Abstract
SET is a multifunctional protein involved in regulating many biological processes of the cell cycle. It is also a regulator of steroidogenesis in the ovary. However, the expression of SET protein in testis, and its function, still remains ambiguous. In this study, we observed the expression of SET in the testes of mice at different developmental stages, and have discussed its potential function in regulating spermatogenesis and androgen production. Forty-eight male mice at different developmental stages (1 week old as the infancy group; 4 weeks old as the prepubertal group; 12 weeks old as the adult group; over 12 months old as the ageing group) were used. Cellular location of SET protein in the testes was observed by immuno-histochemistry. Expression levels of Set mRNA and SET protein were analyzed by quantitative polymerase chain reaction and Western blotting. SET protein was expressed in spermatogonial cells and spermatocytes; the highest level was mainly in haploid and tetraploid cells of the prepubertal and adult groups, and Leydig cells of the adult and ageing groups. There was a low expression in Sertoli cells. Expression of Set mRNA in the prepubertal group was significantly higher than that in the adult group (P < 0.05), while expression of SET protein was at the highest level in the adult group (P < 0.05). SET protein is mainly expressed in spermatogonial cells and spermatocytes, and poorly expressed in Sertoli cells, suggesting that it is involved in spermatogenesis. Expression of SET protein in Leydig cells suggests a possible role in steroidogenesis.
Keywords: androgen production, Leydig cell, Sertoli cell, SET, spermatogenesis
INTRODUCTION
Mammalian spermatogenesis is a unique and complex process of cell division and differentiation.1 Germ cells of the seminiferous epithelium ultimately form spermatozoa through three stages: proliferation of spermatogonia, meiosis of spermatocytes, and spermiogenesis. The whole process of spermatogenesis is controlled by a complete hypothalamic-pituitary-gonadal axis. Gonadotrophin-releasing hormone, one of the more important neuropeptides released from the hypothalamus, regulates reproduction by inducing pulsatile discharges of gonadotrophins, luteinizing hormone (LH) and follicle-stimulating hormone (FSH), from the anterior pituitary gland.2,3,4 LH stimulates testicular Leydig cells to secrete testosterone (T). It is T, not LH directly, that stimulates certain phases of spermatogenesis. FSH with T stimulates mitosis of spermatogonia. Many testicular factors and cytokines are even more importantly involved in regulating spermatogenesis, and Sertoli cells play central roles in this process. Much is unknown on the local regulation of spermatogenesis.
SET (patient “SE translocation”), also termed I2PP2A, PHAP-II, and TAF-1b, an inhibitor of protein phosphatase 2A (PP2A), was first identified in 1992 in a study of leukemia.5 SET has multiple cellular functions, including control of cell cycle, gene transcription, apoptosis, cell migration, and epigenetic regulation. SET has an important role in facilitating cellular growth and proliferation and interacts with pathways that promote tumorigenesis and metastasis. SET protein is expressed at high levels in the developing gonads, gonadal steroid hormone-producing cells, mature oocytes and spermatocytes.6 PP2A, a serine/threonine phosphatase, is a key regulator of diverse signaling pathways.7 SET-mediated PP2A inhibition is an important regulatory mechanism for a number of physiological and pathological processes, including cell differentiation, apoptosis and carcinogenesis.8 One study demonstrated that SET protein in oocytes is essential for the faithful sister chromatid segregation by removing the protection of centromeric cohesion in meiosis II.9 Our evidence suggests that in the ovary SET protein regulates T production by regulating both the promoter activity of CYP17 and the biological activity of P450c17.10,11,12,13 However, it remains still unknown the roles that SET protein plays in testicular spermatogenesis and androgen production.
In this study, we observed the expression of SET protein in the testes of mice at different developmental stages, and have discussed its potential function in regulating spermatogenesis and steroidogenesis. This increases our knowledge of the paracrine and autocrine regulation of testicular functions.
MATERIALS AND METHODS
Animals and protocol
Male ICR mice (named by the Institute for Cancer Research of American) of Specific Pathogen Free Grade II were purchased from Medical Animal Center of Jiangsu Province in Nanjing Medical University. ICR mice have a life span of about 18 months. Mice aged 4 weeks are at the stage of prepubertal development or early maturity. Mice aged 8 weeks have attained sexual maturation; while mice over 12 months are ageing (almost equal to humans over 50 years). Twelve mice aged 1 week were assigned to the infant group (sexual infantilism); 12 mice aged 4 weeks were used as the prepubertal group (early maturity); 12 mice aged 12 weeks were considered as the adult group (sexual maturation) and 12 mice aged over 12 months were used as the ageing group. This study was approved by the Animal and Human Ethics Board of Nanjing Medical University.
After 5 days’ adaptation in the animal house, mice were killed under narcosis with 50 mg kg−1 pentobarbital sodium intraperitoneal (1% (w/v) concentration, 100 μl for 20 g mouse). One testis (left side) from a mouse was fixed in Bouin's solution for pathological exam and immunostaining, another one (right side in general) was freshly washed in ice-cold saline and immediately stored in liquid nitrogen. The fresh testis frozen in liquid nitrogen was then divided into two pieces for RNA isolation and protein extraction, individually. Four testes from the infant group were too small to be divided into two pieces, so two samples were pooled for RNA isolation and other two for protein extraction only. There were 10 samples in the infant group for real-time polymerase chain reaction (RT-PCR) and 10 for Western blot, but 12 samples in other three groups.
Immuno-histochemical analysis
To observe the cellular localization of SET expression and qualitative comparisons, immuno-histochemistry was performed as described previously.14,15 After Bouin-fixation, the paraffin-embedded testicular tissue was cut in 4-μm sections, deparaffinized, and then incubated in 3% (v/v) H2O2 to inhibit endogenous peroxidase activity. The sections were blocked with 5% (w/v) bovine serum albumin and incubated overnight at 4´ with primary antibody to SET (Santa Cruz Biotechnology, Dallas, USA, sc-25564, 1:1000 dilution). Negative control sections were incubated with the corresponding nonimmunized IgG. Following three PBS washes, the sections were incubated with horseradish peroxidase (HRP)-conjugated secondary antibody. Immuno-reactive sites were visualized brown with diamino-benzidine (DAB) and mounted for bright-field microscopy (Axioskop 2 plus; Zeiss, Bremen, Germany). Immuno-histochemical experiments were repeated at least 3 times on different testicular samples from the four groups.
Isolation of total RNA and real-time polymerase chain reaction
Homogenization of the fresh testicular tissues and isolation of total RNA were performed according to the manufacturer's instructions using a Trizol-based commercial kit (Takara Shuzo Co. Ltd., Kyoto, Japan). The purity of each RNA sample was determined by the absorbance ratio at 260 and 280 nm. The integrity of RNA preparations was evaluated by electrophoresis on a 1.2% (w/v) agarose gel containing 0.005% (v/v) a nucleic acid dye, Goldview (Shanghai SaiBaiSheng, Shanghai, China). The extracted RNA, containing ribosomal 28S and 18S RNA with a ratio of absorbance intensity 1.0–1.5 was used for qRT-PCR. In the real-time PCR reaction, cDNAs were used as templates for amplification to quantify the mRNA levels of target genes by using Quanti Tect SYBR Green PCR kits (Takara Shuzo Co Ltd, Kyoto, Japan). The primers were Set sense 5´-CGACGAGACCTCAGAAAAA- GAA-3´ (product size 207 bp) and antisense 5´-TGGTTGACAAATGTTGTTAC- CCA-3´, and β-actin sense 5-CCGTAAAGACCTCTATGCC-3´ and antisense 5´- CTCAGTAACAGTCCGCCTA-3´ (product size 216 bp). Relative quantification of target gene expression was estimated by the 2(−ΔΔCt) method, and qRT-PCR experiments were performed with 10 samples in the infant group and 12 samples in other three groups.
Western blot analysis
Western blots were used for quantitative detection of SET expression in the four groups. Extraction of the total protein of fresh testicular tissue was performed according to the manufacturer's instructions by using the Cell and Tissue Protein Extraction Reagent (Kang Chen KC-415, Shanghai, China), and protein was quantified by using a BCA Protein Assay Kit (Kang Chen KC-430). Ten micrograms of protein was separated by 12% (w/v) sodium dodecyl sulfate-polyacrylamide gel electrophoresis and then transferred onto polyvinylidene difluoride membranes (GE Healthcare, San Francisco, CA, USA). The membrane was blocked in a solution of Tris-buffered saline (TBS) containing 5% (w/v) nonfat milk for 1 h at 37´ and then incubated with rabbit anti-SET antibody (1:1000 dilution, Santa Cruz Biotechnology, sc-25564), or rabbit anti-tubulin antibody (1:10,000 dilution, Abcam, Ab6046-100, Cambridge, MA USA) at 4´ overnight. After being washed in TBS + 0.1% Tween 20 (TBST) thrice for 15 min at 37´ the membrane was incubated for 1 h at 37´ with the appropriate HRP-conjugated secondary antibody (Beijing ZhongShan, Beijing, China). The membrane was washed 3 times for 15 min in TBST. Specific proteins were detected in a darkroom by using an enhanced chemiluminescence detection kit according to the manufacturer's instructions (Amersham Biosciences, London, UK). Western blot experiments were performed with 10 samples in the infant group and 12 samples in other three groups.
Statistical analysis
The original data were shown as the mean ± standard deviation and the results were analyzed using one-way ANOVA test together with the Scheffe’ multiple-range test by the SPSS statistical package 11.0 (SPSS Inc., Chicago, IL, USA). The one-way ANOVA test was used to evaluate generally the difference between groups and the Scheffe’ multiple-range test was used to compare the two groups. P < 0.05 was considered as statistical significance.
RESULTS
Cellular localization and qualitative comparison of SET expression in testes
Results of immuno-histochemical analysis showed that SET protein was mainly located in the cytoplasm and nucleus of spermatogenic cells, as well as in Leydig cells (Figure 1). There was also low expression of SET protein in Sertoli cells. There were different expressions in the testes of mice at different developmental stages, with a consistent cellular localization. SET protein was mainly expressed in spermatogonia and spermatocytes, with the highest expression in haploid and tetraploid cells of 4-week-old mice and 12-week-old mice, and the Leydig cells of 12-week-old mice. Qualitatively, the DAB density in the adult group appeared stronger than those of the infant group and the ageing group (Figure 1).
Figure 1.
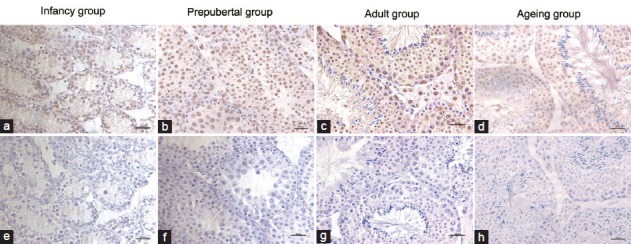
Cellular localization and qualitative comparison of SET expression in mouse testis. There are four groups of the infant (a), prepubertal (b), sexual maturation (c), and ageing (d). Negative control sections are incubated with nonimmunized normal corresponding IgG (e-h). Scale bars = 20 μm.
Expressions of Set mRNA in the four groups
To examine quantitatively the expression levels of the Set gene in the four groups, real-time PCR was performed (Figure 2a). Expression of Set mRNA was very low in the infant group. In general, the prepubertal group had the highest level of Set expression in the four groups. The prepubertal group had significantly higher expression than the infant group (P < 0.05). Expression of Set mRNA in the adult group was maintained at a high level. Interestingly, expression of Set mRNA in the ageing group was significantly lower than that in the adult group (P < 0.05).
Figure 2.
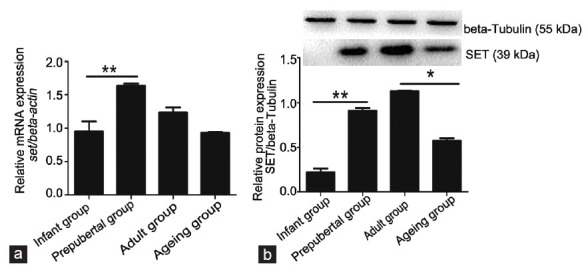
The expression levels of Set mRNA and SET protein in the testes of mice at different developmental stages. There are 10 samples for RNA isolation and 10 samples for protein extraction in the infant group. For other three groups, there are 12 samples for qRT-PCR or Western blot. (a) qRT-PCR analysis of Set mRNA in the four groups. Relative level of SET mRNA is normalized to the reference gene of β-actin. The prepubertal group had the highest level of Set expression in the four groups. The prepubertal group had significantly higher expression than the infant group, while expression of Set mRNA in the ageing group was significantly lower than that in the adult group. (b) Western blot analysis of SET protein in the four groups. Representative autoradiograph of SET protein is normalized using the β-tubulin signal. The expression of SET protein in the prepubertal group was significantly increased than that in the infant group, while the expression in the ageing group was lower than that in the adult group. Differences are determined to be statistically significant if P < 0.05 (*) and P < 0.01 (**).
Expressions of SET protein in the four groups
The different expressions of SET protein in the testicular tissues of the four groups were analyzed by Western blot (Figure 2b). The expression of SET protein in the infant group was very low. The expression of SET protein in the prepubertal group was significantly increased than that in the infant group, with a 4-fold increase (P < 0.05). The expression in the adult group was highest among the four groups (P < 0.05). When compared with the adult group, there was a large decrease in expression of SET protein in the ageing group, with a 2-fold decrease (P < 0.05).
Expression of SET protein in the seminiferous epithelium at different stages
SET protein was expressed in all of the seminiferous epitheliums at different Stages (Figure 3). However, it was mainly expressed in spermatogonia and spermatocytes. There was less expression of SET protein, or even no expression, in testicular spermatozoa.
Figure 3.
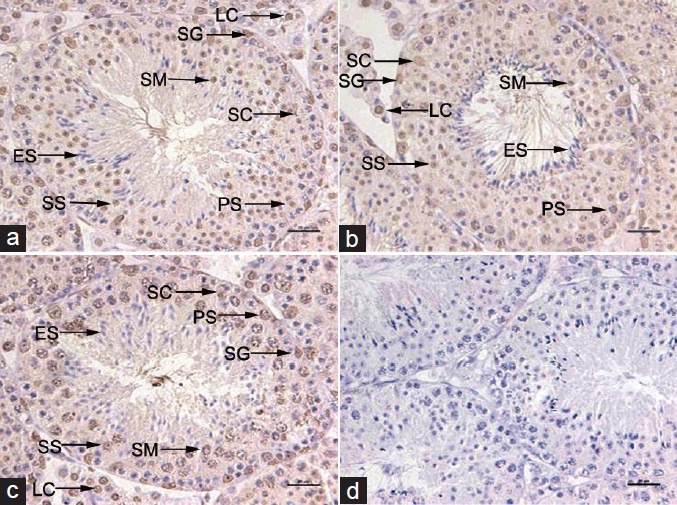
Expression of SET protein in the adult seminiferous epithelium at different stages. The seminiferous epithelium at three stages of the cycle is shown (a) Stages I-VI; (b) Stages VII-VIII; (c) Stages IX-XII; (d) negative control. The immature germ cells lying next to the wall (basement membrane) are spermatogonia. The multi-layered spermatocytes are arranged in the middle of the seminiferous epithelium, while the spermatids and subsequently transformed testicular spermatozoa are arranged near the lumen at the defined stages. SET protein is also expressed at low level in Sertoli cells in the seminiferous epithelium at different stages. Scale bars = 20 μm SG: spermatogonium; PS: primary spermatocyte; SS: secondary spermatocyte; SM: spermatid; ES: elongating spermatid; LC: Leydig cell; SC: Sertoli cell.
DISCUSSION
Testosterone is primarily produced by testicular Leydig cells, which is mainly regulated by LH.16 By binding to the membrane LH receptor, LH induces cAMP synthesis in Leydig cells. cAMP catalyzes PKA synthesis, and PKA transports cholesterol from the cytoplasmic pool to the mitochondria to promote steroidogenesis by steroidogenic enzymes (e.g., CYP11A1, HSD3B) and StAR.17,18 PP2A has been shown to dephosphorylate P450c17; this in turn is regulated by SET, a phosphoprotein, that inhibits its activity. SET is expressed in human NT2 neuronal precursor cells, suggesting a role for it in neuro-steroidogenesis.19 In this study, it was found that SET protein was expressed in the cytoplasm of testicular Leydig cells, suggesting that SET plays a role in regulating androgen production. So it is necessary to investigate the pathway (s) of SET protein in Leydig cells. Binding of bacterially-expressed SET protein to the DNA site at 2418/2399 of rat P450c17 gene trans-activates P450c17 in in vitro cultured Leydig cells.20 The central role of PP2A in SET-mediated regulation of T production was confirmed in our previous study by the findings that SET promoted the lyase activity of P450c17 and that PP2A inhibited its lyase activity.11
The seminiferous tubule exhibits a production line of germ cells; the earlier stages of spermatogenesis (spermatogonia) are found at the basal side of the tubule; towards the tubular lumen, germ cells are in progressively later stages of differentiation.21 Maturing spermatids are nearest the center of the seminiferous tubule, with their tails extending into its lumen. Spermatogenesis in seminiferous tubule from primitive stem cell, the type A spermatogonium, to spermatozoa and matured sperm passes through several complex transformations (Figure 3). In the seminiferous epithelium, the immature germ cells lying next to the wall (basement membrane) are the spermatogonia. These cells multiply by mitosis, and then continue to divide mitotically. Some of them change in appearance, which is called primary spermatocyte. The latter then completes two meiotic divisions to produce haploid cells, the spermatids. As shown in Figure 3, multi-layered spermatocytes were arranged in the middle of seminiferous epithelium, while the spermatides and subsequently transformed spermatozoa (sperm cells) and matured spermatozoa arranged near to the lumen at the defined stages. The cell associations, also called stages of the cycle of the seminiferous epithelium, are most accurately defined by the morphology of the developing acrosomes and of the nuclei of early spermatids.22 Along the seminiferous tubules, these stages follow each other in a wave-like fashion. This makes it possible to study the effect of different regulators on spermatogenesis, by collecting tubular segments from different stages of the cycle.
Pituitary FSH and testicular androgen are the main hormones that regulate spermatogenesis.23,24 Sertoli cells are generally considered the main targets of these hormones in the seminiferous epithelium. In the rat, the various generations of germ cells form the specific associations, labeled I through XIV, follow each other in a fixed sequence in a given area of seminiferous tubules.21 In 1956, Oakberg proposed that the cycle of the mouse seminiferous epithelium was subdivided into 12 stages based on the first 12 steps of the development of spermatids.25 Stages VII and VIII appear to be androgen-dependent.26 The secretory activity of Sertoli cells significantly changes during the cycles, suggesting that Sertoli cells play key roles in the cycle of the seminiferous epithelium. In our previous studies, we found that SET protein was predominantly expressed in theca cells and oocytes in human and mouse ovaries, which located in nucleus, cytoplasm, and chondriosome. Here, we inferred that the role (s) of the expressed SET protein in Sertoli cells was related to metabolism, cell activation, cytokines synthesis and secretion, etc., However, spermatogenic cells are mainly dependent on those factors produced by Sertoli cells under stimulation by FSH.23,24 The local mechanisms are poorly understood. In this study, we found that SET protein was expressed in spermatogonia and spermatocytes of the seminiferous epithelium at different stages. The highest expression of SET protein mainly occurred in haploid and tetraploid cells. There was less expression of SET protein, or even none, in testicular spermatozoa. In recent years, a large number of testicular genes have been claimed to be involved in the chromatin dynamics and DNA repair during mitosis and meiosis. Our findings suggest potential effects of SET protein in spermatogenesis.
The separase-mediated stepwise removal of cohesion, first from chromosome arms and later from the centromere region, is a prerequisite for maintaining sister chromatids together until their separation in meiosis II.27 Centromeric cohesion is protected from separase-dependent removal in meiosis I through the activatin of PP2A-B56 phosphatase, which is recruited to centromeres by shugoshin/MEI-S332 (Sgo).28,29 Matsumoto et al. was the first to study the role of SET protein in chromatin remodeling and transcription.30 Their study suggested that SET protein stimulated both initiation and elongation in DNA replication and transcription of the adenovirus genome as the template-activating factor Ib. SET protein masks the histone tails as a component of the inhibitor of the histone acetyl-transferase complex.31 Recently, SET protein was also found to be a potent activator of chromatin transcription in the early step of transcriptional processes.32 In this study, we found that SET protein was expressed at a lower level in the seminiferous tubule of infantile mice. The increased expression of SET protein in adult mice is in accord with continuous spermatogenesis.
CONCLUSIONS
SET protein is mainly expressed in spermatogonia and spermatocytes of the seminiferous epithelium, as well as Leydig cells. There was low expression of SET protein in Sertoli cells. SET protein has multiple cellular functions, suggesting its direct or indirect regulation of spermatogenesis and androgen production in the testis.
AUTHOR CONTRIBUTIONS
YGC designed the study and revised manuscript. XND and SL carried out the main experiments and wrote the first draft of manuscript. YGC, JYL, CG, LG and LS examined the data, performed statistical analysis, and proofread manuscript. All authors read and approved the final manuscript.
COMPETING INTERESTS
The authors declare no competing interests.
ACKNOWLEDGMENTS
The study was supported by projects from the National Natural Science Foundation of China (NSFC-81370754, 81170559) to YGC and a project funded by the priority academic program development (PAPD) of Jiangsu High Education Institutions.
REFERENCES
- 1.Roosen-Runge EC. The process of spermatogenesis in mammals. Biol Rev Camb Philos Soc. 1962;37:343–77. doi: 10.1111/j.1469-185x.1962.tb01616.x. [DOI] [PubMed] [Google Scholar]
- 2.Zhaohui Z, Yugui C, Yuanming Z, Xuesong W, Xiaobing J, et al. Effect of acupuncture on pubertal development of rats and rabbits at different developmental stages. Neuropeptides. 2007;41:249–61. doi: 10.1016/j.npep.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 3.Zhaohui Z, Jingzhu Z, Guipeng D, Xuesong W, Yuanming Z, et al. Role of neuropeptide Y in regulating hypothalamus-pituitary-gonad axis in the rats treated with electro-acupuncture. Neuropeptides. 2012;46:133–9. doi: 10.1016/j.npep.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 4.Themmen APN, Huhtaniemi IT. Mutations of gonadotropins and gonadotropin receptors: elucidating the physiology and pathophysiology of pituitary-gonadal function. Endocr Rev. 2000;21:551–83. doi: 10.1210/edrv.21.5.0409. [DOI] [PubMed] [Google Scholar]
- 5.von Lindern M, van Baal S, Wiegant J, Raap A, Hagemeijer A, et al. Can, a putative oncogene associated with myeloid leukemogenesis, may be activated by fusion of its 3’ half to different genes: characterization of the set gene. Mol Cell Biol. 1992;12:3346–55. doi: 10.1128/mcb.12.8.3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nagata K, Saito S, Okuwaki M, Kawase H, Furuya A, et al. Cellular localization and expression of template-activating factor I in different cell types. Exp Cell Res. 1998;240:274–81. doi: 10.1006/excr.1997.3930. [DOI] [PubMed] [Google Scholar]
- 7.Trotta R, Ciarlariello D, Dal Col J, Allard J, 2nd, Neviani P, et al. The PP2A inhibitor SET regulates natural killer cell IFN-gamma production. J Exp Med. 2007;204:2397–405. doi: 10.1084/jem.20070419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Madeira A, Pommet JM, Prochiantz A, Allinquant B. SET protein (TAF1beta, I2PP2A) is involved in neuronal apoptosis induced by an amyloid precursor protein cytoplasmic subdomain. FASEB J. 2005;19:1905–7. doi: 10.1096/fj.05-3839fje. [DOI] [PubMed] [Google Scholar]
- 9.Chambon JP, Touati SA, Berneau S, Cladière D, Hebras C, et al. The PP2A inhibitor I2PP2A is essential for sister chromatid segregation in oocyte meiosis II. Curr Biol. 2013;23:485–90. doi: 10.1016/j.cub.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 10.Boqun X, Xiaonan D, Yugui C, Lingling G, Xue D, et al. Expression of SET protein in the ovaries of patients with polycystic ovary syndrome. Int J Endocrinol. 2013;2013:367956. doi: 10.1155/2013/367956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao LL, Liu XQ, Xu BQ, Jiang SW, Cui YG, et al. SET/PP2A system regulates androgen production in ovarian follicles in vitro. Mol Cell Endocrinol. 2013;374:108–16. doi: 10.1016/j.mce.2013.04.012. [DOI] [PubMed] [Google Scholar]
- 12.Xu B, Gao L, Cui Y, Gao L, Dai X, et al. SET protein up-regulated testosterone production in the cultured preantral follicles. Reprod Biol Endocrinol. 2013;11:9. doi: 10.1186/1477-7827-11-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pandey AV, Mellon SH, Miller WL. Protein phosphatase 2A and phosphoprotein SET regulate androgen production by P450c17. J Biol Chem. 2003;278:2837–44. doi: 10.1074/jbc.M209527200. [DOI] [PubMed] [Google Scholar]
- 14.Liu S, Dai X, Cai L, Ma X, Liu J, et al. Effect of Hsp27 on early embryonic development in the mouse. Reprod Biomed Online. 2013;26:491–9. doi: 10.1016/j.rbmo.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 15.Liu S, Dai XN, Wang J, Luo J, Gao C, et al. Expression of calretinin in the testes of rats at different development stages. J Reprod Med. 2012;21:70–6. [Google Scholar]
- 16.Dufau ML. Endocrine regulation and communicating functions of the Leydig cell. Annu Rev Physiol. 1988;50:483–508. doi: 10.1146/annurev.ph.50.030188.002411. [DOI] [PubMed] [Google Scholar]
- 17.Henriksén K, Hakovirta H, Parvinen M. Testosterone inhibits and induces apoptosis in rat seminiferous tubules in a stage-specific manner: In situ quantification in squash preparations after administration of ethane dimethane sulfonate. Endocrinology. 1995;136:3285–91. doi: 10.1210/endo.136.8.7628362. [DOI] [PubMed] [Google Scholar]
- 18.Haider SG. Leydig cell steroidogenesis: unmasking the functional importance of mitochondria. Endocrinology. 2007;148:2581–2. doi: 10.1210/en.2007-0330. [DOI] [PubMed] [Google Scholar]
- 19.Compagnone NA, Zhang P, Vigne JL, Mellon SH. Novel role for the nuclear phosphoprotein SET in transcriptional activation of P450c17 and initiation of neurosteroidogenesis. Mol Endocrinol. 2000;14:875–88. doi: 10.1210/mend.14.6.0469. [DOI] [PubMed] [Google Scholar]
- 20.Zhang P, Compagnone NA, Fiore C, Vigne JL, Culp P, et al. Developmental gonadal expression of the transcription factor SET and its target gene, P450c17 (17alpha-hydroxylase/c17,20 lyase) DNA Cell Biol. 2001;20:613–24. doi: 10.1089/104454901753340604. [DOI] [PubMed] [Google Scholar]
- 21.Roosen-Runge EC, Giesel LO., Jr Quantitative studies on spermatogenesis in the albino rat. Am J Anat. 1950;87:1–30. doi: 10.1002/aja.1000870102. [DOI] [PubMed] [Google Scholar]
- 22.Leblond CP, Clermont Y. Definition of the stages of the cycle of the seminiferous epithelium in the rat. Ann N Y Acad Sci. 1952;55:548–73. doi: 10.1111/j.1749-6632.1952.tb26576.x. [DOI] [PubMed] [Google Scholar]
- 23.Wang C, Swerdloff RS. Male contraception. Best Pract Res Clin Obstet Gynaecol. 2002;16:193–203. doi: 10.1053/beog.2001.0270. [DOI] [PubMed] [Google Scholar]
- 24.O’Donnell L, Pratis K, Wagenfeld A, Gottwald U, Müller J, et al. Transcriptional profiling of the hormone-responsive stages of spermatogenesis reveals cell-, stage-, and hormone-specific events. Endocrinology. 2009;150:5074–84. doi: 10.1210/en.2009-0755. [DOI] [PubMed] [Google Scholar]
- 25.Oakberg EF. A description of spermiogenesis in the mouse and its use in analysis of the cycle of the seminiferous epithelium and germ cell renewal. Am J Anat. 1956;99:391–413. doi: 10.1002/aja.1000990303. [DOI] [PubMed] [Google Scholar]
- 26.Perryman KJ, Stanton PG, Loveland KL, McLachlan RI, Robertson DM. Hormonal dependency of neural cadherin in the binding of round spermatids to Sertoli cells in vitro. Endocrinology. 1996;137:3877–83. doi: 10.1210/endo.137.9.8756560. [DOI] [PubMed] [Google Scholar]
- 27.Petronczki M, Siomos MF, Nasmyth K. Un ménage à quatre: the molecular biology of chromosome segregation in meiosis. Cell. 2003;112:423–40. doi: 10.1016/s0092-8674(03)00083-7. [DOI] [PubMed] [Google Scholar]
- 28.Clift D, Marston AL. The role of shugoshin in meiotic chromosome segregation. Cytogenet Genome Res. 2011;133:234–42. doi: 10.1159/000323793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kitajima TS, Sakuno T, Ishiguro K, Iemura S, Natsume T, et al. Shugoshin collaborates with protein phosphatase 2A to protect cohesin. Nature. 2006;441:46–52. doi: 10.1038/nature04663. [DOI] [PubMed] [Google Scholar]
- 30.Matsumoto K, Nagata K, Ui M, Hanaoka F. Template activating factor I, a novel host factor required to stimulate the adenovirus core DNA replication. J Biol Chem. 1993;268:10582–7. [PubMed] [Google Scholar]
- 31.Seo SB, McNamara P, Heo S, Turner A, Lane WS, Chakravarti D. Regulation of histone acetylation and transcription by INHAT, a human cellular complex containing the set oncoprotein. Cell. 2001;104:119–30. doi: 10.1016/s0092-8674(01)00196-9. [DOI] [PubMed] [Google Scholar]
- 32.Gamble MJ, Erdjument-Bromage H, Tempst P, Freedman LP, Fisher RP. The histone chaperone TAF-I/SET/INHAT is required for transcription in vitro of chromatin templates. Mol Cell Biol. 2005;25:797–807. doi: 10.1128/MCB.25.2.797-807.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]


