Abstract
Magnetotactic bacteria biomineralize ordered chains of uniform, membrane-bound magnetite or greigite nanocrystals that exhibit nearly perfect crystal structures and species-specific morphologies. Transmission electron microscopy (TEM) is a critical technique for providing information regarding the organization of cellular and magnetite structures in these microorganisms. However, conventional TEM can only be used to image air-dried or vitrified bacteria removed from their natural environment. Here we present a correlative scanning TEM (STEM) and fluorescence microscopy technique for imaging viable cells of Magnetospirillum magneticum strain AMB-1 in liquid using an in situ fluid cell TEM holder. Fluorescently labeled cells were immobilized on microchip window surfaces and visualized in a fluid cell with STEM, followed by correlative fluorescence imaging to verify their membrane integrity. Notably, the post-STEM fluorescence imaging indicated that the bacterial cell wall membrane did not sustain radiation damage during STEM imaging at low electron dose conditions. We investigated the effects of radiation damage and sample preparation on the bacteria viability and found that approximately 50% of the bacterial membranes remained intact after an hour in the fluid cell, decreasing to ~30% after two hours. These results represent a first step toward in vivo studies of magnetite biomineralization in magnetotactic bacteria.
Biomineralization is a widespread biological phenomenon occurring in living organisms ranging from single cells to complex multicellular organisms. Biomineralization of inorganic materials in single cell organisms is an ideal system for studying fundamental mechanisms of biomineralization1. Model systems range from prokaryotic organisms, such as magnetite formation in magnetotactic bacteria2,3,4, to eukaryotic organisms, such as silica biomineralization in diatoms5,6. Understanding biomineralization in these organisms in terms of crystal nucleation and growth, as well as the involvement of biological macromolecules and cellular processes, is of fundamental interest to scientists as similar principles can be used to develop synthetic nanomaterials.
Magnetite biomineralization by magnetotactic bacteria is a topic of great interest in nanotechnology7,8,9,10,11,12, functional materials11,12,13,14,15,16, and astrobiology17. Magnetotactic bacteria take up soluble iron species that they use to biomineralize chains of magnetite nanocrystals, known as magnetosomes, in intracellular membrane vesicles2. The nanocrystals have nearly perfect mineral crystal structures with consistent species-specific morphologies, leading to well-defined magnetic properties2,3,9,12,18,19. As a result, magnetotactic bacteria are one of the best model systems for investigating the molecular mechanisms of biomineralization. Magnetite biomineralization in these bacteria is a complex process involving a number of steps including cellular uptake and reduction of ferric ions, complexation of the iron with membrane proteins, and nucleation and growth of mature magnetosomes (cf. Fig. 6 in Faivre and Schuler19). Despite decades of intense research, this process is still not fully understood2,18,20,21. Biological macromolecules, specifically membrane bound proteins in magnetotactic bacteria, play an important role in templating biomineralized magnetosome magnetite crystals, although their exact role remains unclear14,15,16,21.
The variety of processes involved in magnetosome formation, ranging from the bacterial uptake of iron to nucleation and growth of nanoscale magnetite, necessitates the use of various analytical techniques. Materials characterization techniques are typically aimed at direct observations of the magnetite nucleation and growth at high spatial resolution. High resolution TEM is often used to observe formation of magnetosomes in magnetotactic bacteria via cryogenic TEM (cryo-TEM)4,22,23,24,25,26,27,28,29. This technique provides vital information about organization of the cellular structures and development of the magnetosomes20,27. However, the vitreous ice environment of cryo-TEM samples may not be entirely representative of the native hydrated cellular state, and may introduce artifacts due to the preparatory protocols often involving addition of cryo-protectants, thin sectioning, and sample staining. Fluorescence microscopy is often employed in conjunction with TEM imaging to study dynamic biomolecular processes in magnetotactic bacteria, such as localization of membrane proteins during bacteria growth20,26. The spatial resolution of fluorescence microscopy is limited by the diffraction limit to ~200 nm, so establishing connections between biomolecular processes and magnetosome growth mechanisms relies on correlation with cryo-TEM. Correlative cryo-TEM and optical techniques are very complex, often involving computer transfer of sample positions obtained in the optical microscope to the electron microscope via specialized software30.
Fluid cell TEM and scanning TEM (STEM) techniques are unique in that they can be used to directly observe biomacromolecules31,32,33,34,35 and whole cells36,37,38,39,40 in their fully hydrated state. The experimental setup typically consists of a microfluidic chamber comprised of two silicon chips with electron transparent silicon nitride (SiN) windows contained in a hermetically sealed TEM holder41. A thin liquid layer containing the specimen is maintained between the SiN windows; for whole eukaryotic cells the liquid thickness is typically on the order of microns, limiting the spatial resolution to nanometers36,42. Because biological specimens in these thick liquid layers lack sufficient contrast, they are typically incubated with high contrast nanoparticles such as gold36 or CdSe quantum dots38 and imaged with high angle annular dark field (HAADF) STEM, i.e. Z-contrast imaging. De Jonge and co-workers have established correlative fluid cell STEM and fluorescence microscopy as a powerful technique for studying various aspects of eukaryotic cells, such as locating epidermal growth factor receptors in COS7 cells38, exploring the correlation between cellular function and ultrastructure in yeast cells39, and investigating electron beam damage and cell viability in live COS7 cells43. Other electron microscopy techniques exist for imaging whole hydrated cells, such as environmental scanning electron microscopy (ESEM)44, and air SEM (ASEM)45. The cells in these techniques are typically chemically fixed and maintained in a vacuum, making live cell imaging impossible; however, important information about cell receptors can be obtained by incubating the cells with nanoparticles prior to fixation and imaging44.
In this report, we utilize correlative fluorescence and fluid cell STEM imaging to visualize magnetotactic bacteria containing nanoscale biomineralized magnetosomes. We show that HAADF-STEM imaging reveals the bacteria cell membrane in some cases even when magnetite magnetosomes are not present, which is imperative for on-going in vivo magnetite biomineralization studies in the fluid cell. The confined environment of the fluid cell causes a rapid decrease in bacteria viability over a time of two hours, necessitating the use of post-STEM fluorescence microscopy to verify bacteria membrane integrity. We present new criteria for imaging initially live bacteria in the fluid cell, and discuss various radiation damage thresholds and other cell damage mechanisms for fluid cell STEM imaging of bacteria. Correlative STEM and fluorescence imaging of magnetotactic bacteria is a first step in observing biomineralization of magnetite nanocrystals in vivo, and the described approach is expected to be applicable to a broad range of microorganisms that biomineralize various nanomaterials.
Results
Electron microscopy of air-dried magnetotactic bacteria
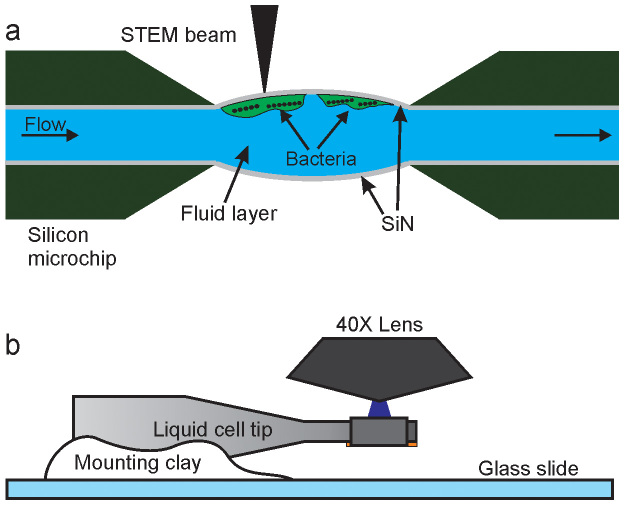
Fig. 1 shows the experimental setup used for the correlative STEM and fluorescence imaging of magnetotactic bacteria. Cells of Magnetospirillum magneticum, strain AMB-1, are attached to the top functionalized 100 nm SiN spacer chip (cf. Methods) and sandwiched with another 100 nm spacer chip, as schematically shown in Fig. 1a. The tip of the liquid cell is removed from the STEM holder platform, mounted on a glass slide and accepted by the fluorescence microscope for imaging with a 40X objective lens, as schematically shown in Fig. 1b.
Figure 1. Schematic of in situ fluid cell STEM and correlative fluorescence microscopy (not to scale).
(a) Fluid cell microfluidic chamber consisting of two silicon microchips supporting two electron transparent SiN membranes. Cells of M. magneticum are attached to the top SiN window and imaged with STEM in the thin liquid layer. (b) The tip of the liquid cell is mounted on a glass slide and subsequently imaged in a fluorescence microscope with a 40X objective lens.
Cells of M. magneticum, strain AMB-1, are helical in shape and typically 1–3 µm in length and 0.4–0.6 µm in diameter46. They produce chains of approximately 10–15 cuboctahedral magnetite magnetosomes, each on the order of 50 nm in size47. A typical HAADF-STEM image of an air-dried, fixed cell of M. magneticum on a grid is shown in Fig. 2a. The magnetosome chains are situated inside the cytoplasmic membrane of the bacterium, held together by networks of actin-like filaments26 (Fig. 2a, white arrow). The term magnetosome refers to the inorganic magnetite nanocrystal along with the enveloping organic vesicle (cf. HRTEM images in Supp. Fig. 1). Other than the magnetosomes, only the cytoplasm of the cell is visible in the HAADF-STEM image; the rest of the cell ultrastructure is not visible due to minor differences in the Z-contrast between various cellular structures. Energy filtered TEM (EFTEM) elemental maps of a single cell of M. magneticum on a grid confirm the presence of oxygen in the cytoplasm (green contrast, Fig. 2b) as well as increased amounts of oxygen in the magnetite magnetosome crystals. An increased iron signal in the nanocrystals (purple contrast) confirms that they consist of an iron oxide species. While imaging magnetotactic bacteria on carbon grids allows for morphological and chemical analysis of the magnetosome structures, glutaraldehyde fixation and the harsh environment of the vacuum makes imaging of the bacteria in their native hydrated state impossible.
Figure 2. Ex situ HAADF-STEM (a) and EFTEM (b) images of cells of M. magneticum prepared on a TEM grid.
(a) HAADF-STEM image of a cell fixed with glutaraldehyde. The magnetite magnetosome chain is denoted with a white arrow. The scale bar is 500 nm. (b) False colored EFTEM elemental map of a cell. Oxygen is shown in green, while iron is shown in purple. The scale bar is 200 nm.
Fluid cell STEM imaging
Briefly, cells of M. magneticum were suspended in growth media and labeled with SYTO 9 and propidium iodide fluorescence dyes and attached to a poly-L-lysine coated 100 nm SiN spacer chip (APTES or BioPlus chips used in other cases will be indicated) and sandwiched with another 100 nm spacer chip (cf. Methods). A flow of fresh media was introduced through the fluid cell prior to STEM imaging to remove excess fluorescence dye; however, there was no fluid flow during STEM imaging (cf. Methods). At the nominal liquid layer thickness of 200 nm the presence of flow could not be confirmed reproducibly, although diffusion of fluorescent dye out of the sample area could not be ruled out. The bacterial cells were kept in a static environment of media during imaging, typically for two hours. Low-loss electron energy loss spectroscopy (EELS) revealed that the liquid layer thickness was typically 500–750 nm at the edges and corners of the SiN window for this spacer configuration48. The HAADF-STEM images in Fig. 3 were acquired with a beam current of 38 pA, yielding a cumulative electron dose of 0.09 electrons · Å−2 for Fig. 3a and 0.34 electrons · Å−2 for Fig. 3b.
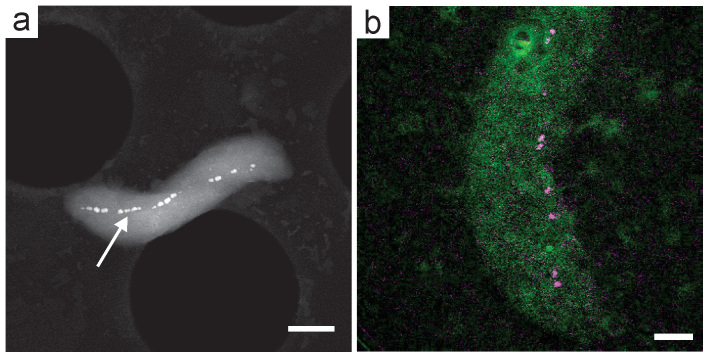
Figure 3. In situ fluid cell HAADF-STEM images of cells of M. magneticum.
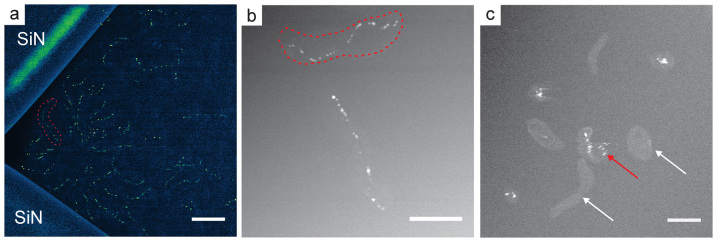
(a) Low magnification image of ~30 bacteria in the corner of the SiN window. The HAADF-STEM image has been background subtracted and false colored so that bacterial magnetosomes appear green on a blue background. The approximate position of the cytoplasm of a cell is highlighted with a dashed red line. (b) HAADF-STEM image of two cells showing the magnetosome chains as well as the cell membranes. The membrane of the top bacteria is highlighted with a dashed red line. (c) STEM image showing bacterial cells with no magnetosomes (white arrows) as well as movement of bacteria indicated by image streaking (red arrow). The scale bar is 2 μm in (a) and 1 μm in (b) and (c).
Fig. 3a shows a low magnification false colored HAADF-STEM image of ~30 cells attached in the corner of the SiN window. The bacterial magnetosomes are clearly visible in green as well as the cytoplasm of the cells, which appear as a light blue contrast above the dark blue background of the fluid (e.g. red dashed line shows the approximate position of the cytoplasm in a single cell). The majority of previous biological cell STEM imaging studies in fluids have employed high-contrast labels, such as gold nanoparticles36 and quantum dots38, to provide contrast in HAADF-STEM images. In our experiments, the biomineralized magnetite magnetosome chains in cells of M. magneticum act as natural high-contrast labels denoting the position of the bacterial cells. Furthermore, alignment of the magnetite nanocrystals in chains is an indicator of the integrity of the bacterium's cellular structure—chains of magnetosomes indicate that the cell membrane and magnetosome membrane vesicles are intact, held together by networks of filaments26. In contrast, lysed bacterial cells would only show collapsed chains or free floating magnetosomes. The bacterial cell wall membrane is typically only visible when the bacterium is near a corner of the SiN window, where the liquid layer is thinnest and the spatial resolution is highest. Fig. 3b shows a higher magnification image of two cells of M. magneticum. In this case the cells were not close to the SiN window corner, so the cell walls are not as visible as in Fig. 3a. The approximate position of the membrane on the upper cell in the figure is outlined with a dashed red line. With poly-L-lysine coated windows, the cells were strongly attached and imaged at magnifications up to M = 28,000 x without detachment of the bacteria from the SiN window. Fig. 3c shows cells of M. magneticum attached to a SiN window without any functionalization. In this experiment, cells were grown in an iron deficient medium so some cells had only partially formed magnetosome chains, while others did not contain any magnetosomes at all (white arrows). There are two noteworthy features in Fig. 3c. Importantly, the cells with no magnetosome chains were still visible in the fluid cell, indicating that imaging of initially “non-magnetic” bacteria for future in vivo studies of magnetite biomineralization is possible. This image also demonstrates the importance of bacterial attachment to the SiN window; with no poly-L-lysine coating on the SiN in this case, some cells were mobile under the electron beam, indicated by streaking in the STEM image (red arrow). In this case, the SiN windows and cells had a negative surface charge, and attachment was much weaker than with the poly-L-lysine treated SiN. It is therefore important to functionalize the SiN window to create a positively charged surface that induces strong electrostatic attachment of the negatively charged bacterial cells. Poly-L-lysine, APTES-functionalized, and BioPlus chips all encouraged consistent attachment of cells, with all the coatings giving qualitatively similar attachment properties.
Electron beam exposure
The effect of the electron beam on bacterial cells in the fluid cell was investigated by exposing them to sequential STEM exposures. Fig. 4 shows two HAADF-STEM images of a single cell exposed to various electron doses indicated in each image. These images were acquired at a magnification of M = 28,000 x, beam current of 38 pA, and pixel dwell time of 8 μs, yielding a cumulative electron dose per image of 2.4 electrons · Å−2. The magnetosome chain in the first STEM image was approximately 1.15 μm in length after being exposed to a cumulative electron dose of 2.4 electrons · Å−2 (Fig. 4a); however, after two more consecutive STEM images and three times the cumulative dose of the first image, the chain length contracted by 9% to 1.05 μm (Fig. 4b). Further contraction of the chain was observed with continued electron beam irradiation. No direct electron beam damage to the magnetite magnetosomes was observed after acquiring three STEM images. Radiolysis damage of the bacterial cytoplasm at these high cumulative electron doses likely led to shrinkage of the entire cell, manifested as a contraction of the magnetosome chain. A single STEM image acquired below a cumulative electron dose of ~0.1 electron ·Å−2 did not induce any observable magnetosome chain shrinkage.
Figure 4. Electron beam damage of cells of M. magneticum by subsequent STEM acquisitions in the fluid cell.
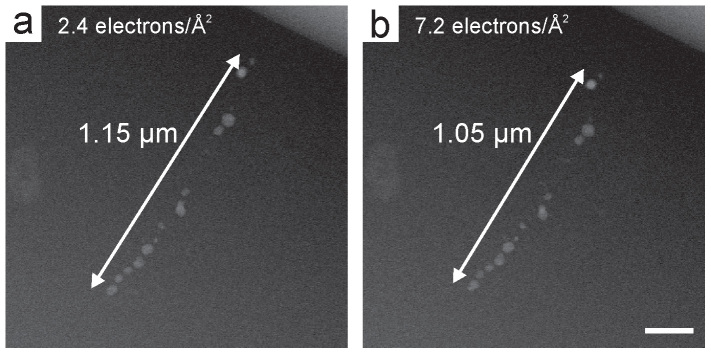
(a) The first and (b) third HAADF-STEM images (cropped) of a cell. The cumulative electron doses and approximate magnetosome chain lengths are indicated in the images. The scale bar in (b) is 200 nm.
Fluorescence Microscopy
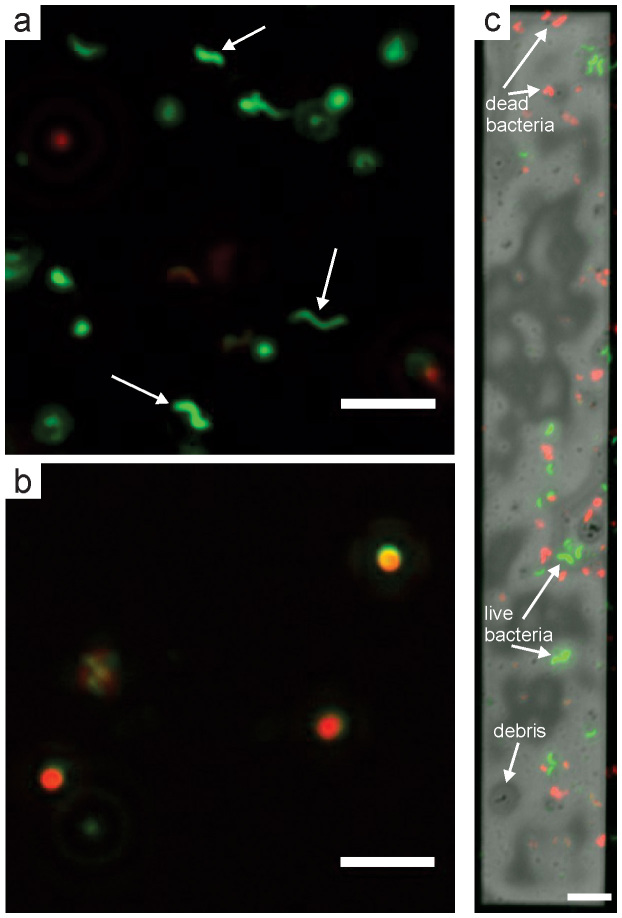
SYTO 9 and propidium iodide fluorescent dyes were used to test the membrane integrity of cells of M. magneticum (cf. Methods). To test the dye protocol for establishing membrane integrity in this specific bacterial strain, we prepared live and dead reference samples on cover slips (Fig. 5). Once bound to a nucleic acid, SYTO 9 fluoresces green while propidium iodide fluoresces red; however, propidium iodide is membrane impermeable. Cells with undamaged membranes will therefore only fluoresce green, while those with damaged membranes will fluoresce both red and green. The reference samples were prepared at the same concentration as a typical fluid cell sample, the dead reference sample was killed with 10% isopropyl alcohol prior to staining. Fig. 5a shows that the live cells of M. magneticum retain their helical shape and show only green fluorescence in the composite image (e.g. white arrows), indicating that they had undamaged cell membranes. On the other hand, killed cells fluoresced both green and red in the composite image, indicating that their membranes were damaged allowing binding of propidium iodide to nucleic acids in the bacteria cytoplasm (Fig. 5b). Many killed cells also had a spherical shape, either as a result of the bacteria rolling up on itself upon exposure to isopropyl alcohol, or due to the lysis of the outer membrane and loss of peptidoglycan. The live and dead reference samples indicate that this two-part staining protocol is a valid method for assessing membrane integrity of cells of M. magneticum in the fluid cell.
Figure 5. Composite fluorescence images of stained cells of M. magneticum prepared on a cover slip and in the fluid cell.
(a) Composite EGFP and rhodamine fluorescence images of cells of M. Magneticum concentrated approximately 20x from the original culture (same as fluid cell preparation). Several bacteria with intact membranes are denoted with white arrows. (b) Cells of M. magneticum killed with 10% isopropyl alcohol. (c) Composite EGFP, rhodamine, and brightfield optical image of bacteria in the fluid cell. The scale bar is 5 μm in (a) and (b) and 10 μm in (c).
Fluorescence imaging of cells of M. magneticum in the fluid cell was performed to identify the positions of viable cells prior to STEM imaging (Fig. 5c). In this experiment, cells were attached to an APTES-coated 100 nm SiN spacer, paired with an uncoated 100 nm SiN spacer. The brightfield contrast on the SiN window is due to thickness variations in the liquid layer due to the presence of debris and cells, both denoted in the image with arrows. Both intact and damaged bacteria can be observed in the fluid cell sample prior to STEM imaging. Approximately half of the cells had intact membranes ~40 minutes after closing the fluid cell. Membrane integrity was typically much higher on cover slip samples (cf. Fig. 5a), so we performed systematic fluorescence microscopy experiments to understand the effect of confinement in the fluid cell on bacterial membrane integrity.
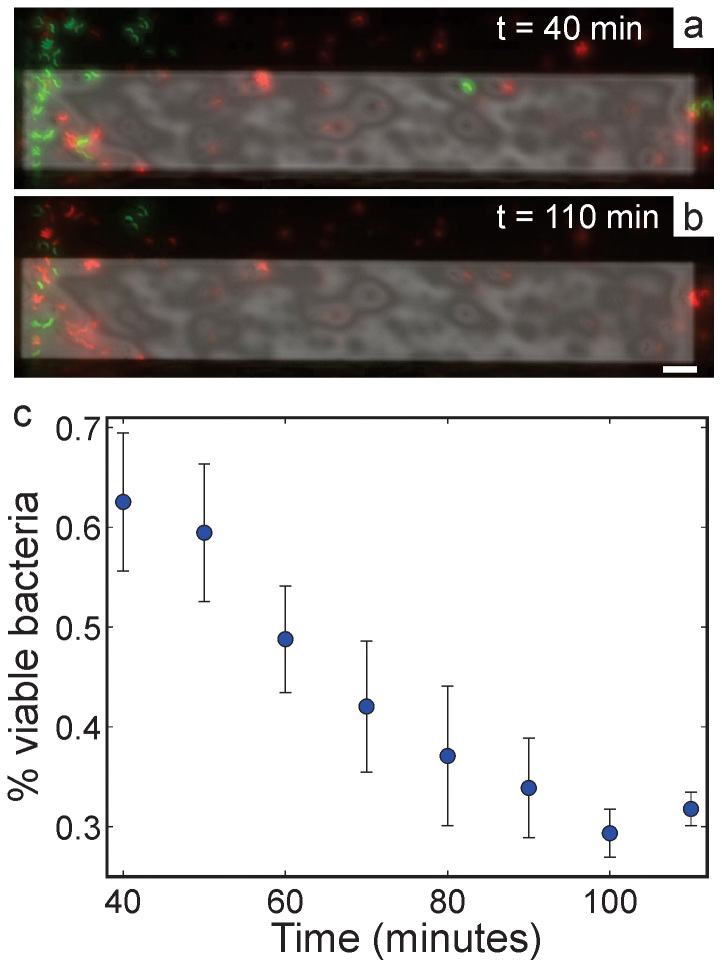
From Fig. 5c, it appears that simply loading the magnetotactic bacteria in the fluid cell drastically decreased their viability even before STEM imaging. We acquired fluorescence images of cells attached to a BioPlus chip every 10 minutes to systematically test bacteria viability as a function of time in the fluid cell (Fig. 6). Figs. 6a and 6b show example fluorescence images taken after 40 and 110 minutes in the fluid cell, respectively. There was a 40 minute lag time between fluid cell assembly and the first fluorescence image to allow for sample preparation and transportation to the fluorescence microscope. Fig. 6c shows a plot of the percent viable bacterial cells as a function of time. After 40 minutes in the fluid cell, approximately half of the bacteria were viable. Over the next 70 minutes, several cells lost their green fluorescence and began to exhibit red fluorescence, with the bacterial viability decreasing at a rate of ~0.5% min−1. After 110 minutes, the bacterial viability decreased to ~30%.
Figure 6. Composite fluorescence images of stained cells of M. magneticum (a) 40 minutes and (b) 110 minutes after sealing the fluid cell tip.
The scale bar in (b) is 10 μm. (c) Percentage of cells with intact membranes (green fluorescence only) as a function of time from fluid cell assembly. Percent viability is determined by dividing green fluorescent bacterial cells by the total number of bacteria cells in the fluid cell. Each data point is the mean of three trials, the error bars are two standard deviations of the mean.
Correlative fluorescence and fluid cell STEM imaging
We combined the STEM and fluorescence techniques described above into a correlative method for imaging viable magnetotactic bacteria in liquid. The correlative imaging method is outlined in general as follows. Fluorescence microscopy of labeled cells of M. magneticum was initially performed to locate viable bacterial cells in the fluid cell. Cells were then immediately imaged in STEM to observe their magnetosome structure. Following STEM imaging, fluorescence microscopy was performed again to confirm viability of the cells in the STEM images. A specific example of this correlative technique is shown in Fig. 7.
Figure 7. Correlative fluorescence and STEM imaging of viable cells of M. magneticum.
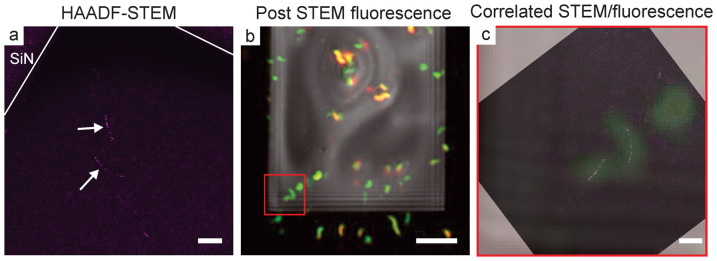
(a) False colored, background subtracted HAADF-STEM image of two bacterial cells near the corner of the SiN window, the magnetosome chains appear in purple and are denoted with white arrows. (b) Post-STEM composite fluorescence image of the same fluid cell sample. (c) Correlated STEM and composite fluorescence image of the bacterial cells highlighted in the red box in (b). The scale bar is 1 μm in (a) and (c) and 10 μm in (b).
Fig. 7 demonstrates the correlation of viable cells between fluorescence and STEM. In this experiment, cells were attached to a BioPlus blank SiN chip paired with a 100 nm spacer chip. Initial fluorescence imaging was performed ~30 min after assembling the fluid cell to locate viable cells for higher resolution STEM imaging (not shown). Fig. 7a shows a HAADF-STEM image of two cells near the corner of the SiN window, indicated by their magnetosome chains (M = 9,900 x, d = 0.17 electrons·Å−2). In this case the bacterial membranes cannot be seen because the fluid layer is too thick. In a typical correlative microscopy technique, fluorescence microscopy is only performed first to determine areas of interest and imaging concludes with electron microscopy30; however, the rapid decrease in bacterial viability in the fluid cell also necessitated fluorescence imaging after STEM. Approximately 20 min after the STEM image was acquired, the fluid cell was imaged in the fluorescence microscope to test bacterial viability (Fig. 7b). We located the same cells that are shown in Fig. 7a in the fluorescence microscope using the SiN window edges and cellular orientation as fiducials (red box). To correlate the STEM and composite fluorescence images, the STEM image was rotated and the composite fluorescence image was cropped, rendered partially transparent, and overlaid (Fig. 7c). While only the magnetite magnetosomes can be observed in the STEM image (Fig. 7a), correlation with the fluorescence image reveals the cytoplasm. Importantly, this image demonstrates that the bacterial cell membranes were not damaged after a single STEM exposure, at least for the cumulative electron dose used in this case.
Discussion
Mitigating radiation damage is critical for electron microscopy of biological samples, especially for fluid cell STEM imaging, which has been shown to have a number of electron beam induced artifacts41,49. Cumulative electron doses on the order of magnitude used here (~0.1 electrons·Å−2) coincide with a recent fluid cell STEM study of gold nanoparticle uptake into eukaryotic cells; these doses were shown to induce only small changes in the cellular structure with a single STEM exposure43. The authors proposed that the first STEM image obtained at the outer cellular region contained information of the ultrastructure of the live cell and subsequent images in other areas likely contained information about the live cell as well. However, for the case of bacteria imaged at the low magnifications in this study, a single STEM exposure irradiates the entire cell at once. Therefore, in this paper, only the first HAADF-STEM images acquired of cells of M. magneticum have been shown as they are the most representative of the initial cellular structure.
While we can use previous fluid cell STEM studies of radiation damage in eukaryotic cells as general guidelines for live imaging of magnetotactic bacteria, the question remains, are the bacterial cells alive following STEM imaging? The death of a bacterial cell is often defined as the inability of the cell to grow into a visible colony in media50. Another commonly used indicator of bacterial viability is cell mobility. In our experiments, it was necessary to attach the cells to the SiN windows to immobilize them for STEM imaging, so we could not evaluate bacterial viability on the basis of either of these criteria. This limits our indicators of radiation damage to cellular function to observations of bacterial shrinkage (Fig. 4) and membrane viability via fluorescence labeling and imaging (Fig. 6), neither of which can be used to directly assess enzymatic or reproductive damage of the cells. Intermediate fluorescence states identified in SYTO 9/propidium iodide labeled cells of Escherichia coli suggested that fluorescent labeling can indicate more than simply membrane integrity, but it was unclear exactly how these states related to specific cell functions and reproductive health of the bacteria50. The cumulative electron doses used in this study are at least four orders of magnitude higher than the lethal electron dose of ~10−5 electrons·Å−2 shown to cause reproductive death and enzyme deactivation in E. coli51,52,53. The lethal electron dose was measured from pulsed electron beam experiments (E = 520 keV) of E. coli on wet Millipore filters52; such a thick sample would absorb more ionizing radiation and facilitate more damage to the bacteria than for fluid cell STEM imaging, which allows for transmission of most of the electrons. Furthermore, calculation of the lethal electron dose assumed TEM irradiation53, which delivers a continuous flux of electrons over a micron sized area, whereas STEM imaging delivers electrons in a nanometer sized probe scanned serially across the sample. Due to these differences, we cannot conclude whether the same lethal electron dose exists for fluid cell STEM imaging of magnetotactic bacteria. Cellular processes in the cells of M. magneticum are likely altered or damaged, but further work on the viability of electron irradiated bacteria in the fluid cell will be needed to establish a lethal electron dose for STEM imaging.
While the electron doses used here have been shown to cause various types of damage in cells of E. coli, they are two or more orders of magnitude below the established damage threshold for cryo-TEM and radiation damage of nucleic acids (~10 electrons·Å−2)30,39,51, and one order of magnitude below the damage threshold for amino acids (~1 electron·Å−2)51. Imaging at electron doses higher than the radiation damage threshold for amino acids induced damage in the form of magnetosome chain shrinkage, likely due to radiation damage of the bacterial cytoplasm (Fig. 4). Importantly, this suggests that as long as the cumulative electron dose for a STEM image is <1 electron·Å−2, the structure of the magnetosome exists in an environment representative of the initial cellular state, even if some of the cellular processes have been altered or arrested due to irradiation. The fact that we image at cumulative doses below the nucleic and amino acid damage thresholds further corroborates our observations of intact bacterial cell membranes following fluid cell STEM imaging. We expect these radiation damage thresholds to be generally applicable to fluid cell electron microscopy imaging of most prokaryotes.
We must also consider changes to the bacterial cells due to fluid cell sample preparation. Potential causes of bacterial membrane damage in the fluid cell include confinement between the two SiN windows, the presence of dimethyl sulfoxide (DMSO) in the fluorescent dye, and non-ideal oxygen conditions in the growth media. Magnetotactic bacteria are cultured under microaerobic conditions (0.25 mbar is optimum for magnetosome biomineralization by M. Magneticum46), and the small amount of media contained between the SiN windows in the fluid cell quickly saturates to atmospheric O2 levels, creating a non-ideal, possibly toxic, environment for the bacteria54. However, this particular strain of M. magneticum is known to be relatively oxygen resistant, and the amount of DMSO in the media is likely below toxic levels, so confinement of the cells is likely the explanation for cell damage. Confinement of the bacterial cells in a hundreds of nanometers thick liquid layer creates compressive stresses on the bacterial cells. Compressive stresses have been shown to increase membrane permeability and damage efflux systems in E. coli, leading to increased absorption of propidium iodide55. High pressures have also been shown to lead to increased rates of cell lysis in Lactobacillus strains56. Our systematic fluorescence imaging showed increased propidium iodide absorption into the cells upon sample preparation and over the two hours in the fluid cell; however, cell lysis was not observed. While the exact cause of the rapid decrease in viability over two hours in the fluid cell is not entirely clear, it necessitates correlative fluorescence imaging after STEM imaging as well, to verify bacterial membrane viability in the electron microscopy images.
Taken together, our bacterial viability experiments and the cumulative electron doses used strongly suggest that some bacterial cells remain alive in the fluid cell up to the point of electron beam irradiation. After STEM imaging, cellular function is altered or arrested due to radiation induced enzyme deactivation and possible reproductive death of the cell. Given these conclusions, we expect in vivo imaging of magnetosome biomineralization in initially live magnetotactic bacteria will be possible using fluid cell STEM imaging. The flow capabilities of the fluid cell will allow for controllable introduction of iron-replete growth media to an iron-deficient bacterial culture. While high resolution microscopy of the magnetosome structure in live bacteria currently may not be possible due to radiation damage, our results demonstrate that STEM imaging at magnifications up to M = 28,000 x allows for visualization of magnetosome structures while retaining an intact bacterial cell membrane. The electron doses used in this study are below that for radiation damage of the bacterial ultrastructure, so imaging of the magnetosome nanostructures in a native cellular environment is possible. High resolution TEM (HRTEM) of intact magnetosome structures in the fluid cell is also possible (cf. Supp. Fig. S1); however, the high electron dose will quickly kill the cells and damage the ultrastructure. One possibility is to perform a fluid cell HRTEM or HRSTEM study where the various steps of magnetosome growth are imaged subsequently in several cells, so that each image of a growing magnetosome is taken from a bacterial cell that is alive up to the moment of imaging. Previous studies have shown that exposure of magnetotactic bacteria to different types of radiation can affect magnetosome shape and size through direct DNA damage57. We didn't observe any direct damage to the magnetosomes by the electron beam in this work and there are no studies on the effect of electrons on magnetosome growth, but possible changes in the magnetosome structure due to high energy electron irradiation should be considered. Another possible route for this technique is in situ chemical imaging of soluble iron uptake by magnetotactic bacteria. We have recently visualized iron binding by micelles of bacterial membrane protein by observing the increase in Z-contrast of the micelles via fluid cell HAADF-STEM imaging58. This study indicates this technique shows promise for visualizing uptake of iron into the bacteria. In situ spectral imaging via EELS59 or energy dispersive x-ray spectroscopy (EDS)60 mapping may provide additional means for visualizing iron uptake, which has only been possible to date in dried bacteria samples using techniques like scanning transmission x-ray microscopy61. Finally, further correlative fluorescence and fluid cell electron microscopy studies utilizing fluorescent proteins could allow investigation of the connections between biomolecular processes and biomineralization of magnetosome magnetite nanocrystals. For instance, fluorescently tagged membrane proteins can be tracked during bacterial cell growth via fluorescence microscopy20, followed by fluid cell STEM imaging of the magnetosome structures to aid in elucidating the role of protein localization in the biomineralization process.
In summary, cells of M. magneticum strain AMB-1 were fluorescently labeled with SYTO 9 and propidium iodide nucleic acid stains and imaged in media via fluid cell STEM and fluorescence microscopy. The magnetite magnetosomes provided high contrast labels to image bacterial cells using HAADF-STEM; in some cases the bacterial cell membrane was resolved when the liquid layer was sufficiently thin. We established the major cellular damage sources to be electron beam irradiation and compression of the bacteria between the SiN windows. The STEM electron dose could be kept sufficiently low to prevent detectable ultrastructure damage to bacteria. However, at high electron doses (>1.0 electron·Å−2 and after multiple STEM exposures), radiation damage in the bacterial cytoplasm was manifested as a contraction of the magnetosome chain length. Correlative fluorescence and STEM imaging indicated that cells of M. magneticum had intact membranes after STEM irradiation in the fluid cell, but could not establish other cell viability criteria such as reproductive ability or enzymatic function. We found that ~50% of the cells typically had intact membranes upon assembling the fluid cell, and approximately half of these bacteria sustained membrane damage due to compressive stresses over the next two hours in the fluid cell. This correlative fluid cell STEM and fluorescence microscopy technique is a first step in directly observing biomineralization of magnetite in viable magnetotactic bacteria. We expect this technique to be generally applicable for in vivo imaging of a wide range of biomineralizing organisms.
Methods
Preparation of cells of M. magneticum
Cells of M. magneticum were grown in a 5 ml flask in standard media at room temperature under microaerobic conditions (approximately 1% O2 in the headspace)46. The original bacterial culture was obtained from the American Type Culture Collection (ATCC); the reference number for Magnetospirillium magneticum is ATCC 700264. A 1 mL aliquot of the culture was taken from a live active culture and concentrated approximately 20x by centrifugation (2 × 2.5 minutes at 7.2 g) and incubated with 5 µM SYTO 9 green fluorescent nucleic acid stain and 55 µM propidium iodide red fluorescent nuclear counterstain (LIVE/DEAD BacLight Viability Kit L-7007, Life Technologies) for 15 min.
Fluid cell assembly and SiN chip functionalization
A continuous flow fluid cell holder platform (Hummingbird Scientific, Lacey, WA, USA) was utilized to image cells of M. magneticum in liquid (cf. schematic in Fig. 1a). Briefly, a thin liquid layer (typically 100–500 nm thick) was formed by sandwiching two SiN coated silicon chips with a 50 × 200 µm opening etched from the center. The 50 nm thick SiN is electron transparent and spans the etch pit in the chip center, creating an imaging window to observe the bacterial cells in the thin liquid layer. The liquid layer and silicon chips are then hermetically sealed to prevent evaporation of the liquid (more details on liquid cell holders for electron microscopy can be found in a previous review paper41).
In a typical experiment, the bacterial cells were attached to one of two types of fluid cell chips: SiN chips with 100 nm thick SU-8 spacers (Hummingbird Scientific, Lacey, WA, USA) or BioPlus biofunctionalized blank SiN chips with no spacers (Dune Sciences and Hummingbird Scientific, Lacey, WA, USA). The BioPlus chips are functionalized with a surface coating that renders them positively charged, encouraging attachment of bacterial cells. In some cases, SiN spacer chips were coated with poly-L-lysine or (3-Aminopropyl)triethoxysilane (APTES) to render them hydrophilic, positively charged, and encourage attachment of the cells62. To coat a SiN window with poly-L-lysine, the chip was floated SiN side down on a film of 0.1% w/v aqueous poly-l-lysine (Sigma-Aldrich) for 5 min, then floated on DI water (Milli-Q grade, 18.2 MΩ) for 1 min, and finally dried with air. To coat a SiN window with APTES (98%, Sigma-Aldrich), the window was cleaned by rinsing in toluene and anhydrous ethanol and then dried under nitrogen. The window was then cleaned in UV/O3 plasma (Bioforce Nanosciences, Ames, IA, USA) for 45 min, followed by APTES functionalization using established protocols63.
To deposit the cells on a SiN spacer chip, the spacer and a blank chip were first treated in UV/O3 plasma for 30 min to clean and render them hydrophilic. A 10 µL drop of the stained concentrated bacteria culture was placed on a Parafilm coated glass slide, and the cleaned spacer chip was placed SiN side down on the drop for 5 min to allow cells to attach. Excess liquid was then removed from the chip using filter paper and the chip was sandwiched on top of a blank chip in the fluid cell holder. This inverted deposition technique encourages preferential attachment of live motile cells, as dead cells likely settle down to the Parafilm by gravity and do not attach. The BioPlus, APTES, and poly-L-lysine chips were used without UV/O3 plasma cleaning to preserve the surface functionality, and cells were deposited by drop casting 0.75 µL of the concentrated bacteria culture on the SiN chip. The cells were allowed to attach for 5 min, the chip was sandwiched with a cleaned spacer chip in the fluid cell holder, and excess liquid was removed with filter paper prior to sealing the tip. For both sample preparations, the cells were attached to the top SiN window to avoid resolution degradation due to STEM beam broadening in the liquid layer (Fig. 1a)42. After the fluid cell was assembled, 100 μL of fresh growth media was allowed to flow through at a rate of 10 μL/min to remove excess fluorescent dye.
Cells of M. magneticum were also imaged air-dried on carbon grids. To prepare a grid sample, the cells were fixed with 2.5% w/v glutaraldehyde (25% w/v in water, Acros Organics), centrifuged, and resuspended in DI water. Cells were then drop cast onto a Quantifoil® carbon grid and allowed to air dry. Images of the dried cells were acquired with HAADF-STEM and energy filtered TEM (EFTEM).
Scanning Transmission Electron Microscopy
Imaging and characterization of the bacteria were performed in a 200 kV FEI Tecnai G2 F20 (S)TEM equipped with a Tridium Gatan image filter. Bacteria in the fluid cell were imaged in STEM mode with a HAADF detector. To minimize electron beam damage to the cells during fluid cell imaging, a beam current of 38 pA and 70 µm condenser lens aperture were used. The approximate electron beam current was measured using the phosphorescent mini-screen. While searching for bacterial cells and focusing images in the fluid cell, a low magnification (M < 14,000 x), short pixel dwell time (2 µs), and small field of view (512 × 512 pixels) were maintained to avoid excessive beam damage, yielding an approximate electron dose rate of d < 0.05 electrons·Å−2·s−1 (cf. Woehl et al.64 for electron dose rate calculation method). HAADF-STEM images were acquired at magnifications ranging from M = 7,000–28,000 x, a pixel dwell time of 8 μs, and image size of 1024 × 1024 pixels, yielding cumulative electron doses of d = 0.1–2.4 electrons·Å−2 for a single STEM scan. Elemental maps of the oxygen K-edge and iron L2,3-edge were acquired in EFTEM mode with a slit width of 30 eV ± 1 eV; EFTEM imaging was performed ex situ on bacterial samples prepared on Quantifoil® carbon grids.
Fluorescence microscopy
The fluid cell was imaged with an upright optical microscope (Zeiss, Axio Imager 2) before and after STEM imaging. To image bacterial cells directly in the fluid cell, the tip of the fluid cell holder was removed and fixed to a glass slide with molding clay (Fig. 1b). Green (EGFP filter, excitation: 470/40 band pass, emission: 525/50 band pass) and red fluorescence (Rhodamine 20 filter, excitation: 546/12 band pass, emission: 607/80 band pass), and transmission brightfield images of the cells in the fluid cell were acquired with a 40x objective lens and monochrome camera. Viable bacterial cells with intact cell membranes only fluoresced green, while those with damaged membranes fluoresced red and green. Typically 30 minutes elapsed between adding the fluorescent dye to the bacteria and initial fluorescence microscopy. ImageJ (NIH) was used to enhance contrast and brightness, false color, and overlay the fluorescence images.
Author Contributions
T.J.W. and T.P. performed fluid cell STEM experiments; T.J.W. carried out fluorescence and correlative microscopy and data analysis. D.T. and T.P.G. cultured bacteria and assisted in establishing specimen loading protocol. S.K. performed preliminary bacterial fluid cell STEM imaging; S.K. and E.F. carried out EFTEM imaging ex-situ. D.F. and D.A.B. helped optimizing the bacterial loading conditions and accessing the cell viability. T.J.W. and T.P. prepared the manuscript.
Supplementary Material
Supplementary Material Submission
Acknowledgments
This work was supported by the U.S. Department of Energy, Office of Basic Energy Science, Division of Materials Sciences and Engineering. The research was performed at the Ames Laboratory, which is operated for the U.S. Department of Energy by Iowa State University under Contract No. DE-AC02-07CH11358. T.P. acknowledges support from the Department of Energy Office of Science Early Career Research Award, Biomolecular Materials Program. The authors would like to thank C. L. Mosher, M. J. Kramer, and T. M. Pepper for informative discussions. This research was supported in D.F's laboratory by the Max Planck Society, and a starting grant from the ERC (Project MB2, no. 256915).
References
- Lowenstan H. A. & Weiner S. (Oxford University Press, New York, 1989).
- Bazylinski D. A. Synthesis of the bacterial magnetosome: the making of a magnetic personality. Int. Microbiol 2, 71–80 (1999). [PubMed] [Google Scholar]
- Bazylinski D. A. & Frankel R. B. Magnetosome formation in prokaryotes. Nat. Rev. Micro. 2, 217–230 (2004). [DOI] [PubMed] [Google Scholar]
- Baumgartner J. et al. Magnetotactic bacteria form magnetite from a phosphate-rich ferric hydroxide via nanometric ferric (oxyhydr)oxide intermediates. Proc. Natl. Acad. Sci. U. S. A. 110, 14883–14888 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumper M. & Brunner E. Learning from diatoms: nature's tools for the production of nanostructured silica. Adv. Funct. Mater. 16, 17–26 (2006). [Google Scholar]
- Sumper M. & Brunner E. Silica biomineralization in diatoms: The model organism Thalassiosira pseudonana. Chem. Bio. Chem. 9, 1187–1194 (2008). [DOI] [PubMed] [Google Scholar]
- Bazylinski D. A. & Schübbe S. in Advances in Applied Microbiology Vol. Volume 62 (eds Sima Sariaslani Allen I. Laskin & M. Gadd Geoffrey) 21–62 (Academic Press., 2007). [DOI] [PubMed]
- Frankel R. B. Biological permanent magnets. Hyperfine Interactions 151/152, 145–153 (2004). [Google Scholar]
- Hergt R. et al. Magnetic properties of bacterial magnetosomes as potential diagnostic and therapeutic tools. J. Magn. Magn. Mater. 293, 80–86 (2005). [Google Scholar]
- Pankhurst Q. A., Thanh N. K. T., Jones S. K. & Dobson J. Progress in applications of magnetic nanoparticles in biomedicine. J. Phys. D Appl. Phys. 42 (2009). [Google Scholar]
- Lang C. & Schüler D. Biogenic nanoparticles: production, characterization, and application of bacterial magnetosomes. J. Phys. Cond. Mat. 18, S2815–S2828 (2006). [Google Scholar]
- Lang C. & Schüler D. Biomineralization of magnetosomes in bacteria: nanoparticles with potential applications. Microb. Bionanotechnol. 107–124 (2006). [Google Scholar]
- Prozorov T., Bazylinski D. A., Mallapragada S. K. & Prozorov R. Novel Magnetic Nanomaterials Inspired by Magnetotactic Bacteria: Topical Review. Mat. Sci. Eng. R. 74, 133–172 (2013). [Google Scholar]
- Prozorov T. et al. Protein-mediated synthesis of uniform superparamagnetic magnetite nanocrystals. Adv. Funct. Mater. 17, 951–957 (2007). [Google Scholar]
- Prozorov T. et al. Cobalt Ferrite Nanocrystals: Out-Performing Magnetotactic Bacteria. ACS Nano 1, 228–233 (2007). [DOI] [PubMed] [Google Scholar]
- Wolff A. et al. Influence of the synthetic polypeptide c25-mms6 on cobalt ferrite nanoparticle formation. J. Nanopart. Res. 14, 1161/1161–1161/1111 (2012). [Google Scholar]
- Thomas-Keprta K. L. et al. Elongated prismatic magnetite crystals in ALH84001 carbonate globules: Potential Martian magnetofossils. Geochim. Cosmochim. Acta 64, 4049–4081 (2000). [DOI] [PubMed] [Google Scholar]
- Frankel R. B. & Bazylinski D. A. Magnetosomes: Nanoscale magnetic iron minerals in bacteria. Nanobiotechnol. 136–145 (2004). [Google Scholar]
- Faivre D. & Schüler D. Magnetotactic Bacteria and Magnetosomes. Chem. Rev. 108, 4875–4898 (2008). [DOI] [PubMed] [Google Scholar]
- Komeili A., Vali H., Beveridge T. J. & Newman D. K. Magnetosome vesicles are present before magnetite formation, and MamA is required for their activation. Proc. Natl. Acad. Sci. U. S. A. 101, 3839–3844 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arakaki A., Webb J. & Matsunaga T. A novel protein tightly bound to bacterial magnetic particles in Magnetospirillum magneticum strain AMB-1. J. Biol. Chem. 278, 8745–8750 (2003). [DOI] [PubMed] [Google Scholar]
- Dey A. & Sommerdijk N. A. J. M. Advanced transmission electron microscopy to explore early stages of bio(mimetic) mineralization. R. S. C. Smart Mater. 4, 74–106 (2013). [Google Scholar]
- Friedrich H., Frederik P. M., de With G. & Sommerdijk N. A. J. M. Imaging of Self-Assembled Structures: Interpretation of TEM and Cryo-TEM Images. Angew. Chem. Int. Ed. 49, 7850–7858 (2010). [DOI] [PubMed] [Google Scholar]
- Baumgartner J. & Faivre D. Magnetite biomineralization in bacteria. Prog. Molec. Subcel. Biol. 52, 3–27(2011). [DOI] [PubMed] [Google Scholar]
- Koernig A. et al. Magnetite Crystal Orientation in Magnetosome Chains. Adv. Funct. Mater. 24, 3926–3932 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komeili A., Li Z., Newman D. K. & Jensen G. J. Magnetosomes are cell membrane invaginations organized by the actin-like protein MamK. Science 311, 242–245 (2006). [DOI] [PubMed] [Google Scholar]
- Scheffel A. et al. An acidic protein aligns magnetosomes along a filamentous structure in magnetotactic bacteria. Nature 441, 248 (2006). [DOI] [PubMed] [Google Scholar]
- Katzmann E., Scheffel A., Gruska M., Plitzko J. M. & Schüler D. Loss of the actin-like protein MamK has pleiotropic effects on magnetosome formation and chain assembly in Magnetospirillum gryphiswaldense. Molec. Microbiol. 77, 208–224 (2010). [DOI] [PubMed] [Google Scholar]
- Wolff A. et al. Oriented attachment explains cobalt ferrite nanoparticle growth in bioinspired syntheses. Beilstein J. Nanotech. 5, 210–218 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartori A. et al. Correlative microscopy: Bridging the gap between fluorescence light microscopy and cryo-electron tomography. J. Struct. Biol. 160 (2007). [DOI] [PubMed] [Google Scholar]
- Evans J. E. et al. Visualizing Macromolecular Complexes with In Situ Liquid Transmission Electron Microscopy. Micron 43, 1085–1090 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashyap S. et al. Visualization of Iron-Binding Micelles in Acidic Recombinant Biomineralization Protein, MamC. J. Nanomater. 7 (2014). [Google Scholar]
- Mirsaidov U. M., Zheng H., Casana Y. & Matsudaira P. Imaging Protein Structure in Water at 2.7 nm Resolution by Transmission Electron Microscopy. Biophys. J. 102 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C. M., Qiao Q., Shokuhfar T. & Klie R. F. High-Resolution Electron Microscopy and Spectroscopy of Ferritin in Biocompatible Graphene Liquid Cells and Graphene Sandwiches. Adv. Mater. 26, 3410–3414 (2014). [DOI] [PubMed] [Google Scholar]
- Gilmore B. L. et al. Visualizing viral assemblies in a nanoscale biosphere. Lab on a Chip 13, 216–219 (2013). [DOI] [PubMed] [Google Scholar]
- de Jonge N., Peckys D. B., Kremers G. J. & Piston D. W. Electron microscopy of whole cells in liquid with nanometer resolution. Proc. Nat. Ac. Sci. USA 106, 2159–2164 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peckys D. B., Veith G. M., Joy D. C. & de Jonge N. Nanoscale Imaging of Whole Cells Using a Liquid Enclosure and a Scanning Transmission Electron Microscope. PLoS One 4 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dukes M. J., Peckys D. B. & de Jonge N. Correlative Fluorescence Microscopy and Scanning Transmission Electron Microscopy of Quantum-Dot-Labeled Proteins in Whole Cells in Liquid. ACS Nano 4, 4110–4116 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peckys D. B., Mazur P., Gould K. L. & de Jonge N. Fully Hydrated Yeast Cells Imaged with Electron Microscopy. Biophys. J. 100, 2522–2529 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang T. W. et al. Self-aligned wet-cell for hydrated microbiology observation in TEM. Lab on a Chip 12, 340–347 (2012). [DOI] [PubMed] [Google Scholar]
- de Jonge N. & Ross F. M. Electron microscopy of specimens in liquid. Nature Nanotech. 6, 695–704 (2011). [DOI] [PubMed] [Google Scholar]
- de Jonge N., Poirier-Demers N., Demers H., Peckys D. B. & Drouin D. Nanometer-resolution electron microscopy through micrometers-thick water layers. Ultramicroscopy 110, 1114–1119 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peckys D. B. & de Jonge N. Liquid Scanning Transmission Electron Microscopy: Imaging Protein Complexes in their Native Environment in Whole Eukaryotic Cells. Microsc. Microanal. 20, 346–365 (2014). [DOI] [PubMed] [Google Scholar]
- Peckys D. B. & de Jonge N. Gold Nanoparticle Uptake in Whole Cells in Liquid Examined by Environmental Scanning Electron Microscopy. Microsc. Microanal. 20, 189–197 (2014). [DOI] [PubMed] [Google Scholar]
- Maruyama Y., Ebihara T., Nishiyama H., Suga M. & Sato C. Immuno EM-OM correlative microscopy in solution by atmospheric scanning electron microscopy (ASEM). J. Struct. Biol. 180, 259–270, 10.1016/jsb.2012.08.006 (2012). [DOI] [PubMed] [Google Scholar]
- Heyen U. & Schüler D. Growth and magnetosome formation by microaerophilic Magnetospirillum strains in an oxygen-controlled fermentor. Appl. Microbiol. Biot. 61, 536–544 (2003). [DOI] [PubMed] [Google Scholar]
- Matsunaga T., Sakaguchi T. & Tadokoro F. Magnetite formation by a magnetic bacterium capable of growing aerobically. Appl. Microbiol. Biot. 35, 651–655 (1991). [Google Scholar]
- Jungjohann K. L., Evans J. E., Aguiar J. A., Arslan I. & Browning N. D. Atomic-Scale Imaging and Spectroscopy for In Situ Liquid Scanning Transmission Electron Microscopy. Microsc. Microanal. 18, 621–627 (2012). [DOI] [PubMed] [Google Scholar]
- Woehl T. J. et al. Experimental procedures to mitigate electron beam induced artifacts during in situ fluid imaging of nanomaterials. Ultramicroscopy 127, 53–63 (2013). [DOI] [PubMed] [Google Scholar]
- Berney M., Hammes F., Bosshard F., Weilenmann H.-U. & Egli T. Assessment and interpretation of bacterial viability by using the LIVE/DEAD BacLight kit in combination with flow cytometry. Appl. Env. Microb. 73, 3283–3290 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimer L. & Kohl H. Transmission Electron Microscopy: Physics of Image Formation. 5th edn, 466–470 (Springer, 2008, New York, NY). [Google Scholar]
- Epp E. R., Weiss H. & Santomasso A. The Oxygen Effect in Bacterial Cells Irradiated with High-Intensity Pulsed Electrons. Radiat. Res. 34, 320–325 (1968). [PubMed] [Google Scholar]
- Stenn K. & Bahr G. F. Specimen damage caused by the beam of the transmission electron microscope, a correlative reconsideration. J. Ultrastruct. Res. 31, 526–550 (1970). [DOI] [PubMed] [Google Scholar]
- Bazylinski D. A. & Williams T. J. in Magnetoreception and Magnetosomes in Bacteria (ed Schuler, D.) 37–75 (Springer, 2007, Berlin). [Google Scholar]
- Benito A., Ventoura G., Casadei M., Robinson T. & Mackey B. Variation in resistance of natural isolates of Escherichia coli O157 to high hydrostatic pressure, mild heat, and other stresses. Appl. Env. Microb. 65, 1564–1569 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone A. S., Shellhammer T. H. & Courtney P. D. Effects of high pressure on the viability, morphology, lysis, and cell wall hydrolase activity of Lactococcus lactis subsp cremoris. Appl. Env. Microb. 68, 4357–4363 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Lin W., Li J. & Pan Y. Changes of cell growth and magnetosome biomineralization in Magnetospirillum magneticum AMB-1 after ultraviolet-B irradiation. Frontiers Microb. 4 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashyap S., Woehl T. J., Liu X., Mallapragada S. K. & Prozorov T. Nucleation of Iron Oxide Nanoparticles Mediated by Mms6 Protein In Situ. ACS Nano 8, 9097–9106 (2014). [DOI] [PubMed] [Google Scholar]
- Holtz M. E. et al. Nanoscale Imaging of Lithium Ion Distribution During In Situ Operation of Battery Electrode and Electrolyte. Nano Let. 14, 1453–1459 (2014). [DOI] [PubMed] [Google Scholar]
- Zaluzec N. J., Burke M. G., Haigh S. J. & Kulzick M. A. X-ray Energy-Dispersive Spectrometry During In Situ Liquid Cell Studies Using an Analytical Electron Microscope. Microsc. Microanal. 20, 323–329 (2014). [DOI] [PubMed] [Google Scholar]
- Kalirai S. S., Lam K. P., Bazylinski D. A., Lins U. & Hitchcock A. P. Examining the chemistry and magnetism of magnetotactic bacterium Candidatus Magnetovibrio blakemorei strain MV-1 using scanning transmission X-ray microscopy. Chem. Geol. 300 (2012). [Google Scholar]
- Peckys D. B. & de Jonge N. Visualizing Gold Nanoparticle Uptake in Live Cells with Liquid Scanning Transmission Electron Microscopy. Nano Lett. 11, 1733–1738 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang A. S., Liang F. Y., Steinbach F. & Caro J. Preparation and separation properties of LTA membranes by using 3-aminopropyltriethoxysilane as covalent linker. J. Membr. Sci. 350, 5–9 (2010). [Google Scholar]
- Woehl T. J., Evans J. E., Arslan L., Ristenpart W. D. & Browning N. D. Direct in Situ Determination of the Mechanisms Controlling Nanoparticle Nucleation and Growth. ACS Nano 6, 8599–8610 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material Submission