Abstract
Individual differences in sleep patterns of children may have developmental origins. Here, two factors known to influence behavioral development, monoamine oxidase A (MAOA) genotype and prenatal iron deficiency, were examined for their influences on sleep in juvenile rhesus monkeys. Sleep was assessed based on a threshold for inactivity as recorded by activity monitors. Pregnant monkeys were fed diets containing either 100 ppm Fe (iron sufficient) or 10 ppm Fe (iron deficient). At 3-4 months of age, male offspring were genotyped for polymorphisms of the MAOA gene that lead to high or low transcription. At one and two years of age, sleep was assessed. Several parameters of sleep architecture changed with age. At one year of age, monkeys with low-MAOA genotype demonstrated a trend toward more sleep at night than the high-MAOA group. When monkeys reached two years of age, prenatal iron deficiency reversed this trend; iron deficiency in the low-MAOA group resulted in sleep fragmentation, more awakenings at night and more sleep episodes during the day, as compared to prenatal iron sufficiency in that genotype. The ability to consolidate sleep during the dark cycle was disrupted by prenatal iron deficiency specifically in monkeys with the low-MAOA genotype.
Keywords: sleep, nonhuman primate, prenatal iron deprivation, MAOA genotype
Introduction
Sleep in primates, including humans, is characterized by a consolidated sleep phase during the dark cycle(1, 2). Sleep locations protected from predators have been seen as the origin of this pattern which contrasts with intermittent sleep periods, peaking in frequency according to the diurnal cycle, in rodents.
Disruption of the consolidated sleep pattern is the basis of common sleep disorders in humans, including delayed onset of sleep, waking during sleep, inability to resume sleep and daytime sleepiness (3). Nonhuman primates, including rhesus monkeys, have been established as valuable models for human sleep based on observation, activity monitoring and electroencephalography(4, 5, 6).
While environmental factors and disease state are becoming known through research as sources for sleep disruption, a developmental origin for sleep regulation variability is less studied. Here we report on the influence of prenatal iron deficiency on sleep patterns in rhesus monkeys 1 and 2 years of age, approximately equivalent developmentally to 4 and 8 years of age in children. Iron deficiency is the most common single nutrient deficiency worldwide with pregnant women and infants most affected. The CDC (Center for Disease Control) reports that 33.8% of pregnant women in the US develop anemia (http://www.cdc.gov/pednss/pnsstables) with the highest frequency in the 3d trimester. Because of the prolonged period of 3d trimester fetal brain development in primates, monkeys, rather than rodents, are the more appropriate model for studying the consequences of 3d trimester iron deficiency for children. To produce a model of 3d trimester iron deficiency, their dams were fed a low iron diet in utero(7).
An additional variable in this study was genotyping for MAOA polymorphisms (high or low expression of the enzyme monoamine oxidase A). A recent study linked MAOA polymorphisms with daytime sleepiness, a possible reflection of sleep fragmentation, in humans(8). Similar MAOA polymorphisms occur in monkeys and have previously shown to interact with prenatal ID to influence behavior in social and cognitive tests in this cohort of monkeys(9, 10). Both ID(11) and MAOA polymorphisms(12, 13, 14) influence monoamine neurotransmitter systems in the brain. We hypothesized that these two factors could interact during fetal brain development to produce long term changes in sleep patterns.
Methods
Compliance with animal research guidelines
Animal husbandry followed the Guide for the Care and Use of Laboratory Animals of the National Research Council. All protocols were approved prior to implementation by the UC Davis Institutional Animal Care and Use Committee. The California National Primate Research Center (CNPRC) is accredited by the Association for Assessment and Accreditation of Laboratory Animal Care. Animal husbandry and veterinary medicine procedures were performed by specialized staff with advanced training in these areas.
Subjects, diets and genotyping
Pregnant rhesus (Macaca mulatta) dams were assigned to the study after screening for reproductive history. Experimental groups were balanced for dam age, weight and parity but were otherwise randomly constructed. Pregnant dams were pair-housed in double cages (120 × 65 × 79cm) in a cage room separate from the rest of the colony. They were fed experimental diet twice a day in premeasured amounts, and water was available ad libitum from an automated system. Their care included daily cleaning of drop pans, twice daily feeding, biweekly cage changes to freshly sterilized cages, automatically controlled light cycles (lights on from 0600 to 1800), and temperature control (20-25°C) and monitoring. Monkeys were observed each morning for health signs and referred to veterinarians for treatment if needed. An individual medical record was maintained for each animal. Monkey dams were time-mated and fed an iron deficient (10 ppm, ID) or an iron sufficient (100 ppm, IS) diet during gestation beginning at pregnancy identification by ultrasound. Pregnancies with male fetuses were identified at this time. Diet composition and feeding schedules have been reported.(7) This dietary regimen has been shown to produce anemia in the 3d trimester (Supplementary Figure S1, available online). After birth, experimental diets were discontinued. Iron repletion was provided to any dams that were anaemic and neonates in the ID group also received iron repletion. Infants were reared by their mothers in double cages with another mother-infant pair until weaning at 5-6 months of age. They were then caged with a like-age peer during a 2 year assessment battery(9) including tests reported here. During the test period, groups did not differ in body weight, an index of growth, or serum haemoglobin, a marker of iron deficiency(9) (Supplementary Figure S2, available online).
Genotyping was obtained from colony records. Genotyping for VNTR polymorphisms in the upstream regulatory region of the rhesus MAOA gene (rhMAOA-LPR) is conducted routinely in 3- to 4- month old rhesus at CNPRC by the Veterinary Genetics Laboratory (VGL) using PCR with MAOA-Forward and MAOA-Reverse primers(9, 15, 16). All PCR reactions included a negative (no DNA template) and two positive (MAOA genotypes 5/5 and 6/7) controls. Genotyping via fragment size analysis used VGL’s STRand software. The overall genotyping error rate at VGL Is <0.5%.
MAOA is an X-linked gene with at least five different VNTR polymorphisms resulting in 13 different female genotypes and 5 different male genotypes in rhesus. Males in our study were identified as hemizygous for low-MAOA (7 VNTR) or hi-MAOA (4, 5 or 6 VNTR) polymorphisms resulting in four groups: hi-MAOA IS (n = 5), hi-MAOA ID (n = 5), low MAOA IS (n = 5) and low-MAOA ID (n = 4). One infant (in the ID group) was excluded from the study due to an ambiguous MAOA VNTR.
Activity monitoring
Actimeters (Actitrac, IM Systems, Baltimore, MD, USA) that record movement were placed in the back of a specially designed vest that the monkeys wore in their home cages for 48 h(17, 18, 19, 20). The home cage environment had a daily 12 h light cycle (lights on 0600 to 1800). The actimeter links to the computer to transfer data to Actitrac software that provides measures of onset, duration and level of each active and inactive period. An inactive period is defined as a 2 min period (epoch) for which that epoch and the epoch prior to and following it average to be below a software-defined activity threshold of 18 counts/2 min. This threshold has been validated as a measure of sleep in children, but not in monkeys. However, studies in monkeys show a good agreement between actimeter and EEG indices of sleep(5).
Subgroups of 6 monkeys, balanced for group, were assessed over the weekend to minimize disturbance. Regular husbandry included daily cage cleaning and twice daily feeding. All entries to the room were recorded for potential exclusion of actimeter readings if necessary. This assessment was performed twice, at one and two years of age (14 months, 22.5 months). The second 24 h of the home cage monitoring period was used for analysis. Sleep parameters selected for analysis were:
time to onset of sleep at night,
number of awakenings at night,
total awake time at night,
number of sleep episodes during the day,
total time asleep during the day.
Statistics and Power Estimates
Selected parameters were analyzed with two-way ANOVA (genotype, ID diet) including the interaction (JMP, SAS, Cary, NC, USA). Planned comparisons looked at the effect of ID diet within genotype. Potential covariates (body weight, cage location, etc.) were screened for relevance to the sleep parameters but none were significant. Data sets were screened for normal distribution prior to analysis.
Group sizes of 10 per diet group were selected for this study based on behavioral effect sizes in a previous cohort. We were not able to estimate effect sizes for the diet × genotype interaction. The effect size for our apical variable, the sleep fragmentation index, comparing the ID and IS groups with the low-MAOA genotype was d=1.74. Small sample nonhuman primate studies are able to detect smaller effects than human studies due to strict environmental control and subject selection.
Results
Diet and genotype did not influence the growth or health of the test cohort as illustrated in Supplementary Figure S2 (available online). At night, about 55% of the time was spent in the sleep (inactive) state at both ages. During the day, monkeys were inactive less than 5% of the time on average. Supplementary Figure S3 (available online) illustrates the pattern of day-night activity as recorded by actimeter.
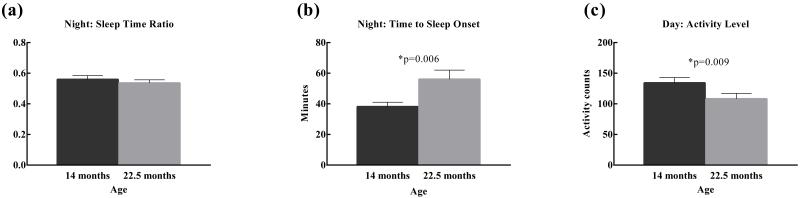
The time spent sleeping at night did not change with age in the test cohort as a whole (Fig 1(a)), but older monkeys took longer to fall asleep after the onset of the dark period (Fig 1(b)), (F(1,19)=9.72, p=.006) and had lower levels of activity during the day (Fig 1(c)), (F(1,19)=8.44, p=.009) compared to younger monkeys.
Figure 1.
Age differences in sleep and activity at one- and two-years of age in male rhesus monkeys. (a) Ratio of sleep to wake time at night, (b) Time to onset of sleep at night. Older animals took longer to fall asleep than younger animals. (c) Activity level during the day. Older animals were less active than younger animals during daytime hours.
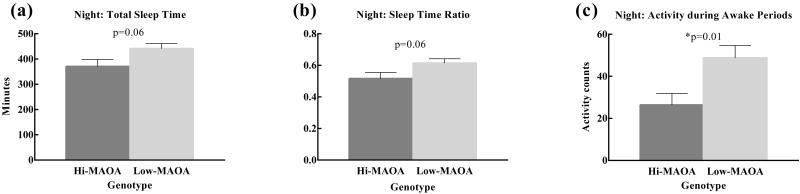
At one year of age, statistical trends suggested that low-MAOA monkeys slept more at night than hi-MAOA monkeys in terms of more total inactive (sleep) time (F(1,15)=4.11, p=.061) and greater percent inactive time (F(1,15)=4.16, p=.059) (Figure 2(a,b)). In addition, the low-MAOA monkeys were more active when they were awake at night (F(1,15)=8.15, p=0.01) (Figure 2(c)). Neither diet nor genotype influenced time to fall asleep, number of awakenings or time awake at night, or daytime sleep periods (data not shown).
Figure 2.
MAOA genotype effects on sleep and activity in one-year-old male rhesus monkeys. Analysis demonstrated a trend toward more sleep at night and a significantly lower level of activity during the day in the low-MAOA genotype group. (a) Night: Total Sleep Time suggests a longer sleep time in low-MAOA monkeys compared to hi-MAOA monkeys. (b) Night: Sleep Time Ratio suggests greater percent sleep at night in low-MAOA monkeys than in hi-MAOA monkeys. (c) Night: Activity during Awake Periods shows more activity in low-MAOA monkeys when they are awake at night compared to hi-MAOA monkeys.
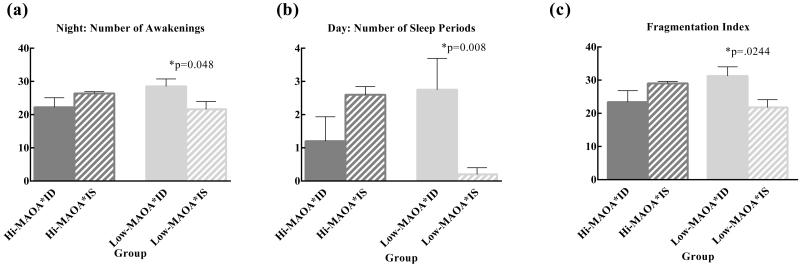
At two years of age, the trend toward greater sleep at night in the low-MAOA monkeys appeared to be reversed by ID. Awakenings at night were more frequent in the low-MAOA ID monkeys than low-MAOA IS monkeys (Figure 3(a)), (interaction F(1,15)=6.35, p=.023, low-MAOA ID>IS p=.048). Conversely, the low-MAOA ID monkeys had more frequent sleep periods during the day (Figure 3(b)) (interaction F(1,15)=11.69, p=.004, low-MAOA ID>IS, p=.008). Total sleep time during the day also showed a significant interaction (F(1,15)=5.69, p=.031) but no post hoc tests were significant. The pattern of means under the iron sufficient condition for both these endpoints suggested a difference between hi-MAOA IS and low-MAOA IS monkeys. Specifically the low-MAOA genotype with adequate prenatal iron had fewer nighttime awakenings (IS low-MAOA<hi-MAOA, p=.133) and fewer daytime sleep periods (IS low-MAOA<hi-MAOA, p=.008) than their hi-MAOA counterparts, but this pattern was reversed by prenatal iron deficiency suggesting a fragmentation of the consolidated sleep pattern. A fragmentation index (number of awakenings at night + number of sleep periods during the day) showed an MAOA × ID interaction (F(1,15)=9.03, p=.01, low-MAOA ID>IS p=.02) (Figure 3(c)). The pattern of means again suggested a difference between iron sufficient high and low-MAOA monkeys (IS low-MAOA<hi-MAOA, p=.053) that was reversed by iron deficiency.
Figure 3.
MAOA × ID effects on sleep and activity in two-year-old male rhesus monkeys. (a) Night: Number of Awakenings shows an MAOA × ID interaction. The low-MAOA ID animals had significantly higher number of awakenings than the low-MAOA IS animals. (b) Day: Number of Sleep Periods shows an MAOA × ID interaction. The low-MAOA ID group had more sleep periods during the day than the low-MAOA IS group. (c) Fragmentation Index, the number of awakenings at night + the number of sleep periods during the day. Low-MAOA ID animals showed more fragmented sleep than low-MAOA IS animals.
Discussion
Research indicates that children differ widely in their sleep patterns. Although environmental conditions can be significant predictors(21), sleep patterns have trait-like stability across childhood(22). Biological origins of trait-like sleep patterns may lie in genetics and/or early developmental influences. This study suggests that MAOA polymorphism and prenatal iron deficiency may play a role in establishing individual differences in sleep patterns.
An effect of prenatal ID on later sleep patterns has not been studied in children. The best known association between ID and sleep has to do with treatment of restless leg syndrome with iron, which is effective in children(23) as well as adults. Ferritin, an index of iron status, was lower in the children who responded to iron therapy for restless legs. Direct links between the current study and the literature on restless leg syndrome and ID are difficult because (1) our subjects were not iron deficient at the time of sleep evaluation, and (2) actimeters were place on the trunk of the monkeys, rather than on a limb, so that limb movements were not recorded. However, links may be established if limb movements can be associated with nighttime awakenings in juvenile monkeys with low-MAOA genotypes and prenatal ID. Also it is possible to suggest that, during prenatal development, the fetus is receptive to low iron environment and programs its future regulation of activity to accommodate a projected lack of iron in the postnatal environment. Rat studies have shown that iron treatments during the developmental period of peak iron uptake in the brain alter expression of iron regulatory genes in the brain throughout the life span(24).
Although prenatal ID has not been studied for an association with sleep regulation, follow-up studies with sleep assessment have been conducted after infant ID, corrected by supplements. At ten years of age, formerly ID children showed more awakenings at night and more leg movements as determined by EMG during monitored overnight sleep(25). Another recent study of activity in 5-year olds after correction of infant ID demonstrated differences from controls on several actimeter measures including more sleep during the day, more activity during sleep during the day, and greater variability in activity during sleep at night(26). An earlier actigraph study in this population as infants recorded more awake time at night and more activity when awake at night in anaemic infants(27). That finding was interpreted as reduced motor inhibition via nigrostriatal dopamine pathways, which are known to be influenced by ID(28). This research supports a potential role of developmental iron deficiency in establishing patterns of sleep regulation in children. A potential interaction of developmental ID with MAOA genotype could also be explored in children.
Rodent studies have shown that iron levels in brain regions follow a diurnal pattern that is disrupted in iron deficiency(29). Iron deficiency also disrupts diurnal patterns of monoamine neurotransmitter systems(30). MAOA was not measured in these studies but indices of dopamine metabolism suggested an impact on activity of this enzyme which could provide a biological basis for the ID*MAOA interaction identified in this study. As anticipated MAOA polymorphisms have been shown to influence monoamine neurotransmitters as reflected in CSF(13). 5HTTLPR polymorphisms, which also impact brain levels of dopamine, as well as serotonin, have recently been shown to influence sleep patterns in adolescents(31).
Recently, a study of college students linked low-MAOA polymorphisms with lower daytime sleepiness as assessed with a questionnaire(8). This result, based on subjective reports of sleepiness, is generally consistent with the pattern of less daytime sleep in the control (iron sufficient) low-MAOA compared to the control hi-MAOA group in the present study (Figure 3b). However, if the low-MAOA infants were iron deprived as fetuses the number of daytime sleep periods increased dramatically.
The interaction pattern of means for sleep parameters suggest that ID differentially affected the two genotypes but did not create abnormal sleep. While sleep fragmentation abnormalities can reach a level that signals sleep disorder, in the current experiment the greater fragmentation in the low-MAOA ID vs. IS monkeys did not lead to a level of fragmentation outside that seen in hi-MAOA IS subjects. MAOA is X-linked so that MAOA-hi and-low polymorphisms as determinants of MAOA transcription are clearly determined in males but vary widely depending on allele combinations in females. Further, the allele distribution of high and low transcribing polymorphisms in the human population is approximately 60/40, hi-/low-MAOA so that both polymorphisms are “normal.” Thus this interaction between MAOA gene transcription and fetal ID is more accurately identified as relevant to individual differences in sleep patterns than to sleep pathology. That said, this interaction in juveniles might be exaggerated in neonates or in aging when sleep patterns are more volatile and sensitive to disruption.
Supplementary Material
Acknowledgements
The authors acknowledge the contribution of the technical staff at CNPRC who assisted with actimeter placements for monitoring. Alicia Bulleri assisted with technical aspects of the project and data management and paper preparation. MSG designed the study and wrote the text. CEH conducted the experimental procedures, developed the actimeter protocols and reviewed the manuscript.
Supported by NIH P01 HD039386200 Betsy Lozoff (PI), Mari Golub PI Project 2. The NIH had no role in the design, analysis or writing of this article.
Footnotes
Conflict of Interest: None
References
- 1.Phillips AJK. Mammalian Sleep Dynamics: How Diverse Features Arise from a Common Physiological Framework. PLOS Computational Biology. 2010;6 doi: 10.1371/journal.pcbi.1000826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fuller PM, Gooley JJ, Saper CB. Neurobiology of the sleep-wake cycle: sleep architecture, circadian regulation, and regulatory feedback. J Biol Rhythms. 2006;21:482–493. doi: 10.1177/0748730406294627. [DOI] [PubMed] [Google Scholar]
- 3.American Academy of Sleep Medicine . International classification of sleep disorders, 2nd ed: Diagostic and coding manual. 2nd ed Westchester, IL: 2005. [Google Scholar]
- 4.Balzamo E, Santucci V, Seri B, et al. Nonhuman primates: laboratory animals of choice for neurophysiologic studies of sleep. Lab Anim Sci. 1977;27:879–886. [PubMed] [Google Scholar]
- 5.Balzamo E, Van Beers P, Lagarde D. Scoring of sleep and wakefulness by behavioral analysis from video recordings in rhesus monkeys: comparison with conventional EEG analysis. Electroencephalogr Clin Neurophysiol. 1998;106:206–212. doi: 10.1016/s0013-4694(97)00152-1. [DOI] [PubMed] [Google Scholar]
- 6.Hsieh KC, Robinson EL, Fuller CA. Sleep architecture in unrestrained rhesus monkeys (Macaca mulatta) synchronized to 24-hour light-dark cycles. Sleep. 2008;31:1239–1250. [PMC free article] [PubMed] [Google Scholar]
- 7.Golub MS, Hogrefe CE, Tarantal AF, et al. Diet-induced iron deficiency anemia and pregnancy outcome in rhesus monkeys. Am J Clin Nutr. 2006;83:647–656. doi: 10.1093/ajcn.83.3.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ojeda DA, Nino CL, Lopez-Leon S, et al. A functional polymorphism in the promoter region of MAOA gene is associated with daytime sleepiness in healthy subjects. J Neurol Sci. 2014;337:176–179. doi: 10.1016/j.jns.2013.12.005. [DOI] [PubMed] [Google Scholar]
- 9.Golub MS, Hogrefe CE, Unger EL. Influence of prenatal iron deficiency and MAOA genotype on response to social challenge in rhesus monkey infants. Genes Brain Behav. 2012;11:278–290. doi: 10.1111/j.1601-183X.2012.00772.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Golub MS, Hogrefe CE. Prenatal iron deficiency and monoamine oxidase A (MAOA) polymorphisms: combined risk for later cognitive performance in rhesus monkeys. Genes Nutr. 2013 doi: 10.1007/s12263-013-0381-3. in revision. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Unger EL, Hurst AR, Georgieff MK, et al. Behavior and monoamine deficits in prenatal and perinatal iron deficiency are not corrected by early postnatal moderate-iron or high-iron diets in rats. J Nutr. 2012;142:2040–2049. doi: 10.3945/jn.112.162198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aklillu E, Karlsson S, Zachrisson OO, et al. Association of MAOA gene functional promoter polymorphism with CSF dopamine turnover and atypical depression. Pharmacogenet Genomics. 2009;19:267–275. doi: 10.1097/FPC.0b013e328328d4d3. [DOI] [PubMed] [Google Scholar]
- 13.Jonsson EG, Norton N, Gustavsson JP, et al. A promoter polymorphism in the monoamine oxidase A gene and its relationships to monoamine metabolite concentrations in CSF of healthy volunteers. J Psychiatr Res. 2000;34:239–244. doi: 10.1016/s0022-3956(00)00013-3. [DOI] [PubMed] [Google Scholar]
- 14.Zalsman G, Huang YY, Harkavy-Friedman JM, et al. Relationship of MAO-A promoter (u-VNTR) and COMT (V158M) gene polymorphisms to CSF monoamine metabolites levels in a psychiatric sample of caucasians: A preliminary report. Am J Med Genet B Neuropsychiatr Genet. 2005;132B:100–103. doi: 10.1002/ajmg.b.30094. [DOI] [PubMed] [Google Scholar]
- 15.Capitanio JP, Del Rosso LA, Calonder LA, et al. Behavioral effects of prenatal ketamine exposure in rhesus macaques are dependent on MAOA genotype. Exp Clin Psychopharmacol. 2012;20:173–180. doi: 10.1037/a0026773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Newman TK, Syagailo YV, Barr CS, et al. Monoamine oxidase A gene promoter variation and rearing experience influences aggressive behavior in rhesus monkeys. Biol Psychiatry. 2005;57:167–172. doi: 10.1016/j.biopsych.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 17.Golub MS, Donald JM. Effect of intrapartum meperidine on behavior of 3- to 12-month-old infant rhesus monkeys. Biol Neonate. 1995;67:140–148. doi: 10.1159/000244155. [DOI] [PubMed] [Google Scholar]
- 18.Golub MS, Hogrefe CE, Germann SL, et al. Neurobehavioral evaluation of rhesus monkey infants fed cow’s milk formula, soy formula, or soy formula with added manganese. Neurotoxicol Teratol. 2005;27:615–627. doi: 10.1016/j.ntt.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 19.Golub MS, Keen CL, Gershwin ME. Behavioral and hematologic consequences of marginal iron-zinc nutrition in adolescent monkeys and the effect of a powdered beef supplement. Am J Clin Nutr. 1999;70:1059–1068. doi: 10.1093/ajcn/70.6.1059. [DOI] [PubMed] [Google Scholar]
- 20.Golub MS, Keen CL, Gershwin ME. Moderate zinc-iron deprivation influences behavior but not growth in adolescent rhesus monkeys. J Nutr. 2000;130:354S–357S. doi: 10.1093/jn/130.2.354S. [DOI] [PubMed] [Google Scholar]
- 21.Lozoff B, Wolf AW, Davis NS. Sleep problems seen in pediatric practice. Pediatrics. 1985;75:477–483. [PubMed] [Google Scholar]
- 22.Jenni OG, Molinari L, Caflisch JA, et al. Sleep duration from ages 1 to 10 years: variability and stability in comparison with growth. Pediatrics. 2007;120:e769–776. doi: 10.1542/peds.2006-3300. [DOI] [PubMed] [Google Scholar]
- 23.Amos LB, Grekowicz ML, Kuhn EM, et al. Treatment of Pediatric Restless Legs Syndrome. Clin Pediatr (Phila) 2013 doi: 10.1177/0009922813507997. epub ahead of print, doi: 10.1177/0009922813507997. [DOI] [PubMed] [Google Scholar]
- 24.Dornelles AS, Garcia VA, de Lima MN, et al. mRNA expression of proteins involved in iron homeostasis in brain regions is altered by age and by iron overloading in the neonatal period. Neurochem Res. 2010;35:564–571. doi: 10.1007/s11064-009-0100-z. [DOI] [PubMed] [Google Scholar]
- 25.Peirano P, Algarin C, Chamorro R, et al. Iron deficiency anemia in infancy exerts long-term effects on the tibialis anterior motor activity during sleep in childhood. Sleep Med. 2012;13:1006–1012. doi: 10.1016/j.sleep.2012.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Angulo-Barroso RM, Peirano P, Algarin C, et al. Motor activity and intra-individual variability according to sleep-wake states in preschool-aged children with iron-deficiency anemia in infancy. Early Hum Dev. 2013;89:1025–1031. doi: 10.1016/j.earlhumdev.2013.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Angulo-Kinzler RM, Peirano P, Lin E, et al. Twenty-four-hour motor activity in human infants with and without iron deficiency anemia. Early Hum Dev. 2002;70:85–101. doi: 10.1016/s0378-3782(02)00092-0. [DOI] [PubMed] [Google Scholar]
- 28.Erikson KM, Jones BC, Beard JL. Iron deficiency alters dopamine transporter functioning in rat striatum. J Nutr. 2000;130:2831–2837. doi: 10.1093/jn/130.11.2831. [DOI] [PubMed] [Google Scholar]
- 29.Unger EL, Earley CJ, Beard JL. Diurnal cycle influences peripheral and brain iron levels in mice. J Appl Physiol. 2009;106:187–193. doi: 10.1152/japplphysiol.91076.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bianco LE, Unger EL, Earley CJ, et al. Iron deficiency alters the day-night variation in monoamine levels in mice. Chronobiol Int. 2009;26:447–463. doi: 10.1080/07420520902820905. [DOI] [PubMed] [Google Scholar]
- 31.Carskadon MA, Sharkey KM, Knopik VS, et al. Short sleep as an environmental exposure: a preliminary study associating 5-HTTLPR genotype to self-reported sleep duration and depressed mood in first-year university students. Sleep. 2012;35:791–796. doi: 10.5665/sleep.1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.