Background: Hitherto, bacterial thiol dioxygenases were only rarely identified and characterized, and the catabolism of mercaptosuccinate is scarcely known.
Results: Mercaptosuccinate dioxygenase catalyzes exclusively the conversion of mercaptosuccinate yielding succinate and sulfite.
Conclusion: Mercaptosuccinate dioxygenase represents a novel thiol dioxygenase and is the key enzyme during mercaptosuccinate degradation.
Significance: The identification of this thiol dioxygenase provides new insights in substrate specificity of cupin motif containing dioxygenases.
Keywords: Bacterial Metabolism, Biodegradation, Dioxygenase, Enzyme, Enzyme Kinetics, Iron, Thiol, Variovorax Paradoxus, Mercaptosuccinate
Abstract
The versatile thiol mercaptosuccinate has a wide range of applications, e.g. in quantum dot research or in bioimaging. Its metabolism is investigated in Variovorax paradoxus strain B4, which can utilize this compound as the sole source of carbon and sulfur. Proteomic studies of strain B4 resulted in the identification of a putative mercaptosuccinate dioxygenase, a cysteine dioxygenase homologue, possibly representing the key enzyme in the degradation of mercaptosuccinate. Therefore, the putative mercaptosuccinate dioxygenase was heterologously expressed, purified, and characterized in this study. The results clearly demonstrated that the enzyme utilizes mercaptosuccinate with concomitant consumption of oxygen. Thus, the enzyme is designated as mercaptosuccinate dioxygenase. Succinate and sulfite were verified as the final reaction products. The enzyme showed an apparent Km of 0.4 mm, and a specific activity (Vmax) of 20.0 μmol min−1 mg−1 corresponding to a kcat of 7.7 s−1. Furthermore, the enzyme was highly specific for mercaptosuccinate, no activity was observed with cysteine, dithiothreitol, 2-mercaptoethanol, and 3-mercaptopropionate. These structurally related thiols did not have an inhibitory effect either. Fe(II) could clearly be identified as metal cofactor of the mercaptosuccinate dioxygenase with a content of 0.6 mol of Fe(II)/mol of enzyme. The recently proposed hypothesis for the degradation pathway of mercaptosuccinate based on proteome analyses could be strengthened in the present study. (i) Mercaptosuccinate is first converted to sulfinosuccinate by this mercaptosuccinate dioxygenase; (ii) sulfinosuccinate is spontaneously desulfinated to succinate and sulfite; and (iii) whereas succinate enters the central metabolism, sulfite is detoxified by the previously identified putative molybdopterin oxidoreductase.
Introduction
The organo-sulfur compound mercaptosuccinate (MS)2 is widely used in diverse fields, e.g. in bioimaging for the detection of markers in breast cancer cells (1, 2) or as stabilizing agent in quantum dots (3). Moreover, as a medical application, hydroxyapatite coupled with poly(allyl methacrylate) and MS can form platin complexes of the anticancer drug cis-diaminedichloroplatinum(II) (cis-platin), which could be applied as nanocarrier for controllable drug delivery to the corresponding cells in the future (4).
Variovorax paradoxus B4 is a Gram-negative, rod-shaped organism belonging to the β-Proteobacteria and was originally isolated due to its ability to utilize MS as sole source of carbon and sulfur (5). The genus Variovorax comprises miscellaneous species, which are able to degrade a remarkable range of substrates and often occur in heavily polluted environments (reviewed by Satola and colleagues (6)). Furthermore, they represent potential plant symbionts, e.g. V. paradoxus 5C-2 in symbiosis with Pivum sativum resulted in improved growth and efficiency of water use for the plant when exposed to water stress (7).
Thiol dioxygenases belong to the cupin superfamily and are characterized by their common β-barrel core as well as their partially conserved cupin motifs (8–10). However, these enzymes exhibit only low overall similarity concerning their amino acid sequences (11). In eukaryotes, cysteine dioxygenase is one of the most important representatives of this family, and it is crucial for the regulation of cysteine levels in the cells (12, 13). It catalyzes the irreversible reaction of cysteine to cysteine sulfinate, which is then transaminated to β-sulfinopyruvate and finally decomposes to form pyruvate and sulfite. Furthermore, cysteine dioxygenase activity is also of importance for the synthesis of taurine in eukaryotic cells (14). In bacteria, only a small number of cysteine dioxygenases has been clearly identified and characterized so far (11, 15). Moreover, cysteine dioxygenase homologues of V. paradoxus TBEA6 and Ralstonia eutropha H16 were identified and characterized as being mercaptopropionate dioxygenases (Mdo) (16). 3-Mercaptopropionate was used as a substrate, whereas the enzymes were incapable of utilizing cysteine (16). These results imply a strong versatility of cysteine dioxygenase homologues concerning the substrate range.
Only recently, another putative novel thiol dioxygenase was identified during proteomic studies with V. paradoxus B4 indicating that this protein might be a mercaptosuccinate dioxygenase and would therefore represent the key enzyme in the degradation of MS in this bacterium (17). Although the putative thiol dioxygenase was originally annotated as a hypothetical protein, further in silico analyses resulted in a hit for the COG5553 domain in the NCBI database comprising metal-dependent enzymes of the double-stranded β helix superfamily (17). Additionally, InterProScan 5 for functional analysis of proteins (EMBL-EBI, Hinxton, United Kingdom) revealed an RmlC-like cupin domain and an RmlC-like jellyroll-fold, representing the aforementioned conserved β-barrel core of thiol dioxygenases (17). These findings supported the assumption that the hypothetical protein might actually be a thiol dioxygenase. In V. paradoxus B4, MS is supposedly converted to sulfinosuccinate by the aforementioned putative MS dioxygenase and then cleaved into succinate and sulfite either by a so far unknown enzyme, by spontaneous hydrolysis, or even by the putative MS dioxygenase itself (17). To verify the postulated reaction of the putative MS dioxygenase and to further unravel the degradation of MS, the enzyme was heterologously expressed and characterized in this study.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Growth Conditions
Bacterial strains, plasmids, and oligonucleotides are listed in Table 1. Strains of Escherichia coli were cultivated in liquid or on solid lysogeny broth (LB) (18) containing ampicillin (75 μg/ml) and chloramphenicol (34 μg/ml).
TABLE 1.
Bacterial strains, plasmids and oligonucleotides (primers) used in this study
| Strain, plasmid, or primer | Relevant characteristic | Source |
|---|---|---|
| Strains | ||
| V. paradoxus B4 | Wild type, MS-degrading | DSM 21786 |
| E. coli Top10 | F− mcrA Δ(mrr-hsdRMS-mcrBC) rpsL nupG Φ80lacZΔM15 ΔlacX74 deoR recA1 araD139 Δ(ara-leu)7697 galU galK endA1 | Invitrogen |
| E. coli BL21(DE3) pLysS | F− ompT hsdSB (rB− mB−) gal dcm (DE3)/pLysS (Cmr) | Novagen |
| Plasmids | ||
| pET-23a(+) | pBR322 ori, f1 ori His6; Apr T7lac | Novagen |
| Primers | ||
| Vp41240_NdeI_fw | AAAACATATGAGCACCCTGTCCGATCCAC | Eurofins MWG Operon, Ebersberg, Germany |
| Vp41240_HindIII_rv | AAAAAGCTTGCGGCGCGTCTGCACG | Eurofins MWG Operon, Ebersberg, Germany |
Cloning, Expression, and Purification of Putative MS Dioxygenase
The putative MS dioxygenase is encoded by the gene with the locus tag NC_0022247.1 and will be designated as msdoB4. Polymerase chain reaction for amplification of msdoB4 was carried out with Phusion High-Fidelity DNA Polymerase (Thermo Scientific, Schwerte, Germany) using total genomic DNA of V. paradoxus B4 and the oligonucleotides listed in Table 1. To extract the desired DNA from an agarose gel, the peqGOLD Gel Extraction Kit (PEQLAB Biotechnologie GmbH, Erlangen, Germany) was applied, following the manufacturer's instructions. Then, DNA and vector pET23a(+) were digested using FastDigest® HindIII and FastDigest® NdeI (both Thermo Scientific, Schwerte, Germany) according to the manufacturer's instructions. Ligation was achieved with T4 DNA ligase (Thermo Scientific). The readily prepared plasmid was used for transformation of CaCl2-competent E. coli Top10 cells (18). For the isolation of plasmid DNA, the peqGOLD Plasmid Miniprep Kit I (PEQLAB Biotechnologie GmbH, Erlangen, Germany) was used according to the manufacturer's instructions. For verification of the correct insert, the isolated plasmid was sequenced, which was carried out by Seqlab (Sequence Laboratories Göttingen GmbH, Göttingen, Germany). After verification of pET23a(+)::msdoB4, competent cells of E. coli BL21(DE3) pLysS (Novagen) were transformed with the plasmid. The main culture was cultivated in 50 ml of LB with antibiotics inoculated with 0.5 ml of a well grown preculture (approximately 16 h, 30 °C, 120 rpm). Induction was achieved by addition of isopropyl 1-thio-β-d-galactopyranoside (final concentration of 400 μm) at an optical density of approximately 0.5 at a wavelength of 600 nm. Then, the culture was incubated at 25 °C for another 24 h at 120 rpm. Cell harvest was carried out in a Universal 320 R centrifuge (Andreas Hettich GmbH & Co. KG, Tuttlingen, Germany) at 4 °C and 9,000 rpm for 15 min. The resulting pellet was stored at −20 °C until further use. For cell disruption, cells were first resuspended in an appropriate amount of binding buffer (50 mm Tris/HCl (pH 7.4), 500 mm NaCl, 20 mm imidazole) and then disrupted by sonication (30 Hz, 50% amplitude, 1 min/ml) (Sonopuls HD2200 MS72, Bandelin Electronic GmbH & Co. KG, Berlin, Germany). The crude extract was then centrifuged at 15,000 × g at 4 °C for 15 min (Centrifuge 5424R, Eppendorf, Hamburg, Germany). For purification, the obtained supernatant was applied to nickel-nitriloacetate (Ni-NTA) affinity chromatography via a His SpinTrap column (GE Healthcare, Uppsala, Sweden) according to the manufacturer's instructions; elution was achieved by a buffer containing 500 mm imidazole instead of 20 mm. The purified enzyme was directly used for further experiments or stored on ice until further use. Protein concentrations were determined according to the method of Bradford (19).
Qualitative Enzyme Assay Using Ellman's Reagent
As a qualitative enzyme assay (20), mixtures with a final volume of 1 ml consisting of 50 mm Tris/HCl (pH 7.4), different concentrations of MS (final concentrations up to 0.1 mm), and 4 μg/ml enzyme were incubated in a cuvette for 30 min at room temperature. Then, Ellman's reagent 5,5′-dithiobis(2-nitrobenzoic acid) (DTNB) was added to a final concentration of 0.2 mm. DTNB reacts with free thiol groups resulting in the release of yellow 2-nitro-5-thiobenzoic acid whose absorption can be followed at 412 nm (ϵ412 nm = 14,150 m−1 cm−1) (21).
Quantitative Activity Assay Applying Oxygen Electrode Measurements
For the quantitative assay the oxygen sensor OXMR connected to picoammeter PA2000 and the software MicOx (all from Unisense, Aarhus, Denmark) were applied according to the manufacturer's instructions.
For calibration, the oxygen electrode was first incubated in a solution containing 0.1 m NaOH and 0.1 m ascorbic acid until a stable value was obtained for these anoxic conditions. To obtain the value for oxygen-saturated conditions, the electrode was incubated in buffer thoroughly aerated with compressed air until a stable value was observed.
The pH optimum was determined applying a broad range buffer composed of (each 40 mm) acetic acid, boric acid, and phosphoric acid (22). The respective pH was adjusted using 0.2 m NaOH solution. Activity was measured at the following pH values: 4.5, 5.0, 5.5, 6.0, 6.5, 7.0, 7.5, 8.0, 8.5, 9.0, 9.5, 10.0, 10.5, 11.0, 11.5, and 12.0.
For determination of kinetic data, substrate specificity, etc. measurements were carried out in 100 mm Tris/HCl buffer (pH 7.4) with different concentrations of mercaptosuccinate or other potential substrates and/or metal cofactors. The reaction was started by addition of 2 μg/ml of enzyme. Control reactions without enzyme or with denatured enzyme were conducted to exclude or detect possible oxygen consumption other than catalyzed by the enzyme. All measurements were carried out in triplicate. To obtain kinetic data, the following MS concentrations were used: 0.05, 0.1, 0.5, 0.75, 1.0, 2.0, 5.0, 7.5, 10, 20, 30, 50, and 75 mm. Nonlinear regression was performed using the Solver add-in of Microsoft Office Excel 2010.
Experimental Setup for Suppression of Disulfide Formation
To obtain enough material for GC and GC/MS analyses, enzyme assays were performed at a larger scale of 4 ml (and were lyophylized) containing 50 mm Tris/HCl (pH 7.4), 5 mm MS, and 2 μg/ml of enzyme. These assays were additionally supplied with oxygen gas via a cannula to ensure sufficient oxygen concentrations for the enzyme reaction. However, the formation of the MS-disulfide was repeatedly observed during GC/MS analyses most probably due to the high oxygen concentrations. To prevent disulfide formation, the enzyme assay setup was modified as described below. A 250-ml Duran bottle (Schott, Mainz, Germany) containing different screw threads (1 × GL 45, 1 × GL 25, 2 × GL 18, and 2 × GL 14) with suitable septa or silicone sealing gaskets was chosen. An amperometric oxygen sensor OxyFerm FDA 225 (Hamilton Messtechnik GmbH, Höchst, Germany) was applied to follow the pO2 concentration online. For calibration, the solution was first vigorously aerated with nitrogen gas (0% oxygen saturation) followed by intense aeration with compressed air (100% oxygen saturation). The probe was guided through the GL 45 lid and the silicone sealing gasket both containing a hole with 2 mm less diameter than the diameter of the probe to ensure leak-tightness of the system. A glass distribution tube (porosity 1) (Sigma) was guided through the GL 25 thread containing a suitable gasket (8 mm hole); this tube was used for controlled oxygen supply during the assay using compressed air. Excess pressure was released through a slightly opened thread. The MS stock solution was freshly prepared in N2-saturated 50 mm Tris/HCl (pH 7.4) buffer to prevent disulfide formation. Additionally, the assay solution was N2-saturated prior to addition of MS and enzyme. The assay was performed in a final volume of 150 ml containing 50 mm Tris/HCl (pH 7.4), 50 mm MS, and 6 μg/ml of enzyme, and was stirred at 25 °C for 8 h. The flow of the compressed air was adjusted keeping pO2 below 20% saturation to prevent disulfide formation. Samples were withdrawn at the beginning of the assay and after 8 h of enzyme reaction, and the samples were subsequently objected to GC and GC/MS analysis as described below.
Determination of Sulfite and/or Sulfate Content
The formation of sulfite in the 150-ml enzyme assay was measured applying a modified protocol of Sörbo (23). Therefore, 100 ml of a solution containing 0.98 g of BaCl2 × 2 H2O and 15 g of PEG-6000 was prepared. Subsequently, 500 μl of a sample were mixed thoroughly with 100 μl of 30% (v/v) hydrogen peroxide, 200 μl of 0.5 m HCl, and 200 μl of the BaCl2-PEG solution. Hydrogen peroxide was used to convert sulfite to sulfate for the assay. This mixture was incubated for 5 min at ambient temperature to allow complete precipitation. The sample was then mixed thoroughly again, and the optical density at 600 nm was determined. For the determination of sulfate formation, the setup was analogous, only hydrogen peroxide was omitted and substituted by the addition of 100 μl of 50 mm Tris/HCl (pH 7.4).
Analytical Size-exclusion Chromatography
To determine the native molecular weight of the putative MS dioxygenase (MsdoB4), an analytical size-exclusion chromatography on a Superdex 200 HP 16/600 column was carried out as described elsewhere (24). Seven milligrams of Ni-NTA-purified enzyme were loaded onto the column.
Metal Cofactor Analysis of Putative MS Dioxygenase
To obtain apoenzyme without metal cofactor, the purified protein was dialyzed. Because the putative MS dioxygenase is a cysteine dioxygenase homologue and therefore contains most probably Fe(II) as metal cofactor, a buffer containing 2 mm 1,10-phenanthroline was chosen to remove all metal ions from the enzyme. Prior to dialysis, the dialysis tube ZelluTrans with a molecular mass cutoff (MWCO) of 12,000–14,000 Da (flat width 45 mm, thickness 20 μm) (Carl Roth GmbH & Co. KG, Karlsruhe, Germany) had to be prepared as follows. The dialysis tube was first placed in H2Odest for 15 min, then incubated for 30 min at 80 °C in a bigger volume of 2% (w/v) NaHCO3 (in H2Odest) on a stirrer for softening the membrane, and finally by transferring the membrane into 10 mm EDTA (in H2OMillipore) for removing possible metal contaminations. The 10 mm EDTA solution was replaced by H2OMillipore, and the membrane was incubated in this solution for another 30 min at 80 °C on a stirrer for total removal of EDTA. After preparation and cooling of the dialysis tube, the enzyme solution was filled into the tube and transferred to a 1000-ml beaker that already contained 50 mm Tris/HCl buffer (pH 7.4) with 2 mm 1,10-phenanthroline. The enzyme solution was dialyzed against this buffer for 3 h at 4 °C, and then dialyzed for approximately 24 h against 50 mm Tris/HCl buffer (pH 7.4) at 4 °C and eventually concentrated applying Vivaspin® 6 with MWCO 10,000 Da (Sartorius AG, Göttingen, Deutschland) following the manufacturer's instructions. Finally, the enzyme was supplied with different metal ions for the activity assay in final concentrations of 0.1 mm: cobalt, copper(II), iron(II), iron(III), magnesium, manganese, nickel, and zinc.
Determination of the Iron Content of the Putative MS Dioxygenase Using Bathophenanthrolinedisulfonic Acid
The determination of the iron content of the enzyme was performed as described elsewhere (25). For the analysis, 100 μl of 100 μm enzyme in 50 mm Tris/HCl buffer (pH 7.4) were applied. The extinction coefficient of the Fe(II)-bathophenanthroline complex is ϵ535 nm = 22,400 m−1 cm−1 (26).
GC and GC/MS Analyses
Enzyme assay mixtures (4 ml with 5 mm MS and 2 μg/ml of enzyme or 1 ml of the 150-ml enzyme assay) were lyophilized. In the second step, the lyophilisates, standards of MS, succinate, and mercaptopropionate were subjected to methylation as described previously (24). The GC analyses were performed with an HP6850 gas chromatograph with a BP21 capillary column (50 m × 0.22 mm; film thickness, 250 nm; SGE, Darmstadt, Germany) and a flame ionization detector as described elsewhere (27). Furthermore, GC/MS analyses were carried out with an HP6890 gas chromatograph equipped with a model 5973 EIMSD mass selective detector (Hewlett-Packard, Waldbronn, Germany) as described previously (24). The identification of peaks was performed using AMDIS software (28) in combination with the NIST database.
Synthesis of Sulfinosuccinate
Synthesis of sulfinosuccinate was performed by adaptation of the instructions of Filby and colleagues (29) in U.S. patent US3950404A. Mercaptosuccinate was methylated prior to synthesis as described above to increase the solubility. The product of the synthesis was analyzed by GC/MS.
Multiple Sequence Alignment of Putative MS Dioxygenase with Other Thiol Dioxygenases
For generating a multiple sequence alignment of the different thiol dioxygenases, the free software BioEdit (Tom Hall, Ibis Biosciences, Carlsbad, CA) was used. The amino acid sequences of the cysteine dioxygenases (Cdos) from human, mouse, and rat were chosen because they represent those investigated in most detail. As representatives for prokaryotic Cdos, the corresponding amino acid sequences of Bacillus subtilis and Cupriavidus necator JMP134 were chosen, because the Cdo of B. subtilis is biochemically characterized and the crystal structure of the Cdo of C. necator JMP134 is available. Furthermore, Advenella mimigardefordensis DPN7T is able to degrade MS (30); therefore, the nearest homologous protein of this strain to MsdoB4 was included in multiple sequence alignment as well. Additionally, the mercaptopropionate dioxygenases of R. eutropha H16 and V. paradoxus TBEA6 were chosen as representatives for thiol dioxygenases other than Cdos. The Cdo of strain B4 was included in the alignment also.
RESULTS
Identification of the Putative MS Dioxygenase
In the course of proteomic studies with V. paradoxus strain B4 (17), several isoforms of a protein with significant up-regulation during growth with MS as carbon source in comparison to growth with gluconate or succinate were identified. This protein was annotated as a hypothetical protein (VAPA_1c41240); however, it showed 46% amino acid similarity to an annotated cysteine dioxygenase of Ralstonia pickettii DTP0602 (17). Further domain searches revealed that the protein might belong to the COG5553 superfamily, comprising metal-dependent enzymes of the double-stranded β helix superfamily (17). This was supported by searches with InterProScan 5 for functional analysis of proteins (EMBL-EBI, Hinxton, United Kingdom) indicating an RmlC-like cupin domain and an RmlC-like jellyroll-fold (17). Cupin domains are characteristic for cysteine dioxygenases and their homologues (11). The amino acid sequence of the putative MS dioxygenase contained the cupin 1 motif (GX5HXHX3–6EX6G) of cysteine dioxygenases, although the conserved glutamate residue is replaced by alanine (Fig. 1) (11). Furthermore, homologies to the cupin 2 motif (GX5–7PXGX2HX3N) were detected, but two of the conserved residues were substituted by others in the MsdoB4 (Fig. 1): instead of the second glycine a glutamate residue occurred and asparagine was substituted by an isoleucine residue. However, all strictly conserved histidine residues required for the coordination of Fe(II) could be identified (Fig. 1) (15). The corresponding gene was cloned and heterologously expressed in E. coli BL21(DE3) pLysS. The protein could be purified to electrophoretic homogeneity applying Ni-NTA affinity chromatography (Fig. 2).
FIGURE 1.
Multiple sequence alignment of amino acid sequences of different cysteine dioxygenases and homologous enzymes from eukaryotic and prokaryotic species. The alignment was generated applying the free software BioEdit (Tom Hall, Ibis Biosciences, Carlsbad, CA). Cupin motifs 1 and 2 are highlighted in gray, strictly conserved amino acid residues among all investigated sequences are highlighted in black, and histidine residues responsible for the coordination of iron are marked by an asterisk.
FIGURE 2.
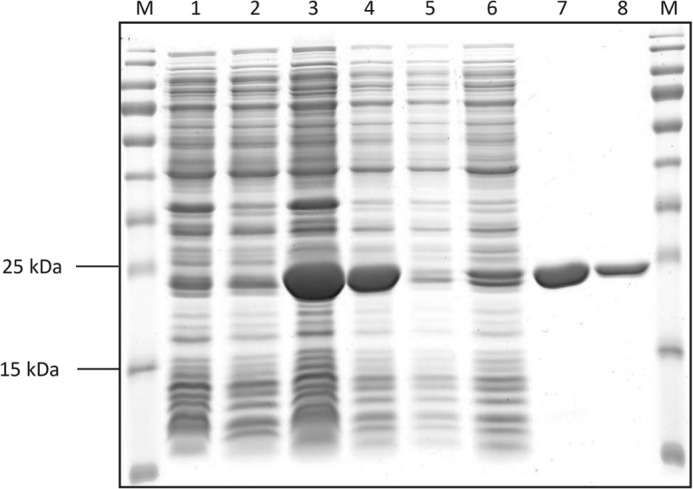
Image of an SDS-polyacrylamide gel after electrophoresis, staining, and de-staining. Lane M, PageRulerTM Prestained Protein Ladder (Thermo Scientific, Schwerte, Germany); lane 1, crude extract of E. coli BL21(DE3) pLysS pET-23a(+) cells as negative control; lane 2, soluble fraction of E. coli BL21(DE3) pLysS pET-23a(+) cells as negative control; lane 3, crude extract of E. coli BL21 (DE3) pLysS pET-23a(+)::msdo cells; lane 4, soluble fraction of E. coli BL21(DE3) pLysS pET-23a(+)::msdo cells; lane 5, flow through after loading of His SpinTrap column; lane 6, washing fraction of His SpinTrap purification; lane 7, eluate (10 μg); lane 8, eluate (5 μg). Expected size for MsdoB4: 23 kDa.
Activity Assays and Kinetic Data
After the successful purification of the putative thiol dioxygenase, an assay to determine the activity of the enzyme had to be established. As a first approach, a qualitative assay applying DTNB was chosen, where the DTNB reacts with free thiol groups of possibly remaining MS after a certain time of incubation resulting in 2-nitro-5-thiobenzoic acid, which exhibits a yellow color observable at 412 nm (ϵ412 nm = 14,150 m−1 cm−1) (20, 21). Testing different concentrations of MS (0.01, 0.05, and 0.1 mm) in the DTNB enzyme assay, it could be shown that the enzyme did indeed consume MS resulting in the absence of color. Measurements with an oxygen electrode applying 2 μg/ml of the enzyme and different concentrations of MS confirmed the oxygen dependence of the catalyzed reaction, which indicates the following reaction.
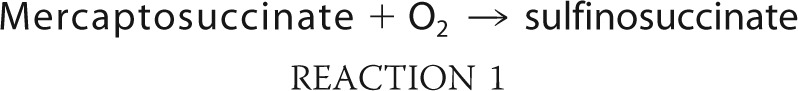 |
The pH optimum of the enzyme was determined revealing a broad range of activity between pH 5.0 and 11.5, with the highest activity observed at pH 7.0 (data not shown). Because the enzyme appeared to be more stable in Tris/HCl buffer and showed higher activity at pH 7.4 than at pH 7.0 using this buffer, 100 mm Tris/HCl (pH 7.4) was chosen for further experiments. An apparent Km value of 0.4 mm, specific activity (Vmax) of 20.0 μmol min−1 mg−1, and a kcat of 7.7 s−1 could be determined (Fig. 3) (Table 2). With denatured protein as a control, no oxygen was consumed. To analyze the products of the reaction, lyophilized enzyme assay reaction mixtures were methylated and then objected to GC/MS analysis. Because the formation of the MS-disulfide was repeatedly observed, which is most probably caused by high oxygen concentrations, the assay was modified as described under “Experimental Procedures” by applying controlled oxygen supply to prevent the disulfide formation. The withdrawn samples were analyzed by GC and GC/MS and their sulfite/sulfate content was determined. Due to the results obtained from GC and GC/MS analyses, succinate could be clearly identified as reaction product (Fig. 4A), whereas no succinate was observed before addition of enzyme (Fig. 4B). This is explainable by the following reaction.
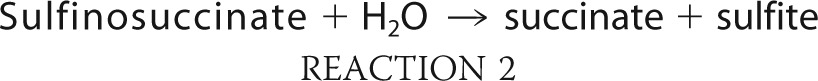 |
During the time course of the assay 17.9 ± 0.7 mm succinate was formed from 50 mm MS as confirmed by GC and GC/MS, whereas 29.0 ± 0.4 mm MS remained in the buffer and the formation of the disulfide could be completely suppressed due to low oxygen concentrations during the assay. Simultaneously, the formation of 15.5 ± 0.4 mm sulfite was determined, whereas no sulfate was detectable.
FIGURE 3.
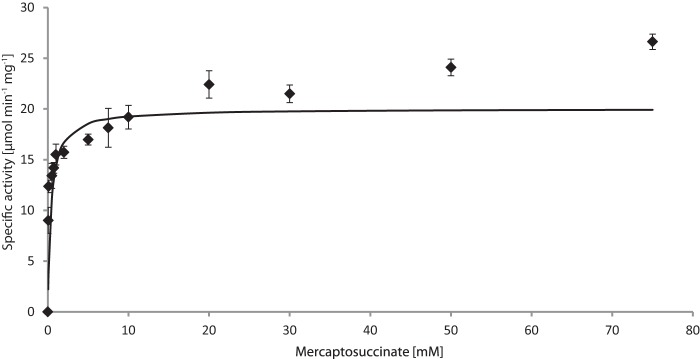
Kinetic data for MsdoB4 were obtained by measuring the oxygen consumed during conversion of MS. Measurements were carried out in triplicate applying different MS concentrations in 100 mm Tris/HCl buffer (pH 7.4). For details please see “Experimental Procedures.”
TABLE 2.
Kinetic data obtained for mercaptosuccinate dioxygenase of V. paradoxus B4 in comparison to bacterial cysteine dioxygenases of B. subtilis, B. cereus, and S. coelicolor
| Thiol dioxygenase | Substrate Km | Specific activity | kcat | kcat/Km |
|---|---|---|---|---|
| mm | μmol min−1 mg−1 | s−1 | mm−1 s−1 | |
| Cdo YubC (B. subtilis) | 3.0a | 1.0a | 0.39a | 0.1a |
| Cdo BC2617 (B. cereus) | 5.7a | 4.4a | 2.0a | 0.4a |
| Cdo SCO3035 (S. coelicolor) | 1.2a | 0.8a | 0.33a | 0.3a |
| Cdo SCO5772 (S. coelicolor) | 3.8a | 0.7a | 0.30a | 0.1a |
| MS dioxygenase (V. paradoxus) | 0.4 | 20.0 | 7.7 | 19.2 |
a Values from Dominy and colleagues (15) obtained via HPLC measurements.
FIGURE 4.
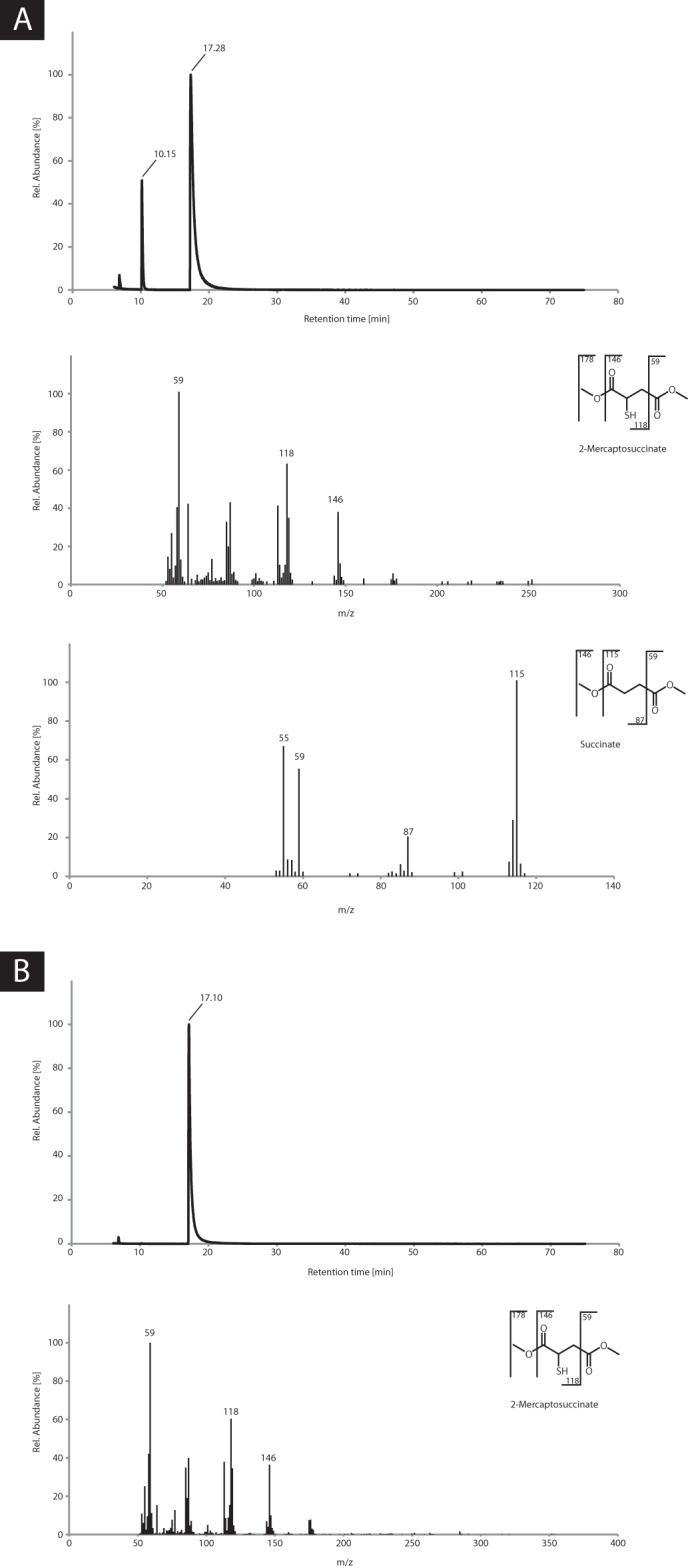
Chromatograms and mass spectra obtained after GC/MS analyses of samples withdrawn (A) at the end of the enzyme assay and (B) before addition of the enzyme. Samples were prepared and analyzed as described under “Experimental Procedures.” A, MS and succinate could clearly be identified; no disulfide of MS was formed. B, only MS was detectable, no disulfide of MS was formed.
To verify if the enzyme catalyzes both reactions, the oxidation of MS as well as the desulfination of the putative intermediate sulfinosuccinate, we aimed at synthesizing the latter compound by the adaption of a method of Filby and colleagues (29), because the synthesis of sulfinosuccinate has not been reported to date. Although metachloroperbenzoic acid was consumed during the reaction resulting in the formation of metachlorobenzoic acid, MS was not completely converted, but nonetheless, succinate was obtained as a reaction product. Furthermore, neither sulfinosuccinate nor sulfosuccinate could be detected as reaction products indicating that sulfinosuccinate might be such instable that it is already hydrolyzed when only traces of water are present.
Substrate Specificity and Structural Features
To elucidate the substrate specificity of the putative MS dioxygenase, different substrates exhibiting thiol groups were applied in enzyme assays with the oxygen electrode. Cysteine, dithiothreitol, 2-mercaptoethanol, and 3-mercaptopropionate were tested (for structures, see Fig. 5) in final concentrations of 0.2, 2, and 20 mm, but no activity was observed with these compounds (data not shown).
FIGURE 5.
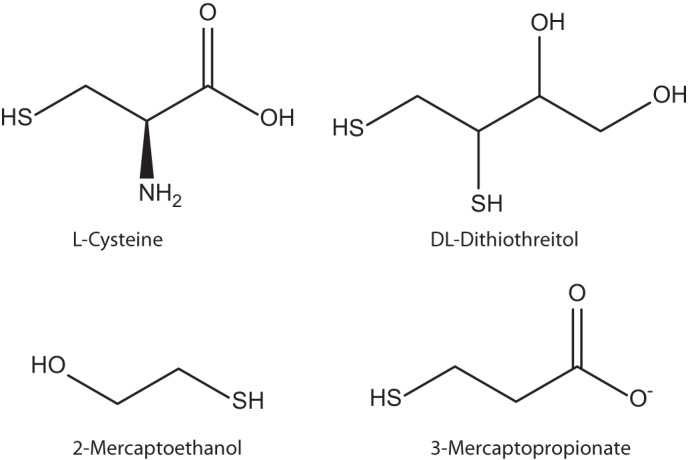
Structures of cysteine, dithiothreitol, 2-mercaptoethanol, and 3-mercaptopropionate, which were applied for investigating the substrate specificity of mercaptosuccinate dioxygenase.
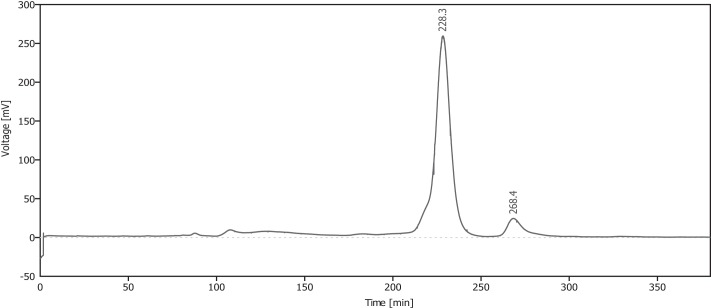
To investigate the subunit composition of the purified enzyme, size-exclusion chromatography, as described under “Experimental Procedures,” was carried out (Fig. 6). The expected size of the monomeric form including the His-tag is about 23 kDa as confirmed by SDS-PAGE of the purified enzyme (Fig. 2). According to the calibration of the size-exclusion chromatography column, the observed retention time of 228 min corresponds to a size of 43 kDa, indicating a homodimeric form of the MsdoB4, which is in accordance to results obtained for the human cysteine dioxygenase (31).
FIGURE 6.
Elution profile of analytical size-exclusion chromatography. A Superdex 200 HP 16/600 column was used and chromatography was carried out as described elsewhere (22). Seven milligrams of Ni-NTA-purified MS dioxygenase was loaded onto the column. The retention time of 228 min corresponds to 43 kDa (expected size for MS dioxygenase dimer including His6 tag: 46 kDa).
Metal Cofactor Analyses
To analyze the metal cofactor of MsdoB4, the Ni-NTA-purified enzyme was dialyzed as described above to finally yield the apoenzyme without its metal cofactor. Because Fe(II) was expected to be the required metal ion for the MS dioxygenase, 1,10-phenanthroline was chosen as a ferrous iron-specific chelator. However, besides Fe(II) other metal ions were tested (cobalt, copper, iron(III), magnesium, manganese, nickel, and zinc) if they could restore enzyme activity. MS readily reacted with the applied metal ions even in the absence of the enzyme forming mostly colored complexes while oxygen was consumed, which was already observed previously (32, 33). Therefore, the apoenzyme was preincubated with the respective metal ion to exclude false-positive results. The obtained result was very distinct: MS dioxygenase showed exclusive activity with Fe(II), and none of the other tested metal ions restored enzyme activity. Additionally, the Fe(II) content was determined applying bathophenanthroline disulfonic acid resulting in 0.6 ± 0.0 mol (n = 3) Fe(II)/mol of enzyme similar to the iron content observed for Cdo by Ye and co-workers (31). A submolar Fe(II) content was also described for the eukaryotic Cdos (12, 31, 34–36). In contrast to the Cdos, MS dioxygenase was active directly after Ni-NTA purification without addition of exogenous iron (12, 15, 34, 35). Moreover, no significant difference in enzyme activity could be observed between (i) the apoenzyme preincubated with iron and (ii) the enzyme used directly after Ni-NTA purification or (iii) Ni-NTA purified enzyme preincubated with iron.
Inhibition of MsdoB4
Enzyme activity assays with MsdoB4 and possible inhibitors were carried out. For this, different thiol compounds besides MS were tested in different concentrations. Neither the addition of cysteine, dithiothreitol, 2-mercaptoethanol, 3-mercaptopropionate nor the reaction products succinate and sulfite showed any significant effect on enzyme activity when added in final concentrations of 0.2, 2, or 20 mm (data not shown). Furthermore, addition of different metal ions (copper, magnesium, manganese, nickel, and zinc) to reaction mixtures with native enzyme did not significantly affect the enzyme activity (data not shown). However, when the native enzyme was preincubated for approximately 10 min on ice with different concentrations of 1,10-phenanthroline, the activity decreased significantly. Concentrations of 0.1, 0.25, 0.5, 1, and 2 mm of the chelator were applied. Preincubation with 0.1 mm 1,10-phenanthroline resulted in approximately 94% activity, 0.25 mm yielded 65% activity, whereas with 0.5 mm only 24% activity was left, which was further decreased to 8% activity with 1 mm, and no activity was detected after preincubation with 2 mm 1,10-phenanthroline.
Comparison of MsdoB4 with Cysteine Dioxygenases and Corresponding Homologues
The multiple sequence alignment (Fig. 1) of the MsdoB4 with Cdos from human, rat, mouse, B. subtilis, C. necator JMP134, and A. mimigardefordensis DPN7T as well as with the Mdos of R. eutropha H16 and V. paradoxus TBEA6 revealed that only four histidine residues, one glycine residue, and one tyrosine residue were strictly conserved among all investigated sequences. Three histidine residues proved to be crucial for the coordination of the metal cofactor as well as for the active site pocket in Cdos (indicated by asterisks) (15). Although the cupin motifs are present in all sequences, the divergence is relatively high, except for the already mentioned strictly conserved amino acids. The MsdoB4 was compared with Mdo of V. paradoxus TBEA6, which is the only other thiol dioxygenase of Gram-negative bacteria that has been thoroughly investigated so far. When comparing the two enzymes, it appears conspicuous that the Mdo activity was dependent on the substrate concentration in a sigmoidal manner (16), whereas the MS dioxygenase followed Michaelis-Menten kinetics, although similar reaction mechanisms of the two enzymes seem conceivable. Until now, only a very low number of bacterial Cdos has been characterized in detail, kinetic data are solely available for Cdos of Gram-positive species. There, enzyme activity was determined by applying HPLC instead of measuring the consumption of dioxygen. A comparison of the available data obtained for Cdos of B. subtilis, Bacillus cereus, two of Streptomyces coelicolor (15), and the MsdoB4 investigated in this study indicates that the latter exhibits a significantly higher specific activity with its substrate than that observed for the Cdos (Table 2). Moreover, MsdoB4 shows the lowest Km value when compared with those of the other Cdos indicating a higher affinity for its substrate (Table 2) (15).
DISCUSSION
In this study, it was clearly demonstrated that the putative MS dioxygenase previously identified during proteomic studies with V. paradoxus B4 does indeed catalyze the conversion of mercaptosuccinate to succinate and sulfite as final reaction products. Although attempts were made to synthesize sulfinosuccinate to verify whether the MsdoB4 also uses this putative intermediate as substrate, these attempts resulted in succinate as product indicating a high chemical instability of sulfinosuccinate. Therefore, it is most likely that the MsdoB4 does only catalyze the conversion of MS to sulfinosuccinate, which then decomposes spontaneously to succinate. After Ni-NTA purification, the ferrous iron content of the enzyme was comparably high with regard to values obtained for Cdos (12, 15, 34, 35), indicating that MsdoB4 might be more robust than the other enzymes and that its active site might be shielded from imidazole more effectively. Furthermore, the enzyme proved to be highly specific for its substrate as could be demonstrated for other Cdos as well (11, 15, 36), because it did not show any activity with the thiols cysteine, dithiothreitol, 2-mercaptoethanol, or 3-mercaptopropionate (Fig. 5). Due to the structural similarity it would be conceivable also that both carboxylic groups are important for the catalytic activity in addition to the thiol group. No significant inhibition could be detected with the tested thiols, either. Additionally, the final reaction products of succinate and sulfite also did not exhibit any inhibitory effect on the activity of the MS dioxygenase.
Hitherto, only very few bacterial Cdos and their homologues have been characterized in detail, kinetic data determined via HPLC are solely available for Cdos of Gram-positive species. A comparison of the available data obtained for the Cdos of B. subtilis, B. cereus, and the two Cdos of S. coelicolor (15) and the MsdoB4 indicated that MS dioxygenase exhibited a significantly higher specific activity with its substrate than that obtained for the Cdos (Table 2), although the Vmax may even be higher due to the observed submolar Fe(II) content of the purified enzyme. Moreover, MS dioxygenase showed the lowest Km value indicating a slightly higher affinity for its substrate (Table 2) (15). The turnover rate of 7.7 s−1 of the MS dioxygenase is also much higher than the rates observed for the other Cdos (Table 2). This kcat value has a significant impact on the comparably high efficiency (kcat/Km) of the enzyme of 19.2 mm−1 s−1.
Aligning the amino acid sequence of the MsdoB4 with characterized Cdos from eukaryotic and prokaryotic organisms, four histidine residues, one glycine residue, and one tyrosine residue are strictly conserved among all aligned enzymes (Fig. 1). As already reviewed by Stipanuk and colleagues (11), three of the conserved histidine residues (indicated by an asterisk in Fig. 1) are important for the coordination of the metal ion cofactor (15). However, the tight binding of the ferrous iron to rat Cdo could only recently be demonstrated by Tchesnokov and co-workers (37). Because the conserved histidine residues are also present in Msdo, it is highly probable that the iron coordination is achieved via these amino acids. Although in eukaryotic Cdos a rare cross-link exists between the conserved tyrosine residue and a cysteine residue (Cys-93) in the active center, this was not observed for prokaryotic Cdos (11, 14, 15, 31, 35). This is probably also true for Mdo of strain TBEA6 and MsdoB4, where the respective cysteine is substituted by an alanine (Fig. 1). For mammalian Cdos, a multitude of crystal structures obtained under different conditions and co-crystallized with different substrates is available, whereas only two crystal structures of prokaryotic Cdos have been deposited to date (14, 35, 38–41).3 Further investigations to elucidate the amino acid residues responsible for substrate specificity and substrate coordination of Msdo are currently in progress.
Taking into account the obtained results, the recent proposal of mercaptosuccinate degradation in V. paradoxus B4 (17) can be supported (Fig. 7). First (i) the conversion of mercaptosuccinate by the MsdoB4 could be confirmed most probably resulting in sulfinosuccinate as intermediate, because (ii) sulfinosuccinate is likely to be highly instable and to spontaneously decompose to succinate and sulfite, and (iii) whereas succinate eventually enters the central metabolism, sulfite is conceivably detoxified by the previously identified putative molybdopterin oxidoreductase.
FIGURE 7.
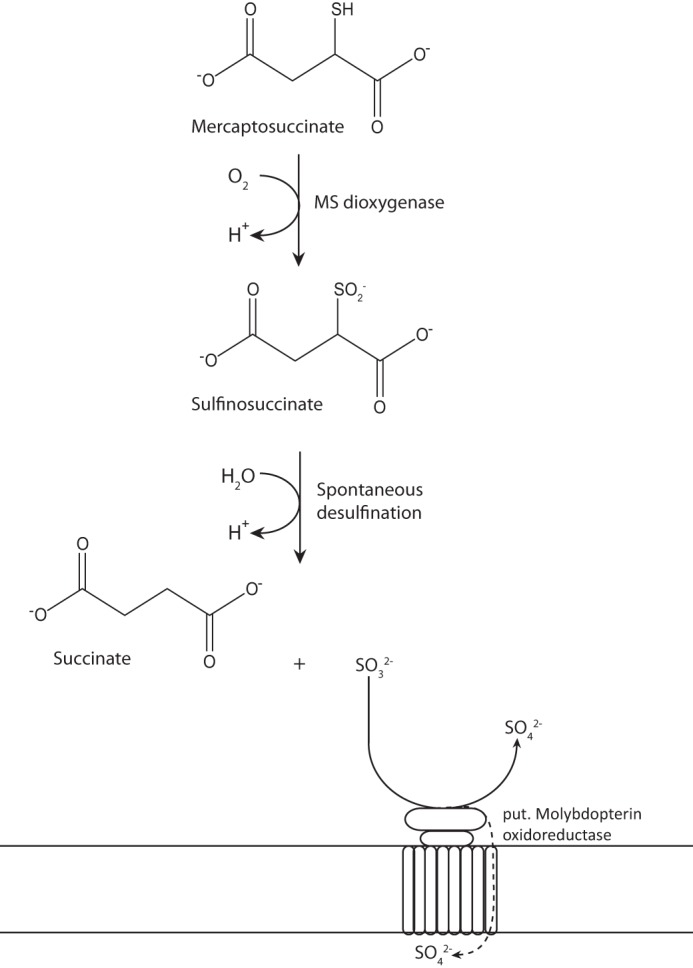
Updated proposal for the degradation pathway of mercaptosuccinate in V. paradoxus B4. First, mercaptosuccinate is converted to sulfinosuccinate by the mercaptosuccinate dioxygenase characterized in this study. Then, sulfinosuccinate spontaneously decomposes to succinate and sulfite. Succinate can eventually enter the central metabolism and serves as carbon source, whereas sulfite has to be detoxified, which is probably achieved by the previously identified molybdopterin oxidoreductase.
G. N. L. Jameson, E. P. Tchesnokov, E. Siakkou, and S. M. Wilbanks, unpublished data.
- MS
- mercaptosuccinate
- Mdo
- mercaptopropionate dioxygenases
- Cdo
- cysteine dioxygenases
- Ni-NTA
- nickel-nitrilotriacetic acid.
REFERENCES
- 1. Ghaderi S., Ramesh B., Seifalian A. M. (2012) Synthesis of mercaptosuccinic acid/mercaptopolyhedral oligomeric silsesquioxane coated cadmium telluride quantum dots in cell labeling applications. J. Nanosci. Nanotechnol. 12, 4928–4935 [DOI] [PubMed] [Google Scholar]
- 2. Rizvi S. B., Yildirimer L., Ghaderi S., Ramesh B., Seifalian A. M., Keshtgar M. (2012) A novel POSS-coated quantum dot for biological application. Int. J. Nanomedicine 7, 3915–3927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fu T., Qin H. Y., Hu H. J., Hong Z., He S. (2010) Aqueous synthesis and fluorescence-imaging application of CdTe/ZnSe core/shell quantum dots with high stability and low cytotoxicity. J. Nanosci. Nanotechnol. 10, 1741–1746 [DOI] [PubMed] [Google Scholar]
- 4. Bach L. G., Islam M. R., Vo T. S., Kim S. K., Lim K. T. (2013) Poly(allyl methacrylate) functionalized hydroxyapatite nanocrystals via the combination of surface-initiated RAFT polymerization and thiol-ene protocol: a potential anticancer drug nanocarrier. J. Colloid Interface Sci. 394, 132–140 [DOI] [PubMed] [Google Scholar]
- 5. Carbajal-Rodríguez I., Stöveken N., Satola B., Wübbeler J. H., Steinbüchel A. (2011) Aerobic degradation of mercaptosuccinate by the Gram-negative bacterium Variovorax paradoxus strain B4. J. Bacteriol. 193, 527–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Satola B., Wübbeler J. H., Steinbüchel A. (2013) Metabolic characteristics of the species: Variovorax paradoxus. Appl. Microbiol. Biotechnol. 97, 541–560 [DOI] [PubMed] [Google Scholar]
- 7. Belimov A. A., Dodd I. C., Hontzeas N., Theobald J. C., Safronova V. I., Davies W. J. (2009) Rhizosphere bacteria containing 1-aminocyclopropane-1-carboxylate deaminase increase yield of plants grown in drying soil via both local and systemic hormone signalling. New Phytol. 181, 413–423 [DOI] [PubMed] [Google Scholar]
- 8. Dunwell J. M., Khuri S., Gane P. J. (2000) Microbial relatives of the seed storage proteins of higher plants: conservation of structure and diversification of function during evolution of the cupin superfamily. Microbiol. Mol. Biol. Rev. 64, 153–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dunwell J. M., Culham A., Carter C. E., Sosa-Aguirre C. R., Goodenough P. W. (2001) Evolution of functional diversity in the cupin superfamily. Trends Biochem. Sci. 26, 740–746 [DOI] [PubMed] [Google Scholar]
- 10. Dunwell J. M., Purvis A., Khuri S. (2004) Cupins: the most functionally diverse protein superfamily? Phytochemistry 65, 7–17 [DOI] [PubMed] [Google Scholar]
- 11. Stipanuk M. H., Simmons C. R., Karplus P. A., Dominy J. E., Jr. (2011) Thiol dioxygenases: unique families of cupin proteins. Amino Acids. 41, 91–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Simmons C. R., Hirschberger L. L., Machi M. S., Stipanuk M. H. (2006) Expression, purification, and kinetic characterization of recombinant rat cysteine dioxygenase, a non-heme metalloenzyme necessary for regulation of cellular cysteine levels. Protein Expr. Purif. 47, 74–81 [DOI] [PubMed] [Google Scholar]
- 13. Stipanuk M. H., Ueki I., Dominy J. E., Jr., Simmons C. R., Hirschberger L. L. (2009) Cysteine dioxygenase: a robust system for regulation of cellular cysteine levels. Amino Acids 37, 55–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Simmons C. R., Liu Q., Huang Q., Hao Q., Begley T. P., Karplus P. A., Stipanuk M. H. (2006) Crystal structure of mammalian cysteine dioxygenase: a novel mononuclear iron center for cysteine thiol oxidation. J. Biol. Chem. 281, 18723–18733 [DOI] [PubMed] [Google Scholar]
- 15. Dominy J. E., Jr., Simmons C. R., Karplus P. A., Gehring A. M., Stipanuk M. H. (2006) Identification and characterization of bacterial cysteine dioxygenases: a new route of cysteine degradation for eubacteria. J. Bacteriol. 188, 5561–5569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bruland N., Wübbeler J. H., Steinbüchel A. (2009) 3-Mercaptopropionate dioxygenase, a cysteine dioxygenase homologue, catalyzes the initial step of 3-mercaptopropionate catabolism in the 3,3′-thiodipropionic acid-degrading bacterium, Variovorax paradoxus. J. Biol. Chem. 284, 660–672 [DOI] [PubMed] [Google Scholar]
- 17. Brandt U., Waletzko C., Voigt B., Hecker M., Steinbüchel A. (2014) Mercaptosuccinate metabolism in Variovorax paradoxus strain B4: a proteomic approach. Appl. Microbiol. Biotechnol. 98, 6039–6050 [DOI] [PubMed] [Google Scholar]
- 18. Sambrook J., Fritsch E. F., Maniatis T. (1989) Molecular cloning: A Laboratory Manual, 2nd Ed., Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 19. Bradford M. M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 20. Ellman G. L. (1959) Tissue sulfhydryl groups. Arch. Biochem. Biophys. 82, 70–77 [DOI] [PubMed] [Google Scholar]
- 21. Riddles P. W., Blakeley R. L., Zerner B. (1983) Reassessment of Ellman's reagent. Methods Enzymol. 91, 49–60 [DOI] [PubMed] [Google Scholar]
- 22. Britton H. T. S., Robinson R. A. (1931) Universal buffer solutions and the dissociation constant of veronal. J. Chem. Soc. 1456–1462 [Google Scholar]
- 23. Sörbo B. (1987) Sulfate: Turbidimetric and nephelometric methods. Methods Enzymol. 143, 3–6 [DOI] [PubMed] [Google Scholar]
- 24. Schürmann M., Deters A., Wübbeler J. H., Steinbüchel A. (2013) A novel 3-sulfinopropionyl coenzyme A (3SP-CoA) desulfinase from Advenella mimigardefordensis strain DPN7T acting as a key enzyme during catabolism of 3,3′-dithiodipropionic acid is a member of the acyl-CoA dehydrogenase superfamily. J. Bacteriol. 195, 1538–1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pierce B. S., Gardner J. D., Bailey L. J., Brunold T. C., Fox B. G. (2007) Characterization of the nitrosyl adduct of substrate-bound mouse cysteine dioxygenase by electron paramagnetic resonance: electronic structure of the active site and mechanistic implications. Biochemistry 46, 8569–8578 [DOI] [PubMed] [Google Scholar]
- 26. Smith G. F., McCurdy W. H., Diehl H. (1952) The colorimetric determination of iron in raw and treated municipal water supplies by use of 4:7-diphenyl-1:10-phenanthroline. Analyst 77, 418–422 [Google Scholar]
- 27. Schürmann M., Wübbeler J. H., Grote J., Steinbüchel A. (2011) Novel reaction of succinyl coenzyme A (succinyl-CoA) synthetase: activation of 3-sulfinopropionate to 3-sulfinopropionyl-CoA in Advenella mimigardefordensis strain DPN7T during degradation of 3,3′-dithiodipropionic acid. J. Bacteriol. 193, 3078–3089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Stein S. E. (1999) An integrated method for spectrum extraction and compound identification from gas chromatography/mass spectrometry data. J. Am. Soc. Mass Spectrom. 10, 770–781 [Google Scholar]
- 29. Filby G., Günther K., Penzhorn R. (April 30, 1974) Method for preparation of sulfinic acids. U. S. Patent US3950404A
- 30. Wübbeler J. H., Lütke-Eversloh T., Van Trappen S., Vandamme P., Steinbüchel A. (2006) Tetrathiobacter mimigardefordensis sp. nov., isolated from compost, a betaproteobacterium capable of utilizing the organic disulfide 3,3′-dithiodipropionic acid. Int. J. Syst. Evol. Microbiol. 56, 1305–1310 [DOI] [PubMed] [Google Scholar]
- 31. Ye S., Wu X., Wei L., Tang D., Sun P., Bartlam M., Rao Z. (2007) An insight into the mechanism of human cysteine dioxygenase: key roles of the thioether-bonded tyrosine-cysteine cofactor. J. Biol. Chem. 282, 3391–3402 [DOI] [PubMed] [Google Scholar]
- 32. Cheney G. E., Fernando Q., Freiser H. (1959) Some metal chelates of mercaptosuccinic acid. J. Phys. Chem. 63, 2055–2057 [Google Scholar]
- 33. Pawelec M., Stochel G., van Eldik R. (2004) Mechanistic information on the copper-catalysed autoxidation of mercaptosuccinic acid in aqueous solution. Dalton Trans. 2, 292–298 [DOI] [PubMed] [Google Scholar]
- 34. Chai S. C., Jerkins A. A., Banik J. J., Shalev I., Pinkham J. L., Uden P. C., Maroney M. J. (2005) Heterologous expression, purification, and characterization of recombinant rat cysteine dioxygenase. J. Biol. Chem. 280, 9865–9869 [DOI] [PubMed] [Google Scholar]
- 35. McCoy J. G., Bailey L. J., Bitto E., Bingman C. A., Aceti D. J., Fox B. G., Phillips G. N., Jr. (2006) Structure and mechanism of mouse cysteine dioxygenase. Proc. Natl. Acad. Sci. U.S.A. 103, 3084–3089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yamaguchi K., Hosokawa Y., Kohashi N., Kori Y., Sakakibara S., Ueda I. (1978) Rat liver cysteine dioxygenase (cysteine oxidase): further purification, characterization, and analysis of the activation and inactivation. J. Biochem. 83, 479–491 [DOI] [PubMed] [Google Scholar]
- 37. Tchesnokov E. P., Wilbanks S. M., Jameson G. N. (2012) A strongly bound high-spin iron(II) coordinates cysteine and homocysteine in cysteine dioxygenase. Biochemistry 51, 257–264 [DOI] [PubMed] [Google Scholar]
- 38. Driggers C. M., Cooley R. B., Sankaran B., Hirschberger L. L., Stipanuk M. H., Karplus P. A. (2013) Cysteine dioxygenase structures from pH 4 to 9: consistent Cys-persulfenate formation at intermediate pH and a Cys-bound enzyme at higher pH. J. Mol. Biol. 425, 3121–3136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Souness R. J., Kleffmann T., Tchesnokov E. P., Wilbanks S. M., Jameson G. B., Jameson G. N. (2013) Mechanistic implications of persulfenate and persulfide binding in the active site of cysteine dioxygenase. Biochemistry 52, 7606–7617 [DOI] [PubMed] [Google Scholar]
- 40. Levin E. J., Kondrashov D. A., Wesenberg G. E., Phillips G. N., Jr. (2007) Ensemble refinement of protein crystal structures: validation and application. Structure. 15, 1040–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Simmons C. R., Krishnamoorthy K., Granett S. L., Schuller D. J., Dominy J. E., Jr., Begley T. P., Stipanuk M. H., Karplus P. A. (2008) A putative Fe2+-bound persulfenate intermediate in cysteine dioxygenase. Biochemistry 47, 11390–11392 [DOI] [PMC free article] [PubMed] [Google Scholar]