Abstract
Purpose of the review
We examine the latest research on the emerging bile acid-gut microbiome axis and its role in health and disease. Our focus revolves around two key microbial pathways for degrading bile salts, and the impact of bile acid composition in the gut on the gut microbiome and host physiology.
Recent findings
Bile acid pool size has recently been shown to be a function of microbial metabolism of bile acids in the intestines. Recent studies have shown potential mechanisms explaining how perturbations in the microbiome affect bile acid pool size and composition. Bile acids are emerging as regulators of the gut microbiome at the highest taxomic levels. The role of bile acids as hormones and potentiators of liver cancer are also emerging.
Summary
The host and microbiome appear to regulate bile acid pool size. The host produces a large, conjugated hydrophilic bile acid pool, maintained through positive-feedback antagonism of FXR in intestine and liver. Members of the microbiome utilize bile acids and their conjugates resulting in agonism of FXR in intestine and liver resulting in a smaller, unconjugated hydrophobic bile acid pool. Hydrophilicity of the bile acid pool is associated with disease states. Reduced bile acid levels in the gut are associated with bacterial overgrowth and inflammation. Diet, antibiotic therapy, and disease states affect the balance of the microbiome-bile acid pool.
Keywords: Bile Salt Hydrolase, bile acid 7α-dehydroxylation, cirrhosis, microbiome
Introduction: The Human Gut Microbiome
The human gut harbors one of the most densely populated and complex ecosystems known. The colon contains approximately 2 to 5 × 1011 bacteria/gram wet weight feces and a total of several hundred grams of bacteria in the adult colon. The vast majority of bacteria colonizing the human gut are mostly obligate anaerobes with lesser numbers of facultative anaerobes, archaea, and yeast [1]. Because the lumen of the gastrointestinal tract is a highly anaerobic environment, microorganisms must carry out fermentative metabolism. The major substrates for growth of gut bacteria include 100-200 grams/wet weight/day of sloughed intestinal cells, plant polysaccharides, starch and cellulose as well as bile components. From these substrates, gut bacteria produce short chain fatty acids (acetate, propionate, butyrate) that can account for up to 10% of the total caloric intake/day [2]. Butyrate is an important energy source and regulatory molecule for colonocytes [3]. Gut bacteria also produce metabolites from the fermentation of amino acids (cresol, phenyacetate, indole) that can be toxic to the host, especially patients with severe liver disease.
In the past, studies of the gut microbiome identified microorganisms by culture-dependent methods along with fluorescence in situ hybridization (FISH), PCR, and gel-based techniques. However, in recent years, high throughput techniques including: 454 pyrosequencing of 16S rRNA genes have dramatically improved the ability to rapidly and cheaply determine the composition of the human gut microbiome [4]. The 16S rRNA gene was chosen because it is the most invariant gene in the bacterial genome and is considered the best phylogenetic marker for molecular taxonomy. Bacterial 16S rRNA genes sharing 97-99% identity is referred to as an operational taxonomic unit (OTU) and represents a “phylotype” [5]. The literature suggests that the human gut microbiota contains more than 1,000 phylotypes. These phylotypes are divided into six divisions/phyla including: Firmicutes, Bacteroidetes, Proteobacteria, Acinetobacteria, Fusobacteria and Verrucomicrobia [1]. Species in the Firmicutes and Bacteroidetes makes up more than 90% of the total gut microbiota [1]. The major genera of obligate anaerobes in the human gut microbiota include: Bacteroides, Bifidobacterium, Clostridium, Eubacterium, Fusobacterium, Peptococcus, Peptostreptococcus, and Rumminococcus. The genera of facultative anaerobic bacteria include: Escherichia, Enterobacter, Enterococcus, Klebsiella, Lactobacillus and Proteus.
Bile acids as Regulators of Gut Microbiome Community Structure
The composition of the human gut microbiota can be altered by diet, age, antibiotics and disease. Bile acids appear to be a major regulator of the gut microbiota. In this regard, a recent report by Kakiyama et al. suggests a connection between the liver health, fecal bile acid concentrations and gut microbiota composition [6]. In this study, levels of fecal bile acids and microbiome community structure as determined by 16S ribosomal gene quantification was compared to control patients and patients with early and advanced cirrhosis. As cirrhosis progressed it was observed that bacterial dysbiosis observed in cirrhosis [7,8] is linked to low bile acid levels entering the intestine [6]. This dysbiosis was characterized by significant reduction in gram-positive members of the normal microbiota such as Blautia, Rumminococcaceae, and [9]. Indeed, there were significant correlations between fecal secondary bile acids and these members of Clostridium cluster XVIa, which include bacteria known to produce them [6]. An increase in pro-inflammatory and potentially pathogenic taxa, Enterobacteriaceae as cirrhosis advances was observed in cirrhotic patients with decreased fecal bile acid levels [6]. Thus, bile acid pool size and composition appear to be important factors in regulating gut microbial community structure in humans.
It is clear that bile acids have both direct antimicrobial effects on gut microbes [10], and indirect effects through FXR-induced antimicrobial peptides [11]. Indeed, the potency of deoxycholic acid (DCA) as an antimicrobial agent, is an order of magnitude greater than cholic acid (CA), owing to its hydrophobicity and detergent properties on bacterial membranes [10]. Complex and significant changes in the gut microbiome are observed when rats are fed bile acids. Islam et al. demonstrated that a medium CA intake (1.25 mmol/kg) and high CA (5 mmol/kg) diet resulted in phylum-level alterations of the gut microbiome with Firmicutes vastly expanding from 54% of the microbiome in control rats to between 93-98% of the microbiome [12]. At the class-level, the Clostridia expanded from 39% in controls to roughly 70% and within the Clostridia, the genus Blautia expanded from 8.3% in control rats to between 55-62% when the mice were fed CA [12]. Blautia includes many species of Clostridium and Ruminococcus spp., many of which are found within Clostridium cluster XIVa, and closely related to human bile acid 7α-dehydroxylating species (resulting in secondary bile acids), which make up a small fraction of this taxonomic group [13]. Total bile acids in feces increase 6-fold and 20-fold in the medium and high CA diet, respectively, with the medium CA diet containing similar levels to those reported in human fecal water on a high-fat diet. These results demonstrate that increased input of bile acids result in significant inhibition of the Bacteroidetes and Actinobacteria, two of the three major phyla reported in the microbiome of humans [12]. Expansion of the Firmicutes results in significant expansion of DCA-producing bacteria. In this regard, we observed a 1,000 fold increase in the levels of bile acid 7α-dehydroxylating bacteria by feeding mice CA [13]. An important question is whether this expansion is a result of bile acid 7α-dehydroxylating bacteria producing a potent antimicrobial agent that reduces competition for nutrients, or if primary bile acids serve as a major fermentative electron acceptor, much in the way pyruvate is used in alcoholic and lactic fermentations.
If bile acid feeding results in expansion of Clostridium cluster XVIa, then we should expect to see a decrease in this taxonomic group when bile acid levels sharply decline. The study by Kakiyama et al suggests precisely this [6]. Levels of bile acids entering the large intestine thus has a profound effect on the major division/phyla level taxa in the lumen of the gut. Bajaj et al. [2012] also demonstrates that the mucosal microbiome community was significantly different from the community in the lumen and these differences correlated with complications of cirrhosis such as hepatic encephalopathy [7]. Taken together, decreasing levels of bile acids in the gut favor gram-negative members of the microbiome, some of which produce potent LPS, and include potential pathogens. Increased bile acid levels in the gut appear to favor gram-positive members of the Firmicutes, including bacteria that 7α-dehydroxylate host primary bile acids to toxic secondary bile acids.
Microbial Bile Acid Products as Promoters of Liver Cancer
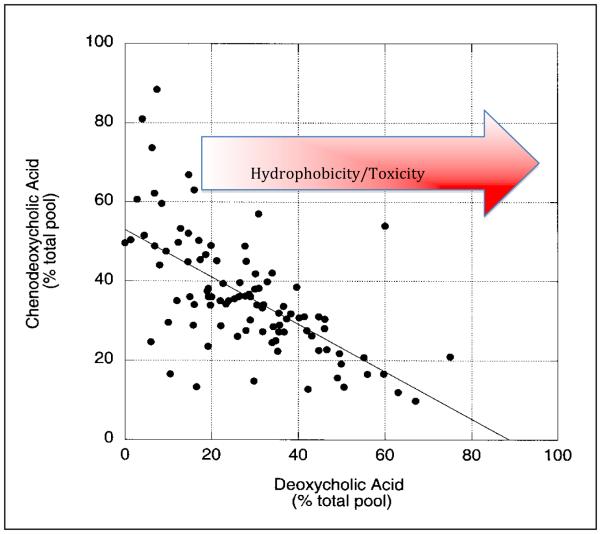
Biotransformation of CA and CDCA/UDCA through bile acid 7α/β-dehydroxylation results in formation of the secondary bile acids, DCA and LCA, respectively. A small population of intestinal species in the genus Clostridium, including C. scindens, C. hiranonis, C. hylemonae (Clostridium cluster XVIa), and C. sordelli (Clostridium cluster XI) are capable of producing secondary bile acids [14]. Unlike rodents, the human liver is incapable of 7α-hydroxylating secondary bile acids returning to the liver via the portal vein, and thus secondary bile acids can accumulate to high levels in the bile of some humans [Figure 1] [14]. Secondary bile acids, particularly DCA are known to accumulate in the BA pool of individuals on a “Western diet” [Figure 1]. Indeed, increased DCA levels in feces, serum, and bile of patients with colon cancer and some cholesterol gallstone disease is widely reported [15]. DCA is known to activate a number of cell-signaling pathways associated with disease phenotypes [16]. Yoshimoto et al. recently reported a novel mechanism by which DCA acts as a major microbial metabolite associated with obesity-associated hepatocellular carcinoma (HCC) [17]. DCA provoked hepatic stellate cells in an animal model to secrete pro-inflammatory and pro-tumorigenic factors in what is known as the senescence-associated secretory pathway (SASP) in the presence of a chemical carcinogen. While DCA was not sufficient to cause cancer in this model, it was necessary, as antibiotics that knocked out DCA production resulted in significant decline in HCC, which was reversed by feeding DCA to antibiotic-fed mice [17].
Figure 1. Accumulation of deoxycholic acid in the bile acid pool of Veterans Affairs patients.
Bile acid 7α-dehydroxylation by intestinal clostridia result in hydrophobic bile acids that can return to the liver via the portal circulation and accumulate in the human bile acid pool. Adapted from Ridlon et al (2006) [14].
The authors performed microbiome analysis and noted significant expansions of Clostridium clusters XI and XVIa in HFD versus normal-chow diet. Clostridium cluster XI was populated by a single species similar to C. sordellii, composing 12% of the gut microbiome in HFD mice. Clostridium cluster XVIa made up only 0.5% of the population in HFD fed mice. These results, as the authors cautiously note, suggest C. sordellii is responsible for the increase in DCA. Previously, our lab quantified bile acid 7α-dehydroxylating activity in a number of human isolates within Clostridium clusters XI and XVIa, including C. sordellii [18]. C. sordellii fell within the “low activity” strains, while species such as C. scindens (cluster XVIa) had 100-fold greater bile acid 7α-dehydroxylating activity in vitro. It is possible that regulation of bile acid 7α-dehydroxylating activity differs in vitro vs. in vivo, and measurements of in vivo rates of bile acid metabolism will be required to settle this issue.
Microbial Control of Bile Acid Pool Size
Recent reports link metabolism of bile salts by gut microbes to bile acid pool size. Observations between germ-free rodents and “conventional” (i.e. acquiring a normal microbiome) revealed striking differences in bile acid pool size. Conventional animals, despite having a greatly reduced BA pool size (decreased ~71%) compared to GF mice, none-the-less have significantly higher levels of BA in the cecum [19]. The increased level of BA in the cecum of GF mice has been reported to be due to the down-regulation of the apical sodium-dependent bile acid transporter (ASBT) which is under control of intestinal FXR [20,21]. Observations of this kind were made decades ago, but only in the past year have some of the mechanisms behind microbial control of the bile acid pool size in animals been elucidated.
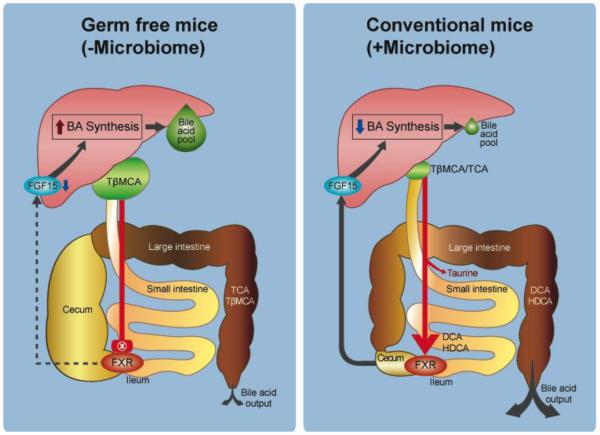
Bile salt deconjugation, carried out by bile salt hydrolase, is wide-spread within the normal microbiota [10,14]. BA synthesis is self-regulated by a feedback loop involving both liver and intestinal expression and activation of the nuclear receptor farnesoid × receptor (FXR) [22]. Intestinal FXR appears to regulate hepatic cholesterol 7α-hydroxylase (CYP7A1), the rate-limiting enzyme in BA synthesis, through a fibroblast growth factor 15 (FGF-15/19)-dependent mechanism [23,24]. BA structure, such as conjugation, extent and orientation of hydroxy groups determine the degree of activation, or indeed inhibition of FXR. In a series of elegant experiments, Sayin et al. demonstrated the mechanism behind the reduction in bile acid pool size when GF mice are conventionalized [19]. The primary bile salt in mice, tauro-β-muricholic acid (T-β-MCA) was shown to be a potent FXR-antagonist, resulting in greater bile acid synthesis and more efficient enterohepatic circulation (EHC) of bile acids [Figure 2] [19]. Hu et al confirmed that expansion of the hydrophilicity of the pool could be achieved by increasing FXR-antagonists (muricholics, UDCA) through elimination of cholic acid in a steroid 12α-hydroxylase knockout mouse model (Cyp8b1−/−) [20]. Interestingly, similar results were obtained in WT mice fed antibiotics: the bile acid pool size increased through a positive-feedback mechanism (FXR antagonism) and reduction in potent microbial bile acid FXR agonists (DCA) [20]. A normal gut microbiome biotransforms T-βMCA through bile salt hydrolase and bile acid 7α-dehydroxlase forming β-MCA and hyodeoxycholic acid, respectively.
Figure 2.
Gut Microbiota Influence bile acid pool size in mice through metabolism of FXR-antagonist, tauro-β-muricholic acid. Panel A: Inhibition of intestinal FXR by T-βMCA increases bile acid pool size through enhanced bile acid synthesis in the liver and efficient enterohepatic circulation due to high levels of apical sodium bile salt transporter (IBAT; SLC10A2) in the ileum. Panel B: Metabolism of T-βMCA, and production of FXR-agonists including secondary bile acids by gut microbiome results in FXR-induced release of FRF-15 in the intestine and inhibition of the rate-limiting enzyme in bile acid synthesis, cholesterol 7α-hydroxylase (CYP7a1) in the liver. Reduction in bile acid synthesis coupled with downregulation of ASBT results in reduction in bile acid pool size.
Dietary components can have effects on the bile acid pool similar to antibiotics due to their effects on bile salt metabolizing members of the microbiome. A recent article by Li et al presented evidence that the antioxidant, tempol, targets intestinal Lactobacilli, a major source of bile salt hydrolase (BSH) activity in the murine gut [25]. By knocking out BSH activity, production of secondary bile acids, potent FXR agonists are down regulated. Additionally, increase in FXR antagonists, such as T-βMCA increase, resulted in increased bile acid pool size. Indeed, tempol feeding resulted in a near inversion of the Firmicutes:Bacteroides ratio. This resulted in a concomitant significant increase in T-βMCA levels in the intestine and inhibition of intestinal FXR signaling that showed similar results to intestinal FXR-KO mice in their study [25]. Taken together, these results pose the following question: Does an increase in hydrophilicity of the human biliary pool lead to an increase in bile acid pool size through similar mechanisms?
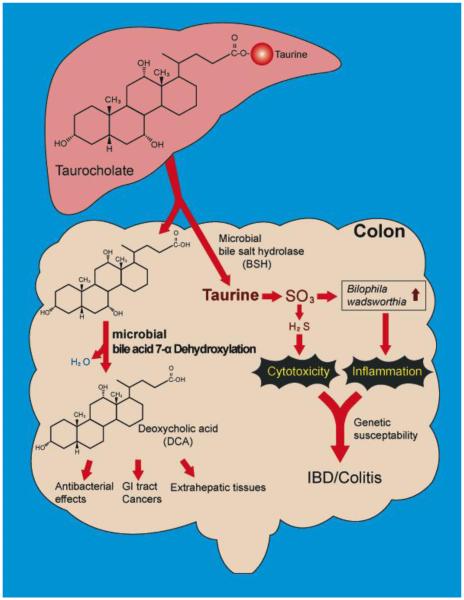
Thus the presence of the microbiome alters the composition of the bile acid pool, which in turn has a profound effect on the bile acid pool size; however, dietary and antimicrobial compounds can affect or even ameliorate this equilibrium. Now, we turn to an oft-overlooked aspect of bile salt metabolism, the taurine and glycine conjugates. Devkota et al presented a truly remarkable study relating diet, taurine-conjugated bile salts, and changes in the microbiome. Mice were fed a high fat diet composed of either milk fat (MF), or polyunsaturated fat and compared with a low-fat chow diet. The authors noted a significant expansion of the Bacteroidetes irrespective of the type of fat, similar to previous results [26]. A member of the deltaproteobacteria, Bilophila wadsworthia, expanded from <1% of the microbiome to ~6% of OTUs detected in the mice fed the MF diet [26]. Previous studies have shown that Bilophila, as its name implies, thrives in the presence of bile, and that indeed, it is able to utilize the sulfite in taurine as a terminal electron acceptor [Figure 3] [27]. This is significant because human dietary studies have shown that a “Western diet” can shift the bile acid pool in humans from glycine to taurine-conjugation [28]. Taken together, these results point to specific host molecules, whose levels are controlled partly by diet, which controls the levels of a specific member of the microbiome.
Figure 3. Metabolism of bile salt, taurocholic acid, by gut microbiome.
Gut microbes in the ileum and large bowel deconjugate bile salts to free bile acids by bile salt hydrolase. Taurine, due to sulfite moiety, can provide pathobionts in the gut with a terminal electron acceptor, allowing for their growth and expansion in the gut [26]. High-fat diet is associated with increased taurine-conjugation in humans [28]. Free primary bile acids are further metabolized to toxic secondary bile acids that can accumulate in the bile acid pool in humans [see Figure 1] and alter host physiology.
Conclusions
We are just beginning to elucidate the complex interactions between the liver, bile acids and the gut microbiome. There are as yet no detailed in vivo studies of bile acid 7α-dehydroxylating bacteria apart from their effects on the bile acid pool. Much of the focus of BSH has been with regard to probiotic organisms and cholesterol-lowering effects of BSH [29] and BSH as a pathogenicity factor [30]. A new area of investigation, as highlighted in this review, is the effect of BSH activity on the size and composition of the bile acid pool and indeed how this affects the structure of the microbiome [19,20,25]. While the composition of the microbiome in adult humans is surely a product of nature [31], it is obvious that nurture, particular diet [26] and disease states [6-8] sculpts the composition of the microbiome. On the one hand, high fat diets increase the levels of bile acids in the gut. This increase in bile acid concentration in the large intestine has effects on the highest taxonomic levels in concert with other dietary variables. What is clear is that increased bile acid feeding leads to blooms of taxa including bile acid 7α-dehydroxylating species [12,13].
Bile acids are now regarded as important hormones [reviewed by 16]. Bacteria in the gut produce secondary bile acids, which appear to bind to and activate a number of host nuclear receptors to a greater extent than host primary bile acids [Table 1][32-37]. Bile acid nuclear receptors are expressed in tissues outside the EHC, such as heart [38], adipose tissue [39] and perhaps kidney [40]. G-protein coupled receptors activated by bile acids include TGR-5 [32], S1PR2 [33] and specific muscarinic receptors [34]. TGR-5 is strongly activated by secondary bile acids microbial products, while S1PR2 and muscarinic receptors are only activated by conjugated bile acids [33-37, 41]. Therefore, the degree of conjugation of the bile acid pool returning from the intestines may significantly alter bile acid signaling in the liver and possible other tissues. The presence of a microbiome has profound effects on the bile acid profile in host tissues; interestingly, bile acid profiles differ between serum and tissues [16]. Unraveling the effects of different bile acid profiles in host tissues will be an important avenue of biomedical research. Approaches to modulate the microbiome-bile acid axis through diet, and antimicrobial agents (antibiotics, small molecule inhibitors of microbial bile acid biotransforming enzymes) appears likely to reduce the risk and/or treat certain diseases of the GI tract and perhaps beyond.
Table 1.
Nuclear Receptors and G-Protein Coupled Receptors Activated by Different Bile Acids
| Nuclear Receptor | Bile Acid Agonist | References |
|---|---|---|
| Farnasoid X Receptor (FXR) | CDCA >LCA = DCA>CA | reviewed by 16 |
| Pregnane-activated receptor (PXR) | LCA>DCA>CA | reviewed by 16 |
| Vitamin D receptor | 3-oxo-LCA>LCA>DCA>CA | 32 |
|
| ||
| G-Protein Coupled Receptors | ||
| TGR-5 | DCA>LCA>CDCA>CA | 33 |
| Sphingosine-1-phosphate Receptor 2 (S1PR2) |
Taurine or Glycine Conjugated Bile Acids |
34 |
| (M2,3) Muscarinic receptors | T-LCA>T-DCA>T-CA | 35-37 |
Key Points.
The global bile acid pool differs markedly between germ-free and conventional mammals
Through bile salt hydrolysis and bile acid 7α-dehydroxylation, the gut microbiome is capable of producing secondary bile acid hormones that affect host physiology
A dynamic equilibrium exists between diet-gut microbiome-bile acid pool size/composition
Perturbations in this equilibrium can result in pathological states (GI cancers, gallstones, liver disease)
Acknowledgements
This work was supported by RO1AA020203 from the National Institute on Alcohol Abuse and Alcoholism, grant RO1DK087913 from the National Institute of Diabetes and Digestive and Kidney Diseases (both to JSB), VA Merit Grant BX001328 to PBH. The authors would like to thank Mikyung Kang for graphic assistance with figures.
References
Papers of particular interest, published within the annual period of review, have been highlighted as:
• Of special interest
•• Of outstanding interest
- ••1.Human Microbiome Project Consortium Structure, function and diversity of the healthy human microbiome. Nature. 2012;486(7402):207–214. doi: 10.1038/nature11234. Culminating work of the Human Microbiome Project which provides an overview of the human microbiome. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Donohoe DR, Garge N, Zhang X, Sun W, O’Connell TM, Bunger MK, Bultman SJ. The microbiome and butyrate regulate energy metabolism and autophagy in the mammalian colon. Cell Metab. 2011;13:517–526. doi: 10.1016/j.cmet.2011.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Christl SU, Eisner HD, Dusel G, Kasper H, Scheppach W. Antagonistic effects of sulfide and butyrate on proliferation of colonic mucosa. Dig Dis Sci. 1996;41:2477–81. doi: 10.1007/BF02100146. [DOI] [PubMed] [Google Scholar]
- 4.Human Microbiome Project Consortium A framework for human microbiome research. Nature. 2012;486(7402):215–221. doi: 10.1038/nature11209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124(4):837–848. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- ••6.Kakiyama G, Pandak WM, Gillevet PM, Hylemon PB, Heuman DM, Daita K, Takei H, Muto A, Nittono H, Ridlon JM, White MB, Noble NA, Monteith P, Fuchs M, Thacker LR, Sikaroodi M, Bajaj JS. Modulation of the fecal bile acid profile by gut microbiota in cirrhosis. J Hepatol. 2013;58(5):949–955. doi: 10.1016/j.jhep.2013.01.003. Study reveals that reduced bile acid concentration in the gut may play an important role in allowing pro-inflammatory microbial taxa to expand. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••7.Bajaj JS, Ridlon JM, Hylemon PB, Thacker LR, Heuman DM, Smith S, Sikaroodi M, Gillevet PM. Linkage of gut microbiome with cognition in hepatic encephalopathy. Am J Physiol Gastrointest Liver Physiol. 2012 Jan 1;302(1):G168–G175. doi: 10.1152/ajpgi.00190.2011. First report linking cognitive disorders resulting from liver disease with specific gut microbial taxa. Study utilized a systems biology approach. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••8.Bajaj JS, Hylemon PB, Ridlon JM, Heuman DM, Daita K, White MB, Monteith P, Noble NA, Sikaroodi M, Gillevet PM. Colonic mucosal microbiome differs from stool microbiome in cirrhosis and hepatic encephalopathy and is linked to cognition and inflammation. Am J Physiol Gastrointest Liver Physiol. 2012;303(6):G675–G685. doi: 10.1152/ajpgi.00152.2012. Demonstrates that gut microbiome research should focus not only on the contents of the colonic lumen, but rather the mucosal microbiota is not only different, but in some disease states may be more relevant. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bajaj JS, Heuman DM, Hylemon PB, Sanyal AJ, White MB, Monteith P, Daita K, Fisher AR, Sikaroodi M, Gillevet PM. The Cirrhosis Dysbiosis Ratio defines Changes in the Gut Microbiome Associated with Cirrhosis and its Complications. J Hepatol. 2013 doi: 10.1016/j.jhep.2013.12.019. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Begley M, Gahan CG, Hill C. The interaction between bacteria and bile. FEMS Microbiol Rev. 2005;29(4):625–51. doi: 10.1016/j.femsre.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 11.Inagaki T, Moschetta A, Lee YK, Peng L, Zhao G, Downes M, Yu RT, Shelton JM, Richardson JA, Repa JJ, Mangelsdorf DJ, Kliewer SA. Regulation of antibacterial defense in the small intestine by the nuclear bile acid receptor. Proc Natl Acad Sci U S A. 2006 Mar;Jul;103(10):3920–3925. doi: 10.1073/pnas.0509592103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••12.Islam KB, Fukiya S, Hagio M, Fujii N, Ishizuka S, Ooka T, Ogura Y, Hayashi T, Yokota A. Bile acid is a host factor that regulates the composition of the cecal microbiota in rats. Gastroenterology. 2011;141(5):1773–1781. doi: 10.1053/j.gastro.2011.07.046. A study demonstrating that increased levels of bile acids reaching the large intestine can significantly alter the composition of the gut microbiome. [DOI] [PubMed] [Google Scholar]
- 13.Ridlon JM, Alves JM, Hylemon PB, Bajaj JS. Cirrhosis, bile acids and gut microbiota: Unraveling a complex relationship. Gut Microbes. 2013;4(5):382–387. doi: 10.4161/gmic.25723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ridlon JM, Kang D, Hylemon PB. Bile salt biotransformations by human intestinal Bacteria. J. Lipid Res. 2006;47:241–259. doi: 10.1194/jlr.R500013-JLR200. [DOI] [PubMed] [Google Scholar]
- 15.Bernstein H, Bernstein C, Payne CM, Dvorakova K, Garewal H. Bile acids as carcinogens in human gastrointestinal cancers. Mutat Res. 2005;589(1):47–65. doi: 10.1016/j.mrrev.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 16.Hylemon PB, Zhou H, Pandak WM, Ren S, Gil G, Dent P. Bile acids as regulatory molecules. J Lipid Res. 2009 Aug;50(8):1509–1520. doi: 10.1194/jlr.R900007-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••17.Yoshimoto S, Loo TM, Atarashi K, Kanda H, Sato S, Oyadomari S, Iwakura Y, Oshima K, Morita H, Hattori M, Honda K, Ishikawa Y, Hara E, Ohtani N. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature. 2013;499(7456):97–101. doi: 10.1038/nature12347. Provides a novel mechanism by which products of gut microbial bile acid 7α-dehydroxylation may promote liver cancer. [DOI] [PubMed] [Google Scholar]
- 18.Doerner KC, Takamine F, LaVoie CP, Mallonee DH, Hylemon PB. Assessment of fecal bacteria with bile acid 7 alpha-dehydroxylating activity for the presence of bai-like genes. Appl Environ Microbiol. 1997;63(3):1185–1188. doi: 10.1128/aem.63.3.1185-1188.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••19.Sayin SI, Wahlström A, Felin J, Jäntti S, Marschall HU, Bamberg K, Angelin B, Hyötyläinen T, Orešič M, Bäckhed F. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 2013;17(2):225–235. doi: 10.1016/j.cmet.2013.01.003. Suggests the important role of FXR-antagonist T-βMCA and gut microbial metabolism of bile acids in regulating bile acid pool size. [DOI] [PubMed] [Google Scholar]
- ••20.Hu X, Bonde Y, Eggertsen G, Rudling M. Muricholic bile acids are potent regulators of bile acid synthesis via a positive feedback mechanism. J Intern Med. 2013 doi: 10.1111/joim.12140. 10.1111/joim.12140. [Epub ahead of print]. Suggests an additional positive-feedback role for MCAs that may work independently of FGF-15. [DOI] [PubMed] [Google Scholar]
- 21.Sinha J, Chen F, Miloh T, Burns RC, Yu Z, Shneider BL. beta-Klotho and FGF-15/19 inhibit the apical sodium-dependent bile acid transporter in enterocytes and cholangiocytes. Am J Physiol Gastrointest Liver Physiol. 2008;295(5):G996–G1003. doi: 10.1152/ajpgi.90343.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sinal CJ, Tohkin M, Miyata M, Ward JM, Lambert G, Gonzalez FJ. Targeted disruption of the nuclear receptor FXR/BAR impairs bile acid and lipid homeostasis. Cell. 2000;102(6):731–744. doi: 10.1016/s0092-8674(00)00062-3. [DOI] [PubMed] [Google Scholar]
- 23.Inagaki T, Choi M, Moschetta A, Peng L, Cummins CL, McDonald JG, Luo G, Jones SA, Goodwin B, Richardson JA, Gerard RD, Repa JJ, Mangelsdorf DJ, Kliewer SA. Fibroblast growth factor 15 functions as an enterohepatic signal to regulate bile acid homeostasis. Cell Metab. 2005;2(4):217–225. doi: 10.1016/j.cmet.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 24.Kim I, Ahn SH, Inagaki T, Choi M, Ito S, Guo GL, Kliewer SA, Gonzalez FJ. Differential regulation of bile acid homeostasis by the farnesoid × receptor in liver and intestine. J Lipid Res. 2007;48(12):2664–2672. doi: 10.1194/jlr.M700330-JLR200. [DOI] [PubMed] [Google Scholar]
- ••25.Li F, Jiang C, Krausz KW, Li Y, Albert I, Hao H, Fabre KM, Mitchell JB, Patterson AD, Gonzalez FJ. Microbiome remodelling leads to inhibition of intestinal farnesoid × receptor signalling and decreased obesity. Nat. Communications. 2013;4(2384) doi: 10.1038/ncomms3384. doi: 10.1038/ncomms3384 Demonstrates how a dietary antioxidant, tempol, ablates lactobacillus spp. in the rodent cecum resulting in reduced BSH activity resulting in FXR antagonism by T-βMCA leading to alteration in bile pool size. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••26.Devkota S, Wang Y, Musch MW, Leone V, Fehlner-Peach H, Nadimpalli A, Antonopoulos DA, Jabri B, Chang EB. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10−/− mice. Nature. 2012;487(7405):104–108. doi: 10.1038/nature11225. Important article linking diet to a host factor (taurine-conjugated bile acids) leading to an expansion of a pathobiont in the human GI tract resulting in colitis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laue H, Denger K, Cook AM. Taurine reduction in anaerobic in anaerobic respiration of Bilophila wadsworthia RZATAU. Appl. Environ. Microbiol. 1997;63:2016–2021. doi: 10.1128/aem.63.5.2016-2021.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hardison WG. Hepatic taurine concentration and dietary taurine as regulators of bile acid conjugation with taurine. Gastroenterology. 1978;75:71–75. [PubMed] [Google Scholar]
- 29.Pavlović N, Stankov K, Mikov M. Probiotics--interactions with bile acids and impact on cholesterol metabolism. Appl Biochem Biotechnol. 2012 Dec;168(7):1880–1895. doi: 10.1007/s12010-012-9904-4. [DOI] [PubMed] [Google Scholar]
- 30.Marchesini MI, Connolly J, Delpino MV, Baldi PC, Mujer CV, DelVecchio VG, Comerci DJ. Brucella abortus choloylglycine hydrolase affects cell envelope composition and host cell internalization. PLoS One. 2011;6(12):e28480. doi: 10.1371/journal.pone.0028480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mäkivuokko H, Lahtinen SJ, Wacklin P, Tuovinen E, Tenkanen H, Nikkilä J, Björklund M, Aranko K, Ouwehand AC, Mättö J. Association between the ABO blood group and the human intestinal microbiota composition. BMC Microbiol. 2012 Jun 6;12:94–106. doi: 10.1186/1471-2180-12-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Khurana S, Raufman JP, Pallone TL. Bile acids regulate cardiovascular function. Clin Transl Sci. 2011 Jun;4(3):210–218. doi: 10.1111/j.1752-8062.2011.00272.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Watanabe M, Houten SM, Mataki C, Christoffolete MA, Kim BW, Sato H, Messaddeq N, Harney JW, Ezaki O, Kodama T, Schoonjans K, Bianco AC, Auwerx J. Bile acids induce energy expenditure by promoting intracellular thyroid hormone activation. Nature. 2006;439(7075):484–489. doi: 10.1038/nature04330. [DOI] [PubMed] [Google Scholar]
- 40.Swann JR, Want EJ, Geier FM, Spagou K, Wilson ID, Sidaway JE, Nicholson JK, Holmes E. Systemic gut microbial modulation of bile acid metabolism in host tissue compartments. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4523–4530. doi: 10.1073/pnas.1006734107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kawamata Y, Fujii R, Hosoya M, Harada M, Yoshida H, Miwa M, Fukusumi S, Habata Y, Itoh T, Shintani Y, Hinuma S, Fujisawa Y, Fujino M. A G protein-coupled receptor responsive to bile acids. J Biol Chem. 2003;278(11):9435–9440. doi: 10.1074/jbc.M209706200. [DOI] [PubMed] [Google Scholar]
- ••33.Studer E, Zhou X, Zhao R, Wang Y, Takabe K, Nagahashi M, Pandak WM, Dent P, Spiegel S, Shi R, Xu W, Liu X, Bohdan P, Zhang L, Zhou H, Hylemon PB. Conjugated bile acids activate the sphingosine-1-phosphate receptor 2 in primary rodent hepatocytes. Hepatology. 2012;55(1):267–276. doi: 10.1002/hep.24681. This article reports activation of cell signaling pathways through TCA activation of G-coupled protein receptor, SIP-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sheikh Abdul Kadir SH, Miragoli M, Abu-Hayyeh S, Moshkov AV, Xie Q, Keitel V, Nikolaev VO, Williamson C, Gorelik J. Bile acid-induced arrhythmia is mediated by muscarinic M2 receptors in neonatal rat cardiomyocytes. PLoS One. 2010 Mar 15;5(3):e9689. doi: 10.1371/journal.pone.0009689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Makishima M, Lu TT, Xie W, Whitfield GK, Domoto H, Evans RM, Haussler MR, Mangelsdorf DJ. Vitamin D receptor as an intestinal bile acid sensor. Science. 2002;296:1313–1316. doi: 10.1126/science.1070477. [DOI] [PubMed] [Google Scholar]
- 36.Raufman JP, Zimniak P, Bartoszko-Malik A. Lithocholyltaurine interacts with cholinergic receptors on dispersed chief cells from guinea pig stomach. Am J Physiol. 1998;274:G997–G1004. doi: 10.1152/ajpgi.1998.274.6.G997. [DOI] [PubMed] [Google Scholar]
- 37.Raufman J-P, Chen Y, Cheng K, Compadre C, Compadre L, Zimniak P. Selective interaction of bile acids with muscarinic receptors: a case of molecular mimicry. Eur J Pharmacol. 2002457:77–84. doi: 10.1016/s0014-2999(02)02690-0. [DOI] [PubMed] [Google Scholar]
- 41.Raufman J-P, Cheng K, Zimniak P. Activation of muscarinic receptor signaling by bile acids. Dig Dis Sci. 2003;48:1431–1444. doi: 10.1023/a:1024733500950. [DOI] [PubMed] [Google Scholar]