast Bni1-FH2 bound to actin (PDB code 1Y64 (38)), human Daam1-FH2 (PDB code 2J1D (45), and the structure of F-actin (46)). The electrostatic potential, calculated using the APBS plugin (53, 54) to PyMOL (Schrödinger, LLC.), is mapped onto the solvent-accessible surface area of the actin minifilament (four subunits; blue ≥ + 3 kBT/e and red ≤ − 3 kBT/e). On the basis of these structures, the positively charged tail (green helix and dashed line) is oriented toward the pointed (−) end and poised to interact favorably with the large region of negative electrostatic potential on the growing filament. B, several possible binding modes through which the tail could contribute to formin processivity. Diagram i is the same structural model as in A, in which an open binding site on the FH2 domain could accept an incoming actin subunit. Diagram ii shows a more open conformation in which only the tail interacts with the terminal subunits. Diagram iii shows a completely free barbed end in which only a single tail is bound near the barbed end. C, the tail mediates several functions of Capu: actin nucleation through G-actin binding, F-actin binding/bundling, processive barbed end association (this work), microtubule binding (19), autoinhibition (13), and Spir binding (15). We hypothesize that the latter two functions involve sequence-specific interactions (solid arrows), whereas the other functions are primarily nonspecific charge-based interactions (dashed arrows). D, because tail binding plays a role in both processive association with the barbed end and filament bundling, we speculate that these functions are mutually exclusive.
On the basis of the DAAM1 structure, the Capu-tail is predicted to overlap with the αT helix of the FH2 domain (Fig. 7A). It has been shown that replacement of this helix with a GGS flexible linker does not disrupt the actin assembly activity of mDia1 FH2-DAD (49). The processivity of the engineered GGS construct was not assayed directly. However, its wild type-like activity in bulk assays indicates that this border region between the FH2 and tail might tolerate conformational changes that allow the tail to maintain contact with the growing filament. We speculate that the tail-binding site on F-actin is different from its binding site on G-actin. The hydrophobic cleft is most likely buried by interprotomer contacts in the filament (50), and, on the basis of the structural model in Fig. 7A, the tail is positioned to interact with the outer corner of subdomain 1.
It is, perhaps, surprising that perturbations that affect the processivity of Capu do not affect the rate of barbed end growth because we would expect these two properties to be intimately related to the translocation cycle of formins. That said, variation in processivity, along with little or no variation in elongation rates, has been observed previously for a series of chimeric formins (48). It is possible that removal of the tail affects barbed end elongation rates for formin FH2 domains other than those of Capu and mDia1 (18). Capu and mDia1 are unusual among formins in that they do not slow barbed end growth in the absence of profilin (Fig. 4 and Ref. 37). Perhaps untethering either of these formins by removing their tails does not increase the rate of growth because they are already functioning at their “speed limits.” It would be interesting to test whether removal of the tail from a formin with a low gating factor such as mDia2 (51) might cause an increase in filament growth rate.
In contrast, we do see a correlation between nucleation activity and processivity (Figs. 2, C and E, and 5, B and C). We speculate that this is because G-actin and F-actin interactions are both mediated by charge/charge interactions with the tail. It is interesting that FMNL1-DAD has a high pI but reduced nucleation activity and processivity. It is possible that not only the overall charge, but also the distribution of charge in the longer DAD sequence, is important for determining how well the tail functions with the FH2 domain. FMNL1-DAD also contains proline and aromatic residues that are not found in the Dia-DAD sequences.
Significance during Drosophila Development
Our data show that Capu-tail has a role in assembling and organizing the actin cytoskeleton. Combined with earlier studies (13, 15, 19), this work establishes the tail as a central hub for the function and regulation of Capu (Fig. 7C). Several of these functions (actin filament nucleation, F-actin binding, and microtubule binding) are most likely dominated by electrostatic interactions that are not dependent on a specific sequence within Capu. This is most directly demonstrated by the approximately wild-type activities of Capu with a scrambled tail sequence (CT-SCR). Despite this, these interactions are dose-dependent and saturable, suggesting that Capu-tail binds to specific sites on F-actin, G-actin, and microtubules. Other functions, such as Spire binding and autoinhibition, are sensitive to the specific sequence within the tail. Point mutations are sufficient to block these interactions. Our data show that Capu is a processive formin with a characteristic run length on the barbed end of ≈310,000 subunits. This is on the high end of formins that have been characterized to date (37, 48, 52). The high processivity of Capu may be important for its function in a large cell like the Drosophila oocyte.
The multifunctionality of formins in general and Capu in particular has led to a variety of models for their in vivo functions. Some have proposed that microtubule and F-actin cross-linking is the major physiological role for Capu in the Drosophila oocyte (40). We found that Capu has a low micromolar affinity for actin filaments (Kd ≈ 4 μm, Fig. 6B). Further, we note that the affinity of Capu-1059 for F-actin is at least 10-fold lower than for microtubules (Kd ≈ 0.12 μm) (19) when measured under similar conditions (50 mm KCl), despite the fact that Capu-tail plays a major role in both interactions. On the basis of these affinities, actin/microtubule cross-linking is unlikely to be relevant in the cell. In addition, it has been shown that the I706A mutation leads to premature cytoplasmic streaming in the oocyte and sterility in Drosophila (20). On the basis of these data and our work showing that CT-1059-706A binds with WT affinity to microtubules (19), we favor a model where filament nucleation and elongation are the essential roles for Capu in the oocyte. We propose that F-actin side binding is most relevant in the context of barbed end elongation and processivity. It is possible that both barbed end elongation and filament bundling are physiologically relevant activities. The importance of Capu-tail for both processivity and bundling supports a model in which those two activities are mutually exclusive (Fig. 7D). We do not yet know what might regulate the switch between the two states.
Currently we do not have a mutant lacking only bundling activity that would allow us to test the role of bundling during development. In general, the multifunctionality of the tail presents challenges for designing genetic experiments that isolate a single function to test the in vivo relevance of that function.
Supplementary Material
Acknowledgments
We thank Andrey Shur for help with protein purification and TIRF reagent preparation, Catherine Leettola for help with molecular biology, Justin Bois for curve-fitting tools, and other members of the Quinlan and Reisler laboratories for discussions.
This work was supported, in whole or in part, by National Institutes of Health National Research Service Award Postdoctoral Fellowship F32GM087857 (to C. L. V.) and National Institutes of Health Grant R01GM096133 (to M. E. Q.). This work was also supported by a Whitcome fellowship (UCLA) (to B. B.), by a Burroughs-Welcome Fund Career Award in the Biomedical Sciences (to M. E. Q.), and by March of Dimes Foundation Grant 1-FY12-442 (to M. E. Q.).

This article contains supplemental Movies 1–7.
- Capu
- Cappuccino
- FH
- formin homology
- TIRF
- total internal reflection fluorescence
- DAD
- Diaphanous autoinhibitory domain
- WH2
- Wiskott-Aldrich syndrom protein homology
- KIND
- kinase noncatalytic C-lobe domain.
REFERENCES
- 1. Goode B. L., Eck M. J. (2007) Mechanism and function of formins in the control of actin assembly. Annu. Rev. Biochem. 76, 593–627 [DOI] [PubMed] [Google Scholar]
- 2. Chesarone M. A., Goode B. L. (2009) Actin nucleation and elongation factors: mechanisms and interplay. Curr. Opin. Cell Biol. 21, 28–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sagot I., Rodal A. A., Moseley J., Goode B. L., Pellman D. (2002) An actin nucleation mechanism mediated by Bni1 and profilin. Nat. Cell Biol. 4, 626–631 [DOI] [PubMed] [Google Scholar]
- 4. Pruyne D., Evangelista M., Yang C., Bi E., Zigmond S., Bretscher A., Boone C. (2002) Role of formins in actin assembly: nucleation and barbed-end association. Science 297, 612–615 [DOI] [PubMed] [Google Scholar]
- 5. Zigmond S. H., Evangelista M., Boone C., Yang C., Dar A. C., Sicheri F., Forkey J., Pring M. (2003) Formin leaky cap allows elongation in the presence of tight capping proteins. Curr. Biol. 13, 1820–1823 [DOI] [PubMed] [Google Scholar]
- 6. Harris E. S., Rouiller I., Hanein D., Higgs H. N. (2006) Mechanistic differences in actin bundling activity of two mammalian formins, FRL1 and mDia2. J. Biol. Chem. 281, 14383–14392 [DOI] [PubMed] [Google Scholar]
- 7. Gaillard J., Ramabhadran V., Neumanne E., Gurel P., Blanchoin L., Vantard M., Higgs H. N. (2011) Differential interactions of the formins INF2, mDia1, and mDia2 with microtubules. Mol. Biol. Cell 22, 4575–4587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Emmons S., Phan H., Calley J., Chen W., James B., Manseau L. (1995) Cappuccino, a Drosophila maternal effect gene required for polarity of the egg and embryo, is related to the vertebrate limb deformity locus. Genes Dev. 9, 2482–2494 [DOI] [PubMed] [Google Scholar]
- 9. Dahlgaard K., Raposo A. A., Niccoli T., St Johnston D. (2007) Capu and Spire assemble a cytoplasmic actin mesh that maintains microtubule organization in the Drosophila oocyte. Dev. Cell 13, 539–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Manseau L. J., Schüpbach T. (1989) Cappuccino and spire: two unique maternal-effect loci required for both the anteroposterior and dorsoventral patterns of the Drosophila embryo. Genes Dev. 3, 1437–1452 [DOI] [PubMed] [Google Scholar]
- 11. Higgs H. N., Peterson K. J. (2005) Phylogenetic analysis of the formin homology 2 domain. Mol. Biol. Cell 16, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Azoury J., Lee K. W., Georget V., Rassinier P., Leader B., Verlhac M. H. (2008) Spindle positioning in mouse oocytes relies on a dynamic meshwork of actin filaments. Curr. Biol. 18, 1514–1519 [DOI] [PubMed] [Google Scholar]
- 13. Bor B., Vizcarra C. L., Phillips M. L., Quinlan M. E. (2012) Autoinhibition of the formin Cappuccino in the absence of canonical autoinhibitory domains. Mol. Biol. Cell 23, 3801–3813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pechlivanis M., Samol A., Kerkhoff E. (2009) Identification of a short Spir interaction sequence at the C-terminal end of formin subgroup proteins. J. Biol. Chem. 284, 25324–25333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Vizcarra C. L., Kreutz B., Rodal A. A., Toms A. V., Lu J., Zheng W., Quinlan M. E., Eck M. J. (2011) Structure and function of the interacting domains of Spire and Fmn-family formins. Proc. Natl. Acad. Sci. U.S.A. 108, 11884–11889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Heimsath E. G., Jr., Higgs H. N. (2012) The C terminus of formin FMNL3 accelerates actin polymerization and contains a WH2 domain-like sequence that binds both monomers and filament barbed ends. J. Biol. Chem. 287, 3087–3098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chhabra E. S., Higgs H. N. (2006) INF2 is a WASP homology 2 motif-containing formin that severs actin filaments and accelerates both polymerization and depolymerization. J. Biol. Chem. 281, 26754–26767 [DOI] [PubMed] [Google Scholar]
- 18. Gould C. J., Maiti S., Michelot A., Graziano B. R., Blanchoin L., Goode B. L. (2011) The formin DAD domain plays dual roles in autoinhibition and actin nucleation. Curr. Biol. 21, 384–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Roth-Johnson E. A., Vizcarra C. L., Bois J. S., Quinlan M. E. (2014) Interaction between microtubules and the Drosophila formin Cappuccino and its effect on actin assembly. J. Biol. Chem. 289, 4395–4404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Quinlan M. E. (2013) Direct interaction between two actin nucleators is required in Drosophila oogenesis. Development 140, 4417–4425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Breitsprecher D., Jaiswal R., Bombardier J. P., Gould C. J., Gelles J., Goode B. L. (2012) Rocket launcher mechanism of collaborative actin assembly defined by single-molecule imaging. Science 336, 1164–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tu D., Graziano B. R., Park E., Zheng W., Li Y., Goode B. L., Eck M. J. (2012) Structure of the formin-interaction domain of the actin nucleation-promoting factor Bud6. Proc. Natl. Acad. Sci. U.S.A. 109, E3424–E3433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Warrens A. N., Jones M. D., Lechler R. I. (1997) Splicing by overlap extension by PCR using asymmetric amplification: an improved technique for the generation of hybrid proteins of immunological interest. Gene 186, 29–35 [DOI] [PubMed] [Google Scholar]
- 24. Lammers M., Rose R., Scrima A., Wittinghofer A. (2005) The regulation of mDia1 by autoinhibition and its release by Rho·GTP. EMBO J. 24, 4176–4187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Stemmer W. P., Crameri A., Ha K. D., Brennan T. M., Heyneker H. L. (1995) Single-step assembly of a gene and entire plasmid from large numbers of oligodeoxyribonucleotides. Gene 164, 49–53 [DOI] [PubMed] [Google Scholar]
- 26. Mouilleron S., Guettler S., Langer C. A., Treisman R., McDonald N. Q. (2008) Molecular basis for G-actin binding to RPEL motifs from the serum response factor coactivator MAL. EMBO J. 27, 3198–3208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Quinlan M. E., Hilgert S., Bedrossian A., Mullins R. D., Kerkhoff E. (2007) Regulatory interactions between two actin nucleators, Spire and Cappuccino. J. Cell Biol. 179, 117–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zuchero J. B. (2007) In vitro actin assembly assays and purification from Acanthamoeba. Methods Mol. Biol. 370, 213–226 [DOI] [PubMed] [Google Scholar]
- 29. Pollard T. D. (1986) Rate constants for the reactions of ATP- and ADP-actin with the ends of actin filaments. J. Cell Biol. 103, 2747–2754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dirks R. M., Bois J. S., Schaeffer J. M., Winfree E., Pierce N. A. (2007) Thermodynamic analysis of interacting nucleic acid strands. SIAM Rev. 49, 65–88 [Google Scholar]
- 31. Bois J. S. (2007) Analysis of Interacting Nucleic Acids in Dilute Solutions, Ph.D. thesis, California Institute of Technology [Google Scholar]
- 32. Smith M. B., Li H., Shen T., Huang X., Yusuf E., Vavylonis D. (2010) Segmentation and tracking of cytoskeletal filaments using open active contours. Cytoskeleton 67, 693–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J.-Y., White D. J., Hartenstein V., Eliceiri K., Tomancak P., Cardona A. (2012) Fiji: an open-source platform for biological-image analysis. Nat. Meth. 9, 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Oliphant T. E. (2007) Python for scientific computing. Comput. Sci. Eng. 9, 10–20 [Google Scholar]
- 35. McGhee J. D., von Hippel P. H. (1974) Theoretical aspects of DNA-protein interactions: co-operative and non-co-operative binding of large ligands to a one-dimensional homogeneous lattice. J. Mol. Biol. 86, 469–489 [DOI] [PubMed] [Google Scholar]
- 36. Kupi T., Gróf P., Nyitrai M., Belágyi J. (2013) Interaction of formin FH2 with skeletal muscle actin: EPR and DSC studies. Eur. Biophys. J. 42, 757–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kovar D. R., Harris E. S., Mahaffy R., Higgs H. N., Pollard T. D. (2006) Control of the assembly of ATP- and ADP-actin by formins and profilin. Cell 124, 423–435 [DOI] [PubMed] [Google Scholar]
- 38. Otomo T., Tomchick D. R., Otomo C., Panchal S. C., Machius M., Rosen M. K. (2005) Structural basis of actin filament nucleation and processive capping by a formin homology 2 domain. Nature 433, 488–494 [DOI] [PubMed] [Google Scholar]
- 39. Paul A. S., Pollard T. D. (2009) Review of the mechanism of processive actin filament elongation by formins. Cell Motil. Cytoskeleton 66, 606–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rosales-Nieves A. E., Johndrow J. E., Keller L. C., Magie C. R., Pinto-Santini D. M., Parkhurst S. M. (2006) Coordination of microtubule and microfilament dynamics by Drosophila Rho1, Spire and Cappuccino. Nat. Cell Biol. 8, 367–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ramabhadran V., Hatch A. L., Higgs H. N. (2013) Actin monomers activate inverted formin 2 by competing with its autoinhibitory interaction. J. Biol. Chem. 288, 26847–26855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chereau D., Kerff F., Graceffa P., Grabarek Z., Langsetmo K., Dominguez R. (2005) Actin-bound structures of Wiskott-Aldrich syndrome protein (WASP)-homology domain 2 and the implications for filament assembly. Proc. Natl. Acad. Sci. U.S.A. 102, 16644–16649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zeth K., Pechlivanis M., Samol A., Pleiser S., Vonrhein C., Kerkhoff E. (2011) Molecular basis of actin nucleation factor cooperativity: crystal structure of the Spir-1 kinase non-catalytic C-lobe domain (KIND)·formin-2 formin SPIR interaction motif (FSI) complex. J. Biol. Chem. 286, 30732–30739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Thompson M. E., Heimsath E. G., Gauvin T. J., Higgs H. N., Kull F. J. (2013) FMNL3 FH2–actin structure gives insight into formin-mediated actin nucleation and elongation. Nat. Struct. Mol. Biol. 20, 111–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lu J., Meng W., Poy F., Maiti S., Goode B. L., Eck M. J. (2007) Structure of the FH2 domain of Daam1: implications for formin regulation of actin assembly. J. Mol. Biol. 369, 1258–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Oda T., Iwasa M., Aihara T., Maéda Y., Narita A. (2009) The nature of the globular- to fibrous-actin transition. Nature 457, 441–445 [DOI] [PubMed] [Google Scholar]
- 47. Paul A. S., Paul A., Pollard T. D., Pollard T. (2008) The role of the FH1 domain and profilin in formin-mediated actin-filament elongation and nucleation. Curr. Biol. 18, 9–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Paul A. S., Pollard T. D. (2009) Energetic requirements for processive elongation of actin filaments by FH1FH2-formins. J. Biol. Chem. 284, 12533–12540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Otomo T., Tomchick D. R., Otomo C., Machius M., Rosen M. K. (2010) Crystal structure of the formin mDia1 in autoinhibited conformation. PLoS ONE 5, e12896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Dominguez R., Holmes K. C. (2011) Actin structure and function. Annu. Rev. Biophys. 40, 169–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Vavylonis D., Kovar D. R., O'Shaughnessy B., Pollard T. D. (2006) Model of formin-associated actin filament elongation. Mol. Cell 21, 455–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Scott B. J., Neidt E. M., Kovar D. R. (2011) The functionally distinct fission yeast formins have specific actin-assembly properties. Mol. Biol. Cell 22, 3826–3839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Baker N. A., Sept D., Joseph S., Holst M. J., McCammon J. A. (2001) Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U.S.A. 98, 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Dolinsky T. J., Czodrowski P., Li H., Nielsen J. E., Jensen J. H., Klebe G., Baker N. A. (2007) PDB2PQR: expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res. 35, W522–W525 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.