ABSTRACT
There is growing evidence that activation of the Notch pathway can result in consequences on cell morphogenesis and behaviour, both during embryonic development and cancer progression. In general, Notch is proposed to coordinate these processes by regulating expression of key transcription factors. However, many Notch-regulated genes identified in genome-wide studies are involved in fundamental aspects of cell behaviour, suggesting a more direct influence on cellular properties. By testing the functions of 25 such genes we confirmed that 12 are required in developing adult muscles, consistent with roles downstream of Notch. Focusing on three, Reck, rhea/talin and trio, we verify their expression in adult muscle progenitors and identify Notch-regulated enhancers in each. Full activity of these enhancers requires functional binding sites for Su(H), the DNA-binding transcription factor in the Notch pathway, validating their direct regulation. Thus, besides its well-known roles in regulating the expression of cell-fate-determining transcription factors, Notch signalling also has the potential to directly affect cell morphology and behaviour by modulating expression of genes such as Reck, rhea/talin and trio. This sheds new light on the functional outputs of Notch activation in morphogenetic processes.
KEY WORDS: Notch, Reck, Talin, Trio, Gene regulation, Myogenesis, Drosophila
INTRODUCTION
Notch signalling is a local cell communication mechanism highly conserved throughout the animal kingdom. It is implicated in a variety of developmental and physiological processes and aberrant Notch activity is linked to many different diseases, including cancers and neurodegenerative disorders (Bolós et al., 2007; Louvi and Artavanis-Tsakonas, 2012). The Notch family of receptors and the Notch ligands, Delta and Serrate (Jagged in vertebrates), are cell surface type I transmembrane proteins. Upon ligand binding, Notch receptors undergo two proteolytic cleavages that lead to the release of the Notch intracellular domain (NICD). NICD is the active form of the receptor and acts in the nucleus as a transcriptional regulator, in cooperation with the DNA-binding protein CSL [also known as CBF1, Su(H), LAG-1] and its co-activator Mastermind (Bray, 2006). Thus, transduction of Notch signalling is relatively simple and primarily results in the regulation of target genes. The identification and characterization of Notch target genes is therefore crucial to fully understand the function of Notch in developmental processes.
The best characterized target genes of Notch encode transcription factors of the HES/E(spl) and Hey/Hesr/Herp gene families (Iso et al., 2003) but more-recent genome-wide studies have uncovered a broader spectrum of Notch-regulated genes (Djiane et al., 2012; Hurlbut et al., 2009; Krejcí et al., 2009; Mazzone et al., 2010; Terriente-Felix et al., 2013; Wang et al., 2011). Among these are genes directly involved in cell shape, cell organization and cell behaviour, whose functional relevance downstream of Notch has not been explored, despite the fact that Notch has been implicated in different morphogenetic processes independently of cell fate. These include the formation of boundaries between different cell populations, such as during somitogenesis and in the Drosophila wing imaginal disc (Becam and Milán, 2008; Major and Irvine, 2006), cell migration (Schober et al., 2005; Wang et al., 2007) and axon guidance [although the latter may involve a non canonical pathway (Le Gall et al., 2008)]. In the majority of contexts, this regulation of morphogenesis involves a transcriptional hierarchy, where activation of Notch results in the expression of a key transcription factor that in turn coordinates the cell behaviours (Niessen et al., 2008; Saad et al., 2010; Schober et al., 2005; Wang et al., 2007). However, it remains plausible that, in some tissues, Notch activity might have a more direct role in co-ordinating the genes that implement cell shape changes, although there is as yet little evidence to support this.
One context where Notch is required for regulating the behaviour of a specified group of cells is in the adult muscle progenitors (AMPs) in Drosophila. As in mammals, Notch activity is required to prevent premature differentiation of the AMPs, which are specified in the embryo and ultimately give rise to the adult muscles of the fly. (Anant et al., 1998; Delfini et al., 2000; Hirsinger et al., 2001). During larval stages, these progenitors proliferate to expand the pool of myoblasts and remain associated with imaginal discs (Bate et al., 1991). Then, at the beginning of metamorphosis the AMPs detach from the epithelium of the imaginal discs and migrate as a ‘swarm’ of associated cells (Roy and VijayRaghavan, 1998). At the target sites, myoblasts fuse with templates formed either from founder myoblasts or, in a few cases, from persistent larval muscles. In doing so, the fusing cells contribute to the differentiating muscle so that it achieves the appropriate size and structure. (Fernandes et al., 1991). Thus, adult myogenesis is a complex morphogenetic process, involving proliferation, migration, cell fusion and differentiation. As Notch signalling is active both in the proliferating AMPs, where it inhibits differentiation, and in the semi-differentiated migrating myoblasts (Bernard et al., 2006; Gildor et al., 2012), it could regulate many aspects of this morphogenetic process. Indeed, a significant fraction of the genes directly regulated by Notch in DmD8 cells encode cytoskeletal regulatory proteins. DmD8 cells are related to AMPs and in particular express the transcription factor Twist, which has been shown to function as a cooperating transcriptional activator for many Notch target genes (Bernard et al., 2010; Krejcí et al., 2009). This suggests that Notch could have a direct role in co-ordinating genes that control cell behaviours in this setting.
Among 25 putative Notch-regulated genes encoding cell-morphogenesis-related proteins that were identified through genome-wide studies (Djiane et al., 2012; Krejci and Bray, 2007; Terriente-Felix et al., 2013), we found that 12 are essential for generating adult flies capable of flight, consistent with appropriate functions in muscle formation. Focusing on three, Reck, rhea/talin (hereafter referred to as talin) and trio, we demonstrate that they are expressed in adult muscle progenitors and exhibit Notch regulation. Thus, these data support the model that Notch activity has the potential to directly regulate genes that coordinate cell morphology, in addition to its more widely accepted role in regulating such characteristics through cell-fate-determining transcription factors.
RESULTS
Identification of Notch target genes involved in adult myogenesis
We first selected a set of genes with known or inferred function in the control of cell shape, organization and behaviour from genome-wide datasets documenting genes associated with Su(H)-bound regions in response to increased Notch signalling in cultured cells and in wing imaginal discs (Djiane et al., 2012; Krejcí and Bray, 2007; Terriente-Felix et al., 2013). Such genes encode different types of proteins including Rho or Rac GTPase exchange factors (e.g. trio), cytoskeleton-binding proteins, a Netrin receptor implicated in cell migration and axon guidance (unc-5; Keleman and Dickson, 2001) and the matrix metalloproteinase inhibitor Reck, an inhibitor of cell invasion in cancer (Takahashi et al., 1998) (Table 1). To test their function in adult myogenesis, a morphogenetic process where Notch signalling is implicated in both cell fate and cell migration (Anant et al., 1998; Gildor et al., 2012), expression of hairpin RNAs targeting the individual genes was directed to the AMPs (using 1151-Gal4 as a driver). Progeny from these crosses were assayed for their ability to fly, as an indication of muscle formation and/or function, and all genes were categorized according to the percentage of flight-deficient individuals observed for each line (Fig. 1A; Table 1; supplementary material Table S1). This approach had the advantage that gene expression was knocked down at all post-embryonic stages of AMP development, from their proliferation in the larvae through to the forming muscles in pupae, but was not disrupted in the larval muscles nor in their embryonic progenitors (Anant et al., 1998). Finally, to avoid false positives from off-target effects or from side effects of RNA interference (RNAi) insertions, where possible we used lines from different sources that contained different types of RNAi constructs.
Table 1. List of candidate genes selected from genome wide studies.
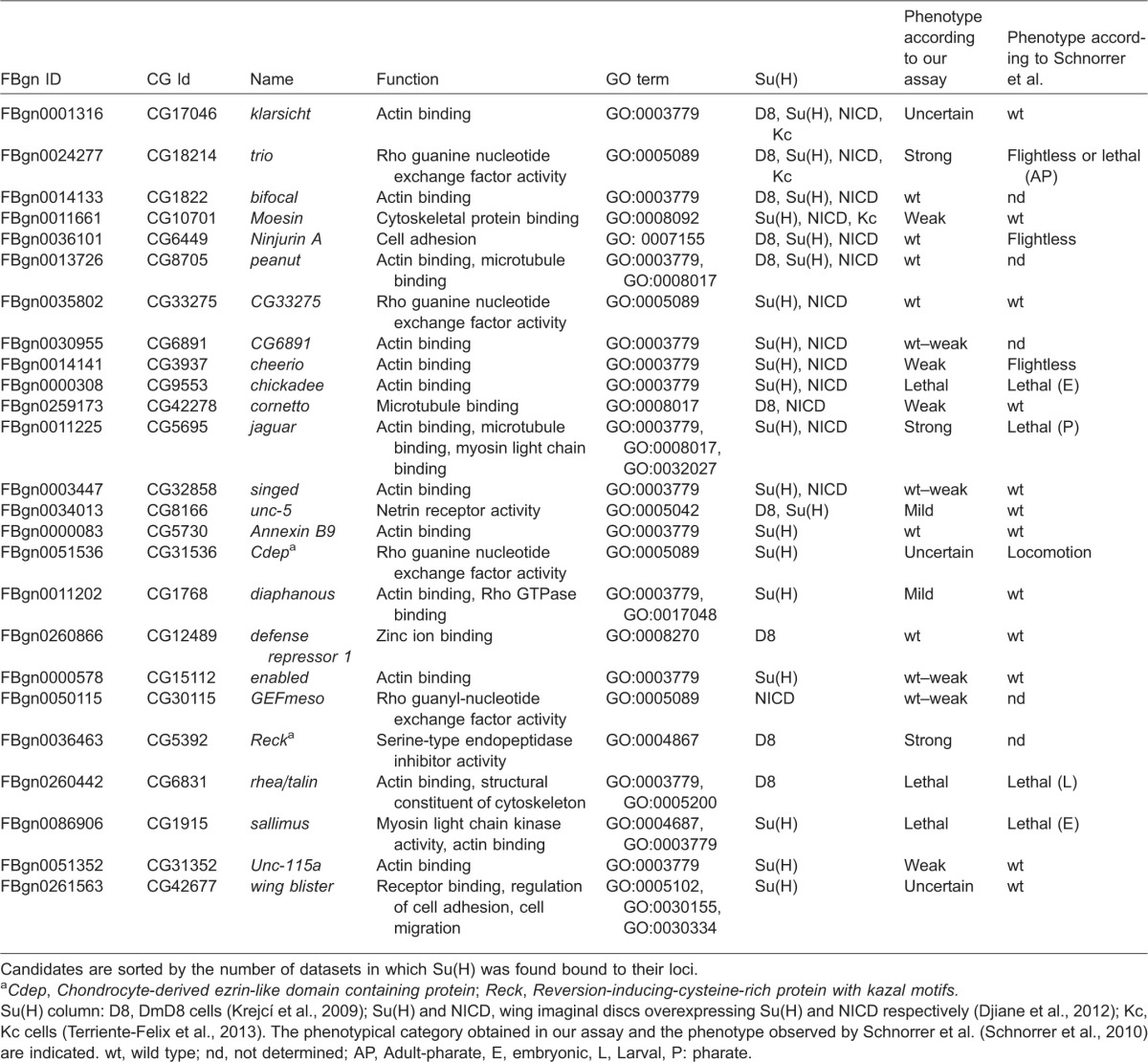
Candidates are sorted by the number of datasets in which Su(H) was found bound to their loci.
Cdep, Chondrocyte-derived ezrin-like domain containing protein; Reck, Reversion-inducing-cysteine-rich protein with kazal motifs.
Su(H) column: D8, DmD8 cells (Krejcí et al., 2009); Su(H) and NICD, wing imaginal discs overexpressing Su(H) and NICD respectively (Djiane et al., 2012); Kc, Kc cells (Terriente-Felix et al., 2013). The phenotypical category obtained in our assay and the phenotype observed by Schnorrer et al. (Schnorrer et al., 2010) are indicated. wt, wild type; nd, not determined; AP, Adult-pharate, E, embryonic, L, Larval, P: pharate.
Fig. 1.
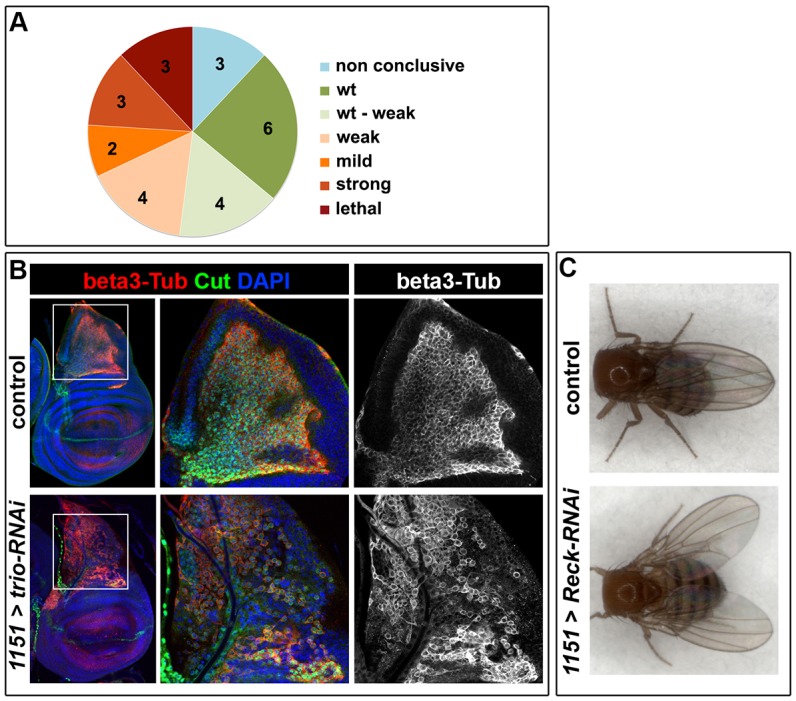
An RNAi assay identifies genes required for muscle formation. (A) Pie chart showing the proportion of genes whose loss of function resulted in the different phenotypical classes (see Materials and Methods for more details about classes). Note that the 1151-Gal4 driver is expressed in AMPs but not in the larval muscles, to avoid any confounding effects from defects in larval musculature (as might occur with mef2-Gal4). (B) AMP organization in wing imaginal discs from control (1151-Gal4) and trio-RNAi (1151-Gal4 UAS-trio-RNAi) larvae detected with β3-tubulin (beta3-Tub, red). Cut (in green) marks the nuclei of AMPs, and DAPI (blue) labels all nuclei. Altered cell morphology and organization is evident in trio-RNAi-expressing flies compared to control (representative examples of intermediate severity are shown). White squares indicate the regions shown at higher magnification. (C) Dorsal view of control (1151-Gal4) and 1151-Gal4-driven Reck-RNAi adult flies. The defective position of the wings observed, a ‘held out wings’ phenotype, has often been associated with flight muscle defects (Greene et al., 2003; Zaffran et al., 1997).
In most cases (18 out of 25) the results were consistent between different RNAi lines targeting the same gene (supplementary material Table S1). For example, lines targeting Reck and trio all induced a high percentage of flightless flies. However, four genes (ena, GEFmeso, singed and CG6891) were classified as ‘wt–weak’ since the results were inconclusive due to variability between lines, although overall there was little indication of any requirement in myogenesis. Furthermore, for three genes (Klar, wb and Cdep) the results were contradictory because one RNAi line induced a severe phenotype, whereas other lines targeting the same gene had no or very little effect. Such differences could be due to differences in the depletion efficiencies or to the inhibition of off target genes, but we have not pursued those genes further.
In total, knockdown of 12 genes gave consistent phenotypes suggesting that they are required for myogenesis (Fig. 1A; Table 1; supplementary material Table S1). Three of these (chic, talin and sls) gave lethality at the pharate stage when inhibited. This is consistent with a deficiency in the adult muscles, and is consistent with previous data showing that talin and sls have roles in myogenesis (Brown et al., 2002; Burkart et al., 2007). Furthermore, in a few instances the adults were found half emerged from the pupal case and, very occasionally a viable flightless fly eclosed. This suggests that the observed lethality was, at least in part, due to the inability to emerge from the puparium rather than to a major developmental defect. In agreement, one of the chic lines gave weaker phenotypes, generating adult flies of which 50% were flightless.
Inhibition of nine other genes, including corn and unc-5, resulted in flight defects with variable penetrance. In all cases the proportion of the progeny that were flightless varied, ranging from completely flightless flies to weak fliers. These appeared to represent a continuum in the severity of the same phenotype, because flightless flies were only observed when the progeny had a high percentage of flight defects, whereas weak fliers were often observed along with normal fliers. Nevertheless these included good candidates to mediate effects from Notch. For example both corn and unc-5 have been shown to be expressed in AMPs, and unc-5 contains a Notch-responsive enhancer (Krejcí et al., 2009).
A previous genome-wide RNAi screen to identify genes with function in muscle formation (Schnorrer et al., 2010) differed from our assay by using mef2-Gal4 as the driver, which is expressed in the muscle lineage from embryonic stages onwards. Despite this difference, the results are broadly consistent (Table 1). Only two (NijA and cher) of the ten genes with no or little effects in our assay were found to give a muscle phenotype with the more widespread depletion used in the Schnorrer et al. study. Likewise, among the six genes with a strong phenotype in our analysis, five had been found to be required for correct muscle formation in the previous screen (Schnorrer et al., 2010). Notably, three genes (chic, talin and sls) whose depletion induced lethality at pharate stage in our assay, gave embryonic or early larval lethality when inhibited from embryonic stage onwards so their role in later development was not considered in the previous study.
Reck, talin and trio are required in AMP lineages
Three genes with a penetrant phenotype obtained with all tested RNAi lines, Reck, talin and trio, were selected for further analysis because they had been shown to be associated with Su(H)-bound regions in response to increased Notch signalling in the AMP-related DmD8 cells (Table 1). In addition, they are well-conserved between Drosophila and vertebrates and have been previously linked to muscle formation: Reck, a metalloproteinase inhibitor, is expressed in developing muscles in the mouse where its expression is regulated by myogenic factors (Echizenya et al., 2005); Talin, a key component of integrin-mediated cell adhesion is involved at different stages of myogenesis in Drosophila (Brown et al., 2002); and Trio, a GTPase exchange factor (GEF) has a role in muscle formation in mammals, where it activates Rac1 to promote myoblast fusion (Charrasse et al., 2007; O'Brien et al., 2000).
Because 1151-Gal4 expression is not specifically restricted to AMPs [for example expression occurs in the salivary gland and some neural cells (Anant et al., 1998)], it was possible that the observed phenotypes were due to Reck, talin and trio inhibition in tissues other than AMPs. We thus first confirmed the requirement of these genes during adult muscle formation by using two other drivers, E(spl)m6-Gal4 and mef2-Gal4, to direct RNAi expression in AMPs (supplementary material Fig. S1). Although small differences were observed in the penetrance obtained with the three drivers (likely due to different expression levels and timing), overall the phenotypes observed for all three genes were very similar (Table 2). As expected, one line targeting talin induced lethality at embryonic or early larval stage when driven by mef2-Gal4. Given that mef-2-Gal4 is also expressed in embryonic and larvae muscles, these results are consistent with the requirement for talin during embryogenesis (Brown et al., 2002; Schnorrer et al., 2010). Similar lethal phenotypes were obtained when Notch-RNAi was expressed under the same conditions. Thus, these results confirmed that Reck, talin and trio are required in the myogenic lineages.
Table 2. Percentage of flies with flight defects obtained with three muscle drivers.
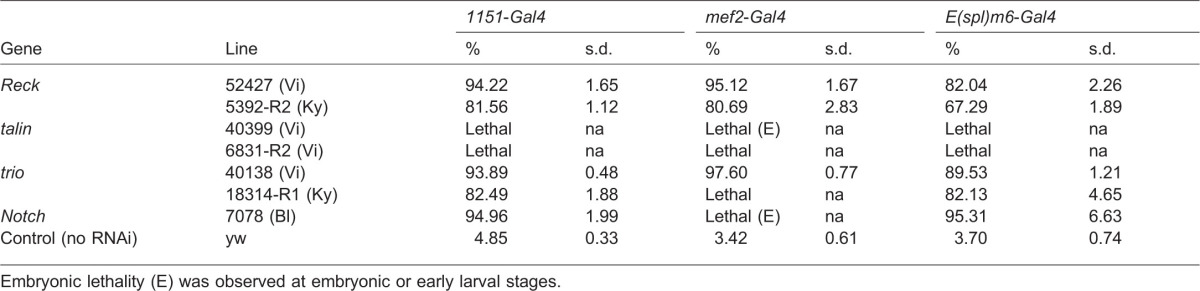
Embryonic lethality (E) was observed at embryonic or early larval stages.
Next, we sought to determine at what stage the three genes are required. To do so, we combined UAS-RNAi lines targeting Reck, talin and trio with the 1151-Gal4 driver in the presence of a thermo-sensitive derivative of Gal80 (Gal80ts), the Gal4 inhibitor (McGuire et al., 2003). RNAi expression was then induced at different stages by incubating larvae at temperatures where Gal80ts was inactivated. As a control, the experiment was first performed with an RNAi line targeting the transcription factor twist, which is required in AMPs but is downregulated when myoblasts differentiate (Anant et al., 1998). In agreement, we observed a severe phenotype when RNAi expression was initiated 2 days before pupation, but not when it was initiated within 12 h after pupation (Table 3). In contrast, inhibition of Notch after pupation resulted in a highly penetrant phenotype (albeit less penetrant and/or severe than when Notch was inhibited prior to pupation), suggesting a longer requirement for Notch compared to Twist. With Reck, talin and trio, the consequences were intermediate between the two. Severe phenotypes were observed when RNAi expression was induced before pupation but the phenotypes were considerably reduced when the animals were shifted within 12 h after pupation. Indeed, with trio-RNAi, phenotypes were already ameliorated with larvae shifted 1 day prior to pupation and were almost non-existent when shifted after pupation. Although it is not possible to distinguish exactly when the gene function will be perturbed, as the precise moment depends on the RNA and protein stability, these results suggest that the three genes are required for myogenesis at the end of larval stages and the beginning of pupation corresponding to the time when the AMPs start to migrate and fuse to form muscle fibres (Fernandes et al., 1991).
Table 3. Percentage of flight defects (or lethality) resulting from the induction of RNAi transcription at different stages.
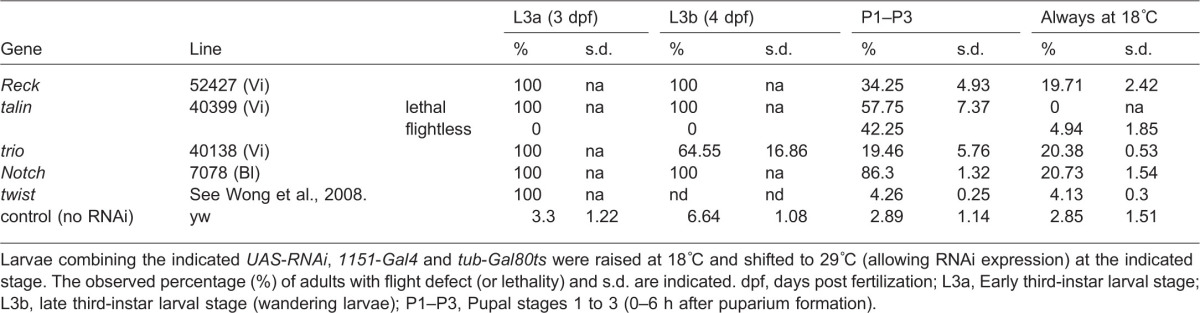
Larvae combining the indicated UAS-RNAi, 1151-Gal4 and tub-Gal80ts were raised at 18°C and shifted to 29°C (allowing RNAi expression) at the indicated stage. The observed percentage (%) of adults with flight defect (or lethality) and s.d. are indicated. dpf, days post fertilization; L3a, Early third-instar larval stage; L3b, late third-instar larval stage (wandering larvae); P1–P3, Pupal stages 1 to 3 (0–6 h after puparium formation).
In order to visualize AMPs shape, morphology and organization we used β3-tubulin (βTub60D), which is highly expressed specifically in the AMPs (Fig. 1B). Consistent with the requirement for Trio during the larval stages, we found that the AMPs were disorganized and had altered morphology when trio expression was inhibited. Indeed, in ∼30% of wing discs, AMP cells were abnormally dispersed, with evident gaps between cells (Fig. 1B). The same phenotype was observed with two independent RNAi constructs (K18214R-1 and V40138, n = 5/15 and 6/20 discs respectively), and suggests that Trio is important in sustaining the integrity of the AMP layer. No such robust changes were detected when Reck or talin expression was perturbed at this stage, although some cells appeared to have lost the expression of the myogenic protein β3-tubulin with one Reck RNAi (data not shown). Furthermore, although individual flies emerging after inhibition of Reck, talin and trio presented a wing posture defect (Fig. 1C; data not shown), using polarized light we did not detect any gross morphological abnormality of indirect flight muscles in these surviving flightless adults. Thus overall it seems likely that the flightless phenotype itself is due to subtler defects in muscle function and that there are also perturbations at earlier developmental stages that correlate to lethality before the flies eclose.
Reck, talin and trio are expressed in AMP cells
Given the implication that Reck, talin and trio are required in AMPs for normal development, we assessed whether they are expressed in these cells, as predicted. Although their overall patterns differed, all three genes were expressed in AMPs. Expression of Reck was largely restricted to the AMPs (Fig. 2A,B; supplementary material Fig. S2, for comparison, see expression of the AMP markers Cut and Mef2 in Fig. 2D,H) where it was present at all stages examined, including young third instar (L3) larvae (Fig. 2A) and early pupae (Fig. 2B). Interestingly the expression levels of Reck seemed to correlate with the AMP maturation process, as higher expression levels were detected at the onset of pupation. In addition, Reck was also detected in the peripodial margin, possibly reflecting a function in disc eversion (Srivastava et al., 2007). In contrast, Talin was broadly expressed throughout the wing disc, with a fairly ubiquitous pattern, as described previously (Brown et al., 2002). However, there was also a clear accumulation in the AMPs, making it plausible that there could be specific regulation of talin in these cells (Fig. 2C,D; supplementary material Fig. S2). Similarly, Trio protein appeared to be expressed widely throughout the wing disc, including in the AMPs (Fig. 2E; supplementary material Fig. S2). Furthermore, in situ hybridizations suggested some differential regulation of trio, with higher expression in the notal region and AMPs (Fig. 2F). Similar enrichment in the AMPs was detected using a trio ‘enhancer trap’ line [corresponding to a P-element insertion at the transcription start of the longest transcript of trio (Bateman et al., 2000)] and this high level of LacZ reporter expression was found to colocalize with the AMP marker Mef2 (Fig. 2G,H; supplementary material Fig. S2). Thus, Reck, talin and trio are all expressed in AMPs, consistent with their proposed function in adult myogenesis.
Fig. 2.
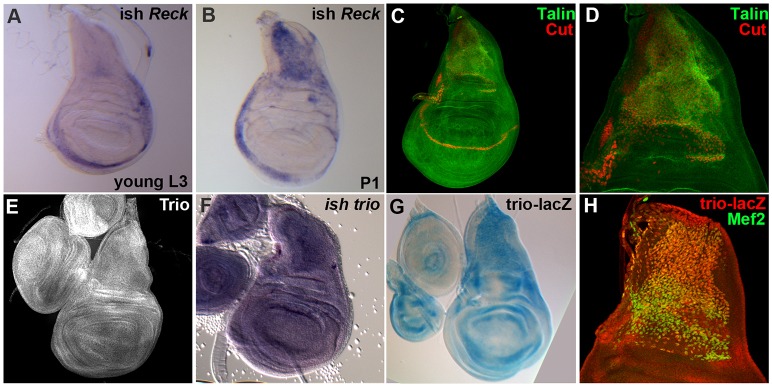
Reck, talin and trio are expressed in AMP cells. Expression profiles of Reck, talin and trio in the wing imaginal disc show that all three genes are expressed in AMPs. (A,B) Reck in situ hybridization (ish) in young L3 larvae (A) and P1 pupae (B). (C,D) talin expression profile revealed by immunostaining and co-stained with the AMP marker Cut (maximum projections of z-stacks from confocal acquisitions are presented; D is shown at twice the magnification of C). (E–H) trio expression profile shown by immunostaining (E, Maximum projections), In situ hybridization (F) and an enhancer-trap reporter line (G) also co-stained with the AMP marker mef2 (H) (H is shown at twice the magnification of G).
Notch regulated enhancers from Reck, talin and trio
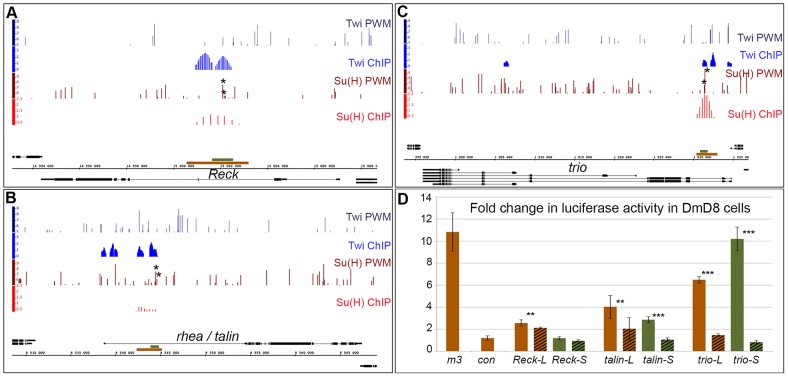
Reck, talin and trio have previously been identified in genome-wide studies as putative direct targets regulated by Notch signalling in the muscle lineage (Krejci et al., 2009). To verify this, we first tested whether the regions occupied by Su(H) in these three loci can function as Notch-responsive enhancers (NREs). DNA fragments corresponding to these Su(H)-bound regions (Fig. 3A–C, brown) were cloned upstream of a minimal luciferase reporter and their ability to respond to the activated form of Notch (NICD) was analysed in co-transfection assays using the DmD8, Twist-expressing, cell line. All three enhancers gave increased luciferase expression in the presence of NICD in DmD8 cells, and this response was compromised when the Su(H)-binding motifs were mutated [Fig. 3D; P<0.001 for trio and P<0.05 for talin and Reck – in each case the two best matches to the Su(H) position weight matrix (PWM) were mutated]. Of the three, the NRE from the trio locus gave the highest fold change in reporter expression in the presence of NICD (6.5 for the trio NRE versus 4.3 and 2.6 for the talin and Reck NRE, respectively) and also gave the most dramatic reduction when the Su(H) motifs were mutated. In contrast, the consequences on the Reck NRE from mutating two Su(H) motifs was modest, although significant. This suggests that additional motifs, with lower Patser scores, might also be important in this NRE. Nevertheless, all of the three identified regions behave as NREs, consistent with the Su(H) occupancy detected by chromatin immunoprecipitation (ChIP).
Fig. 3.
Su(H) ChIP identifies NREs in the Reck, talin and trio loci. (A–C) Genomic region surrounding Drosophila Reck (A), talin (B) and trio (C) genes. Black lines and boxes (exons) represent transcribed regions. Graphs show matches to Su(H) PWM (dark red bars; height of bar indicates Patser score 5–9.79), Twist (Twi) PWM (dark blue bars; Patser score 5–9.3), Su(H) [red; enrichment (AvgM log2)] and Twi (blue) ChIP-enriched regions in DmD8 cells (Bernard et al., 2010; Krejci et al., 2009). Brown and green rectangles represent long (brown) and short (green) fragments tested for their Notch signalling activation sensitivity. Asterisks indicate Su(H)-binding sites mutated in subsequent experiments. (D) Response of the indicated long (brown) and short (green) fragment to Notch signalling activation in transient transfection assays in DmD8 cells. Plain bars represent wild-type fragments and striped bars fragments in which Su(H) sites were mutated. E(spl)m3, which is inducible by Notch in all tested conditions (Krejci and Bray, 2007), was used as positive control. Results are mean±s.d. **P<0.05; ***P<0.001.
To further refine the NREs, smaller regions, centred on the Su(H) motifs, were assayed in a similar manner (∼800 bp, Fig. 3A–C, green). The fold of activation obtained with the short Reck and talin NREs showed a reduced ability to respond to Notch activation compared to their longer counterparts, suggesting that there are important regulatory elements in the surrounding region (Fig. 3D). Indeed, the shortened Reck NRE was no longer responsive to NICD, suggesting there are relevant Su(H) motifs in the larger fragment, which would explain the residual activity of the mutated long Reck NRE. Furthermore, all three long NREs contain Twist-binding regions, which are absent from their shorter counterparts (Fig. 3A–C, ‘Twi ChIP’, blue). Thus, the reduced expression in the short Reck and talin NREs is consistent with the model that, in DmD8, Twist functions as a cooperating transcriptional activator for many Notch target genes (Bernard et al., 2010). Surprisingly, the short trio NRE gave a fold change 50% higher than that of the longer trio NRE, even though it lacks the region previously found to be occupied by Twist. Thus, this trio NRE might bind Su(H) independently of Twist. In agreement, Su(H) was found bound to trio NRE not only in DmD8 cells but also Kc cells which do not express Twist (Terriente-Felix et al., 2013), whereas Reck and talin NRE were only bound to Su(H) in DmD8 cells.
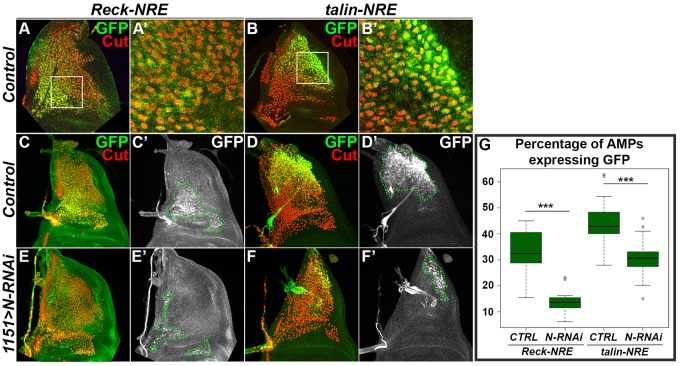
Reck and Talin NRE drive Notch-dependent GFP expression in AMP cells in vivo
To further confirm the relevance of the NREs for Reck, talin and trio expression during adult myogenesis, we tested whether they could drive expression of a reporter in the AMPs. To do this, the identified NRE sequences were cloned upstream of a minimal GFP reporter (Housden et al., 2012) and the resulting expression patterns analysed in wing discs from transgenic flies. Both Reck and talin NREs clearly directed expression of GFP in the AMPs (Fig. 4A–D′,G) although in both cases only in a subset of the AMPs. Interestingly, the subsets differed according to the NRE, which could reflect different stages in the maturation of these cells or different pre-patterns amongst the progenitors. In contrast, the trio NRE was not functional in the AMPs, although it did direct some expression in the wing disc epithelium (data not shown). Therefore, despite its ability to function effectively in transient transfection assays, this fragment appears to lack features that are crucial for activity in vivo. Finally, we tested whether the expression from the Reck and talin NREs were affected when Notch signalling was compromised. Inhibition of Notch in AMPs, using Notch-RNAi driven by 1151-Gal4, caused a clear reduction in the proportion of AMPs expressing the GFP (P<0.01 for both, Fig. 4C–F′,G). Thus the activity of the enhancers appears responsive to Notch signalling in vivo, supporting the hypothesis that Reck and talin are direct Notch targets during adult myogenesis. We also detected weak and variable reductions in expression from the endogenous genes under similar conditions of Notch-RNAi expression (supplementary material Fig. S3).
Fig. 4.
Reck and talin NREs drive Notch-dependent GFP expression in AMPs in vivo. (A–B′) Reck (A, a higher magnification is shown in A′) and talin (B, and higher magnification in B′) long NREs (L-NRE) drive GFP expression in a subset of AMPs as shown by GFP colocalizations with the AMP marker Cut. White squares in A and B indicate regions shown at higher magnification in A′ and B′. Single optical sections from confocal acquisitions are presented. (C–F′) Expression from reporters Reck-L-NRE-GFP (C,C′,E,E′) and talin-L-NRE-GFP (D,D′,F,F′) in wild-type (C–D′) and Notch-depleted conditions (E–F′). Cut expression was used to mark the AMPs. Green dotted lines in C′,D′,E′ and F′ outline Cut- and GFP-expressing cells. Maximum projections of z-stacks from confocal acquisitions are presented. (G) Boxplot representing the percentage of AMPs (Cut-expressing cells) expressing GFP from the Reck-L-NRE-GFP and talin-L-NRE-GFP reporters in wild-type (CTRL) and Notch-depleted (N-RNAi) conditions. The percentage of AMPs expressing GFP was estimated by manually measuring areas occupied by cells expressing Cut or Cut and GFP. The box represents the interquartile range, the middle line the median, and the whiskers show ± 1.5× the interquartile range. ***P<0.001.
Notch can induce ectopic expression of Reck, talin and trio
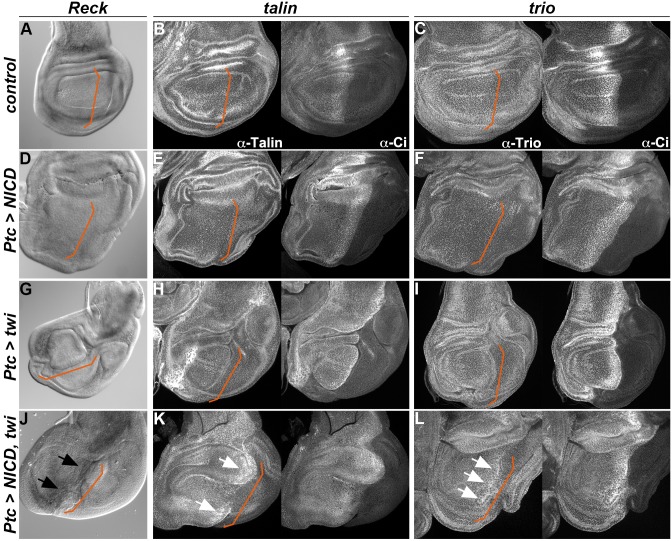
As a further test of whether the three genes can be regulated by Notch activity, we monitored their response to ectopic Notch activity in the wing disc, with and without the co-operating factor Twist. Initially, NICD alone was produced ectopically in a stripe across the wing pouch using the patched-Gal4 driver (ptc-Gal4; Fig. 5A–F). However, none of the genes showed any marked change in expression under these conditions. Subsequently, because many AMP Notch targets require the transcription factor Twist to be induced by Notch (Bernard et al., 2010), we tested the effect of co-expressing NICD and Twist together and of expressing Twist alone. This combination induced ectopic expression of all three genes within the ptc expression domain (Fig. 5J–L), whereas none was clearly upregulated when Twist was expressed alone (Fig. 5G–I). This was even the case for Trio, suggesting that Twist does cooperate with Notch at this target, despite the fact that the NRE binds Su(H) even in cells that lack Twist. Taken together, these results suggest that Reck, talin and trio are regulated by Notch in vivo when Twist is present in the same cells. Although this regulation is likely direct, based on the results from the reporters, it remains possible that there are also indirect mechanisms involved.
Fig. 5.
Notch can induce ectopic expression of Reck, talin and trio in the presence of Twist. (A–L) ptc-Gal4; Tub-Gal80ts was used to drive expression of NICD and/or Twist (Twi) in the wing pouch. The expression profile of Reck (A,D,G,J, in situ hybridization), talin (B,E,H,K, immunostaining) and trio (C,F,I,L, immunostaining) in a wild-type wing pouch (A–C), in NICD-expressing discs (D–F), Twist-expressing discs (G–I) and NICD- plus Twist-expressing discs (J–L). Note the ectopic expression of Reck (J, black arrows) and the upregulation of talin and trio (K,L, white arrows) induced by Notch in the presence of Twist. Ci staining was used to indicate the limit between anterior and posterior domains, along which ptc-Gal4 is expressed (indicated with orange lines).
DISCUSSION
Notch signalling is widely implicated in the control of cell fate during development but also has been shown to influence cell architecture and behaviour in different morphogenetic processes. In most cases, Notch is proposed to coordinate cell morphogenesis by regulating the expression of key transcription factors, rather than by directly regulating the effector genes that implement the cell behaviours (Niessen et al., 2008; Saad et al., 2010; Schober et al., 2005; Wang et al., 2007). One well-characterized example is epithelial-to-mesenchymal transition (EMT), a process that can be triggered by Notch signalling [sometimes in combination with other pathways such as TGFβ (Espinoza and Miele, 2013; Wang et al., 2010)] through its regulation of the key transcription factors Snail and Slug (Niessen et al., 2008; Saad et al., 2010). Our results suggest, however, that genes involved in implementing cell morphology are also directly regulated by Notch. Building on previous genome-wide analyses of Notch-regulated genes, which revealed a wide spectrum of functional targets, we have found that Reck, talin and trio all have some characteristics of direct Notch targets in the muscle progenitors. Genome-wide analysis of CSL (also known as RBPJ) binding in mouse and human T-lymphoblastic leukaemia cells also identified several genes implicated in cell architecture regulation, although those differ from the genes analysed here (Wang et al., 2011). Direct control of genes with roles in co-ordinating cell morphology and behaviour might thus be a general feature of Notch activity in different morphogenetic processes.
Two of the three genes, trio and talin, are very widely expressed. Thus, a large proportion of their expression likely occurs independently of Notch. However, our identification of Su(H)-responsive enhancers associated with each of these genes suggests that Notch activity can modulate their expression at specific stages and/or in specific cell types. This therefore highlights the existence of different categories of Notch-regulated genes. Most commonly the focus is on cell-fate-determining genes, which are specifically switched on in a cell only when Notch activity is present. The regulation of trio and talin suggests that Notch activity also augments the expression of genes that are already transcribed, modifying their expression rather than initiating it de novo. Such subtle changes of gene expression would not have been uncovered by conventional approaches, demonstrating the utility of genome-wide studies in uncovering the full spectrum of target genes. In the AMPs, trio is important for sustaining the normal cell morphology, and might do so by regulating the interaction of AMPs with their niche (Lin et al., 2013) or their transition to a migrating population at the onset of pupation (Kashef et al., 2009; Moore et al., 2013). However, to distinguish the contribution that Notch regulation makes to trio and talin functions it will ultimately be necessary to eliminate the NREs from the endogenous genes.
In contrast, Reck exhibits a much more restricted expression pattern, being specifically upregulated in late-stage AMPs where Notch is active. The identified NRE directs expression in these cells, consistent with Reck expression being controlled by Notch activity in AMPs. Intriguingly, in some mammalian cells (e.g. cortical progenitors,) expression of RECK has been found to activate Notch signalling by directly inhibiting ADAM10-dependent processing of Notch ligands (Muraguchi et al., 2007). As the swarming myoblasts appear to undergo bi-directional Notch signalling, it is possible that Reck could be involved in subtle fine-tuning of signalling between cells. However, so far there is no evidence to indicate that Reck can inhibit Kuzbanian, the Drosophila Adam10 homologue. Instead Reck function has been linked to inhibition of the matrix metalloproteinase MMP1, through its ability, in conjunction with another metalloproteinase inhibitor, to suppress invasion of tumours that had upregulated MMP1 (Srivastava et al., 2007). Similar characteristics are well documented in mammals, where RECK functions as a tumour suppressor by inhibiting migration, invasion, and angiogenesis (Meng et al., 2008; Nagini, 2012; Noda et al., 2003; Takahashi et al., 1998). Although we have not been able to detect any gross defects in the organization of indirect flight muscles, suggesting that AMP migration is not severely affected when Reck is knocked-down, as the RNA is only upregulated in late stage AMPs it is more likely to be involved in positively promoting myoblast migration, similar to its role in zebrafish neural crest cells (Prendergast et al., 2012), than in suppressing migration.
Besides the three genes whose regulation we have analysed in some details, at least nine others appear to be required in adult myogenesis, based on the phenotypes seen when their expression in AMPs is ablated. Two of these, corn and unc-5, have been previously shown to be expressed in AMPs. For unc-5, the Su(H)-bound region has been tested and shown to function as an NRE in the AMPs. Thus, the evidence suggests that other putative targets will be regulated by Notch activity and will contribute to the functional output in regulating the AMPs cell behaviours. The model that emerges is that Notch activity not only regulates transcription factors important in conferring cell fate identify but also directly affects the expression levels of genes encoding proteins that implement cell fates, such as those with roles in regulating cellular architectures and behaviours.
MATERIALS AND METHODS
Drosophila stocks and genetics
Fly stocks used for RNAi experiments are from BDSC (Bloomington, Indiana, IN), DRGC (Kyoto, Japan) or VDRC (Vienna, Austria). Individual line numbers are indicated in supplementary material Table S1. We also used the UAS-twi-RNAi2x line to target twist expression (Wong et al., 2008). Gene ablation was obtained by crossing UAS-RNAi lines with the following drivers: 1151-Gal4 (Anant et al., 1998), mef2-Gal4 (Ranganayakulu et al., 1996) and E(Spl)m6-Gal4 (a gift from Alexis Lalouette, Université Paris-Denis Diderot, France), combined with an UAS-Dcr-2 (BDSC, Bl24650; Dietzl et al., 2007) to enhance the RNAi effect. Crosses were culture at 25°C and progeny was assayed for their ability to fly. In order to limit RNAi expression to a defined period of larval and pupal development, the 1151-Gal4 was combined with a tub-Gal80ts (McGuire et al., 2003). Crosses were cultured at 19°C and individually staged larvae or pupae (see Table 3) were shifted to 29°C (the non-permissive temperature for Gal80ts). Adults were then assayed for their ability to fly.
To assay AMP cell morphology following knockdown of Reck, talin and trio, wing imaginal discs from wandering larvae were dissected and stained for β3-tubulin expression. For the phenotype observed with trio, discs were scored on the basis of whether the AMP cells were abnormally dispersed, with gaps evident.
For Notch and Twist gain-of-function experiments the patched[559.1]-Gal4 driver combined with tub-Gal80ts (ptc-Gal4-Gal80ts) was used to drive UAS-Ni79.2 (NICD) and/or UAS-Twist expression (Baylies and Bate, 1996). Crosses were cultured at 19°C for 7 days, then shifted to 29°C for 48 h before dissection and staining.
The trio-LacZ reporter line (BL 8594) was previously described (Bateman et al., 2000).
Flight assay
For each RNAi line tested, at least 40 adult flies aged 2–8 days were assayed for their ability to fly. For this, flies were dump dropped from their vials at ∼50 cm from the bench and numbers of flies that fell on the bench were scored (i.e. flies that could not fly away). This test was repeated twice from independent crosses and the results were averaged. Depending on the percentage of flies with flight defect, each gene was categorized as wild-type (‘wt’; less than 5%), ‘weak’ (between 5% and 33%), ‘mild’ (between 33% and 66%) and ‘strong’ (more than 66%). When lines targeting the same gene were in different categories, the gene was assigned to the strongest category if all lines were in categories not different by more than one degree (e.g. strong and mild, or mild and weak), except with wt and weak in which case the gene was assigned to the wt–weak category. Finally, genes with lines giving very different results (e.g. weak and strong) were classified as ‘uncertain’.
Muscle preparation
Adult flies were fixed in 4% paraformaldehyde overnight. Thoraces were cut sagittally, mounted in glycerol and viewed under polarized light.
Luciferase experiments and GFP reporters
For luciferase assays, putative NRE fragments from Reck, talin and trio were amplified from Drosophila genomic DNA using primers containing restriction enzyme sequences and cloned into a luciferase vector containing a minimal promoter from the hsp70 gene (pGL3::Min). Genome release 5 coordinates of the cloned fragments were Reck, chr3L: 15000573–15003206 (long) and 15001662–15002553 (short); talin chr3L: 8542368–8545156 (long) and 8543963–8544835 (short); trio chr3L: 1030426–1033023 (long) and 1030865–1031821 (short). Matches to Su(H) motifs were identified using Patser (Hertz and Stormo, 1999) with the position weight matrix described previously (Krejcí et al., 2009).
Mutated Su(H) binding motifs were: in Reck-NRE, CATGGGAA>CATtGttA (at position 15002089) and GTCACACG>GaaAaACG (15002161); in talin-NRE, CATGGGAA>CATtGttA (8544448) and TGGGAGAA>TGGtAttA (8544370); in trio-NRE, TTCCCACG>TaaGaACG (1031441) and GTCCCACA>GaaCaACA (1031363). Cell culture conditions and transfections were as described previously (Nagel et al., 2005; Narasimha et al., 2008). At least three biological replicates were performed in all experiments. Significance of differences in luciferase measurements was assayed with unpaired, two-tailed Welch's t-test using R software (R Core Team, 2013).
To produce GFP reporters, long NREs (as defined above) were cloned in the pGreenRabbit vector (pGR) (Housden et al., 2012). Flies carrying the pGR transgenes were generated by Phi-C31-mediated site-directed integration on the 86Fb platform. To test whether GFP expression was dependent on Notch signalling, reporters were combined with 1151-Gal4 and UAS-Notch-RNAi. Crosses were cultured at 25°C for 4 days, then shifted to 29°C for 48 h before dissection of wandering third-instar larvae and staining. In all, 12–20 wing imaginal discs obtained from independent crosses were imaged. Areas occupied by cells expressing Cut (an AMP marker) or expressing Cut and GFP were measured using ImageJ (Rasband, 1997-2012). Significance of differences in measurements was assayed with unpaired, two-tailed Welch's t-test using R software.
Immunostaining and in situ hybridization
Antibody staining of wing imaginal discs was performed according to standard protocol. The following antibodies from DSHB (Developmental Studies Hybridoma Bank, Iow City, Io, USA) were used: rat anti-ECad (DCAD2, 1∶20), rat anti-Ci (2A1, 1∶20), mouse anti-Cut (2B10. 1∶20), mouse anti β-gal (40-1A, 1∶20) and mouse anti-Trio (9.4A, 1∶20) antibodies. We also used rabbit anti-GFP (1∶500, Life Technologies. Carlsbad, CA, USA), rabbit anti-Talin [1∶500, a gift from Nick Brown (Brown et al., 2002)], rabbit anti-Mef2 [1∶1000, gift of Bruce Paterson (Lilly et al., 1995)] and anti-β3-tubulin [1∶5000, gift from Renate Renkawitz-Pohl (Rudolf et al., 2012)] antibodies. Samples were imaged using a Nikon Eclipse C1 confocal miscroscope. Images were processed with ImageJ and assembled with Adobe Photoshop.
Reck and trio expression were analysed by in situ hybridization with RNA probes synthesised from PCR amplified DNA fragments (∼1.2 kb) corresponding to Reck exon 7 and 8, and trio exon 4 and 5 of the longest isoform. In both cases, the two probes gave the same pattern. In situ hybridization was performed according to standard protocols. Fluorescent in situ hybridization was performed using Tyramide Signal Amplification (Perkin-Elmer, Waltham, MA). Standard colorimetric staining was imaged using a Zeiss Axiophot miscroscope. Fluorescent samples were analysed as above.
Quantitative RT-PCR
Wing imaginal discs from third-instar control (1151-Gal4) and Notch-depleted (1151>N-RNAi) larvae were dissected (20 discs for each genotype). Dorsal halves (corresponding to the notum, where the AMPs are located) were separated from the wing pouch and used for RNA extraction using TRIzol (Life Technologies). Genomic DNA was eliminated using an Ambion DNA-free kit. cDNA was synthesized using random hexamers (Promega. Madisson, WI) and M-MLV reverse transcriptase (Promega). cDNA levels were quantified by real-time (RT)-PCR using QuantiTec Sybr Green PCR mix (Qiagen, Valencia, CA) and the AbiPrism machine. The calibration curve was constructed from serial dilutions of genomic DNA, and values for all genes were normalized to the levels of Elongation factor 2 (Ef2). The following primers were used. Ef2, Fwd 5′-GCCGATCTGCGCTCTAATAC-3′ and Rev 5′-ACGAGTATCCTGGACGATGG-3′ (within exon 5); Notch, Fwd 5′-TGCGATGTTCAGACGATTTC-3′ and Rev CGTATCCCTGGGAGCAGTAG-3′ (within exon 5); Reck, Fwd 5′-TGGACCAAAACTCGACACTG-3′ and Rev 5′-TACTCCTAGGCGGACAATGC-3′ (within exon 8); talin, Fwd 5′-CAGCAGCAGTGAACTTGGAG-3′ and Rev 5′-CTGGGTCATCGAGGTGAGTC-3′ (within exon 15); and trio Fwd 5′-ACCCATGAAAAGGACGTGAC-3′, Rev 5′-CTCTCCTGCTGATCCCTCTG-3′ (within exon 4 of the longest isoform).
Supplementary Material
Acknowledgments
We are grateful to Alexis Lalouette (Institut Jaques Monod, Université Paris-Denis Diderot, France) for the E(spl)m6-Gal4 line, to Renate Renkawitz-Pohl (Philipps-University Marburg, Germany) for the anti-β3-tubulin antibody, to Nick Brown (Gurdon Institute, University of Cambridge, UK) for anti-Talin antibody and to Bruce Paterson (Center for Cancer Research, National Cancer Institute, Bethesda, MD) for anti-Mef2 antibody. We also acknowledge the Bloomington Stock Center (BL), Vienna Drosophila RNAi Center (VDRC), The Kyoto Stock Center (DGRC) and The Developmental Studies Hybridoma Bank (DSHB) for providing Drosophila strains and antibodies. We thank members of the Bray laboratory for valuable discussions.
Footnotes
Competing interests
The authors declare no competing interests.
Author contributions
G.P., H.B. and S.B. conceived and designed the experiments; G.P., H.B. and K.M. performed the experiments; G.P., H.B. and S.B. analysed the data; G.P. and K.M. contributed reagents, materials and/or analysis tools; G.P. and S.B. wrote the paper.
Funding
This work was supported by a programme grant from the Medical Research Council, UK [grant number G0800034 to S.J.B.]; by fellowships to G.P. from Fondation pour la Recherche Medical and from Marie Curie (Intra European Fellowship) [grant number PIEF-GA-2009-236426]; and by an EMBO Long Term Fellowship to H.B. [grant number ALTF 325-2013]. Deposited in PMC for release after 6 months.
Supplementary material available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.151787/-/DC1
References
- Anant S., Roy S., VijayRaghavan K. (1998). Twist and Notch negatively regulate adult muscle differentiation in Drosophila. Development 125, 1361–1369 [DOI] [PubMed] [Google Scholar]
- Bate M., Rushton E., Currie D. A. (1991). Cells with persistent twist expression are the embryonic precursors of adult muscles in Drosophila. Development 113, 79–89 [DOI] [PubMed] [Google Scholar]
- Bateman J., Shu H., Van Vactor D. (2000). The guanine nucleotide exchange factor trio mediates axonal development in the Drosophila embryo. Neuron 26, 93–106. 10.1016/S0896-6273(00)81141-1 [DOI] [PubMed] [Google Scholar]
- Baylies M. K., Bate M. (1996). twist: a myogenic switch in Drosophila. Science 272, 1481–1484. 10.1126/science.272.5267.1481 [DOI] [PubMed] [Google Scholar]
- Becam I., Milán M. (2008). A permissive role of Notch in maintaining the DV affinity boundary of the Drosophila wing. Dev. Biol. 322, 190–198. 10.1016/j.ydbio.2008.07.028 [DOI] [PubMed] [Google Scholar]
- Bernard F., Dutriaux A., Silber J., Lalouette A. (2006). Notch pathway repression by vestigial is required to promote indirect flight muscle differentiation in Drosophila melanogaster. Dev. Biol. 295, 164–177. 10.1016/j.ydbio.2006.03.022 [DOI] [PubMed] [Google Scholar]
- Bernard F., Krejci A., Housden B., Adryan B., Bray S. J. (2010). Specificity of Notch pathway activation: twist controls the transcriptional output in adult muscle progenitors. Development 137, 2633–2642. 10.1242/dev.053181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolós V., Grego-Bessa J., de la Pompa J. L. (2007). Notch signaling in development and cancer. Endocr. Rev. 28, 339–363. 10.1210/er.2006-0046 [DOI] [PubMed] [Google Scholar]
- Bray S. J. (2006). Notch signalling: a simple pathway becomes complex. Nat. Rev. Mol. Cell Biol. 7, 678–689. 10.1038/nrm2009 [DOI] [PubMed] [Google Scholar]
- Brown N. H., Gregory S. L., Rickoll W. L., Fessler L. I., Prout M., White R. A. H., Fristrom J. W. (2002). Talin is essential for integrin function in Drosophila. Dev. Cell 3, 569–579. 10.1016/S1534-5807(02)00290-3 [DOI] [PubMed] [Google Scholar]
- Burkart C., Qiu F., Brendel S., Benes V., Hååg P., Labeit S., Leonard K., Bullard B. (2007). Modular proteins from the Drosophila sallimus (sls) gene and their expression in muscles with different extensibility. J. Mol. Biol. 367, 953–969. 10.1016/j.jmb.2007.01.059 [DOI] [PubMed] [Google Scholar]
- Charrasse S., Comunale F., Fortier M., Portales-Casamar E., Debant A., Gauthier-Rouvière C. (2007). M-cadherin activates Rac1 GTPase through the Rho-GEF trio during myoblast fusion. Mol. Biol. Cell 18, 1734–1743. 10.1091/mbc.E06-08-0766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delfini M. C., Hirsinger E., Pourquié O., Duprez D. (2000). Delta 1-activated notch inhibits muscle differentiation without affecting Myf5 and Pax3 expression in chick limb myogenesis. Development 127, 5213–5224 [DOI] [PubMed] [Google Scholar]
- Dietzl G., Chen D., Schnorrer F., Su K-C., Barinova Y., Fellner M., Gasser B., Kinsey K., Oppel S., Scheiblauer S. et al. (2007). A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448, 151–156. 10.1038/nature05954 [DOI] [PubMed] [Google Scholar]
- Djiane A., Krejci A., Bernard F., Fexova S., Millen K., Bray S. J. (2012). Dissecting the mechanisms of Notch induced hyperplasia. EMBO J. 32, 60–71. 10.1038/emboj.2012.326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echizenya M., Kondo S., Takahashi R., Oh J., Kawashima S., Kitayama H., Takahashi C., Noda M. (2005). The membrane-anchored MMP-regulator RECK is a target of myogenic regulatory factors. Oncogene 24, 5850–5857. 10.1038/sj.onc.1208733 [DOI] [PubMed] [Google Scholar]
- Espinoza I., Miele L. (2013). Deadly crosstalk: Notch signaling at the intersection of EMT and cancer stem cells. Cancer Lett. 341, 41–45. 10.1016/j.canlet.2013.08.027 [DOI] [PubMed] [Google Scholar]
- Fernandes J., Bate M., Vijayraghavan K. (1991). Development of the indirect flight muscles of Drosophila. Development 113, 67–77 [DOI] [PubMed] [Google Scholar]
- Gildor B., Schejter E. D., Shilo B-Z. (2012). Bidirectional Notch activation represses fusion competence in swarming adult Drosophila myoblasts. Development 139, 4040–4050. 10.1242/dev.077495 [DOI] [PubMed] [Google Scholar]
- Greene J. C., Whitworth A. J., Kuo I., Andrews L. A., Feany M. B., Pallanck L. J. (2003). Mitochondrial pathology and apoptotic muscle degeneration in Drosophila parkin mutants. Proc. Natl. Acad. Sci. USA 100, 4078–4083. 10.1073/pnas.0737556100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertz G. Z., Stormo G. D. (1999). Identifying DNA and protein patterns with statistically significant alignments of multiple sequences. Bioinformatics 15, 563–577. 10.1093/bioinformatics/15.7.563 [DOI] [PubMed] [Google Scholar]
- Hirsinger E., Malapert P., Dubrulle J., Delfini M. C., Duprez D., Henrique D., Ish-Horowicz D., Pourquié O. (2001). Notch signalling acts in postmitotic avian myogenic cells to control MyoD activation. Development 128, 107–116 [DOI] [PubMed] [Google Scholar]
- Housden B. E., Millen K., Bray S. J. (2012). Drosophila reporter vectors compatible with ΦC31 integrase transgenesis techniques and their use to generate new notch reporter fly lines. G3 (Bethesda) 2, 79–82. 10.1534/g3.111.001321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurlbut G. D., Kankel M. W., Artavanis-Tsakonas S. (2009). Nodal points and complexity of Notch-Ras signal integration. Proc. Natl. Acad. Sci. USA 106, 2218–2223. 10.1073/pnas.0812024106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iso T., Kedes L., Hamamori Y. (2003). HES and HERP families: multiple effectors of the Notch signaling pathway. J. Cell. Physiol. 194, 237–255. 10.1002/jcp.10208 [DOI] [PubMed] [Google Scholar]
- Kashef J., Köhler A., Kuriyama S., Alfandari D., Mayor R., Wedlich D. (2009). Cadherin-11 regulates protrusive activity in Xenopus cranial neural crest cells upstream of Trio and the small GTPases. Genes Dev. 23, 1393–1398. 10.1101/gad.519409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keleman K., Dickson B. J. (2001). Short- and long-range repulsion by the Drosophila Unc5 netrin receptor. Neuron 32, 605–617. 10.1016/S0896-6273(01)00505-0 [DOI] [PubMed] [Google Scholar]
- Krejcí A., Bray S. (2007). Notch activation stimulates transient and selective binding of Su(H)/CSL to target enhancers. Genes Dev. 21, 1322–1327. 10.1101/gad.424607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krejcí A., Bernard F., Housden B. E., Collins S., Bray S. J. (2009). Direct response to Notch activation: signaling crosstalk and incoherent logic. Sci. Signal. 2, ra1. 10.1126/scisignal.2000140 [DOI] [PubMed] [Google Scholar]
- Le Gall M., De Mattei C., Giniger E. (2008). Molecular separation of two signaling pathways for the receptor, Notch. Dev. Biol. 313, 556–567. 10.1016/j.ydbio.2007.10.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilly B., Zhao B., Ranganayakulu G., Paterson B. M., Schulz R. A., Olson E. N. (1995). Requirement of MADS domain transcription factor D-MEF2 for muscle formation in Drosophila. Science 267, 688–693. 10.1126/science.7839146 [DOI] [PubMed] [Google Scholar]
- Lin G., Zhang X., Ren J., Pang Z., Wang C., Xu N., Xi R. (2013). Integrin signaling is required for maintenance and proliferation of intestinal stem cells in Drosophila. Dev. Biol. 377, 177–187. 10.1016/j.ydbio.2013.01.032 [DOI] [PubMed] [Google Scholar]
- Louvi A., Artavanis-Tsakonas S. (2012). Notch and disease: a growing field. Semin. Cell Dev. Biol. 23, 473–480. 10.1016/j.semcdb.2012.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Major R. J., Irvine K. D. (2006). Localization and requirement for Myosin II at the dorsal-ventral compartment boundary of the Drosophila wing. Dev. Dyn. 235, 3051–3058. 10.1002/dvdy.20966 [DOI] [PubMed] [Google Scholar]
- Mazzone M., Selfors L. M., Albeck J., Overholtzer M., Sale S., Carroll D. L., Pandya D., Lu Y., Mills G. B., Aster J. C. et al. (2010). Dose-dependent induction of distinct phenotypic responses to Notch pathway activation in mammary epithelial cells. Proc. Natl. Acad. Sci. USA 107, 5012–5017. 10.1073/pnas.1000896107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire S. E., Le P. T., Osborn A. J., Matsumoto K., Davis R. L. (2003). Spatiotemporal rescue of memory dysfunction in Drosophila. Science 302, 1765–1768. 10.1126/science.1089035 [DOI] [PubMed] [Google Scholar]
- Meng N., Li Y., Zhang H., Sun X-F. (2008). RECK, a novel matrix metalloproteinase regulator. Histol. Histopathol. 23, 1003–1010 [DOI] [PubMed] [Google Scholar]
- Moore R., Theveneau E., Pozzi S., Alexandre P., Richardson J., Merks A., Parsons M., Kashef J., Linker C., Mayor R. (2013). Par3 controls neural crest migration by promoting microtubule catastrophe during contact inhibition of locomotion. Development 140, 4763–4775. 10.1242/dev.098509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muraguchi T., Takegami Y., Ohtsuka T., Kitajima S., Chandana E. P. S., Omura A., Miki T., Takahashi R., Matsumoto N., Ludwig A. et al. (2007). RECK modulates Notch signaling during cortical neurogenesis by regulating ADAM10 activity. Nat. Neurosci. 10, 838–845. 10.1038/nn1922 [DOI] [PubMed] [Google Scholar]
- Nagel A. C., Krejci A., Tenin G., Bravo-Patiño A., Bray S., Maier D., Preiss A. (2005). Hairless-mediated repression of notch target genes requires the combined activity of Groucho and CtBP corepressors. Mol. Cell. Biol. 25, 10433–10441. 10.1128/MCB.25.23.10433-10441.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagini S. (2012). RECKing MMP: relevance of reversion-inducing cysteine-rich protein with kazal motifs as a prognostic marker and therapeutic target for cancer (a review). Anticancer. Agents Med. Chem. 12, 718–725. 10.2174/187152012802650237 [DOI] [PubMed] [Google Scholar]
- Narasimha M., Uv A., Krejci A., Brown N. H., Bray S. J. (2008). Grainy head promotes expression of septate junction proteins and influences epithelial morphogenesis. J. Cell Sci. 121, 747–752. 10.1242/jcs.019422 [DOI] [PubMed] [Google Scholar]
- Niessen K., Fu Y., Chang L., Hoodless P. A., McFadden D., Karsan A. (2008). Slug is a direct Notch target required for initiation of cardiac cushion cellularization. J. Cell Biol. 182, 315–325. 10.1083/jcb.200710067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda M., Oh J., Takahashi R., Kondo S., Kitayama H., Takahashi C. (2003). RECK: a novel suppressor of malignancy linking oncogenic signaling to extracellular matrix remodeling. Cancer Metastasis Rev. 22, 167–175. 10.1023/A:1023043315031 [DOI] [PubMed] [Google Scholar]
- O'Brien S. P., Seipel K., Medley Q. G., Bronson R., Segal R., Streuli M. (2000). Skeletal muscle deformity and neuronal disorder in Trio exchange factor-deficient mouse embryos. Proc. Natl. Acad. Sci. USA 97, 12074–12078. 10.1073/pnas.97.22.12074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prendergast A., Linbo T. H., Swarts T., Ungos J. M., McGraw H. F., Krispin S., Weinstein B. M., Raible D. W. (2012). The metalloproteinase inhibitor Reck is essential for zebrafish DRG development. Development 139, 1141–1152. 10.1242/dev.072439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team(2013). R: A Language and Environment for Statistical Computing Vienna, Austria: R Foundation for Statistical Computing [Google Scholar]
- Ranganayakulu G., Schulz R. A., Olson E. N. (1996). Wingless signaling induces nautilus expression in the ventral mesoderm of the Drosophila embryo. Dev. Biol. 176, 143–148. 10.1006/dbio.1996.9987 [DOI] [PubMed] [Google Scholar]
- Rasband W. S. (1997-2012). ImageJ Bethesda, MD: US National Institutes of Health [Google Scholar]
- Roy S., VijayRaghavan K. (1998). Patterning muscles using organizers: larval muscle templates and adult myoblasts actively interact to pattern the dorsal longitudinal flight muscles of Drosophila. J. Cell Biol. 141, 1135–1145. 10.1083/jcb.141.5.1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolf A., Buttgereit D., Rexer K. H., Renkawitz-Pohl R. (2012). The syncytial visceral and somatic musculature develops independently of β3-Tubulin during Drosophila embryogenesis, while maternally supplied β1-Tubulin is stable until the early steps of myoblast fusion. Eur. J. Cell Biol. 91, 192–203. 10.1016/j.ejcb.2011.11.002 [DOI] [PubMed] [Google Scholar]
- Saad S., Stanners S. R., Yong R., Tang O., Pollock C. A. (2010). Notch mediated epithelial to mesenchymal transformation is associated with increased expression of the Snail transcription factor. Int. J. Biochem. Cell Biol. 42, 1115–1122. 10.1016/j.biocel.2010.03.016 [DOI] [PubMed] [Google Scholar]
- Schnorrer F., Schönbauer C., Langer C. C. H., Dietzl G., Novatchkova M., Schernhuber K., Fellner M., Azaryan A., Radolf M., Stark A. et al. (2010). Systematic genetic analysis of muscle morphogenesis and function in Drosophila. Nature 464, 287–291. 10.1038/nature08799 [DOI] [PubMed] [Google Scholar]
- Schober M., Rebay I., Perrimon N. (2005). Function of the ETS transcription factor Yan in border cell migration. Development 132, 3493–3504. 10.1242/dev.01911 [DOI] [PubMed] [Google Scholar]
- Srivastava A., Pastor-Pareja J. C., Igaki T., Pagliarini R., Xu T. (2007). Basement membrane remodeling is essential for Drosophila disc eversion and tumor invasion. Proc. Natl. Acad. Sci. USA 104, 2721–2726. 10.1073/pnas.0611666104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi C., Sheng Z., Horan T. P., Kitayama H., Maki M., Hitomi K., Kitaura Y., Takai S., Sasahara R. M., Horimoto A. et al. (1998). Regulation of matrix metalloproteinase-9 and inhibition of tumor invasion by the membrane-anchored glycoprotein RECK. Proc. Natl. Acad. Sci. USA 95, 13221–13226. 10.1073/pnas.95.22.13221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terriente-Felix A., Li J., Collins S., Mulligan A., Reekie I., Bernard F., Krejci A., Bray S. (2013). Notch cooperates with Lozenge/Runx to lock haemocytes into a differentiation programme. Development 140, 926–937. 10.1242/dev.086785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Adam J. C., Montell D. (2007). Spatially localized Kuzbanian required for specific activation of Notch during border cell migration. Dev. Biol. 301, 532–540. 10.1016/j.ydbio.2006.08.031 [DOI] [PubMed] [Google Scholar]
- Wang Z., Li Y., Kong D., Sarkar F. H. (2010). The role of Notch signaling pathway in epithelial-mesenchymal transition (EMT) during development and tumor aggressiveness. Curr. Drug Targets 11, 745–751. 10.2174/138945010791170860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Zou J., Zhao B., Johannsen E., Ashworth T., Wong H., Pear W. S., Schug J., Blacklow S. C., Arnett K. L. et al. (2011). Genome-wide analysis reveals conserved and divergent features of Notch1/RBPJ binding in human and murine T-lymphoblastic leukemia cells. Proc. Natl. Acad. Sci. USA 108, 14908–14913. 10.1073/pnas.1109023108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong M. C., Castanon I., Baylies M. K. (2008). Daughterless dictates Twist activity in a context-dependent manner during somatic myogenesis. Dev. Biol. 317, 417–429. 10.1016/j.ydbio.2008.02.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaffran S., Astier M., Gratecos D., Sémériva M. (1997). The held out wings (how) Drosophila gene encodes a putative RNA-binding protein involved in the control of muscular and cardiac activity. Development 124, 2087–2098 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.