Background: TgMyoA, a Toxoplasma gondii class XIV myosin, is an important parasite virulence factor.
Results: Expression of functional TgMyoA in Sf9 cells requires co-expression with a T. gondii myosin co-chaperone.
Conclusion: TgMyoA can be heterologously expressed and needs two bound light chains to propel actin at fast speeds.
Significance: High yield expression of TgMyoA will enable structure-function studies and drug screening.
Keywords: ATPase, Molecular Chaperone, Molecular Motor, Protein Expression, Protein Folding, Toxoplasma gondii, UCS Proteins, Class XIV Myosin, Myosin Chaperones, Myosin Motor Complex
Abstract
Many diverse myosin classes can be expressed using the baculovirus/Sf9 insect cell expression system, whereas others have been recalcitrant. We hypothesized that most myosins utilize Sf9 cell chaperones, but others require an organism-specific co-chaperone. TgMyoA, a class XIVa myosin from the parasite Toxoplasma gondii, is required for the parasite to efficiently move and invade host cells. The T. gondii genome contains one UCS family myosin co-chaperone (TgUNC). TgMyoA expressed in Sf9 cells was soluble and functional only if the heavy and light chain(s) were co-expressed with TgUNC. The tetratricopeptide repeat domain of TgUNC was not essential to obtain functional myosin, implying that there are other mechanisms to recruit Hsp90. Purified TgMyoA heavy chain complexed with its regulatory light chain (TgMLC1) moved actin in a motility assay at a speed of ∼1.5 μm/s. When a putative essential light chain (TgELC1) was also bound, TgMyoA moved actin at more than twice that speed (∼3.4 μm/s). This result implies that two light chains bind to and stabilize the lever arm, the domain that amplifies small motions at the active site into the larger motions that propel actin at fast speeds. Our results show that the TgMyoA domain structure is more similar to other myosins than previously appreciated and provide a molecular explanation for how it moves actin at fast speeds. The ability to express milligram quantities of a class XIV myosin in a heterologous system paves the way for detailed structure-function analysis of TgMyoA and identification of small molecule inhibitors.
Introduction
Toxoplasma gondii, a member of the phylum Apicomplexa, is a common infectious agent of humans that can cause severe disease in immunocompromised individuals and the developing fetus. This obligate intracellular parasite must invade a host cell and replicate to survive. The invasive stage of the parasite relies on a unique form of substrate-dependent motility called gliding motility, which is driven by a class XIVa myosin motor, TgMyoA.4 The TgMyoA heavy chain, one of 11 myosin heavy chains found in T. gondii (1), is important for efficient parasite motility, invasion, and egress from the host. Parasites lacking TgMyoA are avirulent in a mouse model of infection (2).
The TgMyoA motor is located between the plasma membrane and the inner membrane complex (IMC), a double membrane that is continuous around most of the cell (3) (see Fig. 1). TgGAP50 (a 50-kDa gliding-associated protein), an integral membrane glycoprotein of the IMC, acts as a membrane receptor for the motor (4); TgMyoA is linked indirectly to TgGAP50 through an apicomplexan-specific N-terminal extension of its regulatory light chain, TgMLC1, and TgGAP45 (a 45-kDa gliding-associated protein). Two other membrane-associated proteins, TgGAP40 (a 40-kDa gliding-associated protein) and TgGAP70 (a 70-kDa gliding-associated protein), have recently been identified as additional components of this myosin motor complex (5). The precise mechanism by which the motor complex generates motility is the subject of intense investigation (see Fig. 1 for one model), but there is little debate about the central importance of TgMyoA in the process.
FIGURE 1.
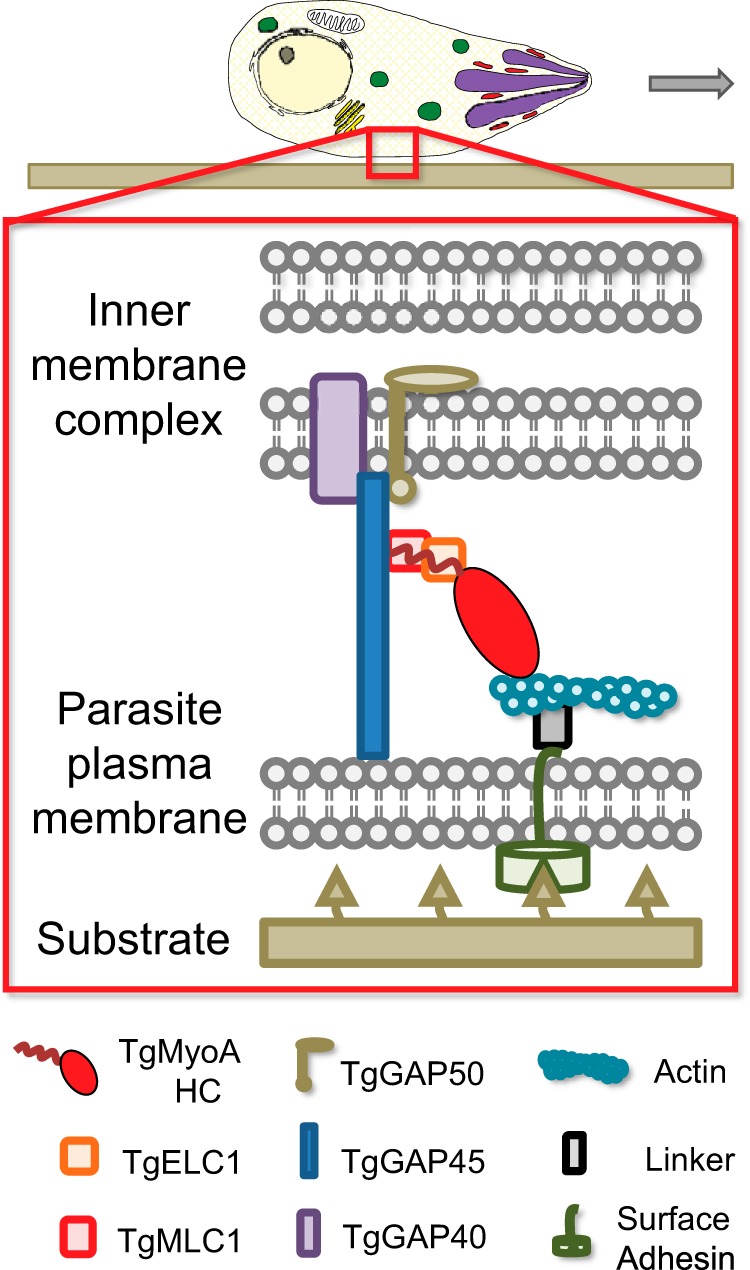
Schematic of T. gondii myosin motor complex. TgMyoA (i.e. TgMyoA heavy chain (HC) with its bound light chains TgMLC1 and TgELC1) and TgGAP45 are anchored to the IMC via transmembrane protein TgGAP50. This multiprotein complex is referred to as the myosin motor complex. Short actin filaments are located between the parasite plasma membrane and the IMC and are thought to be connected to ligands on the host cell surface through linker protein(s) that bind to the cytosolic tails of surface adhesins. TgMyoA (attached to the IMC) displaces the actin filaments (attached to the substrate), causing the parasite to move relative to the substrate. Note that alternative models of parasite motility are emerging (50, 51) in which TgMyoA plays a different but still important role in generating the force required for motility. The figure was adapted from Ref. 9.
TgMyoA is a single-headed motor with several unusual features that distinguish it from other myosin motors. It lacks a highly conserved amino acid in a key actin-binding surface loop that is typically either an acidic residue or a phosphorylatable residue but is a Gln in TgMyoA. It also lacks a conserved Gly in a helix in the motor domain that is thought to act as a pivot point for motion of the light chain binding lever arm (6, 7). The lever arm amplifies small motions at the active site into the larger motions needed to move actin. The light chain-binding region of myosin heavy chains also typically contains one or more α-helical “IQ motifs” (consensus sequence IQXXXRGXXXR), which bind specific myosin light chains and/or calmodulin. The sequence of the IQ motif in TgMyoA is degenerate (6, 7), and thus the number of bound light chains is unclear.
Isolation of a functional motor complex from T. gondii showed that despite these unique features TgMyoA was a bona fide motor able to propel actin at fast speeds of ∼5 μm/s in an in vitro motility assay (7–9). The speed observed in vitro is similar to the speed of parasite motility on two-dimensional surfaces (1–3 μm/s) (10) and in three-dimensional Matrigel (maximum speed, 2 μm/s) (11). The in vitro biophysical characterization of TgMyoA was a tour de force given the limited amounts of protein that could be isolated from the parasite (7). However, it raised some interesting questions. For example, how can a single-headed myosin with only one bound light chain propel actin at such fast speeds? The speed of actin movement by myosin depends on the kinetics of nucleotide binding and release from the motor domain as well as the length of the lever arm, which is determined by the number of bound light chains (12, 13). The unitary step size of TgMyoA was measured to be ∼5.3 nm (7), which is strikingly similar to the step size measured for single-headed subfragment-1 from either fast skeletal muscle myosin or smooth muscle myosin, both of whose lever arms are known to be stabilized by two bound lights chains (for a review, see Ref. 14).
A recent mass spectrometry-based study, which analyzed proteins that bound to the motor complex in the presence and absence of calcium, identified a smaller ∼15-kDa second calmodulin-like light chain in the motor complex immunoprecipitates, which they called the essential light chain (TgELC1) (15). The functional consequences of TgMLC1 and TgELC1 binding to the TgMyoA heavy chain are unknown. To address structure-function questions like these, it is essential to have an expression system that produces sufficient amounts of pure, functional motor protein.
Unfortunately, our previous efforts to express soluble and functional TgMyoA using the baculovirus/Sf9 insect cell expression system were unsuccessful. This was surprising given that many classes of myosin are successfully expressed and properly folded in Sf9 cells including class II smooth (16) and non-muscle myosins (17) and many classes of unconventional myosins including classes V (18–20), VI (21), and VII (22). There is another exception: functional class II striated muscle myosins have not been expressed in Sf9 cells but can be expressed in the C2C12 muscle cell line, implying that proper folding of striated muscle myosins requires factors that are present in muscle cells but absent from Sf9 cells (23–25). Similarly, although active TgMyoA can be purified from T. gondii (7–9), we speculated that a parasite-specific factor was needed for proper folding of TgMyoA in Sf9 cells.
Myosin folding and assembly into filaments in striated muscle is mediated by the general chaperones Hsc70 and Hsp90 and a striated myosin-specific co-chaperone of the UCS protein family, Unc45b (26–28). A second myosin chaperone called Unc45a is expressed in all vertebrate tissues (29). Unc45b forms a stable complex with Hsp90, binds only to the unfolded striated muscle myosin motor domain, and promotes motor domain folding (26–28). The N-terminal region of Unc45 interacts with Hsp90 via three tetratricopeptide (TPR) repeats, a degenerate 34-amino acid motif that is involved in protein-protein interactions (30). The central domain has no known function, and the C-terminal UCS domain of ∼450 amino acids interacts with myosin (26). The UCS domain is named for the three founding members of this family: UNC-45 from Caenorhabditis elegans, CRO1 from the fungus Podospora anserina, and She4p from Saccharomyces cerevisiae.
In the present study, we identified a single unc-45 homolog in T. gondii, referred to as TgUNC, that encodes a protein containing all three domains found in the canonical Unc45 protein. We showed that isolation of functional TgMyoA from Sf9 cells strictly requires co-expression with TgUNC. Using this novel co-expression strategy, we showed that recombinant TgMyoA simultaneously binds both TgMLC1 and TgELC1, similar to the domain structure of conventional class II myosins. The functional consequence of having two bound light chains was a doubling of the speed of actin movement in motility assays compared with that observed when TgMyoA has only a single bound light chain (TgMLC1). The successful expression of TgMyoA will enable structure-function analysis of this unusual class of myosin motor proteins as well as identification of small molecule inhibitors that can serve as leads for new drugs to prevent or treat toxoplasmosis.
EXPERIMENTAL PROCEDURES
Expression Constructs
Full-length (FL) TgUNC (GenBankTM accession number XM_002367241.1) was amplified from T. gondii cosmid number p559; (cosmid generously provided by Dr. M. J. Gubbels) using the primer pair TPRForANcoI (AGCACCATGGAGGATTTGTCAAACGC) and TPRRevAFull MycStpKpnI (CGGCGGTACCTTACAGGTCTTCTTCAGAGATCAGTTTCTGTTCGCTTGAGTCTGGGGTGG). A PCR product coding for TgUNC followed by a Myc tag was cloned into the baculovirus transfer vector pAcSG2 (BD Biosciences). TgUNC pAcSG2 was truncated to make two other constructs. TgUNCΔTPR starts at residue Leu-161, eliminating the TPR region but keeping the central and UCS domains along with the C-terminal Myc tag. The TgUNC UCS-only construct starts with residue Glu-682, thus eliminating both the TPR and central domains.
The plasmid pEB2-FLAG-MyoA5 was used as a template to amplify a product coding for N-terminally FLAG-tagged TgMyoA heavy chain (accession number AAC47724.1) using primer pair EcoR1-FLAG-MyoF1 (GGGGAATTCATGGACTACAAAGACGATGACGACAAGATGGC) and c-TgMyoA-EcoR1 (GGGGGAATTCCTACTACTAGAACGCCGGCTGAACAGTC). The PCR product was cloned into the baculovirus transfer vector pAcUW51 (BD Biosciences). TgMyoA-cBio-cFLAG was constructed by digesting DNA coding for the TgMyoA heavy chain from pVL1392FLAGTgMyoA6 with BamHI/EcoRV and inserting the cleavage fragment into a modified version of pAcSG2 containing both a biotin acceptor site and FLAG tag at the C terminus.
The TgMLC1 gene (GenBank accession number XM_002364961.1) was PCR-amplified from parasite cDNA using primers MLC-F (CATGGAATTCATGAGCAAGGTCGAGAAGA) and MLC-R (CATGAGATCTTGATTACTCCCTTCGCTCGAG) and cloned untagged by ligation of the digested, gel-extracted PCR product at the EcoRI and BglII restriction sites in pAcSG2. The His6-tagged Tg MLC1 gene was amplified from pAcSG2 TgMLC1 using the primer pair 6HisMLCINdeIF (GCACAATCATATGCATCACCACCATCATCACAGCAAGGTCGAGAAGAAATGC) and MLC1BamHIR (GCACAATGGATCCTTACTCCCTTCGCTCGAGCATT). The resulting PCR product was cloned into the bacterial expression vector pET3a (Novagen).
The TgELC1 gene (GenBank accession number XM_002365594) was PCR-amplified from T. gondii RH strain cDNA. The PCR product was ligated downstream of the p10 promoter in vector pAcUW51 for baculovirus expression. In addition, a PCR product coding for TgELC1 with a His6 tag was made using the primer pair 6HisELC1NdeIF (GCACAATCATATGCATCACCACCATCATCACACCTGCCCTCCCCGCGTCCGT) and ELC1BamHIR (GCACAATGGATCCTTATTTCAGCAGCATCTTGACAAAGT) and cloned into the bacterial expression vector pET3a (Novagen).
A dual light chain plasmid was constructed for immunoprecipitation studies. DNA coding for TgELC1with a C-terminal 3xHA tag was cloned into vector pAcUW51 downstream of the p10 promoter, whereas DNA coding for TgMLC1 with an N-terminal 3xMyc tag was cloned downstream of the polH promoter. All constructs were sequenced prior to transfection.
Co-immunoprecipitations
Sf9 cells were harvested 72 h after infection with recombinant baculoviruses (TgMyoA heavy chain and tagged light chains) and lysed with 10 mm imidazole, pH 7.4, 150 mm NaCl, 1 mm EGTA, 5 mm MgCl2, 7% (w/v) sucrose, 3 mm NaN3, 1% (v/v) Nonidet P-40, 1 mm DTT, and 1× protease inhibitor mixture (Sigma-Aldrich, catalogue number P8340). Following addition of 5 mm MgATP, the lysate was spun at 350,000 × g for 20 min at 4 °C. The supernatant was incubated with either rabbit anti-TgMLC1 (a generous gift from Dr. Con Beckers) or rat anti-HA (Roche catalogue number 11867423001) overnight at 4 °C. Rec-Protein A-Sepharose beads (Invitrogen) were added, and the samples were incubated at 4 °C for 60 min. The beads were washed four times with lysis buffer. Bound proteins were eluted by boiling in SDS-PAGE sample buffer, centrifuged at 100 × g for 2 min, and resolved on 4–12% gradient gels. The protein was transferred to Immobilon-FL (Millipore, Bedford, MA) and probed with mouse anti-Myc 9E10 (1:2,000; Developmental Studies Hybridoma Bank, University of Iowa), rat anti-HA (1:400), or rabbit anti-HA (1:2,000; Abcam catalogue number 9110) and mouse anti-FLAG (1:7,500; Sigma-Aldrich). LI-COR Biosciences (Lincoln, NE) secondary antibodies (anti-mouse IRDye680RD, anti-rabbit IRDye800CW, and anti-rat IRDye800CW) were used according to manufacturer's instructions, and blots were scanned using an Odyssey CLx Infrared Imaging System (LI-COR Biosciences).
Protein Expression and Purification
Sf9 cells were co-infected with recombinant baculovirus coding for TgMyoA heavy chain (tagged at the C terminus with a Bio tag and FLAG tag), untagged light chain(s), and the co-chaperone TgUNC. The cells were grown in medium supplemented with 0.2 mg/ml biotin. After 72 h, the cells were lysed by sonication in 10 mm imidazole, pH 7.4, 0.2 m NaCl, 1 mm EGTA, 5 mm MgCl2, 7% (w/v) sucrose, 2 mm DTT, 0.5 mm 4-(2-aminoethyl)benzenesulfonyl fluoride, 5 μg/ml leupeptin, and 5 mm benzamidine. To determine TgMyoA heavy chain solubility, the extracts were centrifuged at 350,000 × g for 20 min. For motor purification, bacterially expressed TgMLC1 and TgELC1 at 25 μg/ml (final concentration) each and 5 mm MgATP were added to the lysate, which was then clarified at 200,000 × g for 30 min. The supernatant was applied to a FLAG affinity resin column (Sigma-Aldrich) and washed with 10 mm imidazole, pH 7.4, 0.2 m NaCl, 1 mm EGTA, and 1 mm NaN3. TgMyoA was eluted from the column with 0.1 mg/ml FLAG peptide in the column buffer. The fractions of interest were combined, concentrated with an Amicon centrifugal filter device (Millipore), and dialyzed against 10 mm imidazole, pH 7.4, 0.2 m NaCl, 50% (v/v) glycerol, 1 mm DTT, and 1 μg/ml leupeptin for storage at −20 °C.
His-tagged light chains (TgELC1 or TgMLC1) in pET3a (Novagen) were expressed in BLR(DE3) competent cells grown in LB broth. The cultures were induced with 0.4 mm isopropyl 1-thio-β-d-galactopyranoside and grown overnight at 27 °C before being pelleted and frozen. The pellets were lysed by sonication in 10 mm sodium phosphate, pH 7.4, 0.3 m NaCl, 0.5% (v/v) glycerol, 7% (w/v) sucrose, 7 mm β-mercaptoethanol, 0.5 mm 4-(2-aminoethyl)benzenesulfonyl fluoride, and 5 μg/ml leupeptin. The cell lysate was clarified at 200,000 × g for 30 min. TgELC1, which is found in the supernatant, was boiled for 10 min in a double boiler and then clarified at 26,000 × g for 30 min. Soluble protein was applied to a HIS-Select® nickel affinity column (Sigma-Aldrich). Nonspecifically bound protein was removed by washing the resin with buffer A (10 mm sodium phosphate, pH 7.4 and 0.3 m NaCl). TgELC1was then eluted from the column with buffer A containing 200 mm imidazole. The protein was dialyzed overnight against 10 mm imidazole, pH 7.4, 150 mm NaCl, 1 mm EGTA, 1 mm MgCl2, and 50% (v/v) glycerol.
Bacterially expressed TgMLC1 is found in the insoluble inclusion bodies. The cell lysate was clarified at 26,000 × g for 10 min. The pellet was dissolved in 20 ml of 8 m guanidine, 150 mm NaCl, 10 mm NaPO4, pH 7.5, and 10 mm DTT and stirred at room temperature until dissolved. It was then clarified at 200,000 × g for 30 min and dialyzed overnight against 2 × 1 liter of buffer A containing 7 mm β-mercaptoethanol and 1 μg/ml leupeptin. The next day the sample was clarified at 26,000 × g for 30 min, and the supernatant was applied to a HIS-Select nickel affinity column. The column was washed with 15 ml of dialysis buffer, and TgMLC1 was eluted with buffer A containing 200 mm imidazole. The protein was dialyzed overnight against 10 mm imidazole, pH 7.4, 150 mm NaCl, 1 mm EGTA, 1 mm MgCl2, and 50% (v/v) glycerol. Both purified light chains were stored at −20 °C.
Gels
Proteins were separated on a 4–12% Bis-Tris NuPAGE gel (Invitrogen) run in MES buffer according to the NuPAGE technical guide.
In Vitro Motility
To prepare the flow cell, 0.2 mg/ml biotinylated BSA in buffer B (150 mm KCl, 25 mm imidazole, pH 7.5, 1 mm EGTA, 4 mm MgCl2, and 10 mm DTT) was added to the nitrocellulose-coated flow cells for 1 min followed by three rinses with 0.5 mg/ml BSA in buffer B. Neutravidin (50 μg/ml; Thermo Scientific) in buffer B was applied for 1 min followed by three rinses with buffer B. Before introduction into the flow cell, TgMyoA was mixed with a 2-fold molar excess of F-actin and 10 mm MgATP in buffer B and centrifuged for 25 min at 350,000 × g to remove ATP-insensitive myosin heads. TgMyoA (0.5 μm) was then introduced into the flow cell. To further block any ATP-insensitive heads, 1 μm vortexed F-actin in buffer C (50 mm KCl, 25 mm imidazole, pH 7.5, 1 mm EGTA, 4 mm MgCl2, and 10 mm DTT) was added for 60 s followed by a 10 mm MgATP wash. Rhodamine-phalloidin-labeled actin was then introduced for 1 min followed by one rinse with buffer C. Three volumes of buffer C, which also contained 5 mm MgATP (unless stated otherwise), 0.5–0.7% methylcellulose, 1 μm TgMLC1, 1 μm TgELC1, 3 mg/ml glucose, 0.125 mg/ml glucose oxidase (Sigma-Aldrich), and 0.05 mg/ml catalase (Sigma-Aldrich) were then flowed into the cell. When assayed with calcium, 1.2 mm calcium was added to this buffer.
Actin movement was observed at 30 °C using an inverted microscope (Zeiss Axiovert 10) equipped with epifluorescence, a Rolera MGi Plus digital camera, and dedicated computer with the Nikon NIS Elements software package. Data were analyzed using a semiautomated filament tracking program described previously (31). The velocities of >600 filaments were determined. Speeds were fit to a Gaussian curve.
Actin-activated ATPase Activity
Assays were performed in 10 mm imidazole, pH 7.0, 5 mm NaCl, 1 mm MgCl2, 1 mm NaN3, and 1 mm DTT at 30 °C. Purified TgMyoA (52 nm) was incubated with various concentrations of skeletal actin. Activity was initiated by the addition of 5 mm MgATP and stopped with SDS every 2 min for 8 min. Inorganic phosphate was determined colorimetrically (32). A low salt concentration was needed to keep the Km values as low as possible. Data were fit to the Michaelis-Menten equation.
Sedimentation Velocity
Sedimentation velocity runs were performed at 20 °C in an Optima XL-I analytical ultracentrifuge (Beckman Coulter) using an An60Ti rotor at 30,000 rpm. The solvent was 20 mm HEPES, pH 7.4, 0.1 m NaCl, and 2 mm DTT. An N-terminally FLAG-tagged TgMyoA heavy chain (no Bio tag) bound to TgMLC1 was used for this experiment. The sedimentation coefficient was determined by curve fitting to one species using the dc/dt program (33).
RESULTS
TgMyoA Heavy Chain Is Insoluble When Expressed in Sf9 Cells
Recombinant baculoviruses encoding TgMyoA heavy chain and its regulatory light chain, TgMLC1, were used to co-infect Sf9 cells. The C terminus of the TgMyoA heavy chain contained two tags: the FLAG tag facilitates purification by affinity chromatography, and the Bio tag, which becomes biotinylated within the Sf9 cells (34), allows the motor to be specifically attached via its C terminus to a streptavidin-coated coverslip for in vitro motility assays (Fig. 2A). Unfortunately, although both TgMyoA heavy chain and TgMLC1 were expressed following infection as detected by Western blotting of the total Sf9 cell lysate, none of the TgMyoA heavy chain was present in the soluble fraction (Fig. 3).
FIGURE 2.
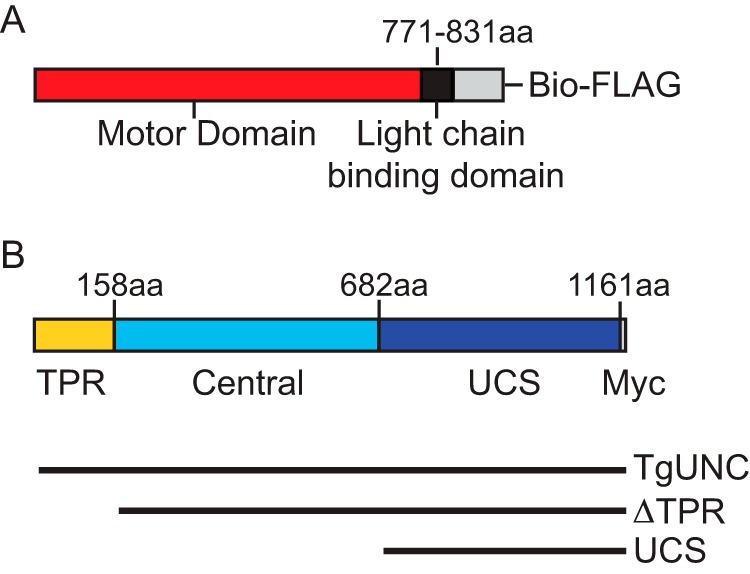
Schematic of the expressed proteins. A, the TgMyoA heavy chain construct contains the motor domain (red) and the light chain-binding region (black) followed by a Bio tag and FLAG tag (gray). B, TgUNC consists of three domains, TPR (yellow), central (cyan), and the UCS region (blue), followed by a Myc tag. The three different TgUNC constructs used during co-expression with TgMyoA heavy chain are shown below the schematic. aa, amino acids.
FIGURE 3.
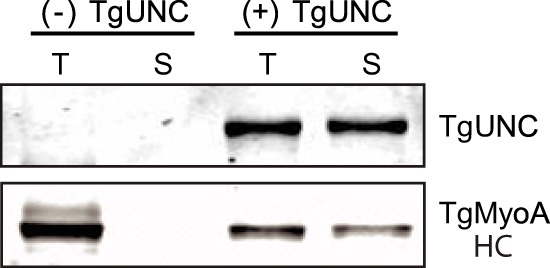
Co-expression of TgMyoA with the chaperone TgUNC in Sf9 cells produces soluble heavy chain. Sf9 cells were co-infected with recombinant baculovirus coding for the TgMyoA heavy chain (HC) and its light chain TgMLC1. The Western blot shows the total (T) and soluble (S) protein fractions after 72-h infection in the absence (left two lanes) or presence (right two lanes) of co-expressed TgUNC. TgUNC was detected using anti-Myc antibody, whereas TgMyoA heavy chain was detected with anti-FLAG antibody.
Identification of a UCS Family Gene in the T. gondii Genome
We speculated that the endogenous Sf9 protein folding machinery was not sufficient to fold this unusual myosin and that a parasite-specific cofactor was required. Potential candidates include proteins in the UCS family of myosin co-chaperones of which UNC-45 from C. elegans is the founding member. Bioinformatics analysis of the T. gondii genome using either full-length Unc45 or the UCS domain only revealed a single candidate, TgUNC (GenBank accession number XM_002367241.1). The TgUNC amino acid sequence was aligned with the three canonical members of the UCS family of myosin chaperones (C. elegans Unc45b (GenBank accession number AAD01976.1), P. anserina CRO1 (GenBank accession number CAA76144.1), and S. cerevisiae She4p (GenBank accession number DAA10818.1)). The percent identity/percent similarity of full-length TgUNC with the other UCS family members is: Unc45b, 17.4/28.7; CRO1, 9.5/16.3; She4p, 9.3/17.4. Comparing only the UCS domains, the percent identity/percent similarity with TgUNC increases: Unc45b, 20.4/31.1; CRO1, 17.4/27.3; She4p,15.3/25.3. A schematic of the domain structure of TgUNC is shown in Fig. 2B.
Production of Soluble TgMyoA in Sf9 Cells Requires Co-expression with TgUNC
In marked contrast to what was observed in the absence of TgUNC, co-expression of TgMyoA heavy chain and TgMLC1 with TgUNC yielded soluble protein (Fig. 3). We then set up a large scale infection for protein purification (500-ml culture; 3 × 109 Sf9 cells) with the three viruses. TgMyoA was purified from the lysate using a FLAG affinity column (see “Experimental Procedures”). Protein that bound to and eluted from the FLAG column showed two predominant bands on SDS gels at the expected sizes of TgMyoA heavy chain and TgMLC1 (Fig. 4A, lane 1). The yield of purified motor was ∼1.5 mg/109 Sf9 cells.
FIGURE 4.
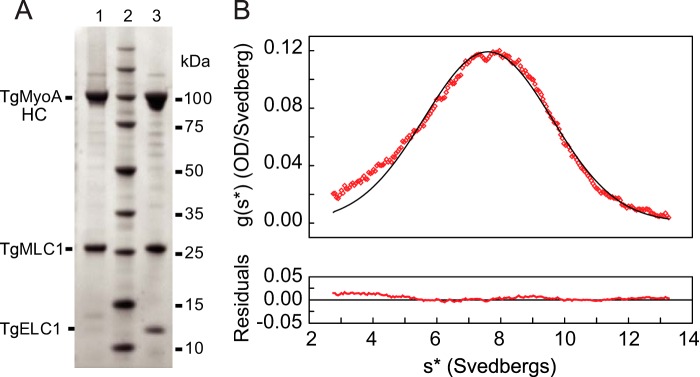
Characterization of purified TgMyoA. A, SDS gel analysis of purified motor proteins. Lane 1, purified protein resulting from co-expression of TgMyoA heavy chain (HC), TgMLC1 light chain, and TgUNC. TgUNC only binds to unfolded protein and does not co-purify with TgMyoA. Lane 2, molecular mass standards. Lane 3, purified protein resulting from co-expression of TgMyoA heavy chain, TgMLC1, TgELC1, and TgUNC. B, sedimentation velocity of TgMyoA heavy chain expressed with TgMLC1. A sedimentation coefficient of 7.7 S was determined by curve fitting to one species. The symmetrical nature of the boundary indicates that a homogeneous species is present. OD, optical density.
The purified motor was analyzed by sedimentation velocity in the analytical ultracentrifuge. The results showed a single symmetrical peak with a sedimentation coefficient of 7.7 S, indicating a homogeneous preparation of protein (Fig. 4B).
In Vitro Motility of Expressed TgMyoA Containing Bound TgMLC1
Solubility does not ensure activity, so an in vitro motility assay was performed to assess function. TgMyoA with bound TgMLC1 was perfused into a chamber containing a streptavidin-coated coverslip. The biotinylated tag at the C terminus of TgMyoA heavy chain ensures that this region will adhere to the coverslip, leaving the motor domain accessible to bind actin. Expressed motor moved rhodamine-phalloidin-labeled skeletal muscle actin at a speed of 1.5 ± 0.2 μm/s (Fig. 5, solid blue triangles). Speeds were calculated using a semiautomated tracking program that allowed thousands of trajectories to be analyzed without selection bias (31). The speeds fit a Gaussian distribution. The speed of actin movement with expressed TgMyoA was slower than that obtained with myosin isolated from parasites (7, 9), which led us to question the reason for the difference.
FIGURE 5.
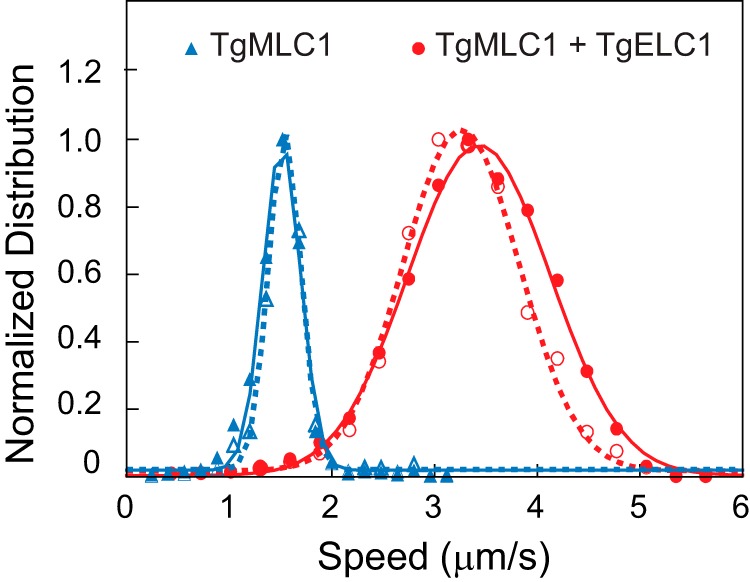
The number of bound light chains determines the speed of actin movement in an in vitro motility assay. The Gaussian fit of speeds for TgMyoA with TgMLC1 only was 1.5 ± 0.2 μm/s (mean ± S.D., n = 2,341 filaments) (solid blue triangles) compared with 3.4 ± 0.7 μm/s (mean ± S.D., n = 4,774 filaments) for TgMyoA with both light chains (solid red circles). Conditions were as follows: 25 mm imidazole, pH 7.5, 50 mm KCl, 1 mm EGTA, 4 mm MgCl2, 10 mm DTT, 5 mm MgATP, 0.5–0.7% (w/v) methylcellulose, and oxygen scavengers at 30 °C. Addition of 1.2 mm calcium (i.e. 0.2 mm free calcium) did not affect motility speed (open symbols). In the presence of calcium, TgMyoA with TgMLC1 only moved actin at 1.5 ± 0.2 μm/s (mean ± S.D., n = 619 filaments), and TgMyoA containing both TgMLC1 and TgELC1 moved actin at 3.3 ± 0.6 μm/s (mean ± S.D., n = 1,729 filaments). The values obtained in calcium versus EGTA were indistinguishable (p > 0.1, Kolmogorov-Smirnov test). For each data set, the bin with the highest speed was normalized to 1 for presentation purposes. Data were obtained from three protein preparations of TgMyoA with both light chains and two protein preparations of TgMyoA with TgMLC1 only.
Binding Both TgMLC1 and TgELC1 to the TgMyoA Heavy Chain Doubles the Speed of Actin Movement
In addition to TgMLC1, an essential light chain called TgELC1 was recently identified as part of the myosin motor complex (15). To test whether TgMLC1 and TgELC1 bind simultaneously to the same heavy chain, we prepared a recombinant baculovirus containing both light chains, each with a different tag. TgMLC1 had an N-terminal 3xMyc tag, whereas TgELC1 had a C-terminal 3xHA tag. Following infection with heavy and light chains, TgMyoA was immunoprecipitated from the Sf9 cell lysate with either an anti-HA antibody (TgELC1) or an anti-TgMLC1 antibody (Fig. 6). The Myc antibody could not be used for TgMLC1 immunoprecipitation because TgUNC was also tagged with Myc. The eluates were analyzed by Western blotting with antibodies to detect TgUNC (anti-Myc), TgMyoA heavy chain (anti-FLAG), TgMLC1 (anti-Myc), and TgELC1 (anti-HA). The results showed that the proteins co-immunoprecipitating with TgMLC1 included TgELC1 and conversely that the proteins co-immunoprecipitating with TgELC1 included TgMLC1. Earlier work showed that individual motor complexes do not physically associate with each other (9), and thus we conclude that both light chains simultaneously bind to the TgMyoA heavy chain.
FIGURE 6.
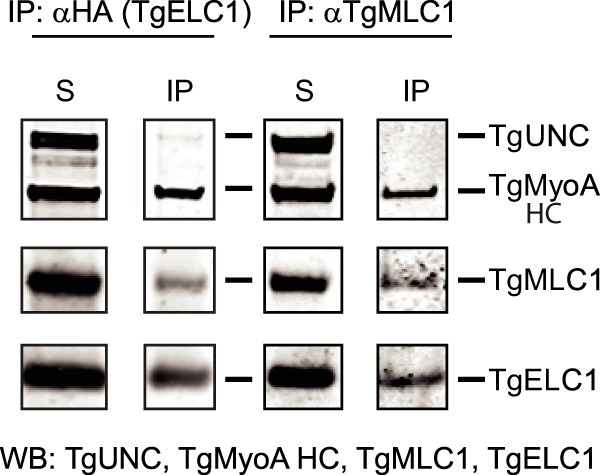
Both TgMLC1 and TgELC1 are bound to the same heavy chain. TgMyoA heavy chain (HC) and TgUNC were co-expressed in Sf9 cells with both 3xMyc-MLC1 and 3xHA-ELC1. The lane labeled S is the total soluble protein after Sf9 cell lysis. TgMyoA was co-immunoprecipitated from cell lysates using either anti-HA antibody (to immunoprecipitate TgELC1; left panels) or anti-TgMLC1 antibody (right panels), confirming that each of the light chains binds to TgMyoA heavy chain. Furthermore, TgMLC1 is present in the TgELC1 pulldowns and vice versa, demonstrating that the two light chains are simultaneously present on the same MyoA heavy chain. TgUNC (∼126 kDa) was detected with anti-Myc antibody, TgMyoA heavy chain (∼104 kDa) was detected with anti-FLAG, 3xMyc-TgMLC1 (∼29 kDa) was detected with anti-Myc in the left panels and anti-TgMLC1 in the right panels, and 3xHA-ELC1 (∼18 kDa) was detected with anti-HA. WB, Western blot; IP, immunoprecipitate.
When TgELC1 was co-expressed with TgMyoA heavy chain, TgMLC1, and TgUNC, both light chains co-purified with the heavy chain, indicating that both TgMLC1 and TgELC1 are tightly bound subunits of TgMyoA (Fig. 4A, lane 3). When TgMyoA containing both light chains was assayed in the in vitro motility assay, it moved actin at 3.4 ± 0.7 μm/s, which is more than twice the speed seen with TgMyoA containing only TgMLC1 (Fig. 5, solid red circles).
Calcium Does Not Regulate Motility Speed in Vitro
To determine whether calcium regulates the speed at which TgMyoA moves actin, an in vitro motility assay was performed in the presence or absence of free calcium. TgMyoA was preincubated in buffer containing either 0.2 mm free calcium or 1 mm EGTA for 1 h before the assay. Whether the myosin contained TgMLC1 only or both light chains, calcium did not affect the speed of actin movement (Fig. 5, open symbols).
Steady-state Actin-activated ATPase Activity
The speed of actin filament movement increased with MgATP concentration in the in vitro motility assay (Fig. 7A). The fit to a rectangular hyperbola defined a maximal speed of 4.6 ± 0.3 μm/s and a Km of 1.3 ± 0.3 mm MgATP. Steady-state actin-activated ATPase assays were thus performed with 5 mm MgATP. Rates of ATP hydrolysis were determined as a function of skeletal actin concentration, and data were fit to the Michaelis-Menten equation. TgMyoA with both light chains bound showed a Vmax of 84 ± 9.5 s−1 and a Km for actin of 136 ± 22 μm at 30 °C (Fig. 7B).
FIGURE 7.
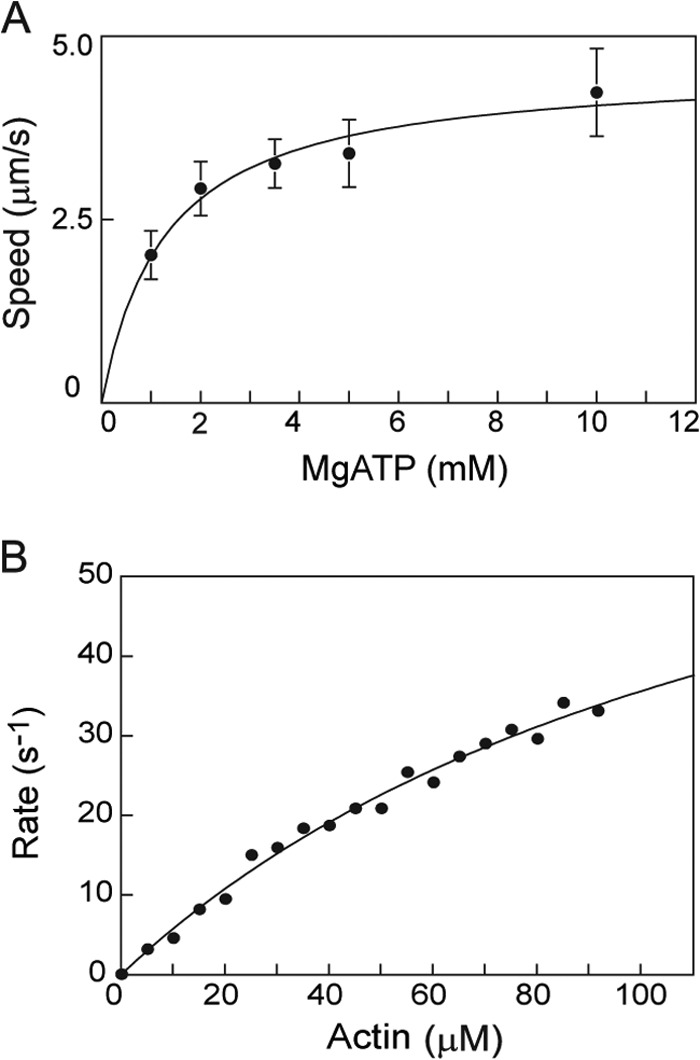
In vitro motility and steady-state actin-activated ATPase assays. A, in vitro motility speed of TgMyoA with both light chains bound as a function of MgATP concentration. The solid line is a fit to a rectangular hyperbola that defines a maximum speed of 4.6 ± 0.3 μm/s, and Km = 1.3 ± 0.3 mm MgATP. Error bars represent S.D. B, steady-state actin-activated ATPase assay of TgMyoA with both light chains bound (52 nm) as a function of skeletal actin concentration. Data were fit to the Michaelis-Menten equation: Vmax = 84 ± 9.5 s−1, and Km = 136 ± 22 μm. Conditions were as follows: 10 mm imidazole, pH 7.0, 5 mm NaCl, 1 mm MgCl2, 1 mm NaN3, 5 mm MgATP, and 1 mm DTT at 30 °C.
In a separate set of experiments, we showed that the actin-activated ATPase activity of TgMyoA with only TgMLC1 bound was indistinguishable from that obtained with TgMyoA containing both light chains (p > 0.1, Student's t test). The values obtained were 55.4 ± 5.4 s−1 (n = 5) for TgMyoA with only TgMLC1 bound and 55.6 ± 10.4 s−1 (n = 6) for TgMyoA with both light chains (at 60 μm actin). These values were obtained at 2 mm MgATP to decrease the ionic strength (relative to Fig. 7B, which was done at 5 mm MgATP). The lower ionic strength decreases the Km and allows the measured values to be nearer Vmax.
The TPR Domain of TgUNC Is Not Required to Express Functional Myosin
We next determined the minimal domain of TgUNC that produces an active motor. In addition to full-length TgUNC, two shorter constructs were cloned (Fig. 2B): one contained the central and UCS domains (ΔTPR), whereas the other contained the UCS domain only. Small scale infections were performed using these three TgUNC constructs, and expression and solubility of TgMyoA were determined by Western blotting (Fig. 8). Total and soluble fractions were probed with either anti-FLAG (top panel) for TgMyoA heavy chain or anti-Myc (bottom panel) for TgUNC and its truncations. The results showed that the TPR domain was dispensable to obtain soluble myosin, whereas the UCS domain alone produced very little soluble myosin.
FIGURE 8.
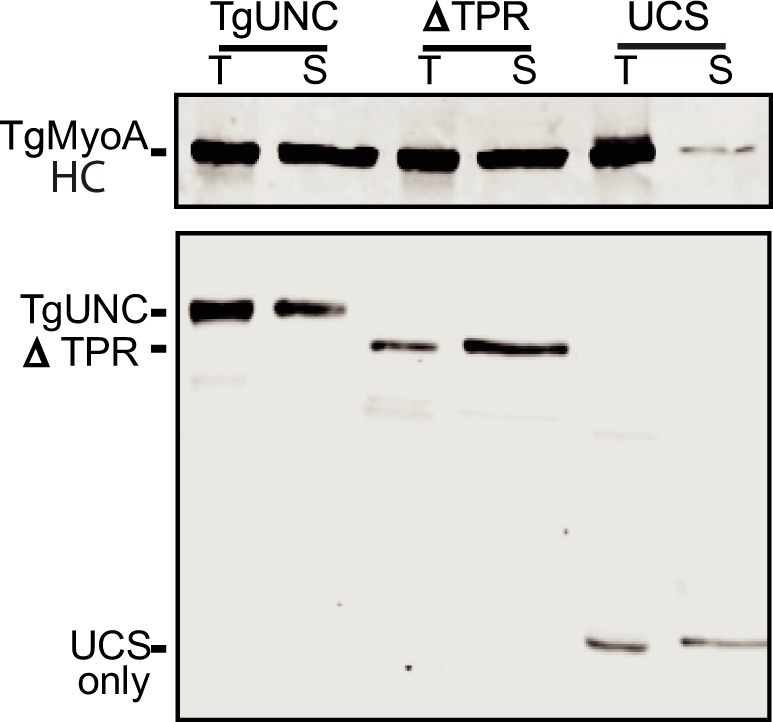
The TPR domain of TgUNC is not required for solubility of TgMyoA. Sf9 cell infections were performed with three TgUNC constructs (see Fig. 2B). Expression and solubility of TgMyoA were determined by Western blotting. Total (T) and soluble (S) fractions were probed with either anti-FLAG (top panel) for TgMyoA heavy chain (HC) or anti-Myc (bottom panel) for TgUNC and its truncations.
To determine the functionality of the TgMyoA co-expressed with these truncated chaperones, large scale co-infections and purifications were done with TgMyoA heavy chain, the two light chains, and each of the TgUNC constructs. All three infections yielded protein, but the yields were reduced for the two shorter TgUNC constructs. Per 109 Sf9 cells, full-length TgUNC yielded ∼1.5 mg of TgMyoA, ΔTPR yielded ∼0.35 mg of TgMyoA, and UCS yielded only ∼0.1 mg of TgMyoA. In the in vitro motility assay, the motor expressed with either full-length TgUNC or ΔTPR generated similar actin sliding speeds (3.4 ± 0.7 and 3.1 ± 0.5 μm/s, respectively; see Fig. 9). In contrast, speeds decreased more than 4-fold to 0.7 ± 0.2 μm/s when TgMyoA was expressed in the presence of the UCS domain alone (Fig. 9).
FIGURE 9.
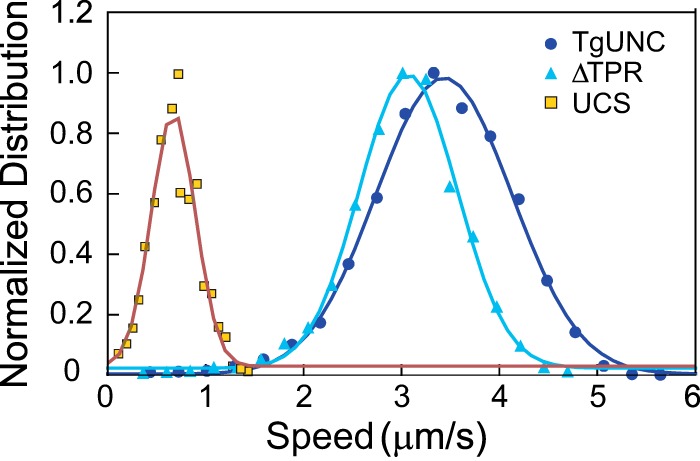
In vitro motility speeds of purified TgMyoA expressed with various TgUNC constructs. When TgMyoA was expressed with either full-length TgUNC or ΔTPR, the speed at which it moved actin was similar: full-length TgUNC, 3.4 ± 0.7 μm/s (mean ± S.D., n = 4,774 filaments) and ΔTPR, 3.1 ± 0.5 μm/s (mean ± S.D., n = 2,048 filaments). Speed decreased when TgMyoA was expressed in the presence of the UCS domain only (0.7 ± 0.2 μm/s, mean ± S.D., n = 1,296 filaments). The bin with the highest speed was normalized to 1 for presentation purposes. Data were obtained from three preparations of TgMyoA co-expressed with TgUNC, two protein preparations of TgMyoA co-expressed with ΔTPR, and one protein preparation of TgMyoA co-expressed with the UCS domain.
DISCUSSION
The first major finding of this study is that when the class XIVa myosin TgMyoA simultaneously binds two light chains, the well established TgMLC1, and a second light chain called TgELC1 (7, 15) the speed at which TgMyoA moves actin in an in vitro motility assay is more than twice that obtained when only TgMLC1 is bound (1.5 versus 3.4 μm/s). TgMyoA had previously been described as containing a motor domain that binds actin and MgATP followed by a short tail. Our data imply that TgMyoA has a fairly conventional lever arm with two bound light chains formed by TgELC1 and TgMLC1 binding to adjacent sites near the C terminus of the TgMyoA heavy chain as suggested previously (15) (Fig. 10). The lever arm amplifies small changes at the active site into the larger motions necessary to propel actin at fast speeds, and the length of the lever arm dictates the speed (for a review, see Ref. 14). Consistent with a number of other studies with skeletal muscle myosin (35), smooth muscle myosin (13), Dictyostelium myosin (36), and myosin V (12), the light chains affect the in vitro motility speed by changing the lever arm length, not by changing the kinetics of the motor domain.
FIGURE 10.
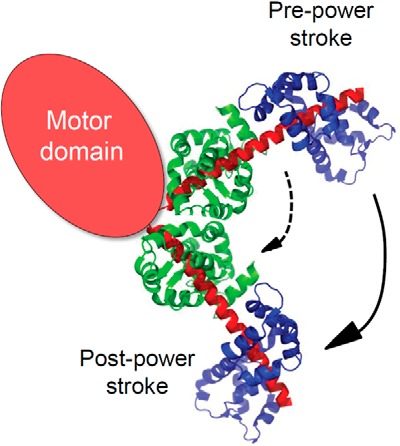
Schematic representation of the domain structure of TgMyoA. We propose that C-terminal to the motor domain TgMyoA has a typical lever arm stabilized by two bound light chains. To illustrate how lever arm length affects step size, a representative lever arm structure (Mlc1p bound to two adjacent motifs of unconventional myosin V; Protein Data Bank code 1N2D) is shown. Rotation of the lever arm upon phosphate release produces a power stroke that moves actin relative to myosin. The distance that myosin can displace actin per cycle of MgATP hydrolysis increases with lever arm length (compare dashed versus solid arrows). The functional impact of having two bound light chains (the equivalent of TgELC1 depicted here in green and TgMLC1 in blue) is a faster speed of movement of actin, consistent with our data.
The faster speed we obtained when both light chains were bound (∼3.4 μm/s) is close to the speed of the motor complex isolated from parasites (∼5 μm/s) (7, 9). The difference in the two values may be due to the fact that we used a semiautomated tracking program that calculates speeds of hundreds of moving filaments without user bias, whereas prior studies tracked a smaller number of individual filaments manually. A more interesting possibility would be that accessory proteins such as GAP45, which were not included in our study, can influence motor speed.
Role of TgMyoA Light Chains
TgELC1 was only recently identified as a component of the TgMyoA motor (15) because with a molecular mass of only ∼15 kDa it can be easily missed depending on how the motor complex components purified from parasites are resolved by SDS-PAGE. Our data and those of Nebl et al. (15) establish that TgELC1 is a bona fide subunit of TgMyoA. In principle, sequence analysis could have predicted that a second light chain would bind, but the region of the heavy chain to which TgELC1 is proposed to bind (15) has at best a very degenerate IQ motif (IQRECLSSWEP).
TgELC1 shows 45% sequence identity to T. gondii calmodulin (GenPept accession number XP_002367258.1), and based on sequence comparisons with calmodulin, there are potential calcium-binding sites in the first and third EF-hands in TgELC1. These two EF-hands in TgELC1 are each composed of a 12-amino acid loop that has a conserved acidic residue (Asp/Glu) at the 12th position necessary for calcium binding (37). These observations suggest that calcium binding to TgELC1 might regulate motor function. However, our in vitro motility data show no calcium-dependent changes in the speed of actin movement, and thus calcium binding does not directly regulate this motor. Because the motility assay is performed in the presence of excess light chains, we cannot rule out the possibility that calcium modulates the affinity of TgELC1 for the heavy chain.
Unlike TgELC1, the association of TgMLC1 with the TgMyoA heavy chain is well established. The TgMLC1 homolog in Plasmodium spp. is called myosin tail-interacting protein (MTIP) (38). TgMLC1 is 42% identical to MTIP from Plasmodium falciparum (PfMTIP). From the crystal structure of PfMTIP bound to the IQ motif from Plasmodium yoelii MyoA, one can predict where TgMLC1 binds to the TgMyoA heavy chain (VQAHIRKKMVA in P. yoelii versus AQAHIRRHLVD in T. gondii) (39). Surprisingly, the seventh amino acid of the IQ motif in the parasite myosins is a long chained basic amino acid (Lys or Arg) instead of the consensus Gly of the IQ motif (IQXXXRGXXXR). Every side chain atom of this Lys residue in P. falciparum MyoA heavy chain contacts MTIP, and the Lys residue appears to be essential for PfMTIP to bind to the heavy chain in a compact conformation (39, 40). Presumably the Arg residue at position 7 of the IQ motif in T. gondii plays a similar role. This residue is part of a dibasic motif shown previously to be important for localizing TgMyoA to the parasite periphery (8) presumably because it is needed for TgMLC1 binding to the heavy chain.
The spacing between the two putative IQ motifs in the T. gondii heavy chain is 36 amino acids, which is much longer than the 21- or 23-residue spacing seen in class V myosins. When a larger spacing between IQ motifs was generated by adding two additional Ala residues between IQ motifs 3 and 4 in myosin V, it took a smaller step because the lever arm became compliant (41). In TgMyoA, it is possible that the ∼70-amino acid N-terminal extension of TgMLC1 binds to some extent to heavy chain residues between the two IQ motifs so that the lever arm remains rigid. In agreement with this idea, isothermal calorimetry data with PfMTIP and heavy chain peptides showed that adding only four additional N-terminal amino acids to the peptide that was used for crystallographic studies of the PfMTIP-heavy chain complex (39) enhanced binding affinity 4-fold (42). The implication of this result is that these heavy chain residues, which lie between the two putative IQ motifs, are engaged in additional interactions with PfMTIP.
Motor Activity
The steady-state ATPase assay shows that TgMyoA has a low affinity (i.e. a high Km) for actin in the presence of MgATP as expected for a single-headed motor. The extrapolated maximal ATPase rate of ∼80 s−1 gives a total time per cycle of MgATP hydrolysis of ∼12 ms. From the measured unitary step size (duni) of 5.3 nm (7) and a speed of actin movement of ∼3.4 μm/s (v), the time the motor spends strongly attached to actin is ∼1.5 ms (ton = duni/v). This is a small percentage of the total cycle time, and thus this motor has a low duty cycle as do most motors designed for speed. Earlier studies also concluded that TgMyoA has a low duty cycle based either on kinetic (7) or in vitro motility data (9). An unusual feature of TgMyoA is the high MgATP concentration (1.3 mm) required to obtain half-maximal speed in the motility assay. This apparent low affinity for substrate is consistent with the Kd of ∼800 μm measured for the dissociation constant of ADP from the actomyosin complex (7).
Folding of TgMyoA into a Functional Motor Requires a Parasite-specific Myosin Co-chaperone
A second major finding of this work is that a T. gondii myosin co-chaperone is required to properly fold TgMyoA heavy chain in the baculovirus/Sf9 insect cell expression system. This is the first demonstration that functional class XIV myosin from an apicomplexan parasite can be expressed in a heterologous system. Although TgMyoA can be purified directly from the parasite, the yields are low, making it difficult to rigorously characterize the parasite-derived motor complex. In contrast, yields of 1 mg of TgMyoA were readily attainable from 1 × 109 infected Sf9 cells (200-ml culture), which is comparable with yields obtained with myosins that do not require co-expression with an exogenous chaperone.
TgUNC has all three domains found in the canonical Unc45 protein, i.e. an N-terminal TPR domain, a central domain, and a C-terminal UCS domain that binds to myosin. The crystal structure of UCS family myosin chaperones from S. cerevisiae (43), Drosophila melanogaster (44), and C. elegans (45) have been solved. She4p, the UCS family protein from budding yeast, lacks a TPR domain, whereas DmUnc45 from Drosophila contains one, but it was disordered and not seen in the crystal structure. The structure of the C. elegans UNC-45 was the first to show the structure of the full-length protein. The UNC-45 structure resembles a checkmark with the shorter arm consisting of the TPR and central domain and the longer arm consisting of the UCS domain (43–45). The structure is composed almost entirely of stacked α-helical motifs known as arginine-rich motif repeats. Forty percent of the arginine-rich motif consensus residues identified in DmUnc45 (44) are identical in TgUNC (supplemental Fig. S1), making it highly likely that their overall structures are similar. Based on sequence homology within the UCS domains of functionally diverse UCS proteins, there is a strikingly high degree of identity at positions that map to a hydrophobic groove near the C terminus of the UCS domain. Based on this, it was proposed that the last five arginine-rich motifs (numbered 17–21) are the sites of interaction with myosin (44).
A novel feature observed with the C. elegans UNC-45 was its ability to form a scaffold of linear protein chains, a desirable feature in that it offers multiple adjacent binding sites for Hsc70, Hsp90, and the client protein myosin (45). It would be interesting to know whether this seemingly advantageous oligomerization is a feature common to all three-domain UCS family proteins including TgUNC.
The N-terminal TPR domain of TgUNC was dispensable for expression of functional TgMyoA in Sf9 cells. Similarly, the TPR domain of C. elegans UNC-45 is not required to rescue lethal unc-45-null mutants arrested in embryonic muscle development (46) even though this domain is typically thought to be essential for recruiting the chaperone Hsp90 to the client protein myosin. The comparable UCS family proteins in both budding (She4p) and fission (Rng3p) yeast also lack a TPR domain, but Rng3p still interacts with fission yeast Hsp90 (Swo1p) by yeast two-hybrid and co-immunoprecipitation analyses (47). This result implies that in fission yeast Hsp90 can bind to another domain of the UCS protein either directly or via another protein. Perhaps TgUNC also has alternative binding sites for Hsp90 that are utilized in the absence of the TPR domain. Alternatively, Hsp90 and TgUNC may act sequentially to fold myosin in the absence of the TPR domain. Given that Unc45 greatly stimulates the activity of Hsp90-mediated motor domain folding (27), this would be a much less efficient folding mechanism than with full-length TgUNC.
Although we have clearly established a requirement for TgUNC in TgMyoA heavy chain folding, many questions still remain about the folding pathway of myosins in general. Why can so many myosins utilize the endogenous folding machinery of Sf9 cells, whereas others such as TgMyoA and vertebrate skeletal and cardiac myosins require specific cofactors? Do other T. gondii myosins (48) require TgUNC for folding? While this paper was under review, it was shown that co-expression with Unc45b and Hsp90 led to the successful purification of mouse myosin 15 from Sf9 cells (49), demonstrating that chaperone-assisted expression may be a useful approach for other myosins. Regardless of the answers to these and other basic mechanistic questions about myosin folding, we have accomplished here the long sought after goal of obtaining milligram quantities of TgMyoA that will for the first time make high throughput screening for drugs against this unusual, virulence-associated motor possible. Furthermore, the genomes of other apicomplexan parasites encode homologs of TgUNC, suggesting that this approach may be similarly useful for the expression of functional class XIV myosins from other apicomplexan parasites of medical or veterinary importance.
Supplementary Material
Acknowledgments
We thank Dr. Marc-Jan Gubbels for providing the T. gondii cosmid number p559 and Dr. Con Beckers for providing the anti-TgMLC1 antibody. We acknowledge the early efforts of Aoife Heaslip and Michelle Shepard in attempting to express functional TgMyoA in Sf9 cells without the chaperone. We thank Susan Lowey for helpful comments on the manuscript. T. gondii genome sequence information was obtained from the Toxoplasma Genome Database. ToxoDB is a component of the Eukaryotic Pathogen Genomics Resource, a Bioinformatics Resource Center supported by the National Institutes of Allergy and Infectious Diseases; we gratefully acknowledge the staff responsible for developing and maintaining this resource.
This work was supported, in whole or in part, by National Institutes of Health Grants GM078097 (to K. M. T.) and AI054961 (to G. E. W.).

This article contains supplemental Fig. S1.
L. Tilley and G. E. Ward, unpublished data.
A. Heaslip and G. E. Ward, unpublished data.
- TgMyoA
- T. gondii myosin A (TgMyoA heavy chain with or without bound light chain(s))
- IMC
- inner membrane complex
- MTIP
- myosin tail-interacting protein
- TgELC1
- T. gondii essential light chain-1
- TgGAP40
- -45, -50, and -70, T. gondii gliding-associated proteins of 40, 45, 50, and 70 kDa, respectively
- TgMLC1
- T. gondii myosin light chain-1
- TPR
- tetratricopeptide repeat
- TgUNC
- T. gondii myosin co-chaperone of the UCS family
- UCS
- a domain conserved in UNC-45/CRO1/She4p proteins.
REFERENCES
- 1. Foth B. J., Goedecke M. C., Soldati D. (2006) New insights into myosin evolution and classification. Proc. Natl. Acad. Sci. U.S.A. 103, 3681–3686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Meissner M., Schlüter D., Soldati D. (2002) Role of Toxoplasma gondii myosin A in powering parasite gliding and host cell invasion. Science 298, 837–840 [DOI] [PubMed] [Google Scholar]
- 3. Sheffield H. G., Melton M. L. (1968) The fine structure and reproduction of Toxoplasma gondii. J. Parasitol. 54, 209–226 [PubMed] [Google Scholar]
- 4. Gaskins E., Gilk S., DeVore N., Mann T., Ward G., Beckers C. (2004) Identification of the membrane receptor of a class XIV myosin in Toxoplasma gondii. J. Cell Biol. 165, 383–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Frénal K., Polonais V., Marq J. B., Stratmann R., Limenitakis J., Soldati-Favre D. (2010) Functional dissection of the apicomplexan glideosome molecular architecture. Cell Host Microbe 8, 343–357 [DOI] [PubMed] [Google Scholar]
- 6. Heintzelman M. B., Schwartzman J. D. (1997) A novel class of unconventional myosins from Toxoplasma gondii. J. Mol. Biol. 271, 139–146 [DOI] [PubMed] [Google Scholar]
- 7. Herm-Götz A., Weiss S., Stratmann R., Fujita-Becker S., Ruff C., Meyhöfer E., Soldati T., Manstein D. J., Geeves M. A., Soldati D. (2002) Toxoplasma gondii myosin A and its light chain: a fast, single-headed, plus-end-directed motor. EMBO J. 21, 2149–2158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hettmann C., Herm A., Geiter A., Frank B., Schwarz E., Soldati T., Soldati D. (2000) A dibasic motif in the tail of a class XIV apicomplexan myosin is an essential determinant of plasma membrane localization. Mol. Biol. Cell 11, 1385–1400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Heaslip A. T., Leung J. M., Carey K. L., Catti F., Warshaw D. M., Westwood N. J., Ballif B. A., Ward G. E. (2010) A small-molecule inhibitor of T. gondii motility induces the posttranslational modification of myosin light chain-1 and inhibits myosin motor activity. PLoS Pathog. 6, e1000720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Håkansson S., Morisaki H., Heuser J., Sibley L. D. (1999) Time-lapse video microscopy of gliding motility in Toxoplasma gondii reveals a novel, biphasic mechanism of cell locomotion. Mol. Biol. Cell 10, 3539–3547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Leung J. M., Rould M. A., Konradt C., Hunter C. A., Ward G. E. (2014) Disruption of TgPHIL1 alters specific parameters of Toxoplasma gondii motility measured in a quantitative, three-dimensional live motility assay. PLoS One 9, e85763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Moore J. R., Krementsova E. B., Trybus K. M., Warshaw D. M. (2004) Does the myosin V neck region act as a lever? J. Muscle Res. Cell Motil. 25, 29–35 [DOI] [PubMed] [Google Scholar]
- 13. Warshaw D. M., Guilford W. H., Freyzon Y., Krementsova E., Palmiter K. A., Tyska M. J., Baker J. E., Trybus K. M. (2000) The light chain binding domain of expressed smooth muscle heavy meromyosin acts as a mechanical lever. J. Biol. Chem. 275, 37167–37172 [DOI] [PubMed] [Google Scholar]
- 14. Tyska M. J., Warshaw D. M. (2002) The myosin power stroke. Cell Motil. Cytoskeleton 51, 1–15 [DOI] [PubMed] [Google Scholar]
- 15. Nebl T., Prieto J. H., Kapp E., Smith B. J., Williams M. J., Yates J. R., 3rd, Cowman A. F., Tonkin C. J. (2011) Quantitative in vivo analyses reveal calcium-dependent phosphorylation sites and identifies a novel component of the Toxoplasma invasion motor complex. PLoS Pathog. 7, e1002222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Trybus K. M. (1994) Regulation of expressed truncated smooth muscle myosins. Role of the essential light chain and tail length. J. Biol. Chem. 269, 20819–20822 [PubMed] [Google Scholar]
- 17. Pato M. D., Sellers J. R., Preston Y. A., Harvey E. V., Adelstein R. S. (1996) Baculovirus expression of chicken nonmuscle heavy meromyosin II-B. Characterization of alternatively spliced isoforms. J. Biol. Chem. 271, 2689–2695 [DOI] [PubMed] [Google Scholar]
- 18. De La Cruz E. M., Wells A. L., Rosenfeld S. S., Ostap E. M., Sweeney H. L. (1999) The kinetic mechanism of myosin V. Proc. Natl. Acad. Sci. U.S.A. 96, 13726–13731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Trybus K. M., Krementsova E., Freyzon Y. (1999) Kinetic characterization of a monomeric unconventional myosin V construct. J. Biol. Chem. 274, 27448–27456 [DOI] [PubMed] [Google Scholar]
- 20. Wang F., Chen L., Arcucci O., Harvey E. V., Bowers B., Xu Y., Hammer J. A., 3rd, Sellers J. R. (2000) Effect of ADP and ionic strength on the kinetic and motile properties of recombinant mouse myosin V. J. Biol. Chem. 275, 4329–4335 [DOI] [PubMed] [Google Scholar]
- 21. Wells A. L., Lin A. W., Chen L. Q., Safer D., Cain S. M., Hasson T., Carragher B. O., Milligan R. A., Sweeney H. L. (1999) Myosin VI is an actin-based motor that moves backwards. Nature 401, 505–508 [DOI] [PubMed] [Google Scholar]
- 22. Yang Y., Kovács M., Xu Q., Anderson J. B., Sellers J. R. (2005) Myosin VIIB from Drosophila is a high duty ratio motor. J. Biol. Chem. 280, 32061–32068 [DOI] [PubMed] [Google Scholar]
- 23. Chow D., Srikakulam R., Chen Y., Winkelmann D. A. (2002) Folding of the striated muscle myosin motor domain. J. Biol. Chem. 277, 36799–36807 [DOI] [PubMed] [Google Scholar]
- 24. Srikakulam R., Winkelmann D. A. (1999) Myosin II folding is mediated by a molecular chaperonin. J. Biol. Chem. 274, 27265–27273 [DOI] [PubMed] [Google Scholar]
- 25. Srikakulam R., Winkelmann D. A. (2004) Chaperone-mediated folding and assembly of myosin in striated muscle. J. Cell Sci. 117, 641–652 [DOI] [PubMed] [Google Scholar]
- 26. Barral J. M., Hutagalung A. H., Brinker A., Hartl F. U., Epstein H. F. (2002) Role of the myosin assembly protein UNC-45 as a molecular chaperone for myosin. Science 295, 669–671 [DOI] [PubMed] [Google Scholar]
- 27. Liu L., Srikakulam R., Winkelmann D. A. (2008) Unc45 activates Hsp90-dependent folding of the myosin motor domain. J. Biol. Chem. 283, 13185–13193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Srikakulam R., Liu L., Winkelmann D. A. (2008) Unc45b forms a cytosolic complex with Hsp90 and targets the unfolded myosin motor domain. PLoS One 3, e2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Price M. G., Landsverk M. L., Barral J. M., Epstein H. F. (2002) Two mammalian UNC-45 isoforms are related to distinct cytoskeletal and muscle-specific functions. J. Cell Sci. 115, 4013–4023 [DOI] [PubMed] [Google Scholar]
- 30. Scheufler C., Brinker A., Bourenkov G., Pegoraro S., Moroder L., Bartunik H., Hartl F. U., Moarefi I. (2000) Structure of TPR domain-peptide complexes: critical elements in the assembly of the Hsp70-Hsp90 multichaperone machine. Cell 101, 199–210 [DOI] [PubMed] [Google Scholar]
- 31. Kinose F., Wang S. X., Kidambi U. S., Moncman C. L., Winkelmann D. A. (1996) Glycine 699 is pivotal for the motor activity of skeletal muscle myosin. J. Cell Biol. 134, 895–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Trybus K. M. (2000) Biochemical studies of myosin. Methods 22, 327–335 [DOI] [PubMed] [Google Scholar]
- 33. Philo J. S. (2000) A method for directly fitting the time derivative of sedimentation velocity data and an alternative algorithm for calculating sedimentation coefficient distribution functions. Anal. Biochem. 279, 151–163 [DOI] [PubMed] [Google Scholar]
- 34. Cronan J. E., Jr. (1990) Biotination of proteins in vivo. A post-translational modification to label, purify, and study proteins. J. Biol. Chem. 265, 10327–10333 [PubMed] [Google Scholar]
- 35. Lowey S., Waller G. S., Trybus K. M. (1993) Skeletal muscle myosin light chains are essential for physiological speeds of shortening. Nature 365, 454–456 [DOI] [PubMed] [Google Scholar]
- 36. Uyeda T. Q., Abramson P. D., Spudich J. A. (1996) The neck region of the myosin motor domain acts as a lever arm to generate movement. Proc. Natl. Acad. Sci. U.S.A. 93, 4459–4464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Beckingham K. (1991) Use of site-directed mutations in the individual Ca2+-binding sites of calmodulin to examine Ca2+-induced conformational changes. J. Biol. Chem. 266, 6027–6030 [PubMed] [Google Scholar]
- 38. Bergman L. W., Kaiser K., Fujioka H., Coppens I., Daly T. M., Fox S., Matuschewski K., Nussenzweig V., Kappe S. H. (2003) Myosin A tail domain interacting protein (MTIP) localizes to the inner membrane complex of Plasmodium sporozoites. J. Cell Sci. 116, 39–49 [DOI] [PubMed] [Google Scholar]
- 39. Bosch J., Turley S., Roach C. M., Daly T. M., Bergman L. W., Hol W. G. (2007) The closed MTIP-myosin A-tail complex from the malaria parasite invasion machinery. J. Mol. Biol. 372, 77–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Turley S., Khamrui S., Bergman L. W., Hol W. G. (2013) The compact conformation of the Plasmodium knowlesi myosin tail interacting protein MTIP in complex with the C-terminal helix of myosin A. Mol. Biochem. Parasitol. 190, 56–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sakamoto T., Wang F., Schmitz S., Xu Y., Xu Q., Molloy J. E., Veigel C., Sellers J. R. (2003) Neck length and processivity of myosin V. J. Biol. Chem. 278, 29201–29207 [DOI] [PubMed] [Google Scholar]
- 42. Douse C. H., Green J. L., Salgado P. S., Simpson P. J., Thomas J. C., Langsley G., Holder A. A., Tate E. W., Cota E. (2012) Regulation of the Plasmodium motor complex: phosphorylation of myosin A tail-interacting protein (MTIP) loosens its grip on MyoA. J. Biol. Chem. 287, 36968–36977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Shi H., Blobel G. (2010) UNC-45/CRO1/She4p (UCS) protein forms elongated dimer and joins two myosin heads near their actin binding region. Proc. Natl. Acad. Sci. U.S.A. 107, 21382–21387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lee C. F., Hauenstein A. V., Fleming J. K., Gasper W. C., Engelke V., Sankaran B., Bernstein S. I., Huxford T. (2011) X-ray crystal structure of the UCS domain-containing UNC-45 myosin chaperone from Drosophila melanogaster. Structure 19, 397–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gazda L., Pokrzywa W., Hellerschmied D., Löwe T., Forné I., Mueller-Planitz F., Hoppe T., Clausen T. (2013) The myosin chaperone UNC-45 is organized in tandem modules to support myofilament formation in C. elegans. Cell 152, 183–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ni W., Hutagalung A. H., Li S., Epstein H. F. (2011) The myosin-binding UCS domain but not the Hsp90-binding TPR domain of the UNC-45 chaperone is essential for function in Caenorhabditis elegans. J. Cell Sci. 124, 3164–3173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mishra M., D'souza V. M., Chang K. C., Huang Y., Balasubramanian M. K. (2005) Hsp90 protein in fission yeast Swo1p and UCS protein Rng3p facilitate myosin II assembly and function. Eukaryot. Cell 4, 567–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Frénal K., Foth B. J., Soldati D. (2008) in Myosins: a Superfamily of Molecular Motors (Coluccio L. M., ed) Vol. 7, pp. 421–440, Springer Verlag, Dordrecht, The Netherlands [Google Scholar]
- 49. Bird J. E., Takagi Y., Billington N., Strub M. P., Sellers J. R., Friedman T. B. (2014) Chaperone-enhanced purification of unconventional myosin 15, a molecular motor specialized for stereocilia protein trafficking. Proc. Natl. Acad. Sci. U.S.A. 111, 12390–12395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Andenmatten N., Egarter S., Jackson A. J., Jullien N., Herman J. P., Meissner M. (2013) Conditional genome engineering in Toxoplasma gondii uncovers alternative invasion mechanisms. Nat. Methods 10, 125–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Egarter S., Andenmatten N., Jackson A. J., Whitelaw J. A., Pall G., Black J. A., Ferguson D. J., Tardieux I., Mogilner A., Meissner M. (2014) The Toxoplasma acto-MyoA motor complex is important but not essential for gliding motility and host cell invasion. PLoS One 9, e91819. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


