SUMMARY
In Arabidopsis, AUXIN RESPONSE FACTOR 3 (ARF3) belongs to the auxin response factor (ARF) family that regulates the expression of auxin-responsive genes. ARF3 is known to function in leaf polarity specification and gynoecium patterning. In this study, we discovered a previously unknown role of ARF3 in floral meristem (FM) determinacy through the isolation and characterization of a mutant of ARF3 that enhanced the FM determinacy defects of agamous (ag)-10, a weak ag allele. Central players in FM determinacy include WUSCHEL (WUS), a gene critical for FM maintenance, and AG and APETALA2 (AP2), which regulate FM determinacy by repression and promotion of WUS expression, respectively. We showed that ARF3 confers FM determinacy through repression of WUS expression, and associates with the WUS locus in part in an AG-dependent manner. We demonstrated that ARF3 is a direct target of AP2 and partially mediates AP2’s function in FM determinacy. ARF3 exhibits dynamic and complex expression patterns in floral organ primordia; altering the patterns spatially compromised FM determinacy. This study uncovered a role for ARF3 in FM determinacy and revealed relationships among genes in the genetic network governing FM determinacy.
Keywords: AUXIN RESPONSE FACTOR 3, floral determinacy, AGAMOUS, APETALA2, WUSCHEL, Arabidopsis
INTRODUCTION
Auxin is a key phytohormone that controls many developmental processes in plants, including embryogenesis, organogenesis, vascular differentiation, tropic growth, root and shoot architecture and senescence (Reinhardt et al., 2000; Vanneste and Friml 2009). Auxin signaling is mediated by two protein families: auxin response factors (ARFs) and Aux/IAA proteins. The 23 ARFs in Arabidopsis function as transcription factors and specifically bind TGTCTC auxin response elements (AuxREs) in the promoters of primary or early auxin response genes (Ulmasov et al., 1995; Guilfoyle et al., 1998; Ulmasov et al., 1999b; Ulmasov et al., 1999a; Guilfoyle and Hagen 2007; Boer et al., 2014). All Arabidopsis ARFs contain a conserved amino-terminal DNA-binding domain (DBD) responsible for AuxRE binding, and all except ARF23 have a middle region that functions as an activation or repression domain. All of the ARFs except ARF3, ARF13 and ARF17 contain a carboxy-terminal dimerization domain (CTD) that interacts with motifs III and IV of Aux/IAA proteins (Liscum and Reed 2002; Tiwari et al., 2003; Guilfoyle and Hagen 2007), which are short-lived nuclear proteins encoded by primary or early auxin response genes. This interaction between motifs III and IV of Aux/IAA proteins and the CTD of ARFs leads to the inhibition of ARF activity (Reed 2001; Tiwari et al., 2003).
Although its encoded protein lacks a CTD, ARF3, which is also known as ETTIN (ETT), nevertheless functions in some auxin-regulated pathways, such as gynoecium morphogenesis, self-incompatibility and de novo organ regeneration (Nemhauser et al., 2000; Tiwari et al., 2003; Tantikanjana and Nasrallah 2012; Cheng et al., 2013). The roles of ARF3 in leaf polarity and floral meristem (FM) and reproductive organ patterning have also been well characterized. ARF3 is expressed in vegetative and developing reproductive tissues (Sessions et al., 1997; Pekker et al., 2005) and can be detected in groups of cells that give rise to new FMs in the inflorescence meristem (IM). In stage 1 and 2 FMs (stages according to (Smyth et al., 1990)), ARF3 is expressed throughout the FM. In stages 3 and 4 FMs, ARF3 RNA is concentrated on the abaxial side of incipient stamen primordia and present in the gynoecium primordium, but is not found in the sepals. In stages 5 to 7 flowers, ARF3 exhibits an abaxial expression pattern in the petal, stamen and gynoecium primordia (Sessions et al., 1997). Consistent with the complex expression pattern of ARF3 and its functions in FM formation and floral organ initiation, arf3/ett mutants display floral organogenesis defects, such as increased sepal and petal number, decreased stamen number and reduced anther formation (Sessions et al., 1997; Sessions 1997).
The establishment of abaxial-adaxial polarity during leaf development involves two small RNA-target pairs (Pulido and Laufs 2010). Although ARF3 is transcribed throughout the leaf, its transcripts and those of the ARF3 homolog ARF4 are restricted to the abaxial side of the leaf by the action of TAS3-derived trans-acting small interfering RNAs (ta-siRNAs) known as tasiR-ARFs. tasiR-ARFs are generated at the adaxial side of the leaf and from a gradient towards the abaxial side, and this gradient effectively restricts ARF3 and ARF4 to the leaf abaxial domain (Williams et al., 2005; Adenot et al., 2006; Garcia et al., 2006; Chitwood et al., 2009). In a similar manner, transcripts from the class III HOMEODOMAIN LEUCINEZIPPER (HD-ZIPIII) genes PHABULOSA (PHB), PHAVULOTA (PHV) and REVOLUTA (REV) are restricted to the leaf adaxial domain by the microRNAs miR165/miR166 (McConnell et al., 2001; Kidner and Martienssen 2004).
The SAM continously produces above-ground plant parts throughout the life of a plant (Kaufmann et al., 2010). In contrast, the FM, which develops from the inflorescence meristem (IM), produces a defined number of floral organs and exhibits determinate growth. The termination of floral organ production is known as FM determinacy and serves as an ideal model for the study of meristem maintenance and termination (Liu et al., 2011). FM determinacy is controlled by the coordinated activities of multiple genes in a molecular framework (Sablowski 2007). WUSCHEL (WUS), which encodes a homeodomain-containing protein critical for SAM establishment and SAM, IM and FM maintenance (Laux et al., 1996; Schoof et al., 2000; Gallois et al., 2004), represents a critical node in this regulatory framework. WUS is expressed in the organizing center (OC), a cluster of cells responsible for maintaining the identity of the stem cells above the OC (Laux et al., 1996; Mayer et al., 1998). During floral development, WUS expression is turned off at stage 6 once the primordium of the gynoecium is produced, resulting in the termination of the floral stem cells (Lenhard et al., 2001). AGAMOUS (AG), a MADS-domain transcription factor that specifies stamen and carpel identity, is required for the temporally regulated repression of WUS expression. In the ag-1 null mutant, prolonged WUS expression beyond stage 6 results in a flowers-in-flower phenotype (Bowman et al., 1989; Lenhard et al., 2001). The delay between the onset of AG expression at stage 3 and the termination of WUS expression by AG at stage 6 suggests that AG regulates WUS indirectly (Lenhard et al., 2001). However, more recent studies have revealed that AG influences WUS expression both directly and indirectly: WUS is directly repressed by AG through Polycomb Group (PcG) recruitment at the WUS locus and indirectly repressed by AG through the AG target gene KNUCKLES (KNU) (Sun et al., 2009; Liu et al., 2011; Sun et al., 2014). APETALA2 (AP2), an AP2-domain containing transcription factor, promotes SAM and FM maintenance by promoting WUS expression and is itself repressed by miR172 (Aukerman and Sakai 2003; Chen 2004; Wurschum et al., 2006; Zhao et al., 2007). The finding that the expression of miR172-resistant AP2 (AP2m3) could dramatically enhance the ag-1 phenotype (Zhao et al., 2007) indicates that AP2 acts independently of AG in terms of FM determinacy. Others studies have shown that AP2 functions as both a transcriptional activator and repressor by directly binding a TTTGTT/AACAAA motif (Yant et al., 2010; Dinh et al., 2012). However, precisely how AP2 promotes FM maintenance and how multiple genes are coordinated to regulate FM determinacy have yet to be determined.
From an ethylmethane sulfonate (EMS) mutagenesis screen of ag-10, a weak ag allele, we isolated and functionally characterized several genes involved in FM determinacy (Ji et al., 2011; Liu et al., 2011; Yumul et al., 2013; Dinh et al., 2014). The present study describes the identification of a previously unknown role of ARF3 in FM determinacy. An arf3 mutant that enhanced the FM determinacy defects of ag-10 was isolated, and molecular genetic studies revealed that ARF3 confers FM determinacy by repressing WUS expression. A chromatin immunoprecipitation assay revealed that ARF3 is associated with the WUS locus in vivo and this association is promoted by AG. ARF3 was also identified as an AP2 target gene and was found to partially mediate the function of AP2 in FM determinacy. Moreover, the complex patterns of ARF3 expression and protein distribution in floral organ primordia were found to be important for its FM determinacy function. These studies not only identify a new player in floral determinacy but also reveal the complex molecular interactions that underlie floral determinacy.
RESULTS
ARF3 is required for floral meristem determinacy
An EMS mutagenesis screen was performed in the ag-10 background to identify FM determinacy regulators (Ji et al., 2011; Liu et al., 2011). While 94% of the siliques in ag-10 plants are morphologically similar to wild-type siliques, 6% are bulged with additional tissue growing inside, indicating a slight FM determinacy defect in ag-10 (Figure 1a,b). Given that the occasional presence of additional organs within ag-10 siliques may be attributable to prolonged stem cell activity, we focused on the isolation of mutants in which all of the siliques were bulged, a phenotype that is reminiscent of, although weaker than, the flowers-in-flower phenotype of the ag-1 mutant.
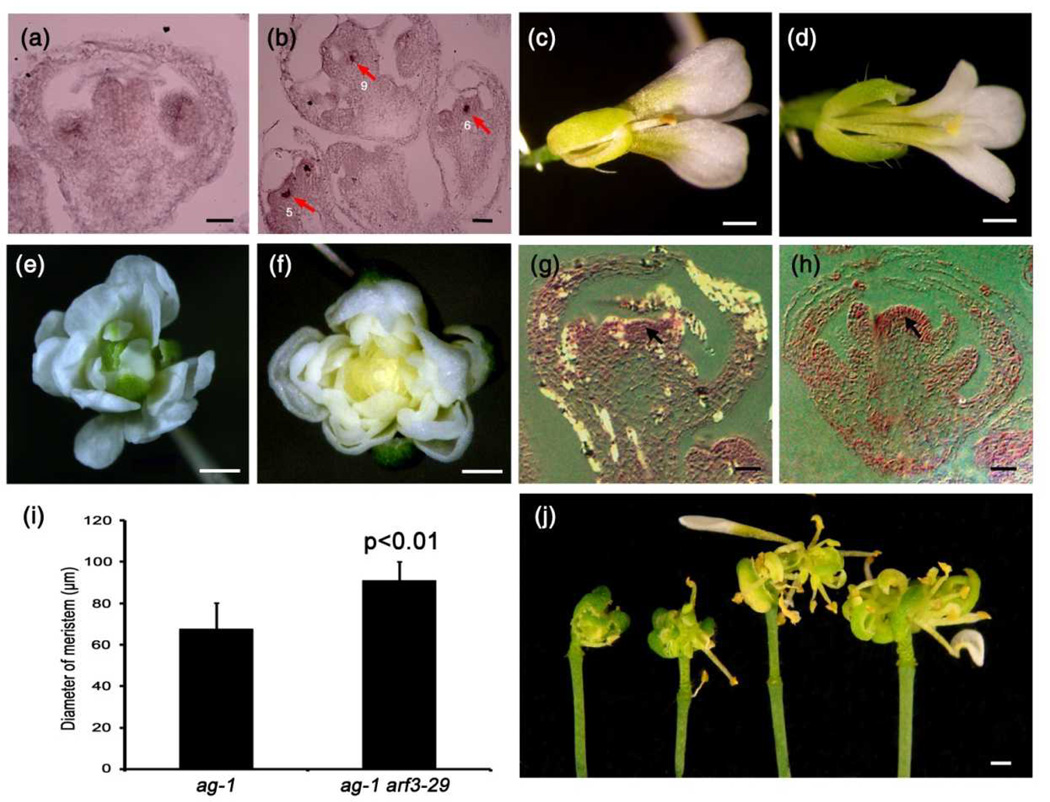
Figure 1. ARF3 is required for FM determinacy.
(a) Wild-type (Ler) siliques.
(b) ag-10 siliques. The one on the right is an example of a bulged silique with internal floral organs.
(c) An ag-10 arf3-29 flower with additional floral organs growing inside of the unfused sepaloid carpels.
(d) An ag-10 ett-3 flower with a similar phenotype as ag-10 arf3-29.
(e) Siliques from two ARF3∷ARF3-GFP transgenic lines (middle and right pairs) and ag-10 arf3-29 siliques as the control (left pair).
(f) An arf3-29 flower (top view).
(g) An arf3-29 flower (side view). The arrows mark the ends of the gynophore.
Bars: 1mm in (a–g).
One such mutant with conspicuously enhanced ag-10 FM determinacy defects had bulged and unfused carpels with organs growing inside despite having normal sepals, petals and stamens (Figure 1c). Map-based cloning revealed a G-to-A mutation at the junction of the eighth exon and eighth intron of ARF3 that disrupted normal splicing and maturation of the mRNA (Supplementary Figure S1a,b). To confirm the effect of the mutation, hereafter referred to as arf3-29, in the ag-10 background, we introduced ett-3, an arf3/ett allele harboring a nonsense mutation in the eighth exon (Sessions et al., 1997) (Supplemental Figure S1a), into ag-10. As with ag-10 arf3-29, the ag-10 ett-3 double mutant also exhibited severe FM determinacy defects characterized by abundant organ growth within the unfused primary carpels (Figure 1d). Introduction of an ARF3∷ARF3-GFP transgene into ag-10 arf3-29 completely rescued the mutant phenotype (Figure 1e), further indicating that the arf3-29 mutation was responsible for the enhanced FM determinacy defects of ag-10 arf3-29.
The ag-10 arf3-29 double mutant was crossed to wild type to obtain the arf3-29 single mutant. arf3-29 was found to be an arf3/ett allele of intermediate strength, and its gynoecium developmental defects resembled those of ett-3, with reduced valves covered by stigma and pronounced medial outgrowths (Figure 1f, g and Supplementary Figure S1c). The extended gynophore of arf3-29 or ett-3 (delimited by arrowheads in Figure 1g and Supplementary Figure S1c), which may be attributed to additional cell layers at the base of the gynoecium at stage 6 (Sessions and Zambryski 1995; Sessions 1997), may also be indicative of a role of ARF3 in regulating floral stem cell activity.
ARF3 confers FM determinacy through the repression of WUS expression
To investigate the molecular basis of the FM determinacy defects of ag-10 arf3-29, we performed in situ hybridization to assess the spatial-temporal expression patterns of WUS (Laux et al., 1996). In wild-type flowers, WUS expression was undetectable at stage 6 of floral development (Supplementary Figure S2a) (Mayer et al., 1998; Ji et al., 2011; Liu et al., 2011; Yumul et al., 2013). While 90% of the flowers (n=13) from ag-10 plants had normal WUS expression patterns as observed in wild type, WUS expression was observed at stage 7 in a small percentage of ag-10 flowers (2/13), consistent with the slight FM determinacy defects of the ag-10 mutant (Supplementary Figure S2b) (Liu et al., 2011). WUS expression beyond stage 6 was not observed in arf3-29 flowers, indicating that the arf3-29 mutation alone did not lead to prolonged WUS expression (Figure 3a). WUS transcript abundance in ag-10 arf3-29 whole inflorescences was not statistically different from that in ag-10 and arf3-29 inflorescences (Supplementary Figure S2c). However, in contrast to ag-10 flowers, all of the ag-10 arf3-29 flowers (9/9) examined had prolonged WUS expression until or beyond stage 7 (Figure 2b). Together, these findings indicate that ARF3 is required for the repression of WUS expression when AG activity is partially compromised.
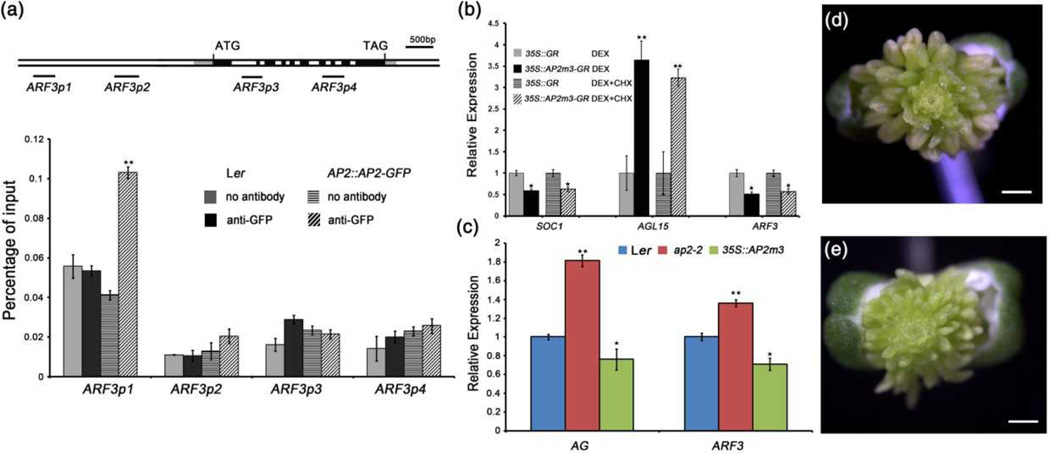
Figure 3. ARF3 is an AP2 target gene.
(a) ChIP-qPCR showing that AP2 binds the ARF3 locus. The tested regions of ARF3 are diagramed in the upper panel. ATG and TAG represent the start and stop codons, respectively. The gray, black and white rectangles represent the 5’ or 3’ untranslated regions, coding regions, and introns or intergenic regions, respectively. The black lines indicate the tested regions. Anti-GFP antibody was used for the analysis, and “no antibody” served as the negative control. Inflorescences containing all unopened flowers were dissected for ChIP assay.
(b) Real-time RT-PCR analysis of ARF3, SOC1 and AGL15 in 35S∷GR and 35S∷AP2m3-GR inflorescences treated with dexamethasone (DEX) for 6 hours with or without cyclohexamide (CHX). Inflorescences containing stage 8 and early flowers were used.
(c) AG and ARF3 expression in Ler, ap2-2 and 35S∷AP2m3 as determined by RT-qPCR Inflorescences containing stage 8 and early flowers were used.
(d) A 35S∷AP2m3 flower. Bars: 0.5mm.
(e) A 35S∷AP2m3 ag-10 arf3-29 flower. Bars: 0.5mm.
Error bars in (a–c) represent SD calculated from three biological replicates. Statistically significant changes are indicated by ★ (p-value < 0.05) and ★★ (p-value < 0.01).
Figure 2. Genetic interactions between arf3-29 and wus-1, ag-1 and knu-1.
(a) WUS expression in a stage 6 arf3-29 flower as examined by in situ hybridization. No WUS signal was detected.
(b) WUS expression in ag-10 arf3-29 flowers as examined by in situ hybridization. Arrows indicate WUS signal, and numbers indicate the floral developmental stage.
(c) A wus-1 flower.
(d) An ag-10 arf3-29 wus-1 flower.
(e) An ag-1 flower.
(f) An ag-1 arf3-29 flower.
(g,h) Longitudinal sections through stage 9 ag-1 (g) and ag-1 arf3-29 (h) flowers. Arrows indicate the floral meristem.
(i) FM diameter of ag-1 and ag-1 arf3-29 flowers. The values indicate means ± SD (n=10). The mean values for ag-1 and ag-1 arf3-29 were significantly different according to a Student’s t-test (p<0.01).
(j) Siliques of ag-10 arf3-29 (left pair) and ag-10 arf3-29 knu-1 (right pair).
Bars: 50µm in (a,b) and (g,h); 1mm in (c–f,j).
To investigate the genetic relationship between ARF3 and WUS, we crossed ag-10 arf3-29/+ plants with the loss-of-function wus-1 mutant (Laux et al., 1996) and generated the ag-10 arf3-29 wus-1 triple mutant. wus-1 plants exhibit premature FM termination, and the incompletely developed flowers of wus-1 have normal sepals and petals, but terminate in one or two stamens (sepal: 3.7±0.6; petal: 3.5±0.9; stamen: 0.8±0.4; n=11) (Figure 2c). The precocious termination of ag-10 arf3-29 wus-1 triple mutant flowers was similar to that of wus-1 flowers, indicating that wus-1 is epistatic to ag-10 arf3-29 in terms of FM determinacy (sepal: 3.8±0.7; petal: 3.6±0.8; stamen: 1.1±0.5; n=15) (Figure 2d).
ARF3 functions partially in the AG pathway in FM determinacy
AG is a key regulator of FM determinacy, and many genes have been found to promote FM determinacy by maintaining AG expression in the center of the meristem (Schultz et al., 1991; Alvarez and Smyth 1999; Carles et al., 2005; Prunet et al., 2008; Das et al., 2009; Maier et al., 2009). To investigate the relationship between ARF3 and AG, we first compared AG transcript levels in early-stage flowers between wild type and arf3-29 and between ag-10 and ag-10 arf3-29. We found that arf3-29 had no effect on AG transcript levels (Supplementary Figure S2c). Moreover, we did not detect ARF3 binding to the AG locus in our chromatin immunoprecipitation (ChIP) assay using ARF3∷ARF3-GFP transgenic plants. Thus, the data indicate that ARF3 does not regulate AG expression.
We next examined whether AG affected ARF3 expression. In ag-10, ARF3 transcript levels were decreased (Supplementary Figure S2d). Additionally, we employed 35S∷AG-GR, a widely used rat glucocorticoid receptor (GR)-induction system in which the AG-GR fusion protein localizes to the nucleus and functions on its target genes after dexamethasone (DEX) treatment (Sablowski and Meyerowitz 1998, Wagner et al. 1999, Ito et al. 2004, William et al. 2004) to evaluate the effects of AG on ARF3 expression., ARF3 expression was increased after 4 days of DEX treatment of 35S∷AG-GR ag-1 inflorescences (Supplementary Figure S2e). However, no enrichment of AG was observed at the ARF3 locus in our ChIP assay using AG∷AG-GFP plants. These results indicate that AG modulates ARF3 expression indirectly, which is consistent with previous reports (Ng et al., 2009).
Next, genetic analysis was performed to determine the molecular pathway in which ARF3 is involved. We separately introduced the ag-1 and knu-1 mutations into ag-10 arf3-29 by crossing. Flowers of the ag-1 null mutant are indeterminate and continually produce new organs in the fourth whorl, resulting in a flowers-in-flower phenotype (Figure 2e) (Bowman et al., 1989). ag-1 arf3-29 flowers appeared morphologically similar to ag-1 flowers, except that all of the floral organs produced in the former after the first whorl sepals were petals, indicating a role of ARF3 in specifying sepal identity in the internal flowers (Figure 2e,f). However, longitudinal sections through ag-1 arf3-29 and ag-1 flowers at the same developmental stage revealed a domelike meristem in ag-1 arf3-29 flowers that was statistically larger than that in ag-1, indicating that arf3-29 enhanced the determinacy defects of ag-1 (Figure 3g–i). KNU is an AG target gene that partly mediates AG’s function in FM determinacy by repressing WUS expression (Sun et al. 2009). Compared to ag-10 arf3-29, the ag-10 arf3-29 knu-1 triple mutant exhibited a more severe floral indeterminacy phenotype with several iterations of stamens and carpels growing from the primary, unfused carpel (Figure 3j). Collectively, these results suggest that ARF3’s function in FM determinacy in at least partly independent of the AG pathway.
ARF3 is an AP2 target gene
AP2 encodes an AP2 domain-containing protein involved in floral organ identity and FM maintenance (Bowman et al., 1991; Zhao et al., 2007). As a bifunctional transcription factor, AP2 can either activate or repress its targets, such as AGL15 and SOC1, respectively (Yant et al., 2010). A previous genome-wide analysis of AP2 binding sites revealed an enrichment of AP2 at the ARF3 locus, indicating that AP2 may directly bind ARF3 (Yant et al., 2010). To determine whether ARF3 is indeed an AP2 target gene, we performed ChIP-qPCR with AP2∷AP2-GFP transgenic plants. Among the ARF3 regions tested, high occupancy of AP2 was observed at a region 2.5kb upstream of the ARF3 transcription start site (TSS), indicating that AP2 binds a specific region of the ARF3 locus (Figure 3a). To assess whether AP2 directly regulates ARF3 expression, we quantified the transcript levels of ARF3 and known AP2 target genes AGL15 and SOC1 in 35S∷AP2m3-GR and 35S∷GR plants treated with either DEX or DEX plus cycloheximide (CHX). Daily DEX treatment of 35S∷AP2m3-GR inflorescences for 1 week led to the induction of the AP2m3 phenotype, thereby demonstrating that the transgene was functional (Zhao et al., 2007; Dinh et al., 2012). After 6 hours of treatment, inflorescences containing stage 8 and younger flowers were micro-dissected, and RT-qPCR was conducted to measure the transcript levels. As expected, AGL15 and SOC1 transcript levels were increased and reduced, respectively, in 35S∷AP2m3-GR compared with 35S∷GR in both DEX and DEX plus CHX treatments. ARF3 transcript levels were reduced by approximately 50% in 35S∷AP2m3-GR relative to 35S∷GR in both treatments (Figure 3b). We also detected higher and lower ARF3 transcript levels in ap2-2 and 35S∷AP2m3 inflorescences, respectively, relative to wild-type, along with changes in AG transcript levels consistent with the repression of AG by AP2 (Figure 3c) (Drews et al., 1991; Zhao et al., 2007; Wollmann et al., 2010). Moreover, we examined ARF3-GFP signals in inflorescences of ARF3∷ARF3-GFP 35S∷AP2m3 and ARF3∷ARF3-GFP. Usually, ARF3-GFP was detected in sepals and the rib zone of floral meristems (see details below and Supplementary Figure S3a). However, reduced GFP signals in sepals and the rib zone of meristems were detected in ARF3∷ARF3-GFP 35S∷AP2m3 compared with ARF3∷ARF3-GFP (compare Supplementary Figure S3a with S3b and Supplementary Figure S3c with Figure 4i). Collectively, these results demonstrated that ARF3 is a direct target of AP2.
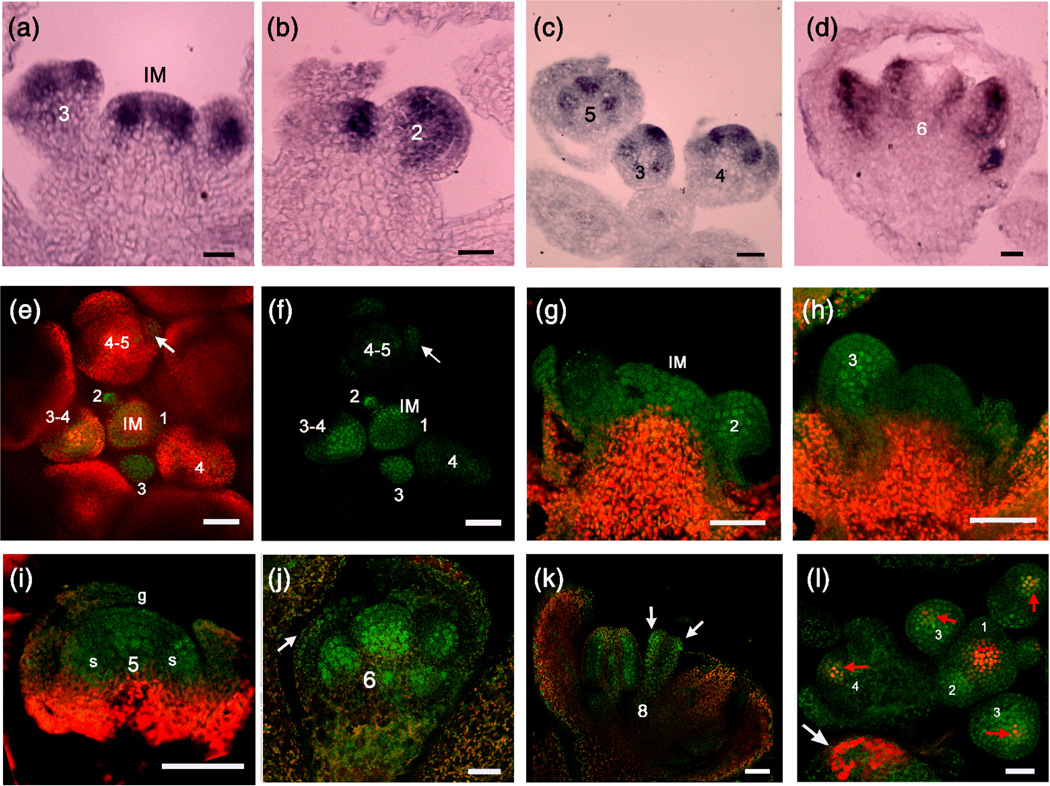
Figure 4. ARF3 RNA and protein distribution in flowers.
(a–d) in situ hybridization with an ARF3 antisense probe. ARF3 expression patterns in the IM and stage 3 (a), stage 2 (b), stage 5 (c) and stage 6 (d) FMs are shown. Numbers indicate the developmental stage.
(e,f) A global view of ARF3-GFP signal distribution in the FM and floral organ promordia during early floral development as observed with a confocal microscope (e: GFP and chlorophyll fluorescence merged channels; f: GFP channel alone). Numbers indicate the developmental stages of the FMs, and the arrow indicates GFP signal in a sepal primordium. Note that the GFP signal is present in the center of the IM but is masked by the strong chlorophyll fluorescence.
(g–k) ARF3-GFP signal in the IM and stage 2 (g), stage 3 (h), stage 5 (i), stage 6 (j) and stage 8 (k) FMs and flowers. Numbers indicate the floral developmental stage. The arrow in (j) indicates GFP signal in a sepal, and the arrows in (k) indicate the abaxial distribution of ARF3-GFP signals. g: gynoecium; s: stamen. Note that in (g) to (i), the regions showing strong chlorophyll fluorescence (red) also had GFP signals, which were masked by the red fluorescence.
(l) Coexpression of ARF3 and WUS in the FM. ARF3∷ARF3-GFP (green) and WUS∷DsRed-N7 (red) were detected in an ARF3∷ARF3-GFP WUS∷DsRed-N7 inflorescence using a confocal microscope, and the signals were merged. Red arrows indicate coexpression regions, and the white arrow indicates chlorophyll autofluorescence. Numbers indicate the floral developmental stage.
Bars: 50µm in (a,b) and (e–j); 25µm in (c,d) and (k,l).
We investigated the genetic relationship between AP2 and ARF3 in terms of FM maintenance by introducing the AP2m3 transgene into ag-10 arf3-29. Consistent with the presence of an indeterminate FM, 35S∷AP2m3 has normal organs in the outer two whorls, numerous stamens produced in a spiral phyllotaxy in the center of the flower and no carpels (Figure 3d) (Chen 2004; Zhao et al., 2007). The floral determinacy phenotypes of ag-10 arf3-29 35S∷AP2m3 resembled those of 35S∷AP2m3 (Figure 3e).
ARF3 expression and protein distribution patterns in the FM and floral organs
One of the major functions of ARF3 is FM and reproductive organ patterning (Sessions et al., 1997). To investigate the molecular basis of this function, we monitored the patterns of ARF3 expression and protein distribution in the FM and floral organs using in situ hybridization and GFP-tagged transgenic plants.
Using an ARF3 antisense probe, strong ARF3 mRNA signals were detected in the peripheral zone of the IM, which produces FMs; no specific signals were found when an ARF3 sense probe was used (Figure 4a and Supplementary Figure 4a). In stage 2 FMs, ARF3 was ubiquitously expressed with high transcript levels in the rib zone of the meristem (Figure 4b). In stages 3 and 5 FMs, ARF3 expression was concentrated in the stamen and carpel primordia and absent in the sepals (Figure 4a–c). From stage 5 to later stages (e.g., stage 8), abaxial ARF3 expression was observed in petals, stamens and the gynoecium (Figure 4d and Supplementary Figure 4b). These results reveal complex patterns of ARF3 expression during early floral development.
The ARF3∷ARF3-GFP transgene fully rescued the floral determinacy and gyneocium developmental defects of ag-10 arf3-29, indicating that the transgene conferred the whole range of ARF3 functions. As a transcription factor, ARF3 localizes in the nucleus (Kelley et al., 2012), and consistently, ARF3-GFP signals were also nuclear (Figure 4e–i) in ARF3∷ARF3-GFP plants. The GFP signal was present in the IM and stages 1 to 8 FMs. The high levels of ARF3 expression during early floral development is consistent with its function in FM and floral organ patterning (Figure 4e,f). Surprisingly, microscopic three-dimensional (3D) imaging of early-stage flowers showed ubiquitous GFP signals in stages 1 to 3 FMs, which differed from the ARF3 mRNA patterns described above (Supplementary Figure 4c, compare Figure 4e,f with Figure 4a,b). Additionally, strong GFP signals were observed in the sepals of stage 4 flowers, although ARF3 mRNA was not detected in sepals by in situ hybridization (compare Figure 4e,f, j and i with Figure 4c). To further investigate the distribution of ARF3-GFP in the floral organs, we embedded and sectioned the tissue then performed fluorescence detection as previously described (Goldshmidt et al., 2008). GFP signal was detected throughout the IM without a gradient (Figure 4e,f ). In stage 1 to early stage 3 FMs, GFP signal was again observed throughout the stem cell population, and was particularly high in the rib zone of the meristem (Figure 4g, h). In late stage 3 FMs, GFP signal was concentrated in groups of cells that would give rise to the sepal primordia (Figure 4i, l). In stage 5 flowers, GFP signal was detected in the center of the FM and the incipient stamen primordia (Figure 4i). In stage 6 flowers, GFP signal was detectable in all organ primordia including sepals and particularly concentrated in the rib zone of the meristem without a gradient (Figure 4j). In stage 6–8 flowers, GFP signal was evenly distributed throughout the gynoecium (Supplementary Figure S4d). After stage 8, ARF3-GFP was restricted to the abaxial side of the gynoecium (Figure 4k). The differences between the patterns of ARF3 transcript and protein distribution suggest that translational or post-translational mechanisms or protein movement may influence the distribution of the protein.
In light of the abundance of ARF3-GFP in the OC, where WUS is expressed, we further investigated whether the expression patterns of ARF3 and WUS overlap by crossing an ARF3∷ARF3-GFP plant with the WUS reporter line WUS∷DsRed-N7 (Gordon et al., 2007). Indeed, a high degree of overlap between GFP and RFP signals was observed, indicating that ARF3 and WUS are coexpressed in early flower development (Figure 4l and Supplementary Figure S4e, f).
The spatial distribution patterns of ARF3 are important for its FM determinacy function
Although ARF3 is transcribed throughout the leaf during leaf development, ARF3 transcripts are restricted to the abaxial leaf domain by tasiR-ARFs, and the resulting spatial pattern of ARF3 expression is critical for the establishment of leaf polarity (Chitwood et al., 2009). To determine whether the spatial patterns of ARF3 expression in the flower similarly influence its FM determinacy function, we generated a WUS∷ARF3m-GFP∷WUS3’ construct. The construct included 3.2kb of the WUS promoter; 1.5kb of the sequence downstream of the WUS coding region; and ARF3m-GFP, which is resistant to regulation by tasiR-ARFs (Hunter et al., 2006; Liu et al., 2011). ag-10 arf3-29/+ plants were transformed with the construct. In the T1 population, all positive transgenic lines were genotyped, and 14 transgenic plants were identified as homozygous for both ag-10 and arf3-29 (i.e., WUS∷ARF3m-GFP∷WUS3’ ag-10 arf3-29). These plants exhibited FM determinacy defects similar to that of ag-10 arf3-29 (Figure 5a, b). As expected, ARF3m-GFP fluorescence was found to be restricted to the OC (Figure 5c). Thus, OC-specific expression of ARF3 failed to rescue the FM determinacy defects of ag-10 arf3-29. We also generated an ARF3∷ARF3m-GFP construct, which is predicted to lead to ubiquitous distribution of ARF3 RNA and an ARF3∷ARF3m-PHB-GFP construct, in which tasiR-ARF-resistant ARF3 is fused to a sequence containing the miR165/166 recognition site from PHB to yield abaxial-adaxial inversion of ARF3 expression. In contrast to ARF3∷ARF3-GFP ag-10 arf3-29 plants (Figure 5d,e), ag-10 arf3-29 plants harboring a single copy insertion of ARF3∷ARF3m-GFP or ARF3∷ARF3m-PHB-GFP displayed only partial complementation: while the FM determinacy defects of later flowers were nearly fully rescued, early flowers clearly exhibited FM determinacy defects (Figure 5f–i). To confirm the expected patterns of ARF3 expression from the transgenes, in situ hybridization was performed in ARF3∷ARF3m-GFP and ARF3∷ARF3m-PHB-GFP plants. As expected, ARF3 was expressed throughout ARF3∷ARF3m-GFP flowers. In contrast, ARF3∷ARF3m-PHB-GFP flowers had stronger ARF3 signal on the adaxial side of floral organs, with the abaxial signal due to endogenous ARF3 transcripts (Supplementary Figure S5a–c).
Figure 5. ARF3’s proper spatial expression is important for its function in FM determinacy.
(a) An ag-10 arf3-29 flower.
(b) A WUS∷ARF3m-GFP∷WUS3’ ag-10 arf3-29 flower.
(c) Fluorescence detection of ARF3m-GFP in a WUS∷ARF3m-GFP∷WUS3’ ag-10 arf3-29 flower. White arrows indicate the GFP signals.
(d,e) An ARF3∷ARF3-GFP ag-10 arf3-29 plant (d) and its siliques (e).
(f,g) An ARF3∷ARF3m-GFP ag-10 arf3-29 plant (f) and its siliques (g).
(h,i) An ARF3∷ARF3m-PHB-GFP ag-10 arf3-29 plant (h) and its siliques (i).
Bars: 1mm in (a,b); 50µm in (c); 1cm in (d,f,h); 1mm in (e,g,i).
AG promotes the binding of ARF3 to the WUS locus in vivo
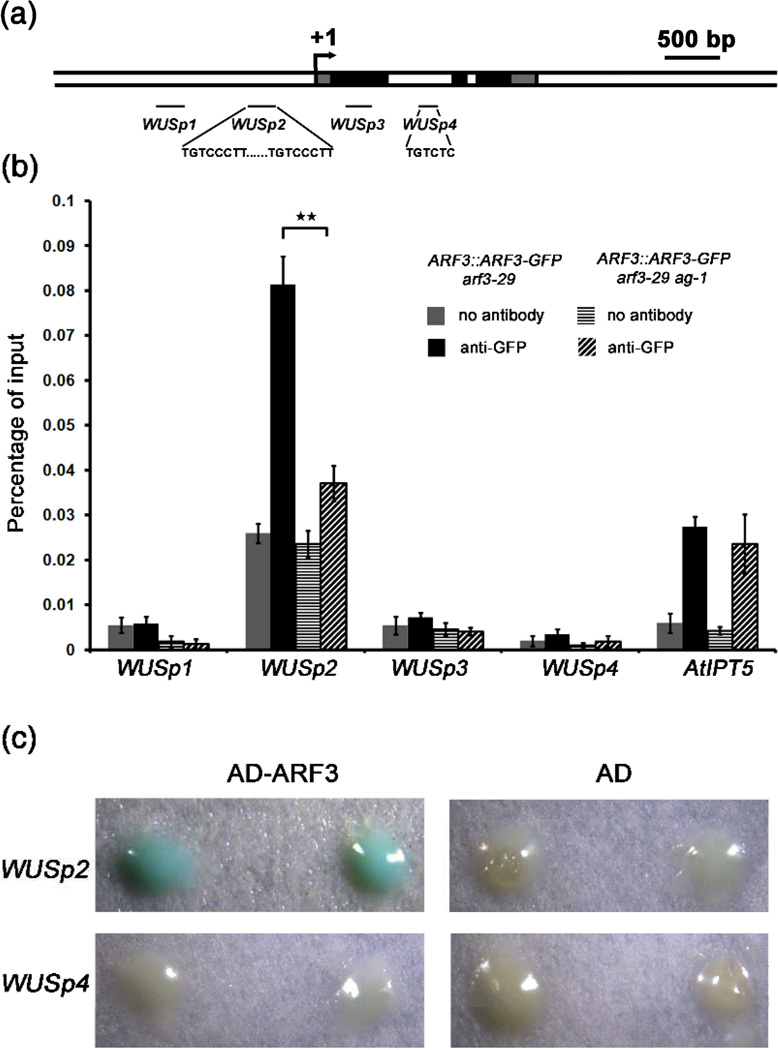
ARF3 and WUS coexpression in the OC raised the possibility that WUS is a target of ARF3. ARF family members specifically bind the AuxRE TGTCTC (Liu et al., 1994; Ulmasov et al., 1995; Liu et al., 1997). In vitro binding analysis revealed that substitutions at position +5 of the TGTCTC element being tolerated by ARF3 (Ulmasov et al., 1999b). We examined the WUS locus sequence and found one TGTCTC motif in the first intron (WUSp4 region) and two tandem TGTCCCTT sequences in the promoter region (WUSp2 region) approximately 500 bp upstream of the TSS (Figure 6a). TGTCCCTT is very similar to the TGTCCCAT cis-acting element found in the auxin-responsive region of the pea PS-IAA4/5 promoter and the soybean SAUR gene (Ballas et al., 1993; Li et al., 1994; Ballas et al., 1995). Using ChIP, we found high ARF3 enrichment at the WUS promoter region containing the TGTCCCTT tandem sequences but not at the first intron of WUS or other regions (Figure 6b).
Figure 6. AG promotes the binding of ARF3 to the WUS locus in vivo.
(a) A diagram of the WUS genomic region with “+1” corresponding to the transcription start site. The gray, black and white rectangles represent the 5’ or 3’ untranslated regions, coding regions and introns or intergenic regions, respectively. The black lines indicate the regions examined in ChIP. The sequences of the putative ARF3 binding sites are also shown.
(b) ARF3 occupancy at WUS and atIPT5 as determined by ChIP-qPCR. Anti-GFP antibody was used for ChIP, and “no antibody” served as the negative control. Error bars represent SD calculated from four biological replicates. Statistically significant changes are indicated by ★★ (p-value < 0.01).
(c) Yeast one-hybrid analysis revealing the direct binding of ARF3 to WUSp2. Yeast strains containing WUSP2∷LacZ or WUSp4∷LacZ reporters were transformed with AD-ARF3 or AD vectors, respectively. Two independent transformants growing on selective medium (SD/-Leu) were selected for β-galactosidase assay.
Next, we investigated whether ARF3 directly binds the WUS locus using yeast one-hybrid analysis. The WUSp2∷LacZ and WUSp4∷LacZ reporters were tested against AD (activation domain)-ARF3 or AD alone in yeast strains. While AD alone was unable to activate either reporter, AD-ARF3 activated the expression of WUSp2∷LacZ but not WUSp4∷LacZ, as shown by aβ-galactosidase assay (Figure 6c), suggesting that ARF3 can directly bind the WUS promoter..
As we previously found that AG also binds the WUS promoter region (Liu et al., 2011), we investigated whether AG affects the binding of ARF3 to WUS. We performed ChIP analysis with ARF3∷ARF3-GFP arf3-29 and ARF3∷ARF3-GFP arf3-29 ag-1 inflorescences. Interestingly, the occupancy of ARF3 at the WUS promoter was dramatically reduced in ARF3∷ARF3-GFP arf3-29 ag-1 compared to ARF3∷ARF3-GFP arf3-29 (Figure 6b). Given that AG promotes ARF3 expression, the reduced ARF3 occupancy at WUS in ag-1 could be due to reduced ARF3 expression. If so, we would expect ARF3 occupancy at its other target genes to be similarly reduced. We examined ARF3 occupancy at the promoter of atIPT5, which was reported to be bound by ARF3 (Cheng et al., 2013). ARF3 occupancy at atIPT5 in ARF3∷ARF3-GFP arf3-29 ag-1 was no different from that in ARF3∷ARF3-GFP arf3-29. Therefore, AG promotes the binding of ARF3 to the WUS promoter (Figure 6b).
DISCUSSION
ARF3 is required for FM determinacy
The roles of ARF3 in FM and floral organ patterning, leaf polarity and gynoecium development have been described (Sessions and Zambryski 1995; Sessions et al., 1997; Fahlgren et al., 2006). arf3 mutants typically exhibit an elongated gynophore, which has been attributed to additional cell layers at the base of the gynoecium primordium in stage 6 arf3 flowers (Sessions and Zambryski 1995; Sessions 1997). Given that floral organ primordia are successively produced from the outer whorls to the inner whorls and that the production of the gynoecium represents the last organogenesis event before the termination of the floral stem cells, the elongated gynophore of arf3 mutants may be indicative of prolonged FM activity. Using forward genetics screening and genetic analysis, we found clear evidence supporting a role of ARF3 in FM determinacy. Specifically, a mutation in ARF3, arf3-29, enhanced the weak FM determinacy defects of ag-10, as does the previously described arf3 mutant ett-3. The double mutants have numerous additional organs growing within the primary unfused sepaloid carpels. Collectively, these results support a role of ARF3 in specifying FM determinacy.
ARF3 partially integrates the AP2 and AG pathways to confer FM determinacy
FM determinacy is a complex process involving the coordination of various regulatory factors during the different floral developmental stages. WUS plays a vital role in SAM and FM maintenance and integrates several genetic pathways that either maintain or terminate the FM (Sablowski 2007; Sun et al., 2009; Ji et al., 2011; Liu et al., 2011; Yumul et al., 2013). The present findings indicate that ARF3, along with AG, promotes FM determinacy by repressing WUS expression within the FM.
Although ARF3 is required for FM determinacy as is AG, our results suggest that ARF3 may largely act independently of the AG pathway. First, in early floral developmental stages, ARF3 was expressed in the IM and FM independently of floral organ identity genes such as AG. While AG-GR increased ARF3 expression, this effect was only observed after hours of DEX treatment, indicating that the induction of ARF3 by AG was indirect. This finding is consistent with a previous report that GIANT KILLER (GIK) mediates the effect of AG on ARF3 expression (Ng et al., 2009). Second, although ag-1 arf3-29 flowers resembled ag-1 flowers in terms of FM determinacy defects (Figure 2e,f), longitudinal sections revealed larger FMs in ag-1 arf3-29 flowers than ag-1 flowers.. Finally, genetic analysis revealed that knu-1 enhanced the ag-10 arf3-29 FM determinacy defects, indicating that ARF3 acts in parallel to KNU, an AG target that represses WUS expression (Sun et al., 2009; Sun et al., 2014). However, it is possible that ARF3 partially mediates the FM determinacy function of AG in light of the finding that AG promotes ARF3 binding to the WUS locus.
AP2 has been implicated in SAM and FM maintenance by promoting WUS expression (Wurschum et al., 2006; Zhao et al., 2007), but the underlying molecular mechanism is unknown. Using ChIP-qPCR and an inducible expression system in the present study, ARF3 was found to be an AP2 direct target, with AP2 repressing ARF3 expression. Genetic analysis indicated that ARF3 mediates the FM determinacy function of AP2 insofar as ag-10 arf3-29 being unable to enhance 35S∷AP2m3. However, comparing the weak FM determinacy defects of arf3-29 and the strong determinacy defects of 35S∷AP2m3, mediation of AP2 function by ARF3 may only be partial.
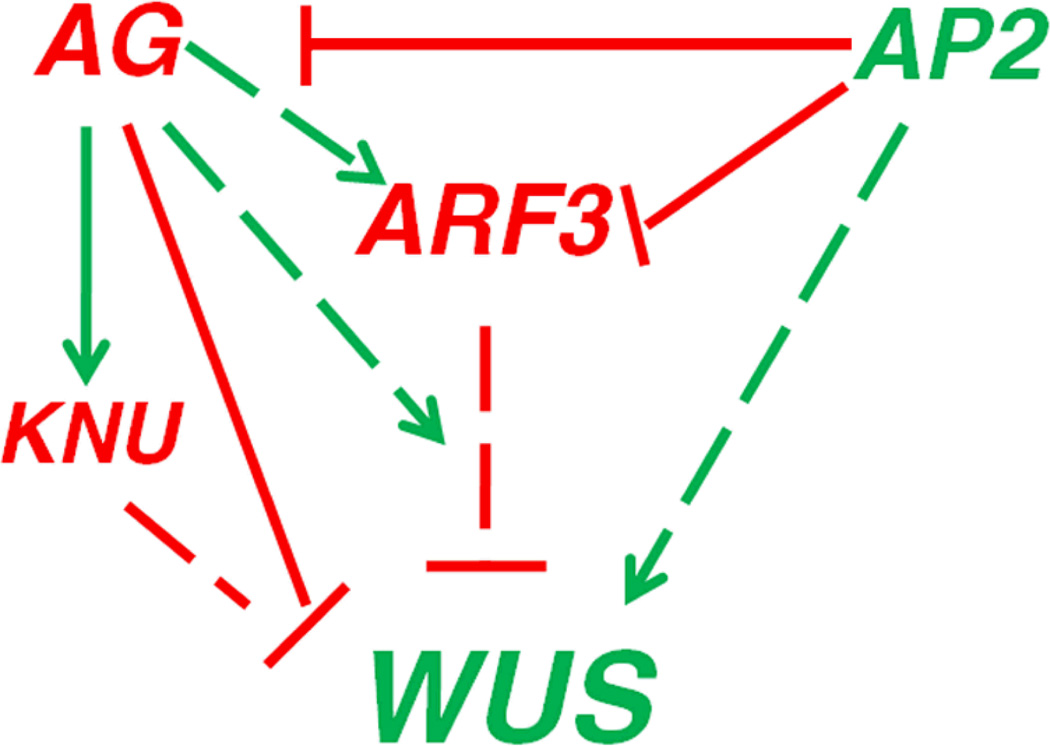
Collectively, the results of the genetic and gene expression analyses led to a working model of ARF3 in regulating FM determinacy (Figure 7). First, ARF3 is probably required for the full function of AG in FM determinacy. Once expressed at stage 3, AG enhances ARF3 expression indirectly and promotes ARF3 binding to the WUS locus, thereby coordinating or enhancing the function of ARF3 at WUS. Second, as a target gene repressed by AP2, ARF3 partially mediates the FM maintenance function of AP2. Therefore, ARF3 partially integrates the functions of both AP2 and AG in FM maintenance and determinacy.
Figure 7. A model of ARF3 in FM determinacy.
ARF3 terminates the floral stem cells by directly or indirectly repressing WUS expression. Besides the direct repression of WUS by AG through the recruitment of PcG to WUS and the indirect repression of WUS through the activation of KNU expression (Sun et al., 2009; Liu et al., 2011), AG indirectly enhances ARF3 expression and promotes ARF3 binding to the WUS locus. As a target gene repressed by AP2, ARF3 partially mediates the FM maintenance function of AP2. Red and green arrows indicate positive and negative effects, respectively. Solid and dotted arrows indicate direct and indirect effects, respectively. ARF3 is likely to have a direct effect on WUS expression, but this has not been definitively proven in vivo, thus a dotted arrow is used.
ARF3 exhibits dynamic and functionally relevant expression patterns
During leaf development, the abaxial distribution of ARF3 transcripts is critical for leaf polarity establishment. Altering the spatial distribution of ARF3 transcripts using tasiR-ARF-resistant ARF3 impairs leaf and gynoecium development (Fahlgren et al., 2006). in situ hybridization analysis in the present study revealed complex expression patterns of ARF3 in the IM and early FMs. ARF3 is expressed in clusters of cells that give rise to new FMs or floral organ primordia, consistent with its functions in FM patterning and floral organ identity. However, differences were observed between the distribution of ARF3 RNA and protein. First, while ARF3 RNA was abaxially distributed in the IM and floral organ primordia of early FMs, ARF3 protein was evenly distributed throughout the IM and gynoecium primordium of early FMs . Second, ARF3 RNA was not detected in sepals by in situ hybridization, but ARF3 protein strongly accumulated there. The latter finding suggests that ARF3 may be involved in sepal organogenesis and is also consistent with the petaloid sepal phenotype of the internal flowers of ag-1 arf3-29 (Figure 2f). The differences in the distribution patterns of ARF3 RNA and protein suggest that the ARF3 protein may be mobile. We note that the patterns of ARF3 RNA distribution are similar to those of auxin in the IM and early stage FMs (Vernoux et al., 2010). This raises the possibility that ARF3 is induced by auxin, although there is currently no evidence for this hypothesis. In turn, the even distribution of ARF3 protein throughout the IM and stages 1–2 FMs may allow ARF3 to coordinate developmental decisions within the meristems.
The abaxial distribution of ARF3 RNA in floral organ primordia was found to contribute to the FM determinacy function of ARF3, as transgenes that altered the ARF3 RNA distribution failed to complement the FM determinacy defects of ag-10 arf3-29. OC-specific expression of ARF3 also failed to rescue the FM determinacy defects of ag-10 arf3-29, although ARF3 was found to bind the WUS promoter region (Figure 6b). These somewhat paradoxical findings suggest that ARF3 exerts both direct and indirect effects on the repression of WUS expression. One possibility is that ARF3 acts via auxin in FM maintenance and determinacy. ARF3 has been found to be involved in SAM induction during de novo organ regeneration through auxin and cytokinin signaling (Cheng et al., 2013). Additionally, physical interactions between ARF3 and KANADI 1 (KAN1) were found to contribute to integument development and polarity determination (Kelley et al., 2012). KAN1 regulates auxin biosynthesis, transport and signaling by binding a specific cis-element as a repressor (Huang et al., 2014). It is possible that ARF3-KAN1 acts outside of the OC to regulate auxin biosynthesis or signaling, which in turn influences FM determinacy.
EXPERIMENTAL PROCEDURES
Plant materials
Plants were grown in soil as previously described by Liu et al. (2011). ag-10, ett-3, ag-1, wus-1, 35S∷AP2m3, knu-1, 35S∷AP2m3-GR,,35S∷GR and 35S∷AG-GR ag-1 were previously described (Sessions et al., 1997; Zhao et al., 2007; Ji et al., 2011; Liu et al., 2011; Dinh et al., 2014).
ChIP and in situ hybridization
The methods used for ChIP and in situ hybridization including information on the WUS probe were previously described (Liu et al., 2011). To generate ARF3 probes for in situ hybridization,, ARF3 sequence was amplified by PCR with ARF3SP6 and ARF3T7 primers and the ARF3∷ARF3m plasmid (see Method S3) as the template. The amplified product was purified and used as the template for in vitro transcription with either T7 or SP6 RNA polymerase to generate the antisense and sense probes, respectively. The primers used are listed in Supplementary Table S1.
Yeast one-hybrid analysis
PCR reactions were performed using WUSp2Y1HF and WUSp2Y1HR or WUSp4Y1HF and WUSp4Y1HR as primers and Ler genomic DNA as the template to obtain the fragments of WUSp2 and WUSp4, respectively. The fragments were cloned into the SmaI site of pLacZi (Clontech), creating pWUSp2∷LacZ and pWUSp4∷LacZ. Each plasmid was linearized by ApaI digestion prior to transformation of the yeast strain YM4271 (Clontech). The full-length cDNA of ARF3 was amplified by PCR from the ARF3∷ARF3m plasmid using primers ARF3Y1HF and ARF3Y1HR and cloned into the pGADT7 vector resulting in the pGADT7-ARF3 plasmid. The pGADT7-ARF3 or pGADT7 plasmids were subsequently transformed into the yeast strain containing the pWUSp2∷LacZ or pWUSp4∷LacZ construct and transformants were selected for growth on selection medium (SD/−Leu). β-galactosidase assay was performed according to the Yeast Protocols Handbook (Clontech, PT3024-1). The primers used are listed in Supplementary Table S1, Method S1, Method S2, Method S3, Method S4.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Dr. Yuval Eshed for the detailed protocol on tissue embedding and slicing. We also thank two anonymous reviewers for their constructive comments. This work was supported by the National Basic Research Program of China (2014CB138100), the National Natural Science Foundation of China (31340056) and the Hundred Talents Program of CAS to X.L. The work was also supported by National Institutes of Health (GM061146) and the Howard Hughes Medical Institute and Gordon and Betty Moore Foundation (GBMF3046) to X.C.
Footnotes
SUPPORTING INFORMATION
Figure S1. Molecular characterization of arf3-29 and the phenotype of ett-3.
Figure S2. WUS, AG and ARF3 expression in various genotypes.
Figure S3. ARF3 protein distribution in 35S∷AP2m3 floral meristems.
Figure S4. ARF3 transcript and protein distribution in FMs and floral organs.
Figure S5. ARF3 expression in various transgenic plants.
Table S1. Primers used in the study.
Method S1. EMS mutagenesis and map-based cloning of ARF3
Method S2. RNA extraction and real-time PCR
Method S3. Plasmid construction
Method S4. Tissue preparation for fluorescence detection
REFERENCES
- Adenot X, Elmayan T, Lauressergues D, Boutet S, Bouche N, Gasciolli V, Vaucheret H. DRB4-dependent TAS3 trans-acting siRNAs control leaf morphology through AGO7. Curr Biol. 2006;16:927–932. doi: 10.1016/j.cub.2006.03.035. [DOI] [PubMed] [Google Scholar]
- Alvarez J, Smyth DR. CRABS CLAW and SPATULA, two Arabidopsis genes that control carpel development in parallel with AGAMOUS. Development. 1999;126:2377–2386. doi: 10.1242/dev.126.11.2377. [DOI] [PubMed] [Google Scholar]
- Aukerman MJ, Sakai H. Regulation of flowering time and floral organ identity by a MicroRNA and its APETALA2-like target genes. Plant Cell. 2003;15:2730–2741. doi: 10.1105/tpc.016238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballas N, Wong LM, Ke M, Theologis A. Two auxin-responsive domains interact positively to induce expression of the early indoleacetic acid-inducible gene PS-IAA4/5. Proc Natl Acad Sci U S A. 1995;92:3483–3487. doi: 10.1073/pnas.92.8.3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballas N, Wong LM, Theologis A. Identification of the auxin-responsive element, AuxRE, in the primary indoleacetic acid-inducible gene, PS-IAA4/5, of pea (Pisum sativum) Journal of molecular biology. 1993;233:580–596. doi: 10.1006/jmbi.1993.1537. [DOI] [PubMed] [Google Scholar]
- Boer DR, Freire-Rios A, van den Berg WA, Saaki T, Manfield IW, Kepinski S, Lopez-Vidrieo I, Franco-Zorrilla JM, de Vries SC, Solano R, Weijers D, Coll M. Structural Basis for DNA Binding Specificity by the Auxin-Dependent ARF Transcription Factors. Cell. 2014;156:577–589. doi: 10.1016/j.cell.2013.12.027. [DOI] [PubMed] [Google Scholar]
- Bowman JL, Smyth DR, Meyerowitz EM. Genes directing flower development in Arabidopsis. Plant Cell. 1989;1:37–52. doi: 10.1105/tpc.1.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman JL, Smyth DR, Meyerowitz EM. Genetic interactions among floral homeotic genes of Arabidopsis. Development. 1991;112:1–20. doi: 10.1242/dev.112.1.1. [DOI] [PubMed] [Google Scholar]
- Carles CC, Choffnes-Inada D, Reville K, Lertpiriyapong K, Fletcher JC. ULTRAPETALA1 encodes a SAND domain putative transcriptional regulator that controls shoot and floral meristem activity in Arabidopsis. Development. 2005;132:897–911. doi: 10.1242/dev.01642. [DOI] [PubMed] [Google Scholar]
- Chen XM. Science. Vol. 303. New York, N.Y.: 2004. A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development; pp. 2022–2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng ZJ, Wang L, Sun W, Zhang Y, Zhou C, Su YH, Li W, Sun TT, Zhao XY, Li XG, Cheng Y, Zhao Y, Xie Q, Zhang XS. Pattern of auxin and cytokinin responses for shoot meristem induction results from the regulation of cytokinin biosynthesis by AUXIN RESPONSE FACTOR3. Plant physiology. 2013;161:240–251. doi: 10.1104/pp.112.203166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitwood DH, Nogueira FT, Howell MD, Montgomery TA, Carrington JC, Timmermans MC. Pattern formation via small RNA mobility. Genes & development. 2009;23:549–554. doi: 10.1101/gad.1770009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das P, Ito T, Wellmer F, Vernoux T, Dedieu A, Traas J, Meyerowitz EM. Floral stem cell termination involves the direct regulation of AGAMOUS by PERIANTHIA. Development. 2009;136:1605–1611. doi: 10.1242/dev.035436. [DOI] [PubMed] [Google Scholar]
- Dinh TT, Girke T, Liu X, Yant L, Schmid M, Chen X. The floral homeotic protein APETALA2 recognizes and acts through an AT-rich sequence element. Development. 2012;139:1978–1986. doi: 10.1242/dev.077073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinh TT, Luscher E, Li S, Liu X, Won SY, Chen X. Genetic Screens for Floral Mutants in Arabidopsis thaliana: Enhancers and Suppressors. Methods in molecular biology. 2014;1110:127–156. doi: 10.1007/978-1-4614-9408-9_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drews GN, Bowman JL, Meyerowitz EM. Negative regulation of the Arabidopsis homeotic gene AGAMOUS by the APETALA2 product. Cell. 1991;65:991–1002. doi: 10.1016/0092-8674(91)90551-9. [DOI] [PubMed] [Google Scholar]
- Fahlgren N, Montgomery TA, Howell MD, Allen E, Dvorak SK, Alexander AL, Carrington JC. Regulation of AUXIN RESPONSE FACTOR3 by TAS3 ta-siRNA affects developmental timing and patterning in Arabidopsis. Curr Biol. 2006;16:939–944. doi: 10.1016/j.cub.2006.03.065. [DOI] [PubMed] [Google Scholar]
- Gallois JL, Nora FR, Mizukami Y, Sablowski R. WUSCHEL induces shoot stem cell activity and developmental plasticity in the root meristem. Genes & development. 2004;18:375–380. doi: 10.1101/gad.291204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia D, Collier SA, Byrne ME, Martienssen RA. Specification of leaf polarity in Arabidopsis via the trans-acting siRNA pathway. Curr Biol. 2006;16:933–938. doi: 10.1016/j.cub.2006.03.064. [DOI] [PubMed] [Google Scholar]
- Goldshmidt A, Alvarez JP, Bowman JL, Eshed Y. Signals derived from YABBY gene activities in organ primordia regulate growth and partitioning of Arabidopsis shoot apical meristems. Plant Cell. 2008;20:1217–1230. doi: 10.1105/tpc.107.057877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon SP, Heisler MG, Reddy GV, Ohno C, Das P, Meyerowitz EM. Pattern formation during de novo assembly of the Arabidopsis shoot meristem. Development. 2007;134:3539–3548. doi: 10.1242/dev.010298. [DOI] [PubMed] [Google Scholar]
- Guilfoyle T, Hagen G, Ulmasov T, Murfett J. How does auxin turn on genes? Plant physiology. 1998;118:341–347. doi: 10.1104/pp.118.2.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilfoyle TJ, Hagen G. Auxin response factors. Curr Opin Plant Biol. 2007;10:453–460. doi: 10.1016/j.pbi.2007.08.014. [DOI] [PubMed] [Google Scholar]
- Huang T, Harrar Y, Lin C, Reinhart B, Newell NR, Talavera-Rauh F, Hokin SA, Barton MK, Kerstetter RA. Arabidopsis KANADI1 Acts as a Transcriptional Repressor by Interacting with a Specific cis-Element and Regulates Auxin Biosynthesis, Transport, and Signaling in Opposition to HD-ZIPIII Factors. Plant Cell. 2014 doi: 10.1105/tpc.113.111526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter C, Willmann MR, Wu G, Yoshikawa M, de la Luz Gutierrez-Nava M, Poethig SR. Trans-acting siRNA-mediated repression of ETTIN and ARF4 regulates heteroblasty in Arabidopsis. Development. 2006;133:2973–2981. doi: 10.1242/dev.02491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji LJ, Liu XG, Yan J, Wang WM, Yumul RE, Kim YJ, Dinh TT, Liu J, Cui X, Zheng BL, Agarwal M, Liu CY, Cao XF, Tang GL, Chen XM. ARGONAUTE10 and ARGONAUTE1 Regulate the Termination of Floral Stem Cells through Two MicroRNAs in Arabidopsis. PLoS genetics. 2011:7. doi: 10.1371/journal.pgen.1001358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann K, Pajoro A, Angenent GC. Regulation of transcription in plants: mechanisms controlling developmental switches. Nature Reviews Genetics. 2010;11:830–842. doi: 10.1038/nrg2885. [DOI] [PubMed] [Google Scholar]
- Kelley DR, Arreola A, Gallagher TL, Gasser CS. ETTIN (ARF3) physically interacts with KANADI proteins to form a functional complex essential for integument development and polarity determination in Arabidopsis. Development. 2012;139:1105–1109. doi: 10.1242/dev.067918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidner CA, Martienssen RA. Spatially restricted microRNA directs leaf polarity through ARGONAUTE1. Nature. 2004;428:81–84. doi: 10.1038/nature02366. [DOI] [PubMed] [Google Scholar]
- Laux T, Mayer KFX, Berger J, Jurgens G. The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development. 1996;122:87–96. doi: 10.1242/dev.122.1.87. [DOI] [PubMed] [Google Scholar]
- Lenhard M, Bohnert A, Jurgens G, Laux T. Termination of stem cell maintenance in Arabidopsis floral meristems by interactions between WUSCHEL and AGAMOUS. Cell. 2001;105:805–814. doi: 10.1016/s0092-8674(01)00390-7. [DOI] [PubMed] [Google Scholar]
- Li Y, Liu ZB, Shi X, Hagen G, Guilfoyle TJ. An auxin-inducible element in soybean SAUR promoters. Plant physiology. 1994;106:37–43. doi: 10.1104/pp.106.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscum E, Reed JW. Genetics of Aux/IAA and ARF action in plant growth and development. Plant Mol Biol. 2002;49:387–400. [PubMed] [Google Scholar]
- Liu XG, Kim YJ, Muller R, Yumul RE, Liu CY, Pan YY, Cao XF, Goodrich J, Chena XM. AGAMOUS Terminates Floral Stem Cell Maintenance in Arabidopsis by Directly Repressing WUSCHEL through Recruitment of Polycomb Group Proteins. Plant Cell. 2011;23:3654–3670. doi: 10.1105/tpc.111.091538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu ZB, Hagen G, Guilfoyle TJ. A G-Box-Binding Protein from Soybean Binds to the E1 Auxin-Response Element in the Soybean GH3 Promoter and Contains a Proline-Rich Repression Domain. Plant physiology. 1997;115:397–407. doi: 10.1104/pp.115.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu ZB, Ulmasov T, Shi X, Hagen G, Guilfoyle TJ. Soybean GH3 promoter contains multiple auxin-inducible elements. Plant Cell. 1994;6:645–657. doi: 10.1105/tpc.6.5.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier AT, Stehling-Sun S, Wollmann H, Demar M, Hong RL, Haubeiss S, Weigel D, Lohmann JU. Dual roles of the bZIP transcription factor PERIANTHIA in the control of floral architecture and homeotic gene expression. Development. 2009;136:1613–1620. doi: 10.1242/dev.033647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer KFX, Schoof H, Haecker A, Lenhard M, Jurgens G, Laux T. Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell. 1998;95:805–815. doi: 10.1016/s0092-8674(00)81703-1. [DOI] [PubMed] [Google Scholar]
- McConnell JR, Emery J, Eshed Y, Bao N, Bowman J, Barton MK. Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature. 2001;411:709–713. doi: 10.1038/35079635. [DOI] [PubMed] [Google Scholar]
- Nemhauser JL, Feldman LJ, Zambryski PC. Auxin and ETTIN in Arabidopsis gynoecium morphogenesis. Development. 2000;127:3877–3888. doi: 10.1242/dev.127.18.3877. [DOI] [PubMed] [Google Scholar]
- Ng KH, Yu H, Ito T. AGAMOUS controls GIANT KILLER, a multifunctional chromatin modifier in reproductive organ patterning and differentiation. PLoS biology. 2009;7:e1000251. doi: 10.1371/journal.pbio.1000251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pekker I, Alvarez JP, Eshed Y. Auxin response factors mediate Arabidopsis organ asymmetry via modulation of KANADI activity. Plant Cell. 2005;17:2899–2910. doi: 10.1105/tpc.105.034876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prunet N, Morel P, Thierry AM, Eshed Y, Bowman JL, Negrutiu I, Trehin C. REBELOTE, SQUINT, and ULTRAPETALA1 function redundantly in the temporal regulation of floral meristem termination in Arabidopsis thaliana. Plant Cell. 2008;20:901–919. doi: 10.1105/tpc.107.053306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulido A, Laufs P. Co-ordination of developmental processes by small RNAs during leaf development. J Exp Bot. 2010;61:1277–1291. doi: 10.1093/jxb/erp397. [DOI] [PubMed] [Google Scholar]
- Reed JW. Roles and activities of Aux/IAA proteins in Arabidopsis. Trends in plant science. 2001;6:420–425. doi: 10.1016/s1360-1385(01)02042-8. [DOI] [PubMed] [Google Scholar]
- Reinhardt D, Mandel T, Kuhlemeier C. Auxin regulates the initiation and radial position of plant lateral organs. Plant Cell. 2000;12:507–518. doi: 10.1105/tpc.12.4.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sablowski R. Flowering and determinacy in Arabidopsis. J Exp Bot. 2007;58:899–907. doi: 10.1093/jxb/erm002. [DOI] [PubMed] [Google Scholar]
- Schoof H, Lenhard M, Haecker A, Mayer KFX, Jurgens G, Laux T. The stem cell population of Arabidopsis shoot meristems is maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell. 2000;100:635–644. doi: 10.1016/s0092-8674(00)80700-x. [DOI] [PubMed] [Google Scholar]
- Schultz EA, Pickett FB, Haughn GW. The FLO10 Gene Product Regulates the Expression Domain of Homeotic Genes AP3 and PI in Arabidopsis Flowers. Plant Cell. 1991;3:1221–1237. doi: 10.1105/tpc.3.11.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessions A, Nemhauser JL, McColl A, Roe JL, Feldmann KA, Zambryski PC. ETTIN patterns the Arabidopsis floral meristem and reproductive organs. Development. 1997;124:4481–4491. doi: 10.1242/dev.124.22.4481. [DOI] [PubMed] [Google Scholar]
- Sessions R. Arabidopsis (Brassicaceae) flower development and gynoecium patterning in wild type and Ettin mutants. American journal of botany. 1997;84:1179. [PubMed] [Google Scholar]
- Sessions RA, Zambryski PC. Arabidopsis gynoecium structure in the wild and in ettin mutants. Development. 1995;121:1519–1532. doi: 10.1242/dev.121.5.1519. [DOI] [PubMed] [Google Scholar]
- Smyth DR, Bowman JL, Meyerowitz EM. Early Flower Development in Arabidopsis. Plant Cell. 1990;2:755–767. doi: 10.1105/tpc.2.8.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun B, Looi LS, Guo S, He Z, Gan ES, Huang J, Xu Y, Wee WY, Ito T. Science. Vol. 343. New York, N.Y: 2014. Timing mechanism dependent on cell division is invoked by Polycomb eviction in plant stem cells; p. 1248559. [DOI] [PubMed] [Google Scholar]
- Sun B, Xu YF, Ng KH, Ito T. A timing mechanism for stem cell maintenance and differentiation in the Arabidopsis floral meristem. Genes & development. 2009;23:1791–1804. doi: 10.1101/gad.1800409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tantikanjana T, Nasrallah JB. Non-cell-autonomous regulation of crucifer self-incompatibility by Auxin Response Factor ARF3. Proceedings of the National Academy of Sciences. 2012;109:19468–19473. doi: 10.1073/pnas.1217343109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari SB, Hagen G, Guilfoyle T. The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell. 2003;15:533–543. doi: 10.1105/tpc.008417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle TJ. Activation and repression of transcription by auxin-response factors. Proc Natl Acad Sci U S A. 1999a;96:5844–5849. doi: 10.1073/pnas.96.10.5844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle TJ. Dimerization and DNA binding of auxin response factors. The Plant journal : for cell and molecular biology. 1999b;19:309–319. doi: 10.1046/j.1365-313x.1999.00538.x. [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Liu ZB, Hagen G, Guilfoyle TJ. Composite structure of auxin response elements. Plant Cell. 1995;7:1611–1623. doi: 10.1105/tpc.7.10.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneste S, Friml J. Auxin: a trigger for change in plant development. Cell. 2009;136:1005–1016. doi: 10.1016/j.cell.2009.03.001. [DOI] [PubMed] [Google Scholar]
- Vernoux T, Besnard F, Traas J. Auxin at the shoot apical meristem. Cold Spring Harb Perspect Biol. 2010;2:a001487. doi: 10.1101/cshperspect.a001487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams L, Carles CC, Osmont KS, Fletcher JC. A database analysis method identifies an endogenous trans-acting short-interfering RNA that targets the Arabidopsis ARF2, ARF3, and ARF4 genes. Proc Natl Acad Sci U S A. 2005;102:9703–9708. doi: 10.1073/pnas.0504029102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollmann H, Mica E, Todesco M, Long JA, Weigel D. On reconciling the interactions between APETALA2, miR172 and AGAMOUS with the ABC model of flower development. Development. 2010;137:3633–3642. doi: 10.1242/dev.036673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurschum T, Gross-Hardt R, Laux T. APETALA2 regulates the stem cell niche in the Arabidopsis shoot meristem. Plant Cell. 2006;18:295–307. doi: 10.1105/tpc.105.038398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yant L, Mathieu J, Dinh TT, Ott F, Lanz C, Wollmann H, Chen X, Schmid M. Orchestration of the floral transition and floral development in Arabidopsis by the bifunctional transcription factor APETALA2. Plant Cell. 2010;22:2156–2170. doi: 10.1105/tpc.110.075606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yumul RE, Kim YJ, Liu XG, Wang RZ, Ding JH, Xiao LT, Chen XM. POWERDRESS and Diversified Expression of the MIR172 Gene Family Bolster the Floral Stem Cell Network. PLoS genetics. 9 doi: 10.1371/journal.pgen.1003218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Kim Y, Dinh TT, Chen X. miR172 regulates stem cell fate and defines the inner boundary of APETALA3 and PISTILLATA expression domain in Arabidopsis floral meristems. The Plant journal : for cell and molecular biology. 2007;51:840–849. doi: 10.1111/j.1365-313X.2007.03181.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.