Abstract
Aims: MiR126 was found to be frequently lost in many types of cancer, including malignant mesothelioma (MM), which represents one of the most challenging neoplastic diseases. In this study, we investigated the potential tumor suppressor function of MiR126 in MM cells. The effect of MiR126 was examined in response to oxidative stress, aberrant mitochondrial function induced by inhibition of complex I, mitochondrial DNA (mtDNA) depletion, and hypoxia. Results: MiR126 was up-regulated by oxidative stress in nonmalignant mesothelial (Met5A) and MM (H28) cell lines. In Met5A cells, rotenone inhibited MiR126 expression, but mtDNA depletion and hypoxia up-regulated MiR126. However, these various stimuli suppressed the levels of MiR126 in H28 cells. MiR126 affected mitochondrial energy metabolism, reduced mitochondrial respiration, and promoted glycolysis in H28 cells. This metabolic shift, associated with insulin receptor substrate-1 (IRS1)-modulated ATP-citrate lyase deregulation, resulted in higher ATP and citrate production. These changes were linked to the down-regulation of IRS1 by ectopic MiR126, reducing Akt signaling and inhibiting cytosolic sequestration of Forkhead box O1 (FoxO1), which promoted the expression of genes involved in gluconeogenesis and oxidative stress defense. These metabolic changes induced hypoxia-inducible factor-1α (HIF1α) stabilization. Consequently, MiR126 suppressed the malignancy of MM cells in vitro, a notion corroborated by the failure of H28MiR126 cells to form tumors in nude mice. Innovation and Conclusion: MiR126 affects mitochondrial energy metabolism, resulting in MM tumor suppression. Since MM is a fatal neoplastic disease with a few therapeutic options, this finding is of potential translational importance. Antioxid. Redox Signal. 21, 2109–2125.
Introduction
MicroRNAs (MiRs) play a crucial role in many biological processes (4, 15), including tumorigenesis (11), and their differential expression in tumors and normal tissues has been documented (10). Switching from profiling studies to the functional role of MiRs resulted in the notion that the aberrant expression of MiRs in cancer plays a causal role in the modulation of the tumorigenic process (10, 58).
We have shown that MiR126 is suppressed in patients with malignant mesothelioma (MM) (52, 60), which is consistent with reports that the down-regulation of MiR126 was observed in tumors (16, 67, 70) and cancer cell lines (23). Its restoration reduced the overall tumor growth and invasiveness of tumor cells (23, 38, 52). MiR126 has been proposed to modulate the PI3K signaling pathway, partly by targeting p85b during colon carcinogenesis (23) and negatively regulating the insulin receptor substrate-1 (IRS1) (67). As an adaptor of the insulin growth factor-1 receptor, IRS1 plays an important role in cell growth and proliferation, primarily via the Akt pathway (5). Mitochondrial dysfunction decreases the expression of IRS1, and MiR126 mediates the repression of the IRS1 protein in response to mitochondrial perturbation (49).
Innovation.
We found that MiR126 expression is regulated by mitochondria-destabilizing stimuli and affects mitochondrial energy metabolism, reducing mitochondrial respiration and up-regulating glycolysis-induced energy in malignant mesothelioma cells. The metabolic re-programming is associated with the inhibition of tumorigenic effects, resulting in the inhibition of tumor growth in an animal model. These data strongly support the tumor suppressor function and therapeutic application of MiR126 in cancers as hard to treat as mesothelioma.
While the molecular mechanism by which MiRs affect the pathogenesis of MM is virtually unknown, their expression in MM is inversely correlated with its severity and prognosis (8). This has been shown, for example, for hsa-MiR29c and MiR31 (44, 30), and the loss of MiR34b/c conferred reduced malignancy in MM cells (34). The level of expression of MiR34b/c is regulated epigenetically (34), a process described for MM (42). The loss of MiR126 in MM patients (52) is in line with the notion of the potential tumor suppressor function of the MiR species (53, 66, 70), and it was proposed as a potential diagnostic marker of MM, in particular in combination with mesothelin (29, 42, 52, 60). However, the molecular mechanism by which MiR126 regulates malignancy of MM has not been examined. Increasing evidence shows the interplay between MiRs and oncogenes/tumor suppressors via key metabolic enzyme effectors (21), and mitochondria play a crucial role in the tumor metabolic re-programming (18, 56). In addition to converting the incoming nutrients into energy in the form of ATP, mitochondria generate intermediates for biosynthetic pathways as well as reactive oxygen species (ROS) that serve as second messengers to modulate signal transduction and metabolism.
Here, we investigated the tumor suppressor effect of MiR126 in response to mitochondria-destabilizing stimuli involved in cancer induction and progression. Our results document that MiR126 is regulated by stress signaling and modulates the response of the cell to the stress. MiR126 affects the IRS1 pathway and the mitochondrial function, resulting in cancer metabolic re-programming. Ectopic MiR126 induced the loss of malignancy and the failure of MM cells to form tumors. This tumor suppressor function of MiR126 was not observed in IRS1-nonresponsive malignant cells. Since MM is a fatal neoplastic disease with hardly any therapeutic options (17, 48, 57, 59), our findings are of potential clinical importance.
Results
MiR126 is regulated by stress and affects the mitochondrial function
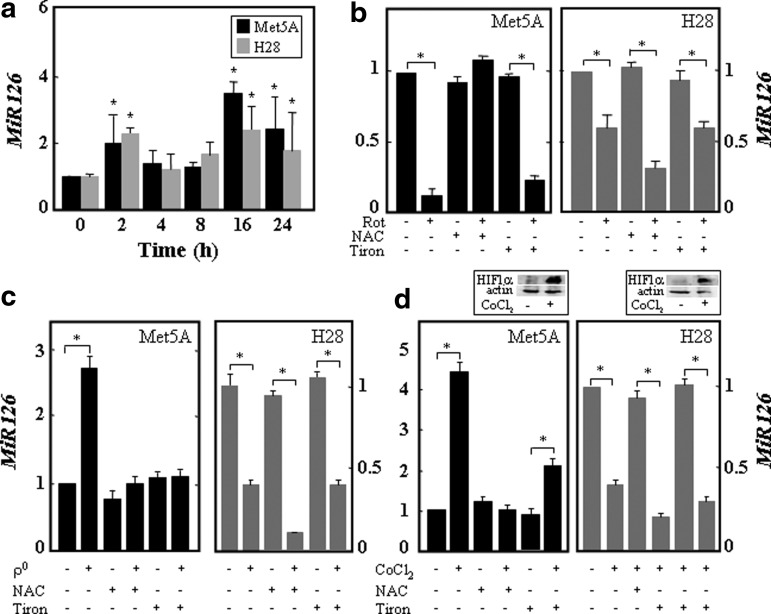
It has been reported that stress alters MiR biogenesis and that MiRs regulate important signaling pathways in mitochondria (36, 37). To assess whether MiR126 is regulated by stress signaling, nonmalignant (Met5A) and MM (H28) cells were exposed to oxidative stress, rotenone, mitochondrial DNA (mtDNA) depletion, and the hypoxia mimetic CoCl2, and the expression of MiR126 was evaluated. To determine a direct relationship between intracellular ROS levels and MiR126 expression mediated by mitochondrial impairment induced by the earlier stimuli, the cells were pretreated with the ROS scavenger N-acetyl cysteine (NAC, 10 mM) or the superoxide scavenger Tiron (10 mM). The dose-dependent curves of the ROS inhibitors on MiR126 level are shown in Supplementary Fig. S1 (Supplementary Data are available online at www.liebertpub.com/ars). MiR126 was transiently induced by oxidative stress in both cell lines, peaking at 2 and 16 h of hydrogen peroxide (200 μM) exposure (Fig. 1a). Rotenone-mediated mitochondrial impairment induced MiR126 down-regulation, which was inhibited by NAC but not by Tiron in Met5A cells (Fig. 1b). Up-regulation of MiR126 caused by mtDNA depletion and by the hypoxia mimetic CoCl2 was suppressed by NAC and Tiron in Met5A cells (Fig. 1c, d), indicating a role for ROS. The various stimuli suppressed the levels of MiR126 in H28 cells, which was rescued neither by NAC nor by Tiron (Fig. 1b–d).
FIG. 1.
Oxidative stress, mitochondrial dysfunction, and hypoxia modulate MiR126 expression. (a) Met5A and H28 cells were exposed over time to hydrogen peroxide (200 μM), and MiR126 level was expressed as fold change versus untreated cells. Met5A and H28 cells were evaluated for MiR126 level after treatment with rotenone (20 μM, 24 h) (b), deletion of mtDNA (c), or exposure to the hypoxia mimetic CoCl2 (100 μM, 5 h) (d) in the presence of NAC (10 mM) or Tiron (10 mM). HIF1α protein levels in Met5A cells and H28 cells treated with CoCl2 (100 μM, 5 h) are shown in panel (d). The results are mean values±S.D. of three experiments performed in duplicate. Comparisons among groups were determined by one-way ANOVA with Tukey post-hoc analysis. The symbol “*” indicates significant differences with p<0.05. ANOVA, analysis of variance; HIF1α, hypoxia-inducible factor-1α; MiR, microRNA; mtDNA, mitochondrial DNA; NAC, N-acetyl cysteine.
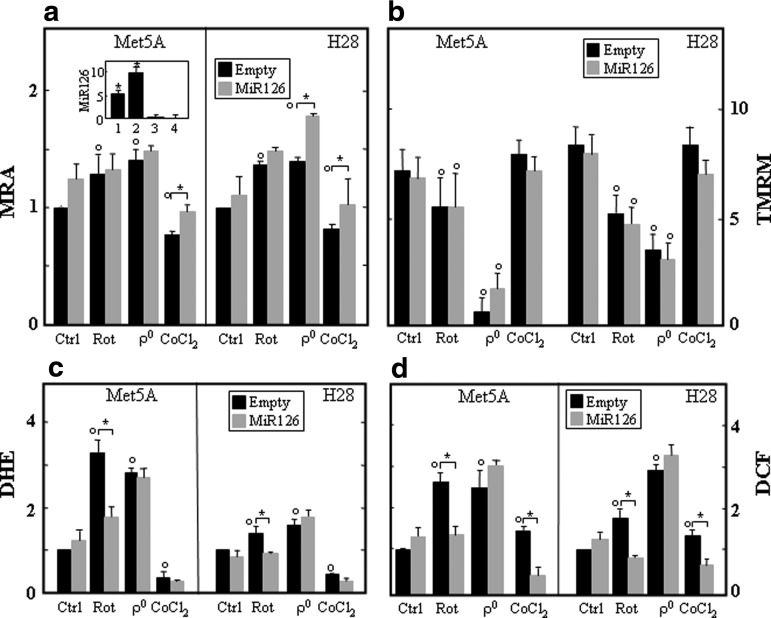
To evaluate whether MiR126 modulates the response of cells to stress stimuli, we generated stable cell lines expressing MiR126 (Met5AMiR126 and H28MiR126 cells) (Fig. 2a, insert), and the mitochondrial function was evaluated as a response to stress. MiR126-transfected cells and their empty plasmid-transfected counterparts (Met5ApCMV-MiR and H28pCMV-MiR cells) were subjected to rotenone exposure, mtDNA depletion, and CoCl2 exposure, and the mitochondrial reducing activity (MRA) was evaluated. Rotenone and mtDNA depletion (ρ0 cells) increased MRA in both parental Met5A and H28 cells, which was further increased by MiR126 in ρ0 H28 cells. MRA reduced in response to CoCl2 and was slightly increased by MiR126 (Fig. 2a). The evaluation of ΔΨm,i revealed a negligible effect of MiR126 in the control and stressed cells (Fig. 2b). As illustrated in Figure 2c and d, MiR126 reduced ROS formation induced by rotenone. Complete loss of mtDNA resulted in higher intracellular levels of ROS in both cell lines, which was not affected by ectopic MiR126. ROS generated in response to CoCl2 and mainly consisted of peroxide-related species, but not superoxide, and their level was reduced in MiR126-transfected cells. No formation of nitric oxide (NO) was observed in parental and MiR126-transfected cell lines exposed to stress (Supplementary Fig. S1d).
FIG. 2.
MiR126 expression affects the mitochondrial function. (a) Mitochondrial reducing activity (MRA) of empty plasmid- and MiR126-transfected Met5A and H28 cells (see inset for MiR126 levels in Met5AMiR126, 1; H28MiR126, 2; Met5Aanti-MiR126, 3; and H28anti-MiR126, 4, relative to their empty plasmid counterparts) was evaluated as the reduction of MTT. The cells were exposed to rotenone (20 μM, 5 h), lacked mtDNA (ρo cells), or were exposed to the hypoxia mimetic CoCl2 (100 μM, 5 h). Formazan production was assessed at 595 nm, normalized to viable cells by crystal violet staining, and the activity of the untreated cells was set as 1. Empty plasmid-transfected Met5A and H28 cells and their MiR126-transfected counterparts treated as described earlier were evaluated for ΔΨm,i using TMRM, expressed as MFI (b), ROS generation using DHE (relative to control cells) (c) or DCF (relative to control cells) (d). The results are mean values±S.D. of three experiments performed in duplicate. Comparisons among groups were determined by one-way ANOVA with Tukey post-hoc analysis. The symbol “*” indicates significant differences, and the symbol “°” significance compared with control with p<0.05. DCF, 2′,7′-dichlorofluorescein diacetate; DHE, dihydroethidinium; MFI, mean fluorescence intensity; MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; ROS, reactive oxygen species; TMRM, tetramethyl rhodamine methyl ester.
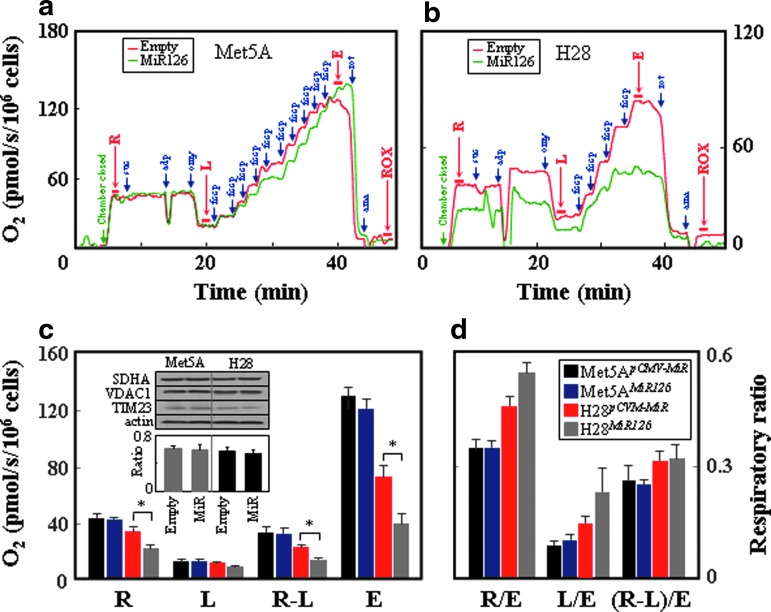
Ectopic MiR126 caused suppression of mitochondrial respiration in H28 but not Met5A cells, with no decrease when the “routine” (R) or “leak” (L) respiration as well as respiration coupled to ATP production (R-L) was related to the total capacity of the uncoupled electron transfer system (ETS, E) (Fig. 3a–d). The reason is due to the fact that MiR126 strongly suppressed, in particular, the maximum respiratory capacity as well as respiration coupled to ATP generation. The lower respiration was not due to different amounts of mitochondria in MiR126- and empty plasmid-transfected cells (Fig. 3c, insert).
FIG. 3.
MiR126 alters respiration in MM cells. Empty plasmid- and MiR126-transfected Met5A (a) and H28 cells (b) were evaluated for respiration using intact cells for routine (R), leak (L), uncoupled (E), and residual (ROX) respiration after additions of succinate (suc; 10 mM), ADP (adp; 3 mM), oligomycin (omy; 1 μg/ml), FCCP (fccp; added at 1 μM aliquots to reach maximum respiration), rotenone (rot; 0.5 μM), and antimycin (ama; 2.5 μM). The individual respiration rates as well as the “netR” value (R-L) are shown in panel (c) as absolute values and in panel (d), they are related to the maximum uncoupled respiration (ETS, E). The inset in panel (c) documents the levels of VDAC1, SDHA, and TIM23 in the empty plasmid- and MiR126-transfected cells and the ratio of mtDNA/nDNA in these cells. The data shown are mean values±S.D. of at least three independent experiments, with the symbol “*” indicating statistically significant differences between empty plasmid and MiR126-transfected cells with p<0.05. The images are representative of three independent experiments. ETS, electron transfer system; MM, malignant mesothelioma; nDNA, nuclear DNA. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
MiR126 induces a glycolytic shift
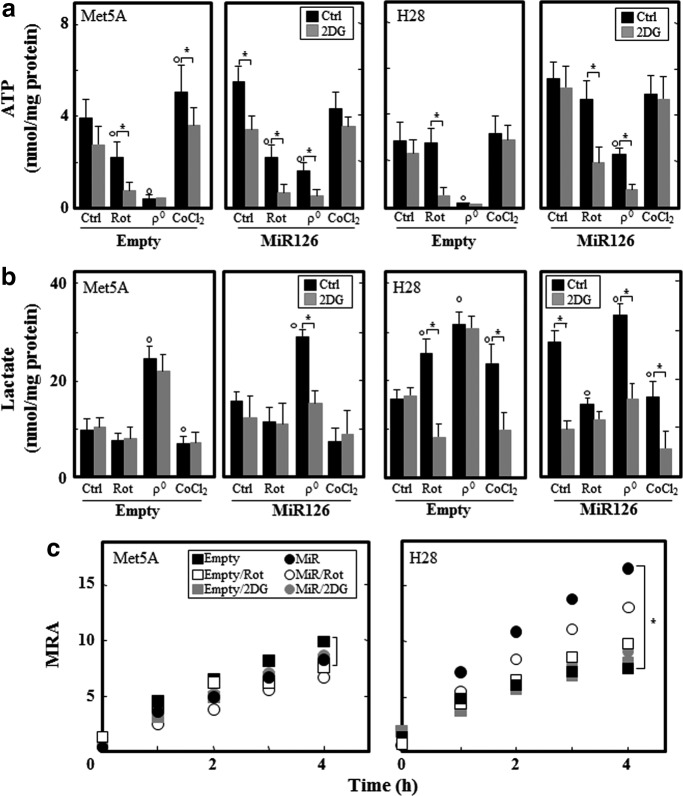
Since ectopic MiR126 lowered the mitochondrial respiration in H28 cells, we expected increased glycolytic activity in H28MiR126 cells to meet their demand for ATP. Intracellular ATP and lactate levels were evaluated in stress-exposed cells in the presence and absence of the glycolytic inhibitor 2-deoxyglucose (2DG). Ectopic MiR126 increased ATP and lactate in both cell lines, which was inhibited by 2DG in Met5AMiR126 cells, suggesting a shift to glycolysis (Fig. 4a, b). Although the H28MiR126 cells showed a glycolytic phenotype, the inhibition of glycolysis or oxidative phosphorylation (OXPHOS) did not affect ATP, which was reduced when 2DG and rotenone were combined, indicating a compensatory mechanism. The depletion of mtDNA inhibited ATP production, which was associated with increased lactate in both cell lines. Unlike in empty plasmid-transfected cells, mtDNA depletion did not completely inhibit ATP in Met5AMiR126 and H28MiR126 cells, in which ATP was derived from glycolysis. The hypoxia mimetic CoCl2 increased ATP, which was generated by glycolysis in Met5A cells. Conversely, the ATP level was not affected by the hypoxia mimetic in H28 cells (Fig. 4a, b). To further confirm the glycolytic shift, MRA, evaluated as the reduction of the mitochondrial cytochrome pool (the resazurin assay), was analyzed in empty plasmid- and MiR126-transfected cells with inhibited OXPHOS or glycolysis. In stark contrast to Met5A cells, MiR126 significantly increased the MRA in H28 cells, which was slightly inhibited by rotenone and suppressed considerably by 2DG (Fig. 4c), suggesting that the mitochondrial function is maintained primarily by glycolysis.
FIG. 4.
MiR126 affects the bioenergetic profile. Empty plasmid- and MiR126-transfected Met5A and H28 cells were evaluated for ATP (nmol/mg protein) (a) and lactate (nmol/mg protein) (b) in presence of rotenone (20 μM, 5 h), after mtDNA depletion (ρ0 cells) of cells exposed to the hypoxic mimetic CoCl2 (100 μM, 5 h), and in the presence of 5 mM 2DG for 5 h. (c) MRA of empty plasmid- and MiR126-transfected Met5A and H28 cells was evaluated in the presence of rotenone (20 μM, 5 h) or 2DG (5 mM, 5 h), and was expressed as MFI per μg protein. The results are mean values±S.D. of three individual experiments performed in triplicate. Comparisons among groups were determined by one-way ANOVA with Tukey post-hoc analysis; the symbol “*” indicates significant differences; symbol “°” significance compared with control with p<0.05. 2DG, 2-deoxyglucose.
MiR126 suppresses the IRS1 pathway and modulates mitochondrial metabolism
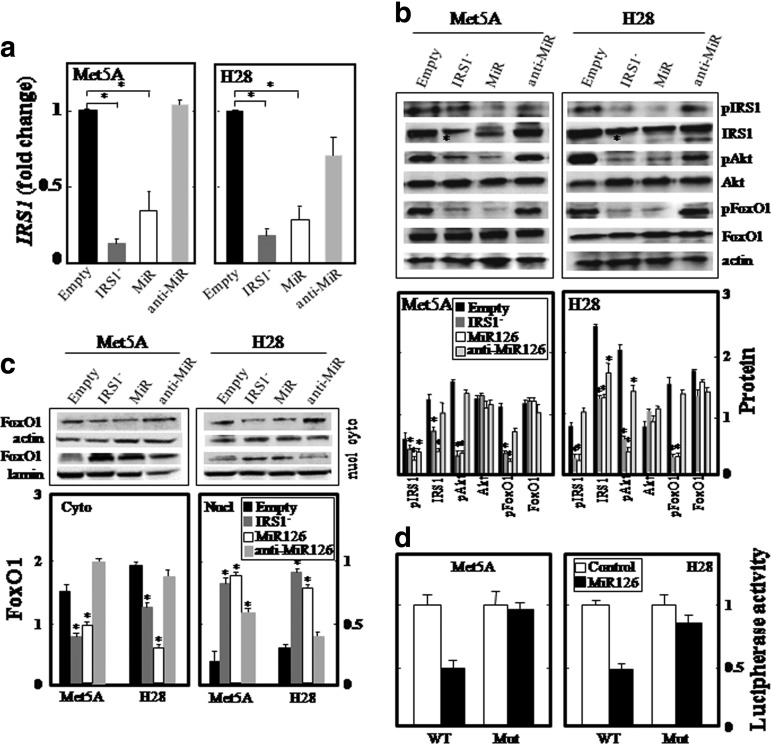
Mitochondrial biogenesis is modulated by IRS1 and IRS2 (49, 50), and MiR126 has been shown to reduce the expression of IRS1 by directly targeting its 3′-UTR (5, 49, 67). Met5AMiR126 and H28MiR126 cells showed strong IRS1 down-regulation (Fig. 5a–c). Targeting of the IRS1 3′-UTR was confirmed using a reporter gene, luciferase, carrying the 3′-UTR of IRS1 containing the MiR126-binding site (IRS1 3Uwt). A mutated 3′-UTR of the IRS1 gene (IRS1 3Umut) lacking of the MiR126 binding site was used as a control. We found that MiR126 significantly inhibited the luciferase activity in both Met5A and H28 cells transfected with IRS1 3Uwt plasmid, whereas luciferase activity with IRS1 3Umut was not suppressed (Fig. 5d). The silencing of IRS1 significantly suppressed Akt activation, confirming its involvement (Fig. 5b). A key Akt pathway down-stream effector, the Forkhead box O1 (FoxO1) transcription factor, is sequestered in the cytoplasm in response to its phosphorylation by Akt. Accordingly, in both IRS1 silencing and MiR126 transfection (Fig. 5b), there was a decrease in the cytoplasm and an increase in the nuclear fraction (Fig. 5c). Restoration of the IRS1-Akt-FoxO1 pathway was observed by blocking the function of MiR126 using antisense MiR126 (anti-MiR) (Fig. 5) (cf also insert in Fig. 2a for MiR126 levels), further confirming the involvement of MiR126.
FIG. 5.
MiR126 affects IRS1 signaling. Empty plasmid-transfected Met5A and H28 cells, and their IRS1-silenced (IRS1−), MiR126-transfected (MiR), and antago-MIR126-transfected (anti-MiR) counterparts were analyzed for the relative level of IRS1 mRNA (a); expression of pIRS1 (S307), IRS1, pAkt, Akt, pFoxO1 (T24), and FoxO1 (b). Lower part of panel (b) documents densitometric evaluation of the bands shown in its upper part related to the level of actin. Cytoplasmic and nuclear expression of the FoxO1 protein (upper panels: western blots of FoxO1, lower panels: their densitographic evaluation) (c) is shown for the different cell types. IRS1-3Uwt or IRS1-3Umut construct was co-transfected with empty plasmid (control) or MiR126 expression plasmid (MiR126) into Met5A and H28 cell lines. Reporter gene assay was performed using dual-luciferase assays kit as described in Methods. The relative luciferase activities were plotted against that of the control, which was set at 1 (d). The data shown are mean values±S.D. of three independent experiments. Comparisons among groups were determined by one-way ANOVA with Tukey post-hoc analysis; the symbol “*” indicates significantly different values compared with empty plasmid-transfected cells with p<0.05. FoxO1, Forkhead box O1; IRS1, insulin receptor substrate-1.
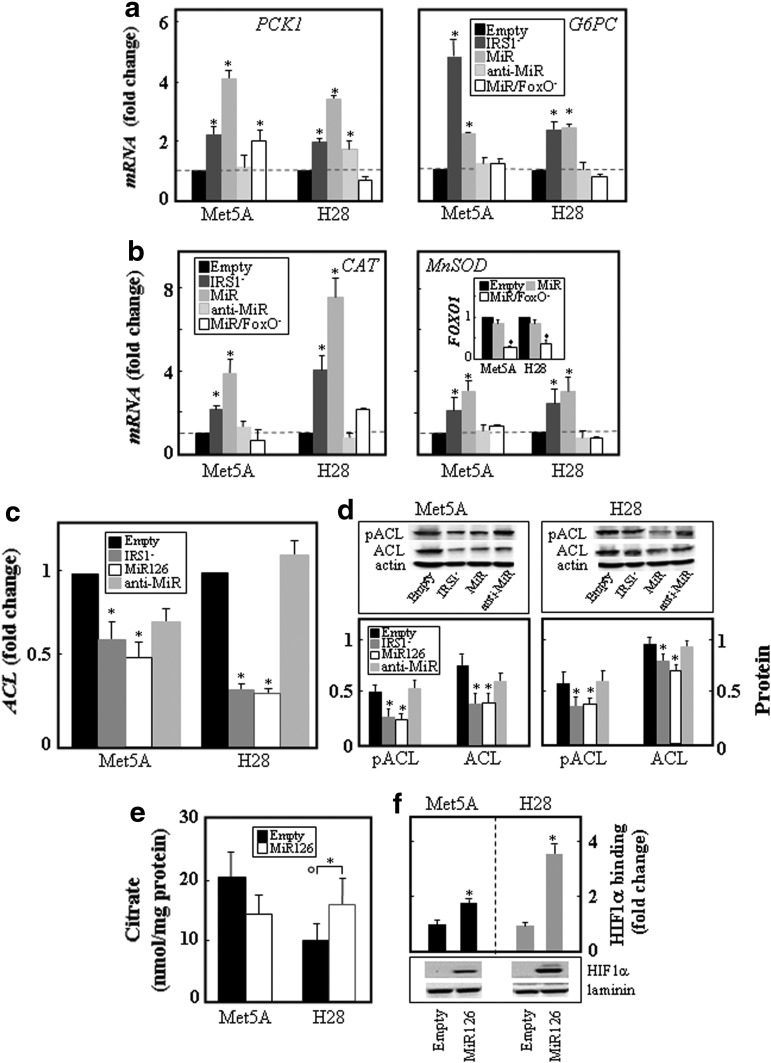
As a result of nuclear localization, FoxO1 caused increased expression of gluconeogenesis genes, including phosphoenolpyruvate carboxykinase 1 (PCK1) and glucose-6-phosphatase catalytic (G6PC), which was consistent with a previous report (68). The elevated expression of PCK1 in MiR126-transfected and IRS1 knock-down cells was normalized on the silencing of FoxO1 and/or MiR126. A similar pattern was observed for G6PC (Fig. 6a). MiR126 as well as knock-down of IRS1 also caused up-regulation of the ROS detoxification enzymes catalase (CAT) and manganese superoxide dismutase (MnSOD). The silencing of FoxO1 as well as the transfection with anti-MiR126 suppressed the MiR126-induced expression of the anti-oxidant enzymes (Fig. 6b).
FIG. 6.
MiR126 alters the IRS1 pathway-dependent mitochondrial function, expression of FoxO1-dependent genes, citrate levels, and HIF1α activation. Empty plasmid-transfected Met5A and H28 cells and their counterparts with silenced IRS1 (IRS1−), and transfected with MiR126 (MiR) and antisense MiR126 (anti-MiR) were evaluated for the level of the mRNA of the gluconeogenesis genes PCK1 and G6PC (a) and oxidative defense genes CAT and MnSOD (b). These cells were further evaluated for the level of ACL mRNA (c) and protein as well as pACL (d; upper panel, western blots, lower panel, densitometric evaluation). Empty plasmid- and MiR126-transfected Met5A and H28 cells were evaluated for intracellular citrate levels (nmol/mg protein) (e) and HIF1α protein level and its DNA binding (f). The data shown are mean values±S.D. derived from three independent experiments; the symbol “*” indicates significant differences compared with empty plasmid-transfected cells, with symbol “°” significance between Met5A and H28 cells with p<0.05. ACL, ATP-citrate lyase; CAT, catalase; G6PC, glucose-6-phosphatase catalytic; MnSOD, manganese superoxide dismutase; PCK1, phosphoenolpyruvate carboxykinase 1.
FoxO1 contributes to glucose homeostasis and lipid metabolism (65), with ATP-citrate lyase (ACL), a key enzyme of de novo lipogenesis being up-regulated and activated in cancer tissue (25). As shown in Figure 6c and d, the ACL protein was expressed more in MM compared with nonmalignant cells. Increased MiR126 as well as suppression of IRS1 resulted in the decrease of ACL at the level of mRNA and protein, as well as its phosphorylation. Anti-MiR126 increased the level and phosphorylation of ACL. Consequently, low citrate levels were found in MM compared with nonmalignant cells, and ectopic expression of MiR126 restored citrate levels, which was associated with hypoxia-inducible factor-1α (HIF1α) stabilization and activation (Fig. 6e, f).
To test the clinical relevance of these results, we analyzed 11 matching pairs of MM tissue and the adjacent normal tissue from MM patients for the level of MiR126 and IRS1, ACL, MnSOD, and CAT mRNA expression. We found that MiR126 levels were inversely correlated with IRS1 and ACL and positively correlated with MnSOD and CAT in the malignant tissue, further supporting the effect of MiR126 on mitochondrial bioenergetics and tumor malignancy (see next) (Supplementary Fig. S2).
Ectopic MiR126 inhibits tumorigenic properties of MM cells and suppresses mesothelioma initiation
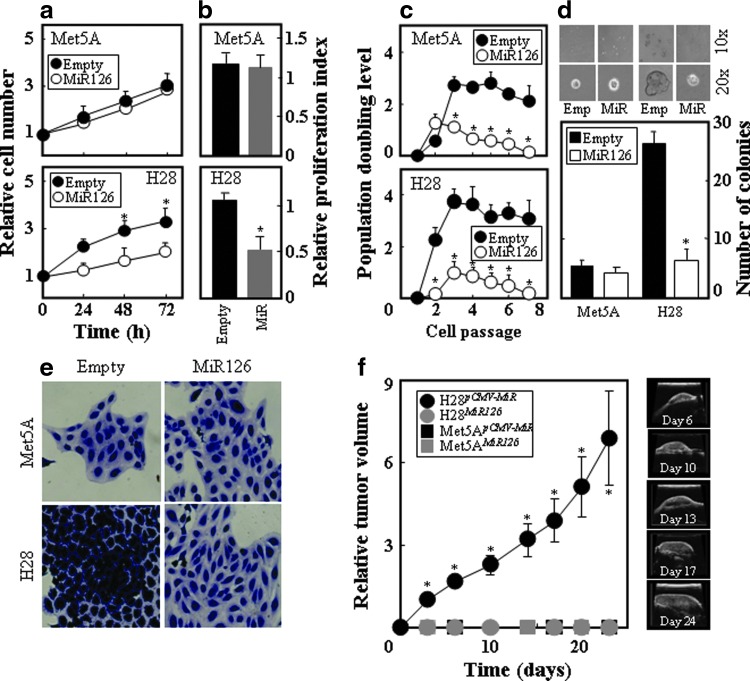
Given that MiR126 affects mitochondrial metabolism in MM, it may regulate tumor progression. Indeed, increased MiR126 expression significantly inhibited H28 cell growth as well as the level of the proliferation marker Ki67. This was not observed in the nonmalignant Met5A cells (Fig. 7a, b). When compared with the empty plasmid-transfected cells, the percentage of H28MiR126 cells in G0/G1 phase increased from 45.6% to 65.9% (p<0.05), whereas the percentage of cells in S phase and G2/M phase decreased from 36.2% to 23.1% (p<0.05) and from 16.4% to 10.6% (p<0.05), respectively. No change in the cell cycle distribution was found for Met5ApCMV-MiR and Met5AMiR126 cells. The MiR126-mediated cell growth inhibition correlated with the reduction in the replication potential, which was evaluated as the population doubling level (PDL), and was more efficiently suppressed in H28 than Met5A cells (Fig. 7c). Similarly, the ability of the cells to form colonies in soft agar was reduced by increased MiR126 in H28 cells but not in Met5A cells (Fig. 7d). In addition, H28MiR126 cells acquired morphological features similar to the nonmalignant Met5A cells (Fig. 7e). To unequivocally confirm the tumor suppressor function of MiR126 in mesothelioma, we tested whether MiR126-transfected cells give rise to tumors in nude mice. Met5A and H28 cells expressing MiR126 and their empty plasmid-transfected counterparts were subcutaneously injected into nude mice and tumor formation was monitored by ultrasound imaging (USI). Figure 7f documents lack of tumor growth when H28MiR126 cells were grafted in the mice, while H28pCMV-MiR cells formed tumors with relatively fast kinetics. As expected, neither Met5ApCMV-MiR nor Met5AMiR126 cells gave rise to tumors.
FIG. 7.
MiR126 suppresses the malignant phenotype. Empty plasmid-transfected H28 and Met5A cells and their MiR126-transfected counterparts were evaluated for cell growth by the MTT assay (a), cell proliferation expressed as Ki67-positive cells (b), PDL (c), and the colony-forming activity (upper panels: microscopic evaluation, lower panels: number of colonies) (d). (e) Empty plasmid- and MiR126-transfected cells were inspected by microscopy for morphology after their staining with crystal violet. (f) Nude mice were injected subcutaneously with H28pCMV-MiR, H28MiR126, Met5A pCMV-MiR, and Met5AMiR126 cells (5×106 cells per mouse in 100 μl of sterile saline). The tumor progression was visualized and quantified by USI (left panel: tumor kinetics, right panels: visualization of a representative tumor). The results are mean values±S.D. of three experiments performed in duplicate; images are representative of three independent experiments. Comparisons among groups were determined by one-way ANOVA with Tukey post-hoc analysis. The symbol “*” indicates significantly different values with p<0.05. Data of tumor volume are mean values±S.E.M. (n=7); the symbol “*” indicates significantly different values with p<0.05. Images are representative of at least three independent experiments. PDL, population doubling level; USI, ultrasound imaging. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
MiR126 is a tumor suppressor in IRS1-responsive MM cells
To see whether the tumor suppressor function of MiR126 is limited to H28 cells, we tested it in the established MM cell line H2452 and the immortalized primary Mes-1 cells. Unlike in the Mes-1 cells, MiR126 increased the level of ATP in the H2452 cells, also shifting them to the glycolytic phenotype (Supplementary Fig. S3a, b). The MiR126-transfected H2452 cells exerted a lower level of respiration unlike Mes-1 cells (Supplementary Fig. S3c, d). MiR126 increased the level of citrate in H2452 but not in the Mes-1 cells, which was associated with ACL down-regulation (Supplementary Fig. S3f) as well as with HIF1α stabilization and activation by MiR126 in the former (Supplementary Fig. S3g). Similarly as for H28 cells, MiR126 in H2452 cells suppressed the level of IRS1, and phosphorylation of Akt as well as of FoxO1 (Supplementary Fig. S4a), Consequently, MiR126 regulated the level of expression of IRS1, CAT, MnSOD, PCK1, G6PC, and ACL genes in H2452 cells (Supplementary Fig. S4b), similarly as observed in H28 cells (cf Figs. 5 and 6). All these changes invoked in H2452 cells by MiR126 cells caused a loss of their malignant phenotype, which was not observed in Mes-1 cells (Supplementary Fig. S4c, d).
To evaluate whether MiR126-induced tumor suppression was linked to binding of the MiR to 3′-UTR of IRS1, Ist-Mes2 MM cells with truncated IRS1 lacking its MiR126 binding site (see sequence in Supplementary Fig. S4e) were transfected with MiR126 plasmid and the tumorigenic effect was evaluated. Small interfering RNA (siRNA) was used as a control to show that the truncated protein was down-regulated by IRS1 siRNA. MM cells carrying IRS1 without the MiR126-binding site at the 3′-UTR were not sensitive to MiR126-induced cell growth arrest and tumor suppression (Supplementary Fig. S4e).
MiR126-induced tumor suppressor is regulated by citrate levels through HIF1α activation
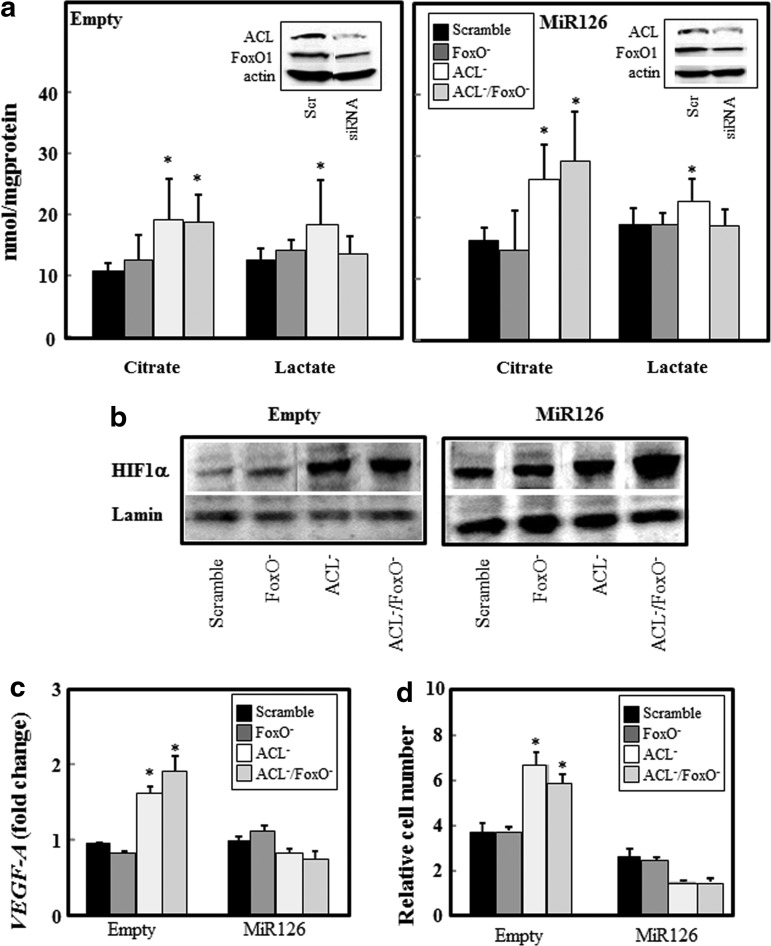
We observed that citrate levels were depleted in MM cells, and MiR126 overexpression rescued citrate by inhibiting ACL, which was associated with HIF1α activation and tumor suppression. To examine the impact of citrate production, HIF1α activation, and glucose metabolism on MiR126-mediated tumor growth inhibition, MiR126- and empty plasmid-transfected MM cells were silenced for ACL, FoxO1 and for both ACL/FoxO1, and the citrate/lactate levels, HIF1α nuclear translocation, HIF1α-responsive vascular endothelial growth factor-A (VEGF-A) gene expression, and cell growth were evaluated. Knocking down ACL (Fig. 8a, insert) induced citrate production in empty-plasmid H28 cells, which was further increased in MiR126-transfected cells (Fig. 8a). The inhibition of FoxO1 (Fig. 8a, insert) did not affect the levels of citrate, even on ACL knockdown (Fig. 8a). Conversely, the silencing of FoxO1 suppressed lactate production induced by inhibiting ACL (Fig. 8a), suggesting its involvement in the MiR126-induced glycolytic shift. The increased levels of citrate mediated by the silencing of ACL induced HIF1α nuclear translocation in both MiR126- and empty plasmid-transfected cells (Fig. 8b). Activation of HIF1α resulted in increased expression of the HIF-responsive VEGF-A gene (Fig. 8c), which was associated with increased cell proliferation in empty plasmid H28 cells (Fig. 8d). Conversely, the accumulation of HIF1α in the nucleus of MiR126 H28 cells resulted in the reduction of the VEGF-A gene expression and cell growth (Fig. 8c, d).
FIG. 8.
Involvement of ACL, FoxO1, and HIF1α in MiR126-induced metabolic changes and MM cell proliferation. MiR126- and empty plasmid-transfected H28 cells were silenced for ACL (ACL−), FoxO1 (FoxO−) or both (ACL−/FoxO−), and citrate/lactate levels (a), HIF1α nuclear translocation (b), HIF1α-responsive VEGF-A gene expression (c), and cell growth evaluated (d). The results are mean values±S.D. of three experiments performed in duplicate; images are representative of three independent experiments. Comparisons among groups were determined by one-way ANOVA with Tukey post-hoc analysis; the symbol “*” indicates significant differences compared with control (scrambled sequences) with p<0.05. VEGF-A, vascular endothelial growth factor-A.
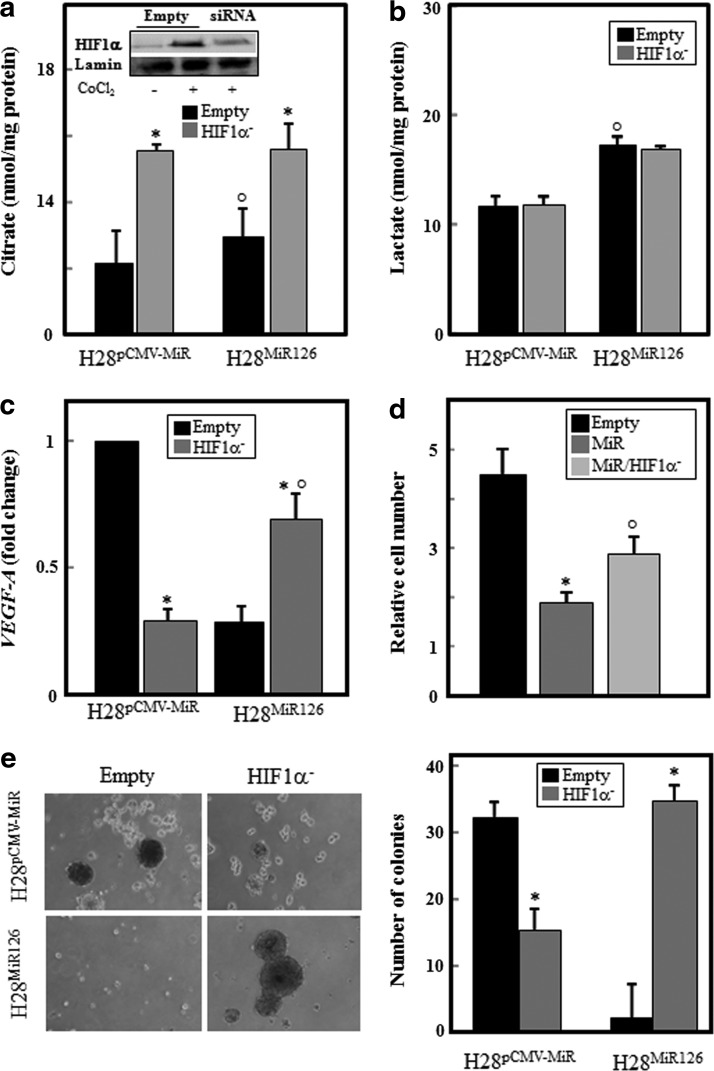
It is established that HIF transactivates several enzymes that are involved in glucose uptake and glycolysis (32). In addition, HIF1α exerts an anti-proliferative effect, and its expression in vitro leads to rapid death of human renal cell carcinoma (32). To gain insights into the role of HIF and the molecular mechanisms underlying the tumor suppression by MiR126, we knocked down HIF1α in MiR126- and empty plasmid-transfected H28 cells, and evaluated the production of citrate and lactate, VEGF-A expression, and cell growth. Silencing HIF1α (Fig. 9a, insert) induced citrate production in both MiR126- and empty plasmid-transfected H28 cells (Fig. 9a) without affecting the lactate levels (Fig. 9b). As expected, HIF1α knock-down considerably reduced VEGF-A gene expression. Likewise, MiR126 overexpression significantly reduced VEGF-A level, which was restored by silencing HIF1α (Fig. 9c). The increased VEGF-A level observed in HIF1α-silenced MiR126-transfected cells leads to increased proliferation of the cells and their ability to form colonies in soft agar (Fig. 9d, e).
FIG. 9.
Silencing of HIF1α restores MiR-126-induced MM cell growth suppression. MiR126- and empty plasmid-transfected H28 cells were silenced for HIF1α (HIF1α−, insert) and citrate/lactate levels (a, b), VEGF-A expression (c), cell growth (d), and soft agar colony forming (e) were evaluated. The results are mean values±S.D. of three experiments performed in duplicate; images are representative of three independent experiments. Comparisons among groups were determined by one-way ANOVA with Tukey post-hoc analysis; the symbol “*” indicates significant differences between silenced and nonsilenced (empty) genes, and the symbol “°” indicates significance between MiR126- and empty plasmid-transfected cells with p<0.05.
Discussion
Although MiR126 has been found down-regulated in different types of cancer, the detailed role of its deregulation in oncogenic transformation remains to be elucidated. Specific MiRs can act as tumor suppressors or oncogenes, depending on the cellular environment (53). Accordingly, it has been reported that MiRs modulate the responses of tissues to physiological and pathological stress (36). In this study, we investigated the role of MiR126 in stress signaling involved in MM development and progression. We have identified a novel link between MiR126, mitochondrial metabolism, and tumor suppression. MiR126 level was induced by oxidative stress in malignant and nonmalignant mesothelioma cells. MiR126 expression increased rapidly after exposure to oxidative insult, with a second rise at prolonged times of exposure. Two oxidation phases have been previously described. A first, an increase of ROS levels was observed in cardiomyocytes after exposure to angiotensin II-induced ROS (phase 1 oxidation), which was followed by phase 2 oxidation, a result of impaired flux through the electron transport chain (46). This raises the hypothesis that the modulation of MiR126 expression by oxidative stress reflects the level of ROS production. Complete loss of mtDNA or exposure to a hypoxia mimetic induced ROS-dependent MiR126 expression in nonmalignant cells while inhibiting MiR126 expression in MM cells, pointing to different cell phenotypes. Conversely, the inhibition of mitochondrial complex I (CI) by rotenone suppressed MiR126 expression in both cell lines (cf Figs. 1 and 2).
A recent study revealed a complex pleiotropic response to rotenone that affects a variety of cellular events, including the modulation of DNA damage response (DDR)-associated genes and the up-regulation of epigenetic regulatory mechanisms. Rotenone induced a DDR repair pathways involving both ROS-dependent and -independent mechanisms (9). In response to damaged chromatin, DNA methyltransferase 1 (DNMT1) was recruited at the site of damage (43); it was reported that methylation of MiR-199a and MiR-125b promoter genes by ROS-mediated DNMT1 was involved in their inhibition (27). Since MiR126 suppression in cancers has been linked to aberrant DNA hypermethylation of its host gene EGFL7 (51), we can postulate that DDR induced by rotenone may inhibit MiR126 expression by DNMT-mediated DNA hypermethylation.
Since MiR126 expression is regulated by mitochondria-destabilizing stimuli (cf Fig. 1), we asked whether MiR126 is able to affect the mitochondrial function. Surprisingly, ectopic MiR126 efficiently reduced ROS generation induced by CI inhibition and hypoxia. In contrast to previous studies, the inactivation of mitochondrial activity (ρ0 cells) led to increased intracellular level of ROS in control and MiR126-transfected cells. It was reported that cells lacking the mitochondrial genome resulted in lower intracellular levels of both superoxide and hydrogen peroxide (28, 45). However, nonmitochondrial sources may also contribute to ROS production, with an example being NADPH oxidases. These enzymes generate superoxide by transferring electrons from NADPH inside the cell across the membrane and coupling them to molecular oxygen to produce superoxide. The increased ROS formation in H28MiR126 ρ0cells was associated with increased MRA and enhanced ATP production (cf Figs. 2 and 4). The mitochondrial dysfunction stimulated 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) reduction that occurs inside the cells mainly by the intracellular FMNH2, FADH2, and NADH/NADPH (7). These intracellular reductants are produced by aerobic glycolysis, and their consumption in the mitochondrial matrix is a consequence of altered homeostasis. It was reported that NADH accumulates in the mitochondrial matrix of highly glycolytic cells in which OXPHOS is dormant (39). This indicates that the increased MRA found in cells with dysfunctional mitochondria could be the result of NADH/NADPH produced via the mitochondrial TCA cycle as a consequence of impaired OXPHOS and/or increased glycolysis.
Constitutively redox-active mitochondria were found in Met5A and H28 cells. However, H28 cells showed higher respiration associated with the glycolytic phenotype compared with Met5A cells. MiR126 suppressed the respiratory activity in MM cells and stimulated glycolysis in response to the inhibition of mitochondrial oxygen consumption by rotenone, indicating a compensatory process. H28MiR126 cells shifted to a more glycolytic phenotype, resulting in the generation of ATP largely via glycolysis, as documented earlier (40). The process is coupled to the glycolytically derived pyruvate that enters a truncated TCA cycle, where citrate is preferentially exported to the cytosol via the tricarboxylate transporter (3, 31). Once in the cytosol, citrate is cleaved by ACL and produces cytosolic acetyl-CoA (AcCoA) that is used for endogenous synthesis of fatty acids, cholesterol, and isoprenoids, as well as for acetylation of proteins. To complete the substrate cycle, ACL-generated oxaloacetate is reduced to malate, which enters mitochondria, recycling carbon units and generating reducing equivalents (65). The coupled conversion of NAD+ to NADH provides a mechanism to preserve the mitochondrial membrane potential and sustain high mitochondrial NADH/NAD+ ratio that maintains the TCA cycle in a repressed state. In our study, we found that MiR126 reduced mitochondrial respiration in MM cells and induced the MRA, which was considerably suppressed by glycolysis inhibition, suggesting that MiR126 modulates the malignant phenotype with maximum ATP synthesis through the TCA cycle coupled to mitochondria (cf Figs. 3 and 4).
The activation of Akt has been shown to alter cellular metabolism and promote the flow of precursors into anabolic pathways (6, 25). Akt activates ACL, promoting the conversion of mitochondria-derived citrate to AcCoA for lipid synthesis. Therefore, the re-programming of mitochondrial citrate metabolism is a central aspect of the PI3K/Akt activity (69). Here, we show that MiR126 repressed Akt activation as a down-stream effector of the adaptor protein IRS1 (cf Fig. 5), a direct target of MiR126 (5, 49, 67). Based on these notions, we can postulate that MiR126 may affect the mitochondrial citrate metabolism by inhibiting the Akt pathway to restore the TCA cycle for the synthesis of ATP via OXPHOS. Indeed, low citrate levels were found in MM, suggesting that these cells use glutamine metabolism via reductive carboxylation (RC) to support efficient carbon utilization for anabolic pathways and growth. Ectopic expression of MiR126 elevated citrate levels by inhibiting ACL (cf Fig. 6). Thus, this mechanism favors glucose oxidation and produces cellular energy rather than converts it to other macromolecules for cellular biosynthesis, as previously reported (20). As a result, a loss of malignancy ensues, featuring the acquisition of a nonmalignant-like cell morphology, lack of anchorage-independent growth, and suppression of tumor initiation and promotion, all results of an increased level of MiR126 (cf Fig. 7 and Supplementary Fig. S4).
A downstream target of Akt is FoxO1, identified as functionally inactive due to its phosphorylation by Akt in a variety of cancers. FoxO1 inactivation favors enhanced cell survival, cell proliferation, and susceptibility to stress, while its activation leads to apoptosis, cell cycle arrest, and stress resistance in various tissues (69). Recent evidence documents that IRS1 is a strong inhibitor of FoxO1 via its Akt-mediated phosphorylation (14, 24). Ectopic MiR126 was found to re-activate FoxO1 via the inhibition of the IRS1/Akt pathway (cf Fig. 5). MiR126 induced nuclear translocation of FoxO1 in Met5A and H28 cells, resulting in the cell cycle blockage and increased expression of genes involved in the glucose metabolism and the mitochondrial function (69). We observed that due to active FoxO1, H28MiR126 cells showed increased expression of genes involved in gluconeogenesis (cf Fig. 6 and Supplementary Fig. S4), which needs to be operational for fast release and utilization of glucose for energy production (14). In MM cells, MiR126 induced the expression of PCK1, which is the main control point for the regulation of gluconeogenesis. Further, MiR126-mediated FoxO1 activation induced the expression of the antioxidant enzymes MnSOD and CAT (cf Fig. 6 and Supplementary Fig. S4), which is consistent with the literature (33, 41). A relationship between MiR126 and the level of expression of the genes it modulates was found in MM patient biopsies (cf Supplementary Fig. S2), further supporting the regulatory role of MiR126.
It is likely that the over-expression of MnSOD and CAT contributes to the reduction of mitochondria-derived ROS induced by mitochondria-destabilizing agents (cf Fig. 2). Enhanced ROS production in cancer drives the onset of aerobic glycolysis, with lactate and ketone production promoting mitochondrial biogenesis and anabolic processes in tumor cells. The alleviation of mitochondrial oxidative stress via enhanced expression of CAT targeted to mitochondria was found to be sufficient to lower tumor severity and to considerably reduce the metastatic tumor burden (55). MnSOD modulates the cellular redox environment and suppresses hypoxia-induced accumulation of the HIF1α protein in MCF7 breast cancer cells. The induction of VEGF due to hypoxia was also suppressed by elevated MnSOD activity, which was paralleled by the HIF1α protein levels (62).
HIF1α plays a key role in the re-programming of cancer metabolism by activating the transcription of genes encoding glucose transporters and glycolytic enzymes, which convert glucose to lactate (32). Several miRNAs that mediate metabolism re-programming can contribute to HIF1α expression and stabilization, including MiR126 (21). The stability and transcriptional activity of HIF1α is regulated by prolyl 4-hydroxylases (PHDs), and certain TCA cycle intermediates and related compounds have recently been reported to inhibit the activity of PHDs (54). Here, we found that the restored citrate levels, induced by ACL inhibition, were linked to HIF1α activation and were not affected by FoxO1 (cf Fig. 8). HIF1α nuclear translocation induced VEGF-A transcription, which resulted in increased cell proliferation. As a consequence of ACL inhibition, higher citrate levels were found in MiR126-transfected cells with regard to their parental cells. The silencing of ACL further increased intracellular citrate, which was associated with HIF1α nuclear translocation, but not with increased VEGF-A gene expression. Although HIF1α affects tumor progression by directly regulating angiogenic target genes, recent evidence indicates that HIF1α also regulates tumor progression by exerting distinct, often opposing effects on critical oncoproteins and tumor suppressors, including MYC, p53, and mTOR (32).
It has been reported that VEGF-A is a target gene of MiR126 (we observed a low level of VEGF-A in H28MiR126 cells; cf Fig. 9C), and down-regulation of MiR126 increases VEGF-A expression in cancer (70). However, other reports indicated that MiR126 is an inducer of angiogenesis by enhancing the pro-angiogenic activity of VEGF-A (19, 63). MiR126 may, therefore, function differently in tumor and stromal cells. Knocking down HIF1α reduced VEGF-A in empty plasmid-transfected MM cells, while restoring VEGF-A levels in MiR126 MM cells. Increased VEGF-A gene expression resulted in enhanced MM cell proliferation and tumorigenesis, supporting the hypothesis that HIF is necessary for MiR126-induced MM cell growth suppression. It is well established that HIF orchestrates and up-regulates several enzymes, ensuring the diversion of pyruvate to lactate production. Indeed, HIF regulates citrate levels, and its increased levels strongly reduce intracellular citrate levels by modulating the reductive flux (20). According to these notions, increased citrate levels were found in HIF1α-silenced MiR126- and empty-plasmid transfected MM cells, and the silencing of HIF1α did not affect lactate levels. Conversely, the inhibition of FoxO1 partially contributed to the reduction of lactate production induced by ACL silencing (cf Fig. 8a), suggesting its involvement in the glycolytic shift.
In this study, we found that the suppression of IRS1 through a direct interaction of MiR126 with the consensus sequences in the 3′-UTR of the IRS1 mRNA is fundamental for tumor inhibition. The suppression of IRS1 as a result of the activity of MiR126 appears pivotal for the regulation of malignancy of MM cells. This is supported by the reporter gene assay showing that MiR126 directly regulates the expression of IRS1 in Met5A and H28 cells. In addition, another MM cell line, H2452, behaves similarly to H28 cells, with strong suppression of IRS1 on MiR126 transfection. On the other hand, Mes-1 cells did not change their malignant status when transfected with MiR126, which is most likely due to the lack of the effect of the MiR on IRS1. This may be reconciled with a previous report documenting that cancer cells may maintain their growth by expressing IRS1 lacking the 3′-UTR (35). This hypothesis was confirmed by using an MM cell line (Ist-Mes2) with truncated IRS1 lacking the MiR126-binding site in the 3′-UTR of IRS1. The transfection of these MM cells with MiR126 did not alter cell growth and tumorigeneis (cf Supplementary Fig. S4e).
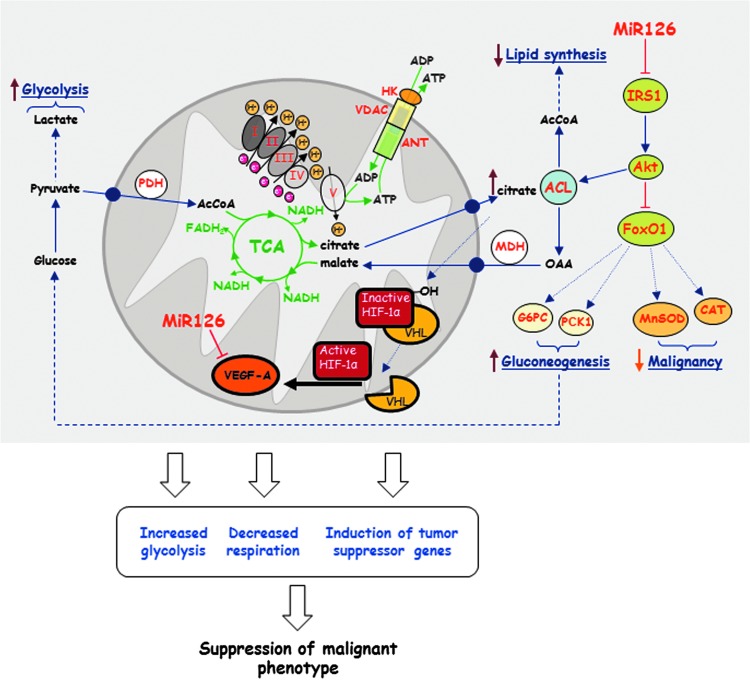
Taken together, we show that MiR126 induces pleiotropic effects in MM cells via the IRS1-Akt axis, branching to FoxO1 and ACL (Fig. 10). The scheme indicates a novel link between MiR126-dependent suppression of IRS1 and the resulting metabolic re-programming that includes the inhibition of RC by ACL and citrate level modulation, with ensuing lack of efficient lipid synthesis, promotion of the TCA cycle, enhanced gluconeogenesis, a glycolytic shift, and enhanced oxidative stress defense. Collectively, these changes, involving HIF1α activation (20), result in the acquisition of a less malignant phenotype that is documented by the loss of the propensity of H28MiR126 cells to form tumors. These data strongly support the therapeutic use of MiRs with the goal to improve the disease response to therapy. Targeting mitochondria as an invariant target by MiRs, as epitomized by MiR126 in MM, is accentuated by recent findings of profound differences in intra- and inter-tumoral mutations, complicating the established therapies (22, 26). A key role of MiR126 in the control of mesothelioma is corroborated by the findings that it is suppressed in MM patients (52, 60).
FIG. 10.
Molecular mechanism of tumor suppressor activity of MiR126. MiR126 suppresses IRS1 by binding to its 3′-UTR, with ensuing inhibition of Akt. This causes the failure of inhibition of FoxO1, which results in the increase in the oxidant defense genes MnSOD and CAT as well as the gluconeogenesis genes G6PC and PCK1 yielding increased glucose accompanied by a glycolytic shift. The TCA cycle is promoted by a lower level and activity of ACL, a result of decreased activity of Akt, and increased activity of FoxO1. The Akt-mediated ACL down-regulation restored citrate levels, which induce the activation and stabilization of HIF1α. HIF1α nuclear translocation is involved in tumor suppression by lowering VEGF-A expression. Collectively, these effects of MiR126 cause a switch of the MM cells to a nonmalignant phenotype. OAA, oxaloacetic acid. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
Materials and Methods
Cell culture and induction of mitochondrial dysfunction
Nonmalignant mesothelial cells (Met5A), sarcomatoid MM cells (H28), and epithelioid MM cells (H2452) were obtained from the ATCC. Immortalized and human primary MM cells (Ist-Mes2 and Mes-1, both epithelioid) were established from a patient and were identified morphologically by a phenotypic analysis (47). All cell lines were grown in the RPMI-1640 medium with antibiotics and 10% FBS. Oxidative stress was induced at different time points by the addition of hydrogen peroxide (200 μM). Mitochondrial dysfunction was induced by the CI inhibitor rotenone (20 μM, 5 or 24 h, mtDNA depletion) (2-week incubation with 0.2 μg/ml of EtBr and 50 μg/ml uridine, resulting in >90% mtDNA depletion) and hypoxia (CoCl2, 100 μM, 5 h). The general ROS scavenger NAC and the superoxide scavenger Tiron (both Sigma) were added at 10 mM 2 h before treatment.
Establishment of stable cell lines expressing MiR126 and siRNA transfection
Cells (2×105) were stably transfected with 1 μg of the pCMV-MiR and/or pRS (OriGene) empty plasmid or with the plasmid carrying the MiR126 sequence 5′-UCG UAC CGU GAG UAA UAA UGC G-3′ (OriGene) and/or shRNA pRS plasmid, with the ACL targeting sequence of CCA TCA CTG AGG TCT TCA AGG AAG AGA TG and the HIF1α targeting sequence of ACA AGA ACC TAC TGC TAA TGC CAC CAC TA using the TransIT-LT1 reagent (Mirus). Selection was carried out with G418 (Sigma) and/or with puromycin (Sigma) added to the cell culture media after transfection at 0.6 mg/ml and 1 μg/ml for 48 h, respectively. G418- and/or puromycin-resistant clones were analyzed for MiR126, ACL, and HIF1α expression. Selected clones were maintained in RPMI with 0.6 mg/ml G418 and/or 1 μg/ml puromycin.
Anti-IRS1 sequence 5′-AGA CCA UCA GCU UCG UGA ATT-3′ (Ambion) and antiFoxO1 sequences 5′-AGC AGU AAA UCA AUG GAA-3′ and 5′-UGG GAU GUU CCA UUG AUU-3′ or scrambled control were used to transiently silence the IRS1 and FoxO1 genes. MiR126 function was blocked with the anti-sense oligonucleotide 5′-GCA UUA UUA CUC ACG GUA CGA-3′ (IDT, Tema Ricerca). Cells (2×105) were transfected with the oligonucleotide (1 μg/well) using TransIT-LT1. After 48 h, the levels of IRS1 and FoxO1 mRNA were evaluated by quantitative RT-PCR (qPCR) and/or western blot analysis.
MTT, cell proliferation, and cell cycle assays
Cells were incubated with 10 μM MTT (5 mg/ml in phosphate-buffered saline [PBS]) at 37°C for 3 h. After removing the media, 200 μl of isopropanol was added to dissolve the crystals. Absorbance was read at 550 nm in an ELISA plate reader (Tecan). For long-term studies, cells were evaluated by the MTT assay at 24, 48, and 72 h. Proliferation was assessed in permeabilized cells using the anti-Ki67 IgG and confocal microscopy (Zeiss; Axiocam MRc5, magnification 60×). The proliferation index was expressed as a percentage of Ki67-positive cells. Cell cycle analysis was performed using the standard propidium iodide assay and flow cytometry (FACS Calibur; Becton Dickinson).
Soft agar colony formation and PDL assay
Cells were seeded in 0.7% low melting point agar in 24-well plates, overlaid with 0.35% low melting point agar, and cultured at 37°C in 5% CO2 for 1 month. Every 7 days, 0.5 ml of fresh medium was added to each well. Colonies containing >50 cells were counted. For the PDL assay, cells were cultured for 6 weeks and counted every week. The PDL value was calculated as log2(D/D0), where D and D0 are cell numbers at the time of harvesting and seeding, respectively (2).
Mitochondrial-reducing activity
The MRA was assessed in live cells as the reduction of MTT or resazurin, with both being based on the mitochondrial metabolic activity (1, 2, 61). The MTT assay was performed as described earlier, and the values were normalized to viable cells evaluated by crystal violet assay (2% crystal violet in 2% ethanol). For the resazurin assay, cells were incubated with resazurin (6 μM) in the presence and absence of rotenone (20 μM, 5 h) and 2-deoxy-glucose (2DG, 5 mM, 5 h). Fluorescence intensity was read at 0–240 min, and the results were normalized to the total protein using the BCA assay (Pierce Biotechnology).
Assessment of ROS/NO generation and mitochondrial membrane potential (ΔΨm,i)
Intracellular superoxide anion and hydrogen peroxide levels were estimated using dihydroethidinium (DHE) and 2′,7′-dichlorofluorescein diacetate (DCF), respectively. Treated and untreated empty plasmid and MiR126-transfected cells were incubated with 20 μM DHE or DCF for 30 min and analyzed by flow cytometry. The level of ROS was expressed as mean fluorescence intensity. NO levels were evaluated in treated and untreated empty plasmid- and MiR126-transfected cells using a kit according to the manufacturer's protocol (Sigma). For ΔΨm,i assessment, cells were incubated with 200 nM tetramethyl rhodamine methyl ester (TMRM; Invitrogen) for 30 min at 37°C and analyzed by flow cytometry for red fluorescence intensity.
Evaluation of lactate, intracellular ATP, citrate, and mitochondrial respiration
Lactate was evaluated using a colorimetric kit (Abcam) according to the manufacturer's protocol. ATP was determined using a luciferase-based assay (CellTiter-Glo® Luminescent Assay; Promega). Citrate was evaluated using a colorimetric kit (Sigma) according to the manufacturer's protocol. The results were normalized to the total protein. Mitochondrial respiration was evaluated using the Oxygraph instrument (Oroboros) according to the manufacturer's instructions. The results were related to the number of cells.
Quantitative RT-PCR analysis
Total RNA from cells and the formalin-fixed, paraffin-embedded (FFPE) tissue samples (10 μg) were obtained using the RNeasy Mini Kit (Qiagen) and the RecoverAll total nucleic acid isolation kit (Ambion), respectively, according to the manufacturer's instructions. The MiR126 first-strand cDNA was synthesized using TaqMan MicroRNA reverse transcription kit (Applied Biosystems). Quantitative RT-PCR (qPCR) was performed using the TaqMan MicroRNA assay (Applied Biosystems) with U6 as the housekeeping gene. The qPCR analysis of mRNA was performed using Prime Time qPCR assay (IDT) and TaqMan gene expression master mix (Applied Biosystems) for IRS1, FoxO1, ACL, MnSOD, CAT, G6PC, PCK1, and VEGF-A. GAPDH was used as the housekeeping gene. The qPCR assays were performed using the Mastercycler EP Realplex (Eppendorf). The results were expressed as ΔCt, and fold changes in relative mRNA expression were calculated using the equation 2−Δ(ΔCt).
Luciferase assay
DNA fragments of IRS1 3′-UTR containing predicted MiR126 binding site (IRS1-3′Uwt) were cloned into the pGL3-promoter plasmid (Promega), and the MiR126 binding sites were replaced with a 4 nt fragment to produce mutated 3′-UTR pGL3 report plasmid (IRS1-3′Umut) as described (49). Briefly, Met5A and H28 cells were plated onto 12-well plates and grown to a 70% confluence. The cells were co-transfected with IRS1-3Uwt or 3Umut and MiR126 expression plasmid along with pRL-SV40 for constitutive expression of Renilla luciferase as an internal control. At 48 h post-transfection, the cells were lysed and the Renilla luciferase activity was assessed by the TECAN Infiniti reader. The results were expressed as firefly luciferase activity to Renilla luciferase activity ratio.
mtDNA copy evaluation
DNA was extracted from empty plasmid- and MiR126-transfected cells using the AquaPure Genomic DNA isolation kit (BioRad). The amount of mtDNA relative to nuclear DNA was determined by qPCR using primers for mitochondrial genes MTRT1, MRT2, MRT3, and MRT4 and nuclear genes MTAIB, MTB2M, and MTBA. The primers are listed in Supplementary Figure S5.
Subcellular fractionation and Western blot analysis
Cells were lysed in the RIPA buffer containing Na3VO4 (1 mM) and protease inhibitors (1 μg/ml). For preparation of the cytoplasmic and nuclear fractions, the cells were washed twice with ice-cold PBS and re-suspended in the cytoplasmic extract (CE) buffer containing 10 mM HEPES, 60 mM KCl, 0.075% NP40, and 1 mM DTT, pH 7.6, and supplemented with protease inhibitors. After 20 strokes in a Dounce homogenizer, the unbroken cells were spun down at 2500 g for 10 min at 4°C, and the supernatant cytoplasmic fraction was collected. The nuclear fraction was washed with the CE buffer and re-suspended in ice-cold hypertonic buffer (20 mM Tris-HCl, 420 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 1 mM PMSF, and 25% glycerol, pH 8.0), mixed, and incubated on ice for 10 min. After centrifugation at 15,000 g for 10 min at 4°C, the supernatant was collected.
The cell lysate proteins were separated using SDS-PAGE and transferred onto nitrocellulose membranes (Protran). After blocking with 5% nonfat milk in PBS-Tween (0.1%), the membranes were incubated with anti-IRS1, anti-HIF1α (both Bethyl), anti-p(S307)-IRS1, anti-Akt, anti-pAkt, anti-FoxO1, anti-p(T24)-FoxO1 (all Cell Signaling), TIM23, VDAC1, SDHA (all Santa Cruz), anti-β-actin, and lamin IgG (both Bethyl). After incubation with the HRP-conjugated secondary IgG (Sigma), blots were developed using the ECL detection system (Pierce Biotechnology). The band intensities were visualized and quantified with ChemiDoc using the Quantity One software (BioRad Laboratories).
HIF1α DNA-binding assay
The binding of HIF1α to DNA was quantified using the TransAM kit (Active Motif) according to the manufacturer's protocol.
Tumor xenograft model
Nude mice (6–7 per group) were injected subcutaneously with H28pCMV-MiR, H28MiR126, Met5ApCMV-MiR, and Met5AMiR126 cells (5×106 cells per mouse in 100 μl of sterile saline). Once tumors appeared, they were visualized and evaluated using the Vevo770 (VisualSonics) USI instrument that was fitted with a 30-μm resolution scan-head as reported (12, 13, 64). Animal studies were performed according to the guidelines of the Australian and New Zealand Council for the Care and Use of Animals in Research and Teaching and were approved by the local Animal Ethics Committee.
Human MM biopsies
FFPE tissue of the subjects affected by MM (n=11) was collected from the Archive of the Pathological Anatomy Unit of the Hospital University of Ancona, Italy. The FFPE samples were cut into 5 μm sections and stored at room temperature until analysis. The adjacent noncancerous tissue was used as normal mesothelium. The patients were not treated with any adjuvant chemotherapy or radiation therapy. The sample collection was carried out according to the Helsinki Declaration, and the samples were processed under the approval of the written consent statement by the ethics committee of the University Hospital of Marche, Italy.
Statistics
All results are expressed as mean±S.D. Comparisons among groups of data were made using one-way analysis of variance with Tukey post-hoc analysis. The two-tailed Student's t-test was used to compare two groups. The associations between MiR126 and gene expression were evaluated by the Spearman's rank correlation test. Differences with p<0.05 were considered statistically significant. All data generated in this study were analyzed using the SPSS software.
Supplementary Material
Abbreviations Used
- 2DG
2-deoxyglucose
- AcCoA
acetyl-CoA
- ACL
ATP-citrate lyase
- CAT
catalase
- CI
complex I
- DCF
2′,7′-dichlorofluorescein diacetate
- DHE
dihydroethidinium
- DNMT1
DNA methyltransferase 1
- ETS
capacity of the electron transfer system
- FFPE
formalin-fixed, paraffin-embedded
- FoxO1
Forkhead box O1
- G6PC
glucose-6-phosphatase catalytic
- HIF1α
hypoxia-inducible factor-1α
- IRS1
insulin receptor substrate-1
- MFI
mean fluorescence intensity
- MiR
microRNA
- MM
malignant mesothelioma
- MnSOD
manganese superoxide dismutase
- MRA
mitochondrial reducing activity
- mtDNA
mitochondrial DNA
- MTT
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- NAC
N-acetyl cysteine
- nDNA
nuclear DNA
- OAA
oxaloacetic acid
- OXPHOS
oxidative phosphorylation
- PCK1
phosphoenolpyruvate carboxykinase 1
- PDL
population doubling level
- qPCR
quantitative RT-PCR
- RC
reductive carboxylation
- ROS
reactive oxygen species
- siRNA
small interfering RNA
- USI
ultrasound imaging
- VEGF-A
vascular endothelial growth factor-A
Acknowledgments
This work was supported by grants from the National Institute against Occupational Injury Insurance to L.S. and M.T., the National Health and Medical Research Council of Australia, the Cancer Council Queensland and Czech Science Foundation (P301/10/1937) to J.N., the Australian Research Council to J.N. and L.F.D., the Czech Science Foundation (P305/12/1708) to J.T., and the Griffith University Fellowship to L.-F.D. M.T. was supported in part by the Apoptosis Research Group, Griffith University, Southport, Queensland, Australia. The project was also supported by the BIOCEV European Regional Development Fund CZ.1.05/1.1.00/02.0109.
Author Disclosure Statement
The authors declare no conflicts of interest.
References
- 1.Abu-Amero KK. and Bosley TM. Detection of mitochondrial respiratory dysfunction in circulating lymphocytes using resazurin. Arch Pathol Lab Med 129: 1295–1298, 2005 [DOI] [PubMed] [Google Scholar]
- 2.Atamna H, Nguyen A, Schultz C, Boyle K, Newberry J, Kato H, and Ames BN. Methylene blue delays cellular senescence and enhances key mitochondrial biochemical pathways. FASEB J 22: 703–712, 2008 [DOI] [PubMed] [Google Scholar]
- 3.Baggetto LG. Deviant energetic metabolism of glycolytic cancer cells. Biochimie 74: 959–974, 1992 [DOI] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297, 2004 [DOI] [PubMed] [Google Scholar]
- 5.Baserga R. The contradictions of the insulin-like growth factor 1 receptor. Oncogene 19: 5574–5581, 2000 [DOI] [PubMed] [Google Scholar]
- 6.Bauer DE, Hatzivassiliou G, Zhao F, Andreadis C, and Thompson CB. ATP citrate lyase is an important component of cell growth and transformation. Oncogene 24: 6314–6322, 2005 [DOI] [PubMed] [Google Scholar]
- 7.Berridge MV, Herst PM, and Tan AS. Tetrazolium dyes as tools in cell biology: new insights into their cellular reduction. Biotechnol Annu Rev 11: 127–152, 2005 [DOI] [PubMed] [Google Scholar]
- 8.Busacca S, Germano S, De Cecco L, Rinaldi M, Comoglio F, Favero F, Murer B, Mutti L, Pierotti M, and Gaudino G. MicroRNA signature of malignant mesothelioma with potential diagnostic and prognostic implications. Am J Respir Cell Mol Biol 42: 312–319, 2010 [DOI] [PubMed] [Google Scholar]
- 9.Cabeza-Arvelaiz Y. and Schiestl RH. Transcriptome analysis of a rotenone model of parkinsonism reveals complex I-tied and -untied toxicity mechanisms common to neurodegenerative diseases. PLoS One 7: e44700, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Calin GA. and Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer 6: 857–866, 2006 [DOI] [PubMed] [Google Scholar]
- 11.Croce CM. and Calin GA. miRNAs, cancer, and stem cell division. Cell 122: 6–7, 2005 [DOI] [PubMed] [Google Scholar]
- 12.Dong LF, Freeman R, Liu J, Zobalova R, Marin-Hernandez A, Stantic M, Rohlena J, Valis K, Rodriguez-Enriquez S, Butcher B, Goodwin J, Brunk UT, Witting PK, Moreno-Sanchez R, Scheffler IE, Ralph SJ, and Neuzil J. Suppression of tumour growth in vivo by the mitocan α-tocopheryl succinate requires respiratory complex II. Clin Cancer Res 15: 1593–1600, 2009 [DOI] [PubMed] [Google Scholar]
- 13.Dong LF, Swettenham E, Eliasson J, Wang XF, Gold M, Medunic Y, Stantic M, Low P, Prochazka L, Witting PK, Turanek J, Akporiaye ET, Ralph SJ, and Neuzil J. Vitamin E analogs inhibit angiogenesis by selective apoptosis induction in proliferating endothelial cells: the role of oxidative stress. Cancer Res 67: 11906–11913, 2007 [DOI] [PubMed] [Google Scholar]
- 14.Dong XC, Copps KD, Guo S, Li Y, Kollipara R, DePinho RA, and White MF. Inactivation of hepatic Foxo1 by insulin signaling is required for adaptive nutrient homeostasis and endocrine growth regulation. Cell Metab 8: 65–76, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Farh KK, Grimson A, Jan C, Lewis BP, Johnston WK, Lim LP, Burge CB, and Bartel DP. The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science 310: 1817–1821, 2005 [DOI] [PubMed] [Google Scholar]
- 16.Feng R, Chen X, Yu Y, Su L, Yu B, Li J, Cai Q, Yan M, Liu B, and Zhu Z. miR-126 functions as a tumour suppressor in human gastric cancer. Cancer Lett 298: 50–63, 2010 [DOI] [PubMed] [Google Scholar]
- 17.Fennell DA, Gaudino G, O'Byrne KJ, Mutti L, and van Meerbeeck J. Advances in the systemic therapy of malignant pleural mesothelioma. Nat Clin Pract Oncol 5: 136–147, 2008 [DOI] [PubMed] [Google Scholar]
- 18.Fiaschi T, Marini A, Giannoni E, Taddei ML, Gandellini P, De Donatis A, Lanciotti M, Serni S, Cirri P, and Chiarugi P. Reciprocal metabolic reprogramming through lactate shuttle coordinately influences tumor-stroma interplay. Cancer Res 72: 5130–5140, 2012 [DOI] [PubMed] [Google Scholar]
- 19.Fish JE, Santoro MM, Morton SU, Yu S, Yeh RF, Wythe JD, Ivey KN, Bruneau BG, Stainier DY, and Srivastava D. miR-126 regulates angiogenic signaling and vascular integrity. Dev Cell 15:272–284, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gameiro PA, Yang J, Metelo AM, Pérez-Carro R, Baker R, Wang Z, Arreola A, Rathmell WK, Olumi A, López-Larrubia P, Stephanopoulos G, and Iliopoulos O. In vivo HIF-mediated reductive carboxylation is regulated by citrate levels and sensitizes VHL-deficient cells to glutamine deprivation. Cell Metab 17: 372–385, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao P, Sun L, He X, Cao Y, and Zhang H. MicroRNAs and the Warburg Effect: new players in an old arena. Curr Gene Ther 12: 285–291, 2012 [DOI] [PubMed] [Google Scholar]
- 22.Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A, Tarpey P, Varela I, Phillimore B, Begum S, McDonald NQ, Butler A, Jones D, Raine K, Latimer C, Santos CR, Nohadani M, Eklund AC, Spencer-Dene B, Clark G, Pickering L, Stamp G, Gore M, Szallasi Z, Downward J, Futreal PA, and Swanton C. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 366: 883–892, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo C, Sah JF, Beard L, Willson JK, Markowitz SD, and Guda K. The noncoding RNA, miR-126, suppresses the growth of neoplastic cells by targeting phosphatidylinositol 3-kinase signaling and is frequently lost in colon cancers. Genes Chromosomes Cancer 47: 939–946, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo S, Copps KD, Dong X, Park S, Cheng Z, Pocai A, Rossetti L, Sajan M, Farese RV, and White MF. The Irs1 branch of the insulin signaling cascade plays a dominant role in hepatic nutrient homeostasis. Mol Cell Biol 29: 5070–5083, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hatzivassiliou G, Zhao F, Bauer DE, Andreadis C, Shaw AN, Dhanak D, Hingorani SR, Tuveson DA, and Thompson CB. ATP citrate lyase inhibition can suppress tumor cell growth. Cancer Cell 8: 311–321, 2005 [DOI] [PubMed] [Google Scholar]
- 26.Hayden EC. Cancer complexity slows quest for cure. Nature 455: 158, 2008 [DOI] [PubMed] [Google Scholar]
- 27.He J, Xu Q, Jing Y, Agani F, Qian X, Carpenter R, Li Q, Wang XR, Peiper SS, Lu Z, Liu LZ, and Jiang BH. Reactive oxygen species regulate ERBB2 and ERBB3 expression via miR-199a/125b and DNA methylation. EMBO Rep 13: 1116–1122, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Higuchi M, Manna SK, Sasaki R, and Aggarwal BB. Regulation of the activation of nuclear factor κB by mitochondrial respiratory function: evidence for the reactive oxygen species-dependent and -independent pathways. Antioxid Redox Signal 4: 945–955, 2002 [DOI] [PubMed] [Google Scholar]
- 29.Hollevoet K, Reitsma JB, Creaney J, Grigoriu BD, Robinson BW, Scherpereel A, Cristaudo A, Pass HI, Nackaerts K, Rodríguez Portal JA, Schneider J, Muley T, Di Serio F, Baas P, Tomasetti M, Rai AJ, and van Meerbeeck JP. Serum mesothelin for diagnosing malignant pleural mesothelioma: an individual patient data meta-analysis. J Clin Oncol 30: 1541–1549, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ivanov SV, Goparaju CM, Lopez P, Zavadil J, Toren-Haritan G, Rosenwald S, Hoshen M, Chajut A, Cohen D, and Pass HI. Pro-tumorigenic effects of miR-31 loss in mesothelioma. J Biol Chem 285: 22809–22817, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaplan RS, Mayor JA, and Wood DO. The mitochondrial tricarboxylate transport protein. cDNA cloning, primary structure, and comparison with other mitochondrial transport proteins. J Biol Chem 268: 13682–13690, 1993 [PubMed] [Google Scholar]
- 32.Keith B, Johnson RS, and Simon MC. HIF1α and HIF2α: sibling rivalry in hypoxic tumour growth and progression. Nat Rev Cancer 12: 9–22, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kops GJ, Dansen TB, Polderman PE, Saarloos I, Wirtz KW, Coffer PJ, Huang TT, Bos JL, Medema RH, and Burgering BM. Forkhead transcription factor FOXO3a protects quiescent cells from oxidative stress. Nature 419: 316–321, 2002 [DOI] [PubMed] [Google Scholar]
- 34.Kubo T, Toyooka S, Tsukuda K, Sakaguchi M, Fukazawa T, Soh J, Asano H, Ueno T, Muraoka T, Yamamoto H, Nasu Y, Kishimoto T, Pass HI, Matsui H, Huh NH, and Miyoshi S. Epigenetic silencing of microRNA-34b/c plays an important role in the pathogenesis of malignant pleural mesothelioma. Clin Cancer Res 17: 4965–4974, 2011 [DOI] [PubMed] [Google Scholar]
- 35.La Rocca G, Badin M, Shi B, Xu SQ, Deangelis T, Sepp-Lorenzinoi L, and Baserga R. Mechanism of growth inhibition by MicroRNA 145: the role of the IGF-I receptor signaling pathway. J Cell Physiol 220: 485–491, 2009 [DOI] [PubMed] [Google Scholar]
- 36.Leung AK. and Sharp PA. MicroRNA functions in stress responses. Mol Cell 40: 205–215, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li P, Jiao J, Gao G, and Prabhakar BS. Control of mitochondrial activity by miRNAs. J Cell Biochem 113: 1104–1110, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Musiyenko A, Bitko V, and Barik S. Ectopic expression of miR-126*, an intronic product of the vascular endothelial EGF-like 7 gene, regulates protein translation and invasiveness of prostate cancer LNCaP cells. J Mol Med 86: 313–322, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nadege B, Patrick L, and Rodrigue R. Mitochondria: from bioenergetics to the metabolic regulation of carcinogenesis. Front Biosci 14: 4015–4034, 2009 [DOI] [PubMed] [Google Scholar]
- 40.Nakashima RA, Paggi MG, and Pedersen PL. Contributions of glycolysis and oxidative phosphorylation to adenosine 5′-triphosphate production in AS-30D hepatoma cells. Cancer Res 44: 5702–5706, 1984 [PubMed] [Google Scholar]
- 41.Nemoto S. and Finkel T. Redox regulation of forkhead proteins through a p66shc-dependent signaling pathway. Science 295: 2450–1452, 2002 [DOI] [PubMed] [Google Scholar]
- 42.Nocchi L, Tomasetti M, Amati M, Neuzil J, Santarelli L, and Saccucci S. Thrombomodulin is silenced in malignant mesothelioma by a poly(ADP-ribose)polymerase-1-mediated epigenetic mechanism. J Biol Chem 286: 19478–19488, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O'Hagan HM, Wang W, Sen S, Destefano Shields C, Lee SS, Zhang YW, Clements EG, Cai Y, Van Neste L, Easwaran H, Casero RA, Sears CL, and Baylin SB. Oxidative damage targets complexes containing DNA methyltransferases, SIRT1, and polycomb members to promoter CpG Islands. Cancer Cell 20: 606–619, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pass HI, Goparaju C, Ivanov S, Donington J, Carbone M, Hoshen M, Cohen D, Chajut A, Rosenwald S, Dan H, Benjamin S, and Aharonov R. hsa-miR-29c* is linked to the prognosis of malignant pleural mesothelioma. Cancer Res 70: 1916–1924, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rasmussen AK, Chatterjee A, Rasmussen LJ, and Singh KK. Mitochondria-mediated nuclear mutator phenotype in Saccharomyces cerevisiae. Nucleic Acids Res 31: 3909–3917, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ricci C, Pastukh V, Leonard J, Turrens J, Wilson G, Schaffer D, and Schaffer SW. Mitochondrial DNA damage triggers mitochondrial-superoxide generation and apoptosis. Am J Physiol Cell Physiol 294: C413–C422, 2008 [DOI] [PubMed] [Google Scholar]
- 47.Rippo MR, Moretti S, Vescovi S, Tomasetti M, Orecchia S, Amici G, Catalano A, and Procopio A. FLIP overexpression inhibits death receptor-induced apoptosis in malignant mesothelial cells. Oncogene 23: 7753–7760, 2004 [DOI] [PubMed] [Google Scholar]
- 48.Robinson BWS, Musk AW, and Lake AR. Malignant mesothelioma. Lancet 366: 397–408, 2005 [DOI] [PubMed] [Google Scholar]
- 49.Ryu HS, Park SY, Ma D, Zhang J, and Lee W. The induction of microRNA targeting IRS-1 is involved in the development of insulin resistance under conditions of mitochondrial dysfunction in hepatocytes. PLoS One 6: e17343, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sadagurski M, Cheng Z, Rozzo A, Palazzolo I, Kelley GR, Dong X, Krainc D, and White MF. IRS2 increases mitochondrial dysfunction and oxidative stress in a mouse model of Huntington disease. J Clin Invest 121: 4070–4081, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saito Y, Friedman JM, Chihara Y, Egger G, Chuang JC, and Liang G. Epigenetic therapy upregulates the tumor suppressor microRNA-126 and its host gene EGFL7 in human cancer cells. Biochem Biophys Res Commun 379: 726–731, 2009 [DOI] [PubMed] [Google Scholar]
- 52.Santarelli L, Strafella E, Staffolani S, Amati M, Emanuelli M, Sartini D, Pozzi V, Carbonari D, Bracci M, Pignotti E, Mazzanti P, Sabbatini A, Ranaldi R, Gasparini S, Neuzil J, and Tomasetti M. Association of MiR-126 with soluble mesothelin-related peptides, a marker for malignant mesothelioma. PLoS One 6: e18232, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schetter AJ, Okayama H, and Harris CC. The role of microRNAs in colorectal cancer. Cancer J 18: 244–252, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Selak MA, Armour SM, MacKenzie ED, Boulahbel H, Watson DG, Mansfield KD, Pan Y, Simon MC, Thompson CB, and Gottlieb E. Succinate links TCA cycle dysfunction to oncogenesis by inhibiting HIF-α prolyl hydroxylase. Cancer Cell 7: 77–85, 2005 [DOI] [PubMed] [Google Scholar]
- 55.Sotgia F, Martinez-Outschoorn UE, and Lisanti MP. Mitochondrial oxidative stress drives tumor progression and metastasis: should we use antioxidants as a key component of cancer treatment and prevention? BMC Med 9: 62, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sotgia F, Whitaker-Menezes D, Martinez-Outschoorn UE, Salem AF, Tsirigos A, Lamb R, Sneddon S, Hulit J, Howell A, and Lisanti MP. Mitochondria “fuel” breast cancer metabolism: fifteen markers of mitochondrial biogenesis label epithelial cancer cells, but are excluded from adjacent stromal cells. Cell Cycle 11: 4390–4401, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stapelberg M, Gellert N, Swettenham E, Tomasetti M, Witting PK, Procopio A, and Neuzil J. α-Tocopheryl succinate inhibits malignant mesothelioma by disrupting the FGF autocrine loop: the role of oxidative stress. J Biol Chem 280: 25369–25376, 2005 [DOI] [PubMed] [Google Scholar]
- 58.Tavazoie SF, Alarcón C, Oskarsson T, Padua D, Wang Q, Bos PD, Gerald WL, and Massagué J. Endogenous human microRNAs that suppress breast cancer metastasis. Nature 451: 147–152, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tomasetti M, Gellert N, Procopio A, and Neuzil J. A vitamin E analogue suppresses malignant mesothelioma in a pre-clinical model: a prototype of a future drug against a fatal neoplastic disease? Int J Cancer 109: 641–642, 2004 [DOI] [PubMed] [Google Scholar]
- 60.Tomasetti M, Staffolani S, Nocchi L, Neuzil J, Strafella E, Manzella N, Mariotti L, Bracci M, Valentino M, Amati M, and Santarelli L. Clinical significance of circulating miR-126 quantification in malignant mesothelioma patients. Clin Biochem 45: 575–581, 2012 [DOI] [PubMed] [Google Scholar]
- 61.van de Loosdrecht AA, Beelen RH, Ossenkoppele GJ, Broekhoven MG, and Langenhuijsen MM. A tetrazolium-based colorimetric MTT assay to quantitate human monocyte mediated cytotoxicity against leukemic cells from cell lines and patients with acute myeloid leukemia. J Immunol Methods 174: 311–320, 1994 [DOI] [PubMed] [Google Scholar]
- 62.Wang M, Kirk JS, Venkataraman S, Domann FE, Zhang HJ, Schafer FQ, Flanagan SW, Weydert CJ, Spitz DR, Buettner GR, and Oberley LW. Manganese superoxide dismutase suppresses hypoxic induction of hypoxia-inducible factor-1α and vascular endothelial growth factor. Oncogene 24: 8154–8166, 2005 [DOI] [PubMed] [Google Scholar]
- 63.Wang X, Tang S, Le SY, Lu R, Rader JS, Meyers C, and Zheng ZM. Aberrant expression of oncogenic and tumor-suppressive microRNAs in cervical cancer is required for cancer cell growth. PLoS One 3: e2557, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang XF, Birringer M, Dong LF, Veprek P, Low P, Swettenham E, Stantic M, Yuan LH, Zobalova R, Wu K, Ledvina M, Ralph SJ, and Neuzil J. A peptide adduct of vitamin E succinate targets breast cancer cells with high erbB2 expression. Cancer Res 67: 3337–3344, 2007 [DOI] [PubMed] [Google Scholar]
- 65.Ward PS. and Thompson CB. Metabolic reprogramming: a cancer hallmark even Warburg did not anticipate. Cancer Cell 21: 297–308, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, Calin GA, Liu CG, Croce CM, and Harris CC. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell 9: 189–198, 2006 [DOI] [PubMed] [Google Scholar]
- 67.Zhang J, Du YY, Lin YF, Chen YT, Yang L, Wang HJ, and Ma D. The cell growth suppressor, mir-126, targets IRS-1. Biochem Biophys Res Commun 377: 136–140, 2008 [DOI] [PubMed] [Google Scholar]
- 68.Zhang W, Patil S, Chauhan B, Guo S, Powell DR, Le J, Klotsas A, Matika R, Xiao X, Franks R, Heidenreich KA, Sajan MP, Farese RV, Stolz DB, Tso P, and Koo SH. FoxO1 regulates multiple metabolic pathways in the liver: effects on gluconeogenic, glycolytic, and lipogenic gene expression. J Biol Chem 281: 10105–10117, 2006 [DOI] [PubMed] [Google Scholar]
- 69.Zhang X, Tang N, Hadden TJ, and Rishi AK. Akt, FoxO and regulation of apoptosis. Biochim Biophys Acta 1813: 1978–1986, 2011 [DOI] [PubMed] [Google Scholar]
- 70.Zhu N, Zhang D, Xie H, Zhou Z, Chen H, Hu T, Bai Y, Shen Y, Yuan W, Jing Q, and Qin Y. Endothelial-specific intron-derived miR-126 is down-regulated in human breast cancer and targets both VEGFA and PIK3R2. Mol Cell Biochem 351: 157–164, 2011 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.