ABSTRACT
The protein nephrocystin-4 (NPHP4) is widespread in ciliated organisms, and defects in NPHP4 cause nephronophthisis and blindness in humans. To learn more about the function of NPHP4, we have studied it in Chlamydomonas reinhardtii. NPHP4 is stably incorporated into the distal part of the flagellar transition zone, close to the membrane and distal to CEP290, another transition zone protein. Therefore, these two proteins, which are incorporated into the transition zone independently of each other, define different domains of the transition zone. An nphp4-null mutant forms flagella with nearly normal length, ultrastructure and intraflagellar transport. When fractions from isolated wild-type and nphp4 flagella were compared, few differences were observed between the axonemes, but the amounts of certain membrane proteins were greatly reduced in the mutant flagella, and cellular housekeeping proteins >50 kDa were no longer excluded from mutant flagella. Therefore, NPHP4 functions at the transition zone as an essential part of a barrier that regulates both membrane and soluble protein composition of flagella. The phenotypic consequences of NPHP4 mutations in humans likely follow from protein mislocalization due to defects in the transition zone barrier.
KEY WORDS: CEP290, Chlamydomonas, Cilia, Flagella, Nephrocystin-4, Transition zone
INTRODUCTION
Nephronophthisis (NPHP) is a recessive form of cystic kidney disease that is the most frequent genetic cause of chronic renal failure in children (Wolf and Hildebrandt, 2011). To date, 18 causative human genes for NPHP have been reported (Wolf and Hildebrandt, 2011; Failler et al., 2014; Renkema et al., 2014); one of these is NPHP4, which encodes nephrocystin-4 (NPHP4). Mutations in NPHP4 lead to juvenile NPHP type 4 as well as retinitis pigmentosa; both disease features occur together in Senior–Løken syndrome (Hoefele et al., 2005; Mollet et al., 2002; Otto et al., 2002; Schuermann et al., 2002).
NPHP and Senior–Løken syndrome are ciliopathies (diseases caused by defects in cilia), and there is abundant evidence from model systems that loss of NPHP4 causes a ciliary phenotype. A study of Nphp4-mutant mice has shown that Nphp4 is necessary for normal photoreceptor formation and maintenance as well as sperm development (Won et al., 2011). In zebrafish, nphp4 morphants exhibit classic ciliary phenotypes, including abnormal body curvature and pronephric cysts (Burcklé et al., 2011; Slanchev et al., 2011). In Caenorhabditis elegans, RNA interference (RNAi)-mediated knockdown of NPHP-4 causes defects in male mating behavior, which is mediated by sensory cilia (Wolf et al., 2005), and a mutant null for NPHP-4 has mild defects in cilia-dependent sensory functions (Winkelbauer et al., 2005). Cilia of the same mutant are stunted, misshapen and have axonemal ultrastructural defects (Jauregui et al., 2008).
These phenotypes most likely result from defective function of NPHP4 at the ciliary transition zone, a specialized region between the basal body and the cilium proper (Shiba and Yokoyama, 2012; Czarnecki and Shah, 2012; Reiter et al., 2012). Several studies have reported that NPHP4 localizes to the transition zone. LAP-tagged NPHP4 in IMCD3 cells is present at sites of cell–cell contact and at the base of the primary cilium, specifically in the transition zone (Sang et al., 2011). Others have reported the presence of NPHP4 in the transition zone of cultured mouse renal epithelial cells (Shiba et al., 2010) and in the connecting cilium – a greatly elongated transition zone – of mouse photoreceptor cells (Roepman et al., 2005; Patil et al., 2012; Won et al., 2011). In Chlamydomonas reinhardtii, NPHP4 was identified in a proteomic study of basal bodies, which included the transition zone (Keller et al., 2005). In C. elegans, NPHP4 has been found to localize to the transition zone of the sensory cilia (Winkelbauer et al., 2005; Jauregui et al., 2008; Williams et al., 2011).
Evidence is accumulating that the transition zone functions as the ‘pore’ or gate proposed to exist at the base of the cilium to control access to the ciliary compartment (Rosenbaum and Witman, 2002). A highly conserved feature of the transition zone is the presence of Y-shaped links (Y-links) that connect the transition zone doublet microtubules to the overlying membrane (Fisch and Dupuis-Williams, 2011). In C. reinhardtii, these links are partially disrupted in the absence of CEP290, another transition zone protein, resulting in abnormal flagellar protein composition (Craige et al., 2010). The mutant also has defects in intraflagellar transport (IFT), which is used by nearly all organisms to build cilia and flagella (Pedersen and Rosenbaum, 2008; Ishikawa and Marshall, 2011). These results provide experimental evidence that the transition zone functions to control protein and IFT particle entry into the cilium, and they indicate that CEP290 is important for this function. NPHP4 also might be involved in this activity; cilia of the C. elegans nphp-4 mutant accumulate the membrane-associated proteins RGI-2 and TRAM-1a, which normally are excluded from the cilium (Williams et al., 2011).
Despite this progress, we still do not know the precise location of NPHP4 in the transition zone nor how its loss affects overall ciliary composition. In this study, we have used C. reinhardtii to learn more about NPHP4. C. reinhardtii has many advantages for studying cilia and ciliary components. Particularly relevant for this study, its flagella can be isolated to determine the biochemical consequences of loss of NPHP4. NPHP4 is highly conserved (C. reinhardtii to human BLASTP E value = 1e−85), so conclusions from studying C. reinhardtii NPHP4 are likely to be applicable to humans and other organisms. We found that NPHP4 is located at the periphery of the distal transition zone, close to the membrane and distal to CEP290. In contrast to CEP290, NPHP4 at the transition zone does not undergo rapid turnover. We identified an NPHP4-null mutant; the mutant has full-length flagella and generally has normal transition zone and axonemal ultrastructure. We isolated the flagella of the nphp4 mutant and found that a subset of the membrane-associated proteins that are present in wild-type flagella were greatly decreased in amount; conversely, the flagella contained many large cytosolic housekeeping proteins that normally are excluded from wild-type flagella. The results indicate that NPHP4 is a crucial component of the selective gate that functions at the transition zone to control the movement of both soluble and membrane-associated proteins between the flagellar and cytoplasmic compartments. It is likely that the various phenotypic consequences of NPHP4 mutations in humans and other organisms all follow from protein mislocalization due to defects in the transition zone barrier.
RESULTS
NPHP4 loss has minor effects on cell motility but slows flagellar assembly
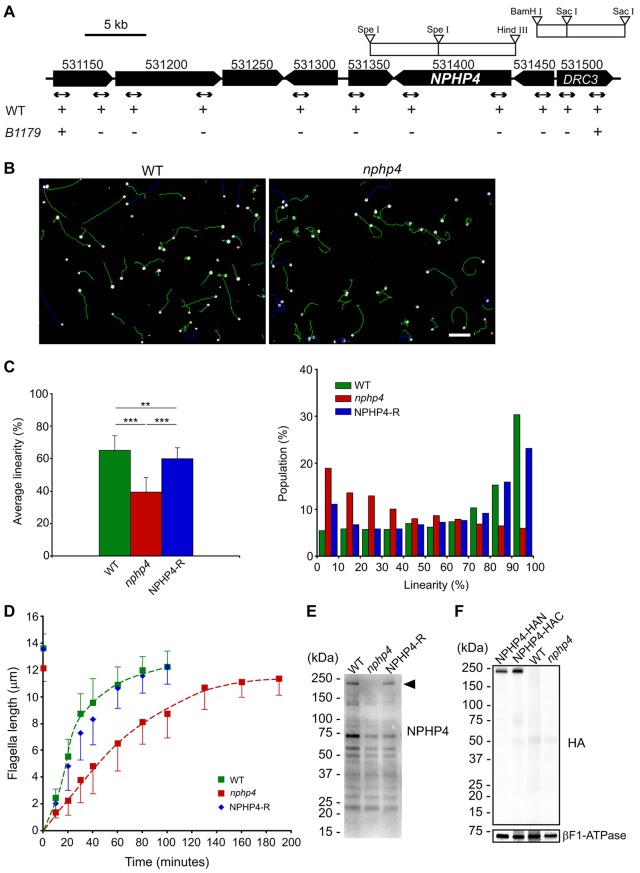
To identify a C. reinhardtii nphp4 mutant, genomic DNA from our collection of insertional mutants (Pazour et al., 1995; Pazour et al., 1998) was screened by real-time PCR with primer pairs specific to NPHP4. In one strain (B1179), no product was amplified. Further analysis by PCR revealed that the mutation, termed nphp4-1, deleted ∼40 kbp around NPHP4, including the entire NPHP4 gene, several predicted genes that have no known association with the flagellum, and a part of DRC3 (Fig. 1A). DRC3 was first identified in our flagellar proteomic study as FAP134 (Pazour et al., 2005) and was later shown to be a component of the nexin–dynein regulatory complex (Lin et al., 2011). To ensure that the phenotype being analyzed in the studies that follow was not compromised by the absence of DRC3, B1179 was backcrossed twice to a wild-type strain, and some of the mutant progeny were then transformed with a DNA fragment containing the DRC3 gene (Fig. 1A). One of the resulting transformants, rescued for DRC3, was used in this study as the nphp4 strain.
Fig. 1.
Characterization of the C. reinhardtii nphp4 mutant. (A) Map of C. reinhardtii genome near the NPHP4 locus. Numbers above each locus correspond to gene IDs in Phytozome version 9.1 (http://www.phytozome.net). Arrows indicate the positions of PCR products used to delimit the deleted region; plus and minus marks indicate whether the PCR products were amplified. Genome fragments from bacterial artificial chromosome (BAC) clones used for knock-in of full-length NPHP4 and DRC3 are indicated by white rectangles. WT, wild type. (B) Representative images showing swimming paths of wild-type and nphp4 cells. White dots show cells where CASA began to monitor their tracks; green lines are the swimming paths analyzed. Blue tracks were not analyzed because the cells swam outside the microscope field before recording was completed. Cells marked with red dots were immobilized by attachment to the coverslip. The swimming paths of nphp4 cells are more erratic than those of wild-type cells. Scale bar: 50 µm. (C) Bar graph (left) showing the mean±s.d. of linearity </emph>and histogram (right) summarizing the distribution of linearity in swimming paths of populations of wild-type, nphp4 and NPHP4-R cells (see supplementary material Fig. S1). For each strain, the population was calculated from the sum of values from a total of ten fields in each of five independent experiments. Statistical significance was determined by the Tukey–Kramer method: **P<0.01; ***P<0.001. (D) Flagella length prior to deflagellation and as a function of time after deflagellation of wild-type, nphp4 and NPHP4-R cells. Flagellar length at each time-point is the mean±s.d. of 30 flagella. (E) Western blot of whole cells probed with an antibody against the C-terminus of NPHP4. The antibody recognized a band (arrowhead) of the predicted size for NPHP4 (210 kDa) in wild-type cells and the NPHP4-R strain. (F) Western blot of wild-type, nphp4, NPHP4–HAN and NPHP4–HAC whole cells probed with a monoclonal antibody against the HA peptide. βF1-ATPase was used as a loading control.
The nphp4-mutant cells are motile and can undergo phototaxis (data not shown), but their swimming paths are slightly more erratic than those of wild-type cells (Fig. 1B). To assess this objectively, motility was analyzed using computer-aided sperm analysis (CASA) (Fig. 1C) (Mortimer, 2000). Swimming speeds of wild-type cells and the nphp4 mutant were similar (supplementary material Fig. S1B, VCL), but linearity was much lower for the mutant (Fig. 1C; see supplementary material Fig. S1A for definition of terms). Transformation of the nphp4 mutant with the wild-type NPHP4 gene to generate strain NPHP4-R almost completely restored linearity (Fig. 1C), confirming that the phenotype is due to the absence of NPHP4. Consistent with the ability to swim nearly normally, steady-state flagella of nphp4 mutant cells are of near-normal length and appear to be ultrastructurally normal (Fig. 1D; Fig. 2A; data not shown). Following flagellar amputation, the nphp4 mutant regenerated new flagella more slowly than did wild-type cells; normal flagellar regeneration was completely restored in the NPHP4-R strain (Fig. 1D). Therefore, NPHP4 is needed to build the flagella with normal kinetics.
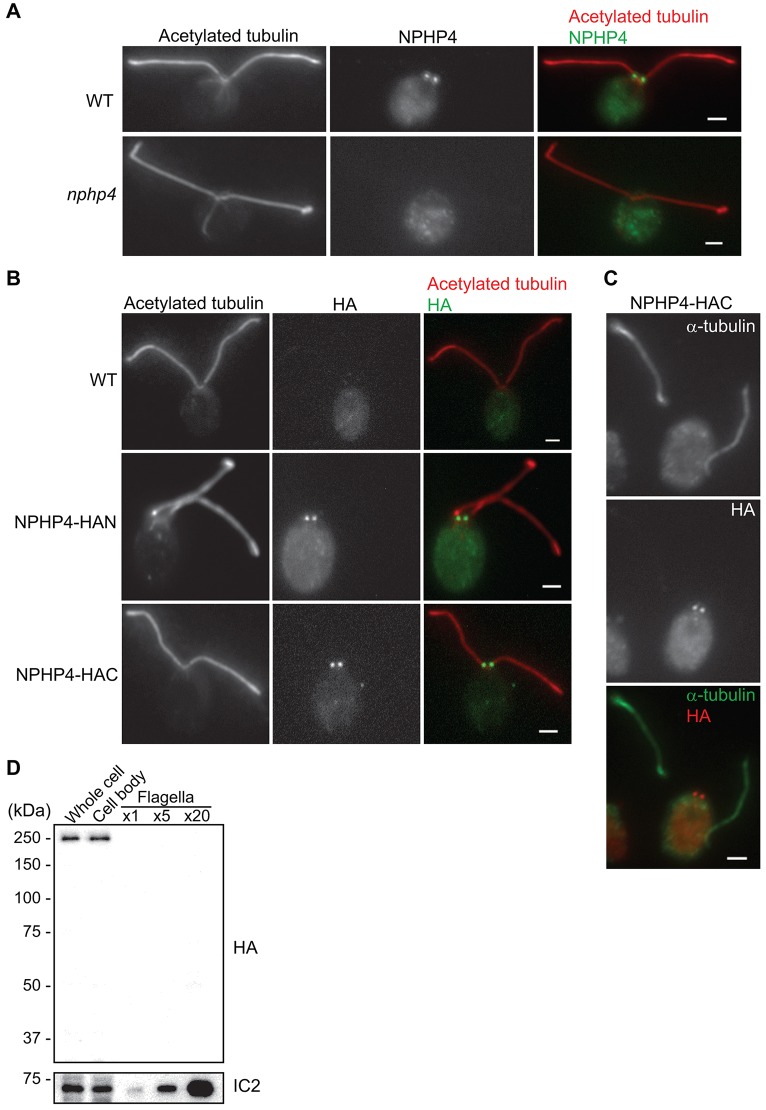
Fig. 2.
NPHP4 is localized to the base of flagella. (A) Wild-type (WT) and nphp4 cells were labeled with antibodies against acetylated tubulin (a marker for flagellar microtubules) and NPHP4; merged images are on the right. (B) Wild-type, NPHP4–HAN and NPHP4–HAC cells were labeled with anti-acetylated tubulin and anti-HA; merged images are on the right. (C) Detached flagella and cell body of an NPHP4–HAC cell fixed and labeled with antibodies against α-tubulin and the HA peptide. Scale bars: 2 µm. (D) NPHP4–HAC whole cells, cell bodies and isolated flagella were analyzed on a western blot probed with the anti-HA antibody; the axonemal protein IC2 was used as a loading control. ×1, ×5 and ×20 indicate that ∼1, 5 or 20 flagella pairs were loaded per cell body.
NPHP4 is located in the transition zone
To determine where NPHP4 is located in C. reinhardtii, we made an antibody against the C-terminus of NPHP4. The antibody recognized a band of the expected size in western blots of wild-type but not nphp4 whole cells; the band was restored in the NPHP4-R strain (Fig. 1E). These results confirmed that NPHP4 is missing in the mutant. We also rescued the mutant with constructs expressing a 3xHA tag at either the N- or C-terminus of NPHP4 to generate NPHP4–HAN and NPHP4–HAC cells, respectively; the anti-HA antibody recognized the tagged protein with high specificity in the rescued cells (Fig. 1F). In immunofluorescence microscopy, the anti-NPHP4 antibody strongly labeled the bases of the flagella of wild-type cells but not nphp4 cells (Fig. 2A), indicating that this localization is specific to NPHP4. Similarly, the anti-HA antibody localized specifically to the bases of flagella in both NPHP4–HAN and NPHP4–HAC cells but not in wild-type cells, indicating that the localization is specific to HA-tagged NPHP4 (Fig. 2B).
NPHP4 has been found by immunofluorescence microscopy to localize to primary cilia of MDCK and mouse kidney cells (Mollet et al., 2005; Patil et al., 2012). Therefore, we carefully determined the distribution of NPHP4 in C. reinhardtii flagella versus cell bodies. When cells are deflagellated, the flagella detach at the junction of the transition zone and axoneme proper (Sanders and Salisbury, 1989). NPHP4–HAC cells were deflagellated with dibucaine and the resulting cell bodies and detached flagella were stained with anti-HA; NPHP4–HAC was found to stay with the cell bodies (Fig. 2C). To confirm this result, whole cells, cell bodies and isolated flagella were compared by western blotting; all of the NPHP4–HAC remained within the cell body and none could be detected in the isolated flagella (Fig. 2D). The results indicate that C. reinhardtii NPHP4 is located in either the transition zone or the basal body, but not in the flagella.
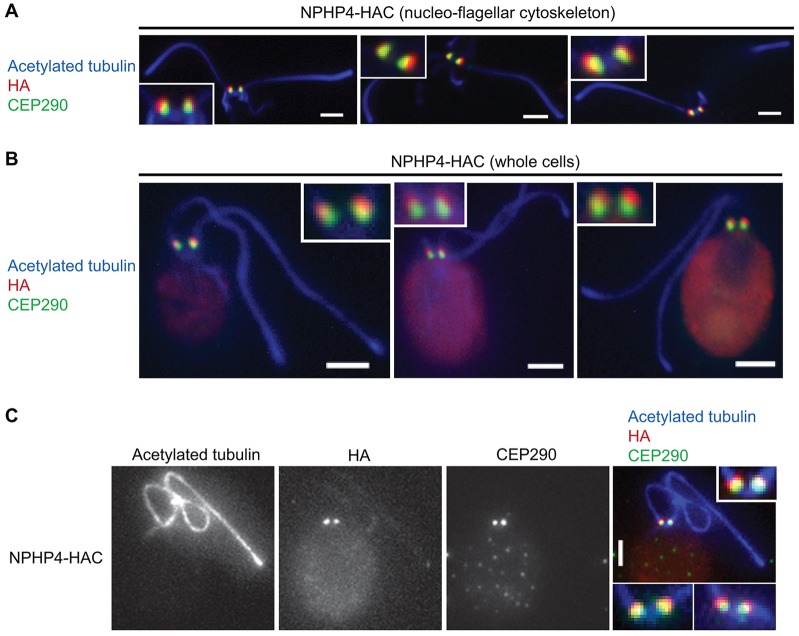
To further narrow down the location of NPHP4, both nucleoflagellar apparatuses and whole cells of the NPHP4–HAC strain were labeled with antibodies against the HA peptide and CEP290, which is located in the transition zone (Craige et al., 2010). By both wide-field immunofluorescence microscopy (Fig. 3A,B) and total internal reflection fluorescence/epi-fluorescence structured light microscopy (TESM) (Fig. 3C), the NPHP4–HAC label was observed to be distal to the CEP290 label, indicating that NPHP4 is located in the transition zone. Analysis of TESM images indicated that the mean peak-to-peak distance between CEP290 and the C-terminus of NPHP4–HAC is ∼137 nm (n = 130).
Fig. 3.
NPHP4 is localized to the transition zone. Isolated nucleoflagellar apparatuses of NPHP4–HAC cells (A) or whole NPHP4–HAC cells (B,C) were labeled with antibodies against acetylated tubulin, HA peptide and CEP290. Images were acquired by conventional widefield immunofluorescence microscopy (A,B) or TESM (C). All the images in A and B and the image in the right panel of C are merged from separate channels (shown only in C). The insets in A and B and the upper inset in C are enlargements of the transition zones shown in the panels; the lower insets in C show two additional examples. Scale bars: 2 µm.
NPHP4 is located at the periphery of the distal transition zone
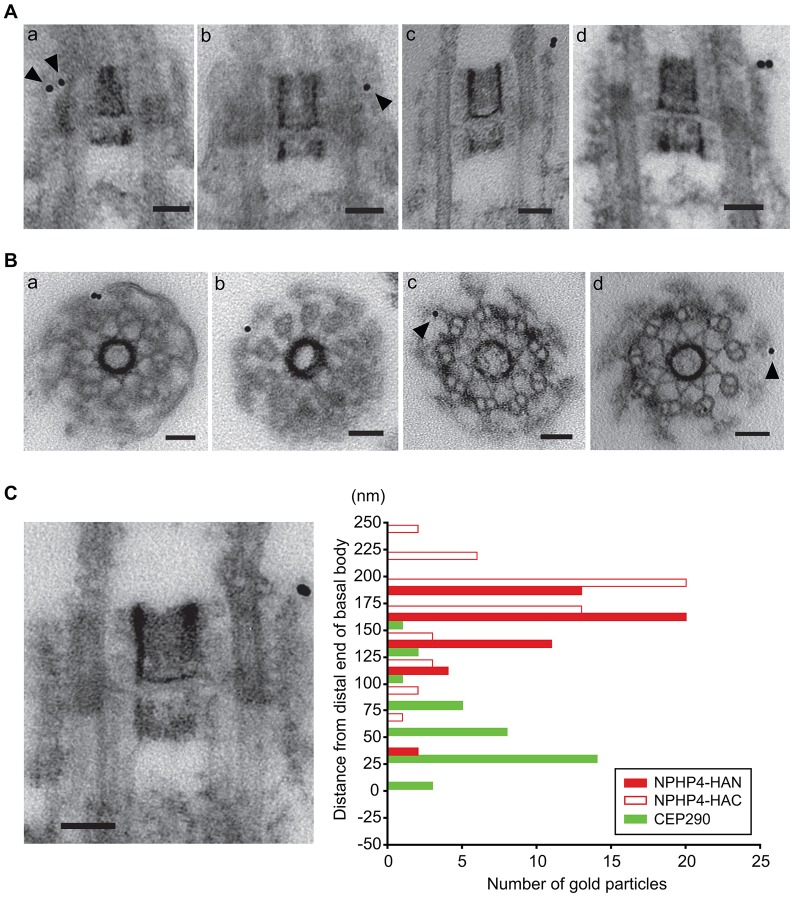
To determine the location of NPHP4 within the transition zone, we carried out pre-embedding immunoelectron microscopy of nucleoflagellar apparatuses from wild-type, NPHP4–HAN and NPHP4–HAC cells. As expected from the immunofluorescence microscopy results (Fig. 2B), gold particles were associated with the transition zone in both NPHP4–HAN and NPHP4–HAC specimens (Fig. 4A,B). Most gold particles in longitudinal sections and all particles in cross-sections of the transition zone were located at the transition zone periphery, in close association with remnants of the transition zone membrane, which is resistant to treatment with nonionic detergent (Kamiya and Witman, 1984). We observed no gold particles associated with the wild-type transition zones, indicating that the immunogold labeling was specific to HA-tagged NPHP4.
Fig. 4.
NPHP4 is located in the distal transition zone. Isolated nucleoflagellar apparatuses of NPHP4–HAN or NPHP4–HAC cells were incubated with rat IgG directed against the HA peptide, followed by incubation with anti-rat-IgG conjugated to 10-nm gold particles. (A) Typical longitudinal sections of NPHP4–HAN (a,b) and NPHP4–HAC (c,d) transition zones labeled with gold particles (arrowheads). (B) Cross-sections of NPHP4–HAN (a,b) and NPHP–HAC (c,d) transition zones labeled with gold particles (arrowheads). All gold particles were found at the periphery of the transition zone, in close association with the transition zone membrane. (C) Distribution of gold particles associated with longitudinal sections of NPHP4–HAN and NPHP4–HAC transition zones; 50 gold particles were scored for each. The electron micrograph shows a typical example matched to the scale of the y-axis; ‘0’ on the y-axis indicates the boundary (visible in the longitudinal section) between the basal body and the transition zone. The C-terminus of NPHP4 might be located slightly more distally than the N-terminus. The green bars show the distribution of CEP290 replotted from Craige et al. (Craige et al., 2010). Scale bars: 50 nm.
The locations, relative to the proximal end of the transition zone, of a total of 50 gold particles from longitudinal sections of both NPHP4–HAN and NPHP4–HAC specimens were measured and compared to that determined for CEP290 by Craige et al. (Craige et al., 2010) (Fig. 4C). Consistent with our immunofluorescence microscopy observations (Fig. 3), the distribution of both NPHP4–HAN and NPHP4–HAC gold particles had sharp peaks in the distal transition zone. The average distance of gold particles from the proximal end of the transition zone was 173±37 nm (±s.d.) for NPHP4–HAC and 155±31 nm for NPHP4–HAN, compared to 61±38 nm for CEP290; the average distance between NPHP4–HAC and CEP290 gold particles was 111 nm, in good agreement with the separation (∼137 nm) measured by TESM.
NPHP4 and CEP290 localize to the transition zone independently of each other
To explore if C. reinhardtii NPHP4 can be assembled at the transition zone independently of CEP290 and vice versa, we carried out immunofluorescence microscopy of cep290- and nphp4-mutant cells with antibodies against NPHP4 and CEP290, respectively. Localization of NPHP4 appeared to be normal in the cep290 cells, and localization of CEP290 was normal in the nphp4 cells (supplementary material Fig. S2A,B). Therefore, these two proteins are transported to and assemble into the transition zone independently of each other.
NPHP4 does not turn over rapidly
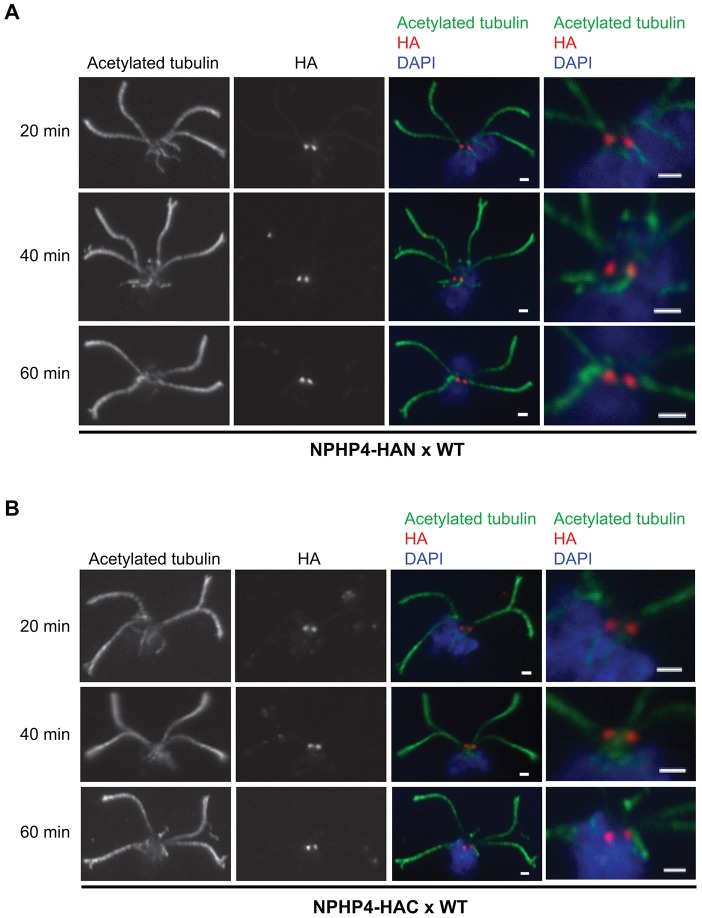
We reported previously that CEP290 at the transition zone is highly dynamic (Craige et al., 2010). To determine if NPHP4 is similarly dynamic, we mixed wild-type gametes with NPHP4–HAN or NPHP4–HAC gametes to form quadriflagellated zygotes in which two transition zones contained untagged NPHP4 and two contained HA-tagged NPHP4 in a common cytoplasm. Nucleoflagellar apparatuses were prepared at 20, 40 and 60 min after initiation of the mating reaction and labeled with anti-HA (Fig. 5). Even after 60 min, only two transition zones were labeled by the antibody, indicating that little, if any, HA-tagged NPHP4 was incorporated into the wild-type transition zones over this time period. Moreover, the label was as bright at 60 min as at 20 min, indicating that little, if any, HA-tagged NPHP4 was replaced with wild-type NPHP4. Therefore, in contrast to CEP290, NPHP4 is static at the transition zone.
Fig. 5.
NPHP4 at the transition zone is static. Wild-type (WT) gametes were mixed with NPHP4–HAN (A) or NPHP4–HAC (B) gametes to initiate the mating reaction. At the indicated times after mixing, nucleoflagellar apparatuses of the resulting quadriflagellated zygotes were prepared, fixed and stained with anti-acetylated tubulin, anti-HA and DAPI. The right panels are enlargements of the transition zones shown in the merged images. Scale bars: 1 µm.
Ultrastructure of the transition zone in the nphp4 mutant
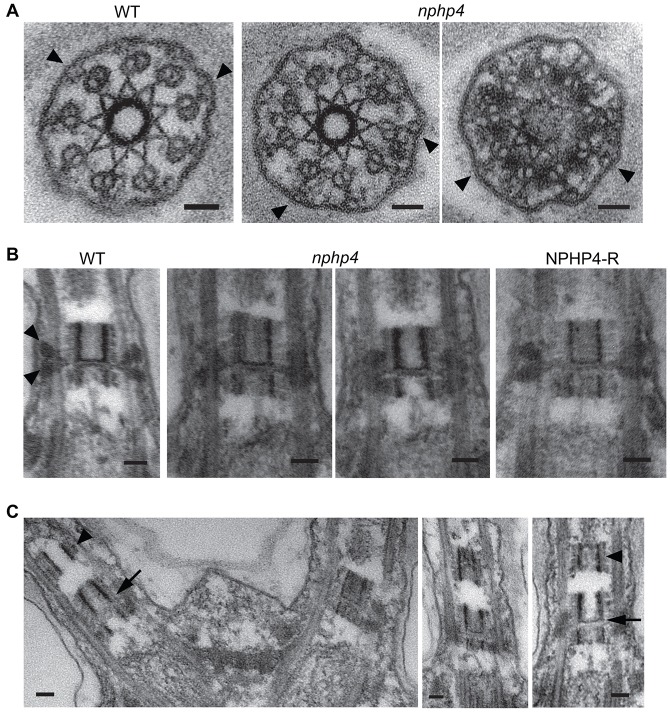
To determine whether loss of NPHP4 caused structural abnormalities in the transition zone, we compared the transition zones of wild-type and nphp4-mutant cells by electron microscopy. In cross sections, the Y-links between the transition zone doublet microtubules and membrane, which are disrupted in the cep290 mutant (Craige et al., 2010), were present and appeared normal (Fig. 6A). In longitudinal sections, the wedge-shaped connectors, which extend between the transition zone doublets and membrane in wild-type cells, appeared to be missing or collapsed in the nphp4 mutant (Fig. 6B). However, the wedge-shaped structures are similarly altered in transition zones of cells lacking the IFT protein IFT46 (Hou et al., 2007). To clarify whether disruption of this structure is correlated with lack of NPHP4, we performed immunofluorescence microscopy of ift46 cells and found that NPHP4 was normally located at the base of their flagella (supplemental material Fig. S2C). Thus, the morphological variation in this structure is not specifically related to the loss of NPHP4. Moreover, because the Y-links are present in the nphp4 mutant, we can conclude that they and the wedge-shaped structures are not the same. Finally, in ∼30% of longitudinal sections of nphp4 flagella, we observed, just distal to the transition zone, the apparently ectopic localization of dense material resembling the central cylinders of the C. reinhardtii transition zone (Fig. 6C).
Fig. 6.
Transition zone structure in the nphp4 mutant. (A) Cross-sections through transition zones of wild-type (WT) and nphp4 cells. The Y-links (arrowheads) appear normal in the nphp4 mutant. (B) Longitudinal sections through transition zones of wild-type, nphp4 and NPHP4-R cells. The electron-opaque wedge-shaped structures (arrowheads) are present in wild-type and NPHP4-R transition zones, but are missing, deformed or obscured in the nphp4 transition zones. (C) About one-third of nphp4 flagella contain ectopic cylindrical structures (arrowheads) just distal to the transition zone; these structures resemble the normal transition zone cylinders (arrows). Scale bars: 50 nm.
Protein composition of the flagellar membrane and matrix is drastically altered by loss of NPHP4
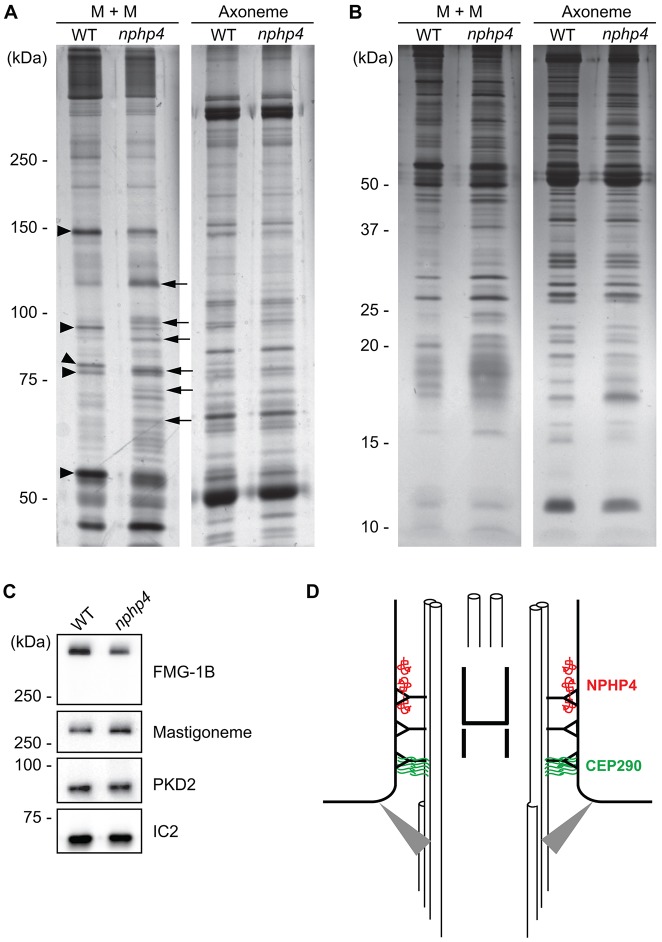
To compare the protein compositions of wild-type versus nphp4 flagella, flagella were isolated and treated with NP-40 to separate proteins into a detergent-soluble membrane-plus-matrix fraction and an axonemal fraction. In SDS-polyacrylamide gels of the axoneme fractions, very few differences were observed between wild-type and nphp4 samples (Fig. 7A,B). However, numerous differences were observed in the membrane-plus-matrix fractions. The levels of some proteins were decreased in the nphp4 mutant (arrowheads, Fig. 7A), whereas others were increased (arrows, Fig. 7A). Normal flagellar protein composition was restored in NPHP4-R (supplementary material Fig. S3), confirming that the differences were due to loss of NPHP4. Importantly, most differences between the wild-type and nphp4 membrane-plus-matrix fractions were observed in the mass range above 50 kDa; few differences were observed below 50 kDa (Fig. 7B).
Fig. 7.
Flagella of nphp4 cells have abnormal protein composition. 6% (A) or 12% (B) SDS-polyacrylamide gels were used to compare wild-type (WT) versus nphp4 membrane-plus-matrix (M+M) and axonemal fractions. Arrowheads indicate some proteins that decreased in amount in the nphp4 membrane-plus-matrix fraction; arrows indicate some proteins that increased in amount in the mutant flagella. (C) Western blot of isolated wild-type and nphp4 flagella probed with antibodies against FMG-1B, the mastigoneme protein and PKD2. The outer arm dynein intermediate chain IC2 served as a loading control. (D) Diagram illustrating the distribution of NPHP4 and CEP290 in the transition zone. NPHP4 (red) is located in close association with the membrane at the distal end of the transition zone. CEP290 (green) is located in the space between the doublet microtubules and membrane at the proximal end of the transition zone. Gray triangles represent transition fibers connecting the basal body to the plasma membrane.
To identify the proteins that were decreased or increased in the nphp4 flagella, sets of slices containing the discordant bands were excised from gels of the membrane-plus-matrix fraction of wild-type, nphp4 and NPHP4-R flagella (supplementary material Fig. S3). The slices were then subjected to analysis by mass spectrometry. Multiple proteins were identified in all slices (supplementary material Table S1). To conclude that the amount of a protein was altered in the mutant flagella, we used the following criteria: (1) the number of peptides recovered had to be increased or decreased by more than ten compared to wild type, or the number of peptides recovered had to be greater than five when the protein was not found in the other slice (wild type or nphp4) being compared; and (2) the change was specifically rescued in the NPHP4-R cells. Based on these criteria, the proteins that were found to be increased or decreased in amount in the nphp4-mutant flagella are listed in Table 1 and supplementary material Table S1 (where blue or red font indicates increased or decreased proteins, respectively). Except for ATPase V1 complex-subunit A, all proteins that increased in amount were housekeeping proteins that presumably are abundant in the cell bodies. Valyl- and phenylalanyl-tRNA synthetase, ADP ribosylglycohydrolase and RNA-binding protein were not found in the C. reinhardtii flagellar proteome (Pazour et al., 2005). All increased proteins are predicted to be larger than 50 kDa (Table 1). We conclude that large housekeeping proteins are specifically increased in nphp4-mutant flagella.
Table 1. Proteins decreased or increased in nphp4 flagella.
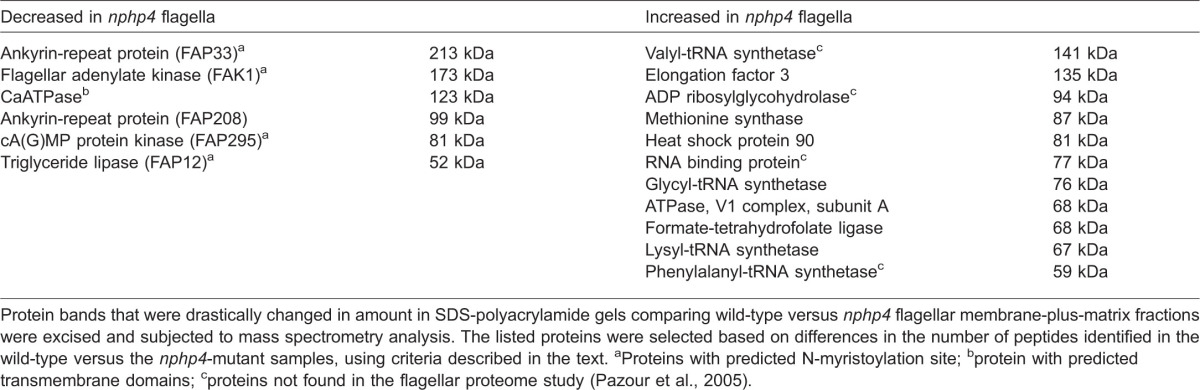
Protein bands that were drastically changed in amount in SDS-polyacrylamide gels comparing wild-type versus nphp4 flagellar membrane-plus-matrix fractions were excised and subjected to mass spectrometry analysis. The listed proteins were selected based on differences in the number of peptides identified in the wild-type versus the nphp4-mutant samples, using criteria described in the text. aProteins with predicted N-myristoylation site; bprotein with predicted transmembrane domains; cproteins not found in the flagellar proteome study (Pazour et al., 2005).
By contrast, all proteins that decreased in amount (other than FAP208) are predicted to be membrane associated – the calcium ATPase is predicted to have a transmembrane domain, and flagellar adenylate kinase, FAP33, FAP295 and FAP12 have N-myristoylation sites. Additionally, when we examined other proteins also predicted to be membrane associated but not meeting the above criteria (these proteins were identified by less than five peptides in wild-type flagella, were not detected at all in the nphp4 mutant and were restored in the rescued strain), all were decreased in the mutant flagella. These included the large flagellar membrane glycoprotein FMG1-B, a predicted ion channel, another calcium transport ATPase, an ABC transporter and FAP29 – all of which have transmembrane domains – and ankyrin-repeat protein and apyrase, which contain N-myristoylation sites (yellow font in supplementary material Table S1). Nearly all proteins that decreased in amount were previously identified in the C. reinhardtii flagellar proteome (Pazour et al., 2005). Thus, proteins normally associated with the flagellar membrane are either not transported into or are not retained in the flagella in the absence of NPHP4.
To determine whether all flagellum-associated membrane proteins are reduced in amount in nphp4 flagella, we probed western blots of isolated flagella from wild-type and the nphp4-mutant cells with antibodies against two well-characterized flagellar membrane proteins not identified in our dataset. The flagellar mastigoneme protein (Nakamura et al., 1996) and PKD2 (a homolog of human polycystin-2; Huang et al., 2007) were not decreased in the nphp4 flagella (Fig. 7C). We also probed for FMG-1B, which appeared to be decreased in nphp4 flagella in our mass spectrometry analysis. The western blot confirmed that it was reduced in the mutant flagella. We conclude that only a subset of proteins normally associated with the flagellar membrane is decreased in the absence of NPHP4.
IFT in the nphp4 mutant
Several IFT proteins were identified by mass spectrometry, and the levels of most of these proteins did not seem to be altered in the mutant flagella (supplementary material Table S1). To confirm this, we examined the amounts of IFT components in isolated flagella of wild-type and nphp4 cells by western blotting. The levels of IFT motors, of IFT proteins of both complex A and B, and of BBS4, a subunit of the BBSome, were unaltered or slightly elevated in the nphp4-mutant flagella (supplementary material Fig. S4A). To more directly assess whether IFT is affected by lack of NPHP4, IFT-particle movement in steady-state flagella was analyzed (supplementary material Fig. S4B). IFT frequency was not affected, whereas IFT velocity was slightly decreased in the absence of NPHP4 (supplementary material Fig. S4C).
DISCUSSION
NPHP4 and CEP290 define distinct regions of the transition zone
In agreement with previous studies in C. elegans and mammals (Winkelbauer et al., 2005; Jauregui et al., 2008; Shiba et al., 2010; Sang et al., 2011; Williams et al., 2011), we found that C. reinhardtii NPHP4 is located in the transition zone. However, our studies provide much more precise information on the distribution and localization of NPHP4. Using both immunofluorescence microscopy and immunoelectron microscopy, we determined that NPHP4 is located in the distal part of the transition zone, distal to CEP290. The NPHP4 and CEP290 peaks observed by TESM were separated by a minimum of ∼140 nm, and immunoelectron microscopy revealed that there is little overlap between the two proteins. Thus, NPHP4 and CEP290 define distinct structural domains of the transition zone (Fig. 7D). Immunoelectron microscopy also indicated that the N- and C-termini of NPHP4 are located at the periphery of the transition zone, in close association with the transition zone membrane. Thus, NPHP4 is well positioned to control entry and/or exit of membrane proteins from the flagellum. This apparent localization of NPHP4 to the extreme periphery of the transition zone contrasts with that of CEP290, which is distributed throughout the space between the doublet microtubules and the membrane (Craige et al., 2010); however, we cannot rule out that the middle part of NPHP4 extends into the space between the membrane and microtubules. Finally, our biochemical studies of isolated flagella indicate that there is no detectable NPHP4 in the flagellum proper.
Consistent with NPHP4 and CEP290 being located in different regions of the transition zone, we found that NPHP4 and CEP290 each assembled normally into the transition zone in the absence of the other. Both the localization and assembly results indicate that C. reinhardtii NPHP4 and CEP290 are integrated into different complexes in the transition zone. These findings agree with those of Sang et al. (Sang et al., 2011), who concluded that mammalian NPHP4 and CEP290 were components of distinct modules, based on immunoprecipitation of LAP-tagged proteins from whole-cell lysates.
We also observed that NPHP4 at the transition zone undergoes little or no turnover during a 1-h period; this is in sharp contrast to CEP290, which is at least 50% replaced in the same time span (Craige et al., 2010). Therefore, NPHP4 is static at the transition zone, as might be expected for a structural protein. By contrast, CEP290 appears to be much more dynamic. Although highly speculative, the results raise the possibility that CEP290 has a role in shuttling IFT or other ciliary proteins into the transition zone.
NPHP4 and CEP290 loss have different consequences for transition zone structure
A highly conserved and defining feature of the transition zone is the occurrence of Y-links between the doublet microtubules and membrane as observed in electron microscopy cross-sections of various organisms (Gilula and Satir, 1972; Perkins et al., 1986; Ringo, 1967). Craige et al. (Craige et al., 2010) observed that the Y-links were missing or disrupted in C. reinhardtii CEP290 mutants. We found that loss of NPHP4, by contrast, did not affect the Y-links. Therefore, NPHP4 is not required for Y-link formation. The same is true in C. elegans, where loss of NPHP4 alone did not cause disruption of the Y-links (Jauregui et al., 2008). However, when mutations in NPHP4 and members of the MKS/B9 module were combined in C. elegans, the Y-links were lost (Williams et al., 2011).
Although no specific structural abnormalities were observed in nphp4 transition zones, the flagella often contained, distal to the transition zone, what appeared to be a partial duplication of the central cylinder that normally occupies the lumen of the transition zone of C. reinhardtii. Interestingly, a similar duplication of the central cylinder has been observed in uniflagellar mutants of this organism (Huang et al., 1982; Piasecki et al., 2008). The duplication might occur because the cell detects abnormalities associated with the absence of NPHP4 and responds by moving additional central cylinder precursors into the flagellum.
A size-dependent barrier to ciliary entry of large soluble housekeeping proteins is breached in nphp4 cells
Because of the ease with which C. reinhardtii flagella can be purified, we were able to assess the biochemical consequences of loss of NPHP4 for the flagella. Strikingly, nearly all of the differences observed were in the membrane-plus-matrix fraction, not the axonemal fraction. The amounts of many proteins were increased in the nphp4 flagella, and, with the possible exception of subunit A of the V1-ATPase, all of these proteins were soluble housekeeping proteins. Most of these proteins were not detected at all in the membrane-plus-matrix fraction from wild-type flagella, suggesting that their presence in the nphp4 flagellum resulted from the breakdown of a mechanism that normally keeps them out of the organelle.
Importantly, abnormal entry of soluble cytoplasmic proteins into the nphp4 flagellum was observed only for proteins that migrated above 50 kDa in SDS-PAGE, indicating that there is an NPHP4-dependent barrier at the transition zone that excludes soluble proteins of ∼50 kDa or larger. Consistent with this, Kee et al. (Kee et al., 2012) observed that when fluorescently labeled protein A (41 kDa) and bovine serum albumin (BSA) (67 kDa) were microinjected into hTERT RPE cells, the former entered the ciliary compartment, whereas the latter did not. Similarly, Breslow et al. (Breslow et al., 2013), working with permeabilized IMCD3 cells, found that the rate of entry of soluble proteins into the cilium progressively slowed as the proteins increased in size from a Stokes radius of 3.1 nm to 4.3 nm (corresponding to ∼30 to ∼70 kDa); no entry was observed for larger proteins. Taken together, these results indicate that there is a size-dependent barrier to diffusion of soluble proteins into the cilia of organisms ranging from protists to humans and that this barrier is dependent upon NPHP4 in the transition zone.
Using a chemically inducible diffusion trap approach, Lin et al. (Lin et al., 2013) also obtained evidence for a size-dependent diffusion barrier at the base of primary cilia. However, they observed slow entry of proteins as large as ∼600–650 kDa, and suggested that “the steady-state distribution of proteins in cilia is unlikely to be regulated by the diffusion barrier.” Because the differences that we observed were in steady-state flagella that were at least 3 h old, the C. reinhardtii NPHP4-dependent diffusion barrier appears to be more impermeable to the diffusion-based entry of large soluble proteins.
NPHP4 is essential to a transition zone barrier that retains a subset of membrane proteins in the flagella
Although the levels of large soluble housekeeping proteins increased in nphp4 flagella, levels of many membrane-associated proteins decreased. However, the levels of some other flagellar membrane proteins, such as PKD2 and the mastigoneme protein, were not affected in nphp4 flagella. C. reinhardtii PKD2 is likely to be anchored to the axoneme because most PKD2 peptides were found in axonemal fractions in the flagellar proteome (Pazour et al., 2005) and >90% of GFP-tagged PKD2 is stationary in the flagella (Huang et al., 2007). Similarly, 75% of peptides from the mastigoneme protein identified in the flagellar proteome were found in the fraction resistant to detergent, implying that this protein also is anchored to the axoneme (Pazour et al., 2005). These results agree well with reports that PKD2 and an odorant receptor are normally located in C. elegans nphp-4 cilia (Jauregui et al., 2008; Williams et al., 2011). We further observed that the level of FMG-1B was decreased; in the C. reinhardtii flagellar proteome, peptides from FMG-1B were evenly distributed between the membrane and axonemal fractions (Pazour et al., 2005), and at least some of this protein is moved in the plane of the flagellar membrane by IFT (Guilford and Bloodgood, 2013). Taken together, these results suggest that a subset of flagellar membrane proteins – specifically those presumed to be mobile in the plane of the membrane – are present at lower levels in nphp4 flagella, whereas those that are anchored to the axoneme exhibit no reduction. Because PKD2 and the mastigoneme protein that bind to the axoneme appear to be delivered normally to the nphp4 flagella, it is possible that most or all flagellar membrane proteins are targeted normally to the flagella, but those that are free to move in the plane of the membrane subsequently diffuse out of the organelle in the absence of an NPHP4-dependent barrier at the transition zone.
Previous studies have provided evidence for a membrane protein barrier at the base of the cilium that is consistent with our findings. Studies with mammalian cells showed that disruption of members of the MKS/B9 complex, which localizes to the transition zone, resulted in a decrease in the amount of several GFP-tagged membrane proteins in primary cilia (Garcia-Gonzalo et al., 2011; Chih et al., 2012). NPHP4 is not part of the MKS/B9 complex, but studies in C. elegans indicate that NPHP4 and proteins of the MKS/B9 complex genetically interact in functionally redundant pathways that are important for ciliary development (Williams et al., 2008; Williams et al., 2010; Williams et al., 2011; Huang et al., 2011; Warburton-Pitt et al., 2012). Therefore, NPHP4 and the MKS/B9 proteins are likely to function together to prevent ciliary membrane proteins from diffusing out of the cilium. NPHP4 is located close to or at the transition zone membrane and several of the MKS/B9 proteins are predicted to be transmembrane proteins, so it will be of interest to determine precisely where the MKS/B9 proteins are located relative to NPHP4 and CEP290 in the transition zone.
Loss of NPHP4 has mild effects on flagellar motility
Interestingly, the only behavioral defect that we observed in the nphp4 mutant was erratic swimming. The lack of a stronger motility defect is consistent with our finding that there is little, if any, change in the protein composition of the flagellar axoneme, which constitutes the motile machinery. However, changes in intraflagellar Ca2+ are crucial for controlling flagellar waveform (Bessen et al., 1980; Kamiya and Witman, 1984), so the erratic swimming that we observed is entirely consistent with the loss of membrane proteins such as the Ca2+-transport ATPases, which are likely to be involved in maintaining normal intraflagellar Ca2+ levels.
NPHP4 and CEP290 define different structural and functional domains in the transition zone
Although nphp4 cells are able to assemble full-length flagella, the kinetics of flagellar regeneration following flagellar amputation are significantly impaired. Presently, the reason for this is unclear. IFT frequency is normal and velocity is only slightly reduced in steady-state nphp4 flagella, suggesting that loss of NPHP4 does not directly affect the IFT machinery. These results are generally consistent with those in C. elegans, where mutation of NPHP-4 has modest (Jauregui et al., 2008) or no (Williams et al., 2011) effect on IFT. The slower regeneration kinetics that we observed might reflect reduced IFT cargo loading, impaired flagellar signaling necessary for normal regeneration or altered intraflagellar ion concentrations (see above section). In any case, the very slight effect of NPHP4 loss on IFT is in contrast to the effect of CEP290 loss, which results in severe IFT abnormalities and greatly impaired flagellar formation (Craige et al., 2010). Therefore, although NPHP4 and CEP290 are both essential to the transition zone barrier, CEP290 has an additional function in IFT. We conclude that NPHP4 and CEP290 define different functional as well as structural domains in the transition zone.
MATERIALS AND METHODS
Strains and growth medium
C. reinhardtii wild-type strains 137c (nit1, nit2, mt+) and CC124 (nit1, nit2, mt−) were obtained from the Chlamydomonas Resource Center (University of Minnesota, St Paul, MN). B1179 (nphp4-1, drc3-1, arg7, ARG7, mt+) was generated by insertional mutagenesis of H11 (arg7, mt+) with the pARG7.8 plasmid (Debuchy et al., 1989). The nphp4 strain 79D8 (nphp4-1, drc3, DRC3, mt+), rescued for DRC3, was produced by crossing B1179 sequentially to the wild-type strains CC124 and 137c, and then transforming selected progeny with DRC3. NPHP4-R (nphp4-1, NPHP4, drc3, DRC3, mt+) was made by transformation of 79D8 with NPHP4. NPHP4-HAN (nphp4-1, NPHP4-HAN, drc3, DRC3, mt+) was generated by transformation of 79D8 with NPHP4-HAN. NPHP4-HAC (nphp4-1, NPHP4-HAC, drc3, DRC3, mt−) was made by transformation of B1179 with NPHP4-HAC followed by two crosses to wild-type strains (CC124 and 137c, sequentially) and transformation of selected offspring with DRC3. Cells were grown in M medium I (Sager and Granick, 1953) modified with 2.2 mM KH2PO4 and 1.7 mM K2HPO4 (Witman, 1986) or TAP medium (Gorman and Levine, 1965).
Real-time PCR
The deletion site of B1179 was determined by real-time PCR using a thermal cycler (Opticon, MJ Research), the QuantiTect SYBR Green PCR kit (Qiagen) and primers listed in supplementary material Table S2.
Gene cloning and transformation
To clone the full-length NPHP4 gene, a 7159-bp HindIII–SpeI fragment and a 6357-bp SpeI fragment were cut from BAC clone 18L19 containing NPHP4 and inserted sequentially into pBluescript II SK- (Stratagene). To generate a construct encoding NPHP4–HAN [containing three consecutive HA peptides (3xHA tag) between the first and second amino acids of NPHP4], a SmaI–NaeI fragment of the p3xHA plasmid (Silflow et al., 2001) was inserted into the AfeI site of NPHP4. To generate a construct encoding NPHP4–HAC (containing a 3xHA tag between the penultimate and last amino acids), a StuI–SmaI fragment from the p3xHA plasmid was subcloned into the PsiI site of NPHP4. To clone DRC3, a 2225-bp BamHI–SacI fragment and a 5144-bp SacI fragment were cut from BAC clone 34B19 and inserted into pBluescript II SK-. All BAC clones were purchased from Clemson University Genomics Institute.
All transformations were performed by the glass beads method (Kindle, 1990). For transformation with wild-type and HA-tagged NPHP4, cells were co-transformed with the ble gene as a selectable marker (Stevens et al., 1996). Colonies on TAP medium plates containing zeomycin were randomly selected and screened by immunofluorescence microscopy with anti-NPHP4 or anti-HA antibodies. For transformation with DRC3, the aph7″ gene was used as the selectable marker (Berthold et al., 2002).
Analysis of motility
Cultures of 137c, nphp4 and NPHP4-R cells at 104–105 cells/ml were diluted 5–10 times with modified M medium for motility analysis. To make a 100-µm-deep chamber for observation of the cells, 22×22-mm and 24×30-mm No. 1 coverslips were glued to opposite ends of a 25×75-mm slide glass so that the two coverslips were separated by ∼23 mm. An aliquot (∼20 µl) of the cell culture was then placed between the coverslips and covered with a 2X-CEL cover glass (Hamilton Thorne Research). The chamber was inserted into an IVOS sperm analyzer (version 12; Hamilton Thorne Research) and cell movement was recorded for 3.3 s using dark-field optics and a 4× objective.
Antibodies
The last exon of C. reinhardtii NPHP4 was amplified by PCR with primers NPHP4Ex28-N and -C linked to BamHI and SalI sites, respectively. The amplified DNA fragment was cut with those enzymes and cloned into pMAL-cRI (New England Biolabs) to express the last exon of NPHP4 fused to maltose-binding protein (MBP). The fusion proteins were purified with amylose resin and used to produce rabbit polyclonal antibodies (Covance). The NPHP4 antibodies were purified by affinity chromatography with the antigen attached to agarose resin. Bound antibodies were eluted and applied to an agarose column containing MBP to remove anti-MBP antibodies; the flow-through fraction was used in this study. Other antibodies used were anti-βF1-ATPase (Atteia et al., 2006); anti-HA peptide (3F10, Roche); anti-acetylated tubulin (Sigma-Aldrich); anti-α-tubulin (Silflow and Rosenbaum, 1981); anti-CEP290 (Craige et al., 2010); anti-FMG1-B (Bloodgood et al., 1986); anti-mastigoneme (Nakamura et al., 1996); anti-PKD2 (Huang et al., 2007); anti-KAP (Mueller et al., 2005); anti-DHC1b (Pazour et al., 1999); anti-IC2 (King et al., 1985); anti-BBS4 (Lechtreck et al., 2009); anti-IFT46 (Hou et al., 2007); and anti-IFT57, anti-IFT81, anti-IFT139 and anti-IFT172 (Cole et al., 1998).
Immunofluorescence microscopy
Cells were applied for 1–3 min to a coverslip coated with polyethyleneimine, the excess cell suspension was removed by blotting, and the coverslip was immersed in chilled methanol (−20°C) for 15 min. The coverslip was then completely dried before treating with blocking solution (3% fish gelatin, 1% BSA in PBS pH 7.0) for 30 min at room temperature. Primary antibody diluted in the blocking solution was applied to the sample overnight at 4°C, followed by four washes with PBS. Secondary antibodies diluted in the blocking buffer were then added to the coverslip for 1 h at room temperature. For double staining, secondary antibodies conjugated to Alexa Fluor 488, 568 or 594 (Life Technologies) were used. For tricolor labeling, secondary antibodies conjugated to Alexa Fluor 350, 488 and 594 were used. Finally, the coverslip was given three 10-min rinses with PBS and mounted on a glass slide with ProLong Gold Antifade Reagent (Life Technologies).
To prepare nucleoflagellar apparatuses, NPHP4–HAC cells were treated with autolysin, resuspended in ice-cold microtubule-stabilizing (MTSB) buffer (30 mM HEPES-KOH pH 7.4, 5 mM MgSO4, 15 mM KCl, 2 mM EGTA) and mixed with an equal volume of 1% NP-40 (Calbiochem) in MTSB buffer. The solution was left on ice for 15 min and then mixed with an equal volume of 6% paraformaldehyde in MTSB buffer. After incubation on ice for 60 min, the nucleoflagellar apparatuses were collected by centrifugation at 1100 g for 10 min. The cytoskeletons were attached to a coverslip coated with poly-L-lysine and dried. The samples were blocked, labeled and mounted as described above for whole cells.
The microscope, camera and software used to take fluorescence images were as described previously (Craige et al., 2010). Gamma adjustment and creation of merged images were performed with Photoshop CS2; all images mounted together were processed similarly.
TESM
TESM images were taken using the custom-built TESM system of our Biomedical Imaging Group (Navaroli et al., 2012). Cells triple labeled with Alexa Fluor 488, 594 and 647 and mounted in ProLong Gold Antifade Reagent were imaged using three color epi-fluorescence illumination. The pixel size corresponded to 0.083 µm. Z-stacks consisted of 100 images with a z separation of 100 nm. At each z-plane, an exposure of 25 ms was taken in each emission bandpass.
Tetraspec beads (200 nm) were used to measure chromatic aberration in the microscope system. Point spread functions fitted to the beads in each color revealed a chromatic aberration between the green and the red and far red channels of 1.6 pixels in the x direction. This measured offset was used to correct the experimental data.
Electron microscopy
Whole cells were pre-fixed in 1% glutaraldehyde in modified M medium for 15 min at room temperature, collected by centrifugation, resuspended with 1% glutaraldehyde in 100 mM sodium cacodylate-NaOH pH 7.2 (cacodylate buffer) and left for 2 h at room temperature. Specimens were then processed for ultra-thin sectioning as described previously (Craige et al., 2010).
For pre-embedding immunogold electron microscopy, nucleoflagellar apparatuses of 137c, NPHP4–HAN and NPHP4–HAC cells were prepared as described for immunofluorescence microscopy. After fixation with 3% paraformaldehyde, the samples were washed three times with PBS+0.02% Tween-20 (PBST) and blocked with 5% skimmed milk, 4% BSA in PBST for 1 h at room temperature. The cytoskeletons were then resuspended with the anti-HA antibody diluted 1∶400 in PBST and incubated overnight at 4°C. After three washes with PBST, the samples were next incubated with 1% BSA in PBST containing anti-rat-IgG conjugated to 10-nm gold particles (Aurion). The cytoskeletons were then washed three times in MTSB buffer and fixed with 1% OsO4 in MTSB buffer for 30 min at rom temperature. The pellets were then washed once with MTSB buffer, rinsed three times with distilled water and stained with 1% uranyl acetate overnight at 4°C. Dehydration and embedding was performed as for whole cells, except that post-sectioning staining was omitted.
Generation of gametes and dikaryons
To produce gametes, 30 ml of 137c, CC124, NPHP4–HAN and NPHP4–HAC cultures grown to ∼105 cells/ml in M medium I were collected and resuspended twice with nitrogen-free M medium. The second suspensions were transferred to 250-ml flasks and agitated under constant light overnight. Quadriflagellated zygotes were generated by mixing 137c with NPHP4–HAC and CC124 with NPHP4–HAN gametes. At various times after initiation of the mating reaction, aliquots of the mixture were removed and nucleoflagellar apparatuses were prepared as described for immunofluorescence microscopy.
Flagella isolation, SDS-PAGE and western blotting
Flagella were isolated by the dibucaine method (Witman, 1986) with the following modifications: DTT was omitted from the HMDS solution. The isolated flagella were resuspended with HMDEK buffer containing 30 mM HEPES pH 7.4, 5 mM MgSO4, 1 mM DTT, 1 mM EGTA and 25 mM KCl. To obtain axonemal and membrane-plus-matrix fractions, NP-40 was added to the flagellar suspension to a final concentration of 1% and the suspension left on ice for 10 min. The suspension was then centrifuged (26,890 g for 10 min at 4°C) and the supernatant was collected as the membrane-plus-matrix fraction. The pellet containing axonemes was resuspended in HMDEK buffer. For SDS-PAGE, each fraction was mixed with one-fourth volume of 5× SDS-sample solution (0.3 M Tris-HCl pH 6.8, 5 mM EDTA, 10% SDS, 25% sucrose and 0.01% Bromophenol Blue), and DTT was added to a final concentration of 50 mM. The mixture was then incubated at 75°C for 20 min. Protein bands were visualized with silver stain (Silver Stain Plus; Bio-Rad). To prepare whole flagella for western blotting, a suspension of isolated flagella was mixed with SDS-sample solution and DTT as described above. For whole cells, cells were collected by centrifugation, lysed with the 5× SDS-sample solution and incubated at 75°C for 20 min in the presence of 50 mM DTT. Proteins in SDS-polyacrylamide gels were transferred onto Immobilon-P (Millipore). For western blots of whole cells, antibodies were diluted with HIKARI (Nacalai Tesque). The chemiluminescence or fluorophore signals from the secondary antibodies were captured with a FluorChem Q equipped with a CCD camera (ProteinSimple).
Mass spectrometry
Protein bands of interest were excised from silver-stained gels and subjected to trypsin digestion as described previously (Spiess et al., 2011). Mass spectrometry analysis was performed as described previously (Wagner et al., 2014) with the following modifications: the length of the LC column was 15 cm, and the flow rate of sample injection was 250 nl/min. The resulting spectra were searched against the C. reinhardtii Augustus 5 protein database (Erik Hom, personal communication; based on Assembly 4 of the C. reinhardtii genome from the Joint Genome Institute: http://genome.jgi-psf.org/Chlre4/Chlre4.home.html) using SEQUEST (Bioworks software, v3.3.1; Thermo Electron).
Flagella regeneration
To determine the kinetics of flagellar regeneration, the pH of mid-log phase cultures of 137c, nphp4 and NPHP4-R cells was lowered to 4.3 with 0.5 M acetic acid. As soon as deflagellation was confirmed by microscopy, the pH was raised to 6.9 with 0.5 M KOH. To fix cells at each time-point, a small aliquot of the cell suspension was mixed with an equal volume of M medium containing 2% glutaraldehyde. Flagella lengths were measured with the Segmented Line tool in ImageJ (National Institutes of Health) and mean values at each point were obtained from 30 cells.
Observation of IFT
137c and nphp4 cells at ∼104 cells/ml were used to observe IFT as described previously (Craige et al., 2010). Kymographs were made by ImageJ with the MultipleKymograph plugin.
Supplementary Material
Acknowledgments
We are grateful to all those who generously provided antibodies, to the University of Massachusetts Medical School (UMMS) Core Electron Microscopy Facility for expert help with electron microscopy and to the Vermont Genetics Network (VGN) Proteomics Facility for mass spectrometry. We thank Eric Coleman Johnson (UMMS, MA) for identifying the nphp4 mutant. We also thank Erik Hom (University of Mississippi, University, MS) for providing C. reinhardtii Augustus 10 gene models, and Branch Craige (UMMS, MA) for critical discussion.
Footnotes
Competing interests
The authors declare no competing interests.
Author contributions
J.A. designed and performed most experiments. S.T. performed the initial characterization of the nphp4 mutant. C.S., K.D.B. and K.E.F. acquired data with TESM. K.F.L. assisted with some experiments. J.A. and G.B.W. prepared the manuscript. G.J.P and G.B.W. supervised and funded this study and helped to interpret the data.
Funding
This research was supported by the National Institutes of Health [grant numbers GM060992 (to G.J.P.) and GM030626 (to G.B.W.)]; by the Robert W. Booth Endowment at UMMS (to G.B.W.); and by the National Institute of General Medical Sciences Institutional Development Award (IDeA) [grant number P20 GM103449 (to the VGN)]. Deposited in PMC for release after 12 months.
Supplementary material available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.155275/-/DC1
References
- Atteia A., van Lis R., Gelius-Dietrich G., Adrait A., Garin J., Joyard J., Rolland N., Martin W. (2006). Pyruvate formate-lyase and a novel route of eukaryotic ATP synthesis in Chlamydomonas mitochondria. J. Biol. Chem. 281, 9909–9918. 10.1074/jbc.M507862200 [DOI] [PubMed] [Google Scholar]
- Berthold P., Schmitt R., Mages W. (2002). An engineered Streptomyces hygroscopicus aph 7″ gene mediates dominant resistance against hygromycin B in Chlamydomonas reinhardtii. Protist 153, 401–412. 10.1078/14344610260450136 [DOI] [PubMed] [Google Scholar]
- Bessen M., Fay R. B., Witman G. B. (1980). Calcium control of waveform in isolated flagellar axonemes of Chlamydomonas. J. Cell Biol. 86, 446–455. 10.1083/jcb.86.2.446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloodgood R. A., Woodward M. P., Salomonsky N. L. (1986). Redistribution and shedding of flagellar membrane glycoproteins visualized using an anti-carbohydrate monoclonal antibody and concanavalin A. J. Cell Biol. 102, 1797–1812. 10.1083/jcb.102.5.1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breslow D. K., Koslover E. F., Seydel F., Spakowitz A. J., Nachury M. V. (2013). An in vitro assay for entry into cilia reveals unique properties of the soluble diffusion barrier. J. Cell Biol. 203, 129–147. 10.1083/jcb.201212024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burcklé C., Gaudé H. M., Vesque C., Silbermann F., Salomon R., Jeanpierre C., Antignac C., Saunier S., Schneider-Maunoury S. (2011). Control of the Wnt pathways by nephrocystin-4 is required for morphogenesis of the zebrafish pronephros. Hum. Mol. Genet. 20, 2611–2627. 10.1093/hmg/ddr164 [DOI] [PubMed] [Google Scholar]
- Chih B., Liu P., Chinn Y., Chalouni C., Komuves L. G., Hass P. E., Sandoval W., Peterson A. S. (2012). A ciliopathy complex at the transition zone protects the cilia as a privileged membrane domain. Nat. Cell Biol. 14, 61–72. 10.1038/ncb2410 [DOI] [PubMed] [Google Scholar]
- Cole D. G., Diener D. R., Himelblau A. L., Beech P. L., Fuster J. C., Rosenbaum J. L. (1998). Chlamydomonas kinesin-II-dependent intraflagellar transport (IFT): IFT particles contain proteins required for ciliary assembly in Caenorhabditis elegans sensory neurons. J. Cell Biol. 141, 993–1008. 10.1083/jcb.141.4.993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craige B., Tsao C. C., Diener D. R., Hou Y., Lechtreck K. F., Rosenbaum J. L., Witman G. B. (2010). CEP290 tethers flagellar transition zone microtubules to the membrane and regulates flagellar protein content. J. Cell Biol. 190, 927–940. 10.1083/jcb.201006105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czarnecki P. G., Shah J. V. (2012). The ciliary transition zone: from morphology and molecules to medicine. Trends Cell Biol. 22, 201–210. 10.1016/j.tcb.2012.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debuchy R., Purton S., Rochaix J. D. (1989). The argininosuccinate lyase gene of Chlamydomonas reinhardtii: an important tool for nuclear transformation and for correlating the genetic and molecular maps of the ARG7 locus. EMBO J. 8, 2803–2809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Failler M., Gee H. Y., Krug P., Joo K., Halbritter J., Belkacem L., Filhol E., Porath J. D., Braun D. A., Schueler M. et al. (2014). Mutations of CEP83 cause infantile nephronophthisis and intellectual disability. Am. J. Hum. Genet. 94, 905–914. 10.1016/j.ajhg.2014.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisch C., Dupuis-Williams P. (2011). Ultrastructure of cilia and flagella – back to the future! Biol. Cell 103, 249–270. 10.1042/BC20100139 [DOI] [PubMed] [Google Scholar]
- Garcia-Gonzalo F. R., Corbit K. C., Sirerol-Piquer M. S., Ramaswami G., Otto E. A., Noriega T. R., Seol A. D., Robinson J. F., Bennett C. L., Josifova D. J. et al. (2011). A transition zone complex regulates mammalian ciliogenesis and ciliary membrane composition. Nat. Genet. 43, 776–784. 10.1038/ng.891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilula N. B., Satir P. (1972). The ciliary necklace. A ciliary membrane specialization. J. Cell Biol. 53, 494–509. 10.1083/jcb.53.2.494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman D. S., Levine R. P. (1965). Cytochrome f and plastocyanin: their sequence in the photosynthetic electron transport chain of Chlamydomonas reinhardi. Proc. Natl. Acad. Sci. USA 54, 1665–1669. 10.1073/pnas.54.6.1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilford W. H., Bloodgood R. A. (2013). Laser trap measurements of flagellar membrane motility. Methods Enzymol. 525, 85–107. 10.1016/B978-0-12-397944-5.00005-5 [DOI] [PubMed] [Google Scholar]
- Hoefele J., Sudbrak R., Reinhardt R., Lehrack S., Hennig S., Imm A., Muerb U., Utsch B., Attanasio M., O'Toole J. F. et al. (2005). Mutational analysis of the NPHP4 gene in 250 patients with nephronophthisis. Hum. Mutat. 25, 411. 10.1002/humu.9326 [DOI] [PubMed] [Google Scholar]
- Hou Y., Qin H., Follit J. A., Pazour G. J., Rosenbaum J. L., Witman G. B. (2007). Functional analysis of an individual IFT protein: IFT46 is required for transport of outer dynein arms into flagella. J. Cell Biol. 176, 653–665. 10.1083/jcb.200608041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B., Ramanis Z., Dutcher S. K., Luck D. J. (1982). Uniflagellar mutants of Chlamydomonas: evidence for the role of basal bodies in transmission of positional information. Cell 29, 745–753. 10.1016/0092-8674(82)90436-6 [DOI] [PubMed] [Google Scholar]
- Huang K., Diener D. R., Mitchell A., Pazour G. J., Witman G. B., Rosenbaum J. L. (2007). Function and dynamics of PKD2 in Chlamydomonas reinhardtii flagella. J. Cell Biol. 179, 501–514. 10.1083/jcb.200704069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L., Szymanska K., Jensen V. L., Janecke A. R., Innes A. M., Davis E. E., Frosk P., Li C., Willer J. R., Chodirker B. N. et al. (2011). TMEM237 is mutated in individuals with a Joubert syndrome related disorder and expands the role of the TMEM family at the ciliary transition zone. Am. J. Hum. Genet. 89, 713–730. 10.1016/j.ajhg.2011.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H., Marshall W. F. (2011). Ciliogenesis: building the cell's antenna. Nat. Rev. Mol. Cell Biol. 12, 222–234. 10.1038/nrm3085 [DOI] [PubMed] [Google Scholar]
- Jauregui A. R., Nguyen K. C., Hall D. H., Barr M. M. (2008). The Caenorhabditis elegans nephrocystins act as global modifiers of cilium structure. J. Cell Biol. 180, 973–988. 10.1083/jcb.200707090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya R., Witman G. B. (1984). Submicromolar levels of calcium control the balance of beating between the two flagella in demembranated models of Chlamydomonas. J. Cell Biol. 98, 97–107. 10.1083/jcb.98.1.97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee H. L., Dishinger J. F., Blasius T. L., Liu C. J., Margolis B., Verhey K. J. (2012). A size-exclusion permeability barrier and nucleoporins characterize a ciliary pore complex that regulates transport into cilia. Nat. Cell Biol. 14, 431–437. 10.1038/ncb2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller L. C., Romijn E. P., Zamora I., Yates J. R., III and Marshall W. F. (2005). Proteomic analysis of isolated chlamydomonas centrioles reveals orthologs of ciliary-disease genes. Curr. Biol. 15, 1090–1098. 10.1016/j.cub.2005.05.024 [DOI] [PubMed] [Google Scholar]
- Kindle K. L. (1990). High-frequency nuclear transformation of Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 87, 1228–1232. 10.1073/pnas.87.3.1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King S. M., Otter T., Witman G. B. (1985). Characterization of monoclonal antibodies against Chlamydomonas flagellar dyneins by high-resolution protein blotting. Proc. Natl. Acad. Sci. USA 82, 4717–4721. 10.1073/pnas.82.14.4717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechtreck K. F., Johnson E. C., Sakai T., Cochran D., Ballif B. A., Rush J., Pazour G. J., Ikebe M., Witman G. B. (2009). The Chlamydomonas reinhardtii BBSome is an IFT cargo required for export of specific signaling proteins from flagella. J. Cell Biol. 187, 1117–1132. 10.1083/jcb.200909183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J., Tritschler D., Song K., Barber C. F., Cobb J. S., Porter M. E., Nicastro D. (2011). Building blocks of the nexin-dynein regulatory complex in Chlamydomonas flagella. J. Biol. Chem. 286, 29175–29191. 10.1074/jbc.M111.241760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y. C., Niewiadomski P., Lin B., Nakamura H., Phua S. C., Jiao J., Levchenko A., Inoue T., Rohatgi R., Inoue T. (2013). Chemically inducible diffusion trap at cilia reveals molecular sieve-like barrier. Nat. Chem. Biol. 9, 437–443. 10.1038/nchembio.1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollet G., Salomon R., Gribouval O., Silbermann F., Bacq D., Landthaler G., Milford D., Nayir A., Rizzoni G., Antignac C. et al. (2002). The gene mutated in juvenile nephronophthisis type 4 encodes a novel protein that interacts with nephrocystin. Nat. Genet. 32, 300–305. 10.1038/ng996 [DOI] [PubMed] [Google Scholar]
- Mollet G., Silbermann F., Delous M., Salomon R., Antignac C., Saunier S. (2005). Characterization of the nephrocystin/nephrocystin-4 complex and subcellular localization of nephrocystin-4 to primary cilia and centrosomes. Hum. Mol. Genet. 14, 645–656. 10.1093/hmg/ddi061 [DOI] [PubMed] [Google Scholar]
- Mortimer S. T. (2000). CASA – practical aspects. J. Androl. 21, 515–524 [PubMed] [Google Scholar]
- Mueller J., Perrone C. A., Bower R., Cole D. G., Porter M. E. (2005). The FLA3 KAP subunit is required for localization of kinesin-2 to the site of flagellar assembly and processive anterograde intraflagellar transport. Mol. Biol. Cell 16, 1341–1354. 10.1091/mbc.E04-10-0931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura S., Tanaka G., Maeda T., Kamiya R., Matsunaga T., Nikaido O. (1996). Assembly and function of Chlamydomonas flagellar mastigonemes as probed with a monoclonal antibody. J. Cell Sci. 109, 57–62 [DOI] [PubMed] [Google Scholar]
- Navaroli D. M., Bellvé K. D., Standley C., Lifshitz L. M., Cardia J., Lambright D., Leonard D., Fogarty K. E., Corvera S. (2012). Rabenosyn-5 defines the fate of the transferrin receptor following clathrin-mediated endocytosis. Proc. Natl. Acad. Sci. USA 109, E471–E480. 10.1073/pnas.1115495109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto E., Hoefele J., Ruf R., Mueller A. M., Hiller K. S., Wolf M. T., Schuermann M. J., Becker A., Birkenhäger R., Sudbrak R. et al. (2002). A gene mutated in nephronophthisis and retinitis pigmentosa encodes a novel protein, nephroretinin, conserved in evolution. Am. J. Hum. Genet. 71, 1161–1167. 10.1086/344395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil H., Tserentsoodol N., Saha A., Hao Y., Webb M., Ferreira P. A. (2012). Selective loss of RPGRIP1-dependent ciliary targeting of NPHP4, RPGR and SDCCAG8 underlies the degeneration of photoreceptor neurons. Cell Death Dis. 3, e355. 10.1038/cddis.2012.96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour G. J., Sineshchekov O. A., Witman G. B. (1995). Mutational analysis of the phototransduction pathway of Chlamydomonas reinhardtii. J. Cell Biol. 131, 427–440. 10.1083/jcb.131.2.427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour G. J., Wilkerson C. G., Witman G. B. (1998). A dynein light chain is essential for the retrograde particle movement of intraflagellar transport (IFT). J. Cell Biol. 141, 979–992. 10.1083/jcb.141.4.979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour G. J., Dickert B. L., Witman G. B. (1999). The DHC1b (DHC2) isoform of cytoplasmic dynein is required for flagellar assembly. J. Cell Biol. 144, 473–481. 10.1083/jcb.144.3.473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour G. J., Agrin N., Leszyk J., Witman G. B. (2005). Proteomic analysis of a eukaryotic cilium. J. Cell Biol. 170, 103–113. 10.1083/jcb.200504008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen L. B., Rosenbaum J. L. (2008). Intraflagellar transport (IFT) role in ciliary assembly, resorption and signalling. Curr. Top. Dev. Biol. 85, 23–61. 10.1016/S0070-2153(08)00802-8 [DOI] [PubMed] [Google Scholar]
- Perkins L. A., Hedgecock E. M., Thomson J. N., Culotti J. G. (1986). Mutant sensory cilia in the nematode Caenorhabditis elegans. Dev. Biol. 117, 456–487. 10.1016/0012-1606(86)90314-3 [DOI] [PubMed] [Google Scholar]
- Piasecki B. P., LaVoie M., Tam L. W., Lefebvre P. A., Silflow C. D. (2008). The Uni2 phosphoprotein is a cell cycle regulated component of the basal body maturation pathway in Chlamydomonas reinhardtii. Mol. Biol. Cell 19, 262–273. 10.1091/mbc.E07-08-0798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter J. F., Blacque O. E., Leroux M. R. (2012). The base of the cilium: roles for transition fibres and the transition zone in ciliary formation, maintenance and compartmentalization. EMBO Rep. 13, 608–618. 10.1038/embor.2012.73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renkema K. Y., Stokman M. F., Giles R. H., Knoers N. V. (2014). Next-generation sequencing for research and diagnostics in kidney disease. Nat. Rev. Nephrol. 10, 433–444. 10.1038/nrneph.2014.95 [DOI] [PubMed] [Google Scholar]
- Ringo D. L. (1967). Flagellar motion and fine structure of the flagellar apparatus in Chlamydomonas. J. Cell Biol. 33, 543–571. 10.1083/jcb.33.3.543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roepman R., Letteboer S. J., Arts H. H., van Beersum S. E., Lu X., Krieger E., Ferreira P. A., Cremers F. P. (2005). Interaction of nephrocystin-4 and RPGRIP1 is disrupted by nephronophthisis or Leber congenital amaurosis-associated mutations. Proc. Natl. Acad. Sci. USA 102, 18520–18525. 10.1073/pnas.0505774102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum J. L., Witman G. B. (2002). Intraflagellar transport. Nat. Rev. Mol. Cell Biol. 3, 813–825. 10.1038/nrm952 [DOI] [PubMed] [Google Scholar]
- Sager R., Granick S. (1953). Nutritional studies with Chlamydomonas reinhardi. Ann. N. Y. Acad. Sci. 56, 831–838. 10.1111/j.1749-6632.1953.tb30261.x [DOI] [PubMed] [Google Scholar]
- Sanders M. A., Salisbury J. L. (1989). Centrin-mediated microtubule severing during flagellar excision in Chlamydomonas reinhardtii. J. Cell Biol. 108, 1751–1760. 10.1083/jcb.108.5.1751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang L., Miller J. J., Corbit K. C., Giles R. H., Brauer M. J., Otto E. A., Baye L. M., Wen X., Scales S. J., Kwong M. et al. (2011). Mapping the NPHP-JBTS-MKS protein network reveals ciliopathy disease genes and pathways. Cell 145, 513–528. 10.1016/j.cell.2011.04.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuermann M. J., Otto E., Becker A., Saar K., Rüschendorf F., Polak B. C., Ala-Mello S., Hoefele J., Wiedensohler A., Haller M. et al. (2002). Mapping of gene loci for nephronophthisis type 4 and Senior-Løken syndrome, to chromosome 1p36. Am. J. Hum. Genet. 70, 1240–1246. 10.1086/340317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiba D., Yokoyama T. (2012). The ciliary transitional zone and nephrocystins. Differentiation 83, S91–S96. 10.1016/j.diff.2011.11.006 [DOI] [PubMed] [Google Scholar]
- Shiba D., Manning D. K., Koga H., Beier D. R., Yokoyama T. (2010). Inv acts as a molecular anchor for Nphp3 and Nek8 in the proximal segment of primary cilia. Cytoskeleton 67, 112–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silflow C. D., Rosenbaum J. L. (1981). Multiple alpha- and beta-tubulin genes in Chlamydomonas and regulation of tubulin mRNA levels after deflagellation. Cell 24, 81–88. 10.1016/0092-8674(81)90503-1 [DOI] [PubMed] [Google Scholar]
- Silflow C. D., LaVoie M., Tam L. W., Tousey S., Sanders M., Wu W., Borodovsky M., Lefebvre P. A. (2001). The Vfl1 Protein in Chlamydomonas localizes in a rotationally asymmetric pattern at the distal ends of the basal bodies. J. Cell Biol. 153, 63–74. 10.1083/jcb.153.1.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slanchev K., Pütz M., Schmitt A., Kramer-Zucker A., Walz G. (2011). Nephrocystin-4 is required for pronephric duct-dependent cloaca formation in zebrafish. Hum. Mol. Genet. 20, 3119–3128. 10.1093/hmg/ddr214 [DOI] [PubMed] [Google Scholar]
- Spiess P. C., Deng B., Hondal R. J., Matthews D. E., van der Vliet A. (2011). Proteomic profiling of acrolein adducts in human lung epithelial cells. J. Proteomics 74, 2380–2394. 10.1016/j.jprot.2011.05.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens D. R., Rochaix J. D., Purton S. (1996). The bacterial phleomycin resistance gene ble as a dominant selectable marker in Chlamydomonas. Mol. Gen. Genet. 251, 23–30 [DOI] [PubMed] [Google Scholar]
- Wagner D. E., Bonenfant N. R., Sokocevic D., DeSarno M. J., Borg Z. D., Parsons C. S., Brooks E. M., Platz J. J., Khalpey Z. I., Hoganson D. M. et al. (2014). Three-dimensional scaffolds of acellular human and porcine lungs for high throughput studies of lung disease and regeneration. Biomaterials 35, 2664–2679. 10.1016/j.biomaterials.2013.11.078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburton-Pitt S. R., Jauregui A. R., Li C., Wang J., Leroux M. R., Barr M. M. (2012). Ciliogenesis in Caenorhabditis elegans requires genetic interactions between ciliary middle segment localized NPHP-2 (inversin) and transition zone-associated proteins. J. Cell Sci. 125, 2592–2603. 10.1242/jcs.095539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams C. L., Winkelbauer M. E., Schafer J. C., Michaud E. J., Yoder B. K. (2008). Functional redundancy of the B9 proteins and nephrocystins in Caenorhabditis elegans ciliogenesis. Mol. Biol. Cell. 19, 2154–2168. 10.1091/mbc.E07-10-1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams C. L., Masyukova S. V., Yoder B. K. (2010). Normal ciliogenesis requires synergy between the cystic kidney disease genes MKS-3 and NPHP-4. J. Am. Soc. Nephrol. 21, 782–793. 10.1681/ASN.2009060597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams C. L., Li C., Kida K., Inglis P. N., Mohan S., Semenec L., Bialas N. J., Stupay R. M., Chen N., Blacque O. E. et al. (2011). MKS and NPHP modules cooperate to establish basal body/transition zone membrane associations and ciliary gate function during ciliogenesis. J. Cell Biol. 192, 1023–1041. 10.1083/jcb.201012116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkelbauer M. E., Schafer J. C., Haycraft C. J., Swoboda P., Yoder B. K. (2005). The C. elegans homologs of nephrocystin-1 and nephrocystin-4 are cilia transition zone proteins involved in chemosensory perception. J. Cell Sci. 118, 5575–5587. 10.1242/jcs.02665 [DOI] [PubMed] [Google Scholar]
- Witman G. B. (1986). Isolation of Chlamydomonas flagella and flagellar axonemes. Methods Enzymol. 134, 280–290. 10.1016/0076-6879(86)34096-5 [DOI] [PubMed] [Google Scholar]
- Wolf M. T., Hildebrandt F. (2011). Nephronophthisis. Pediatr. Nephrol. 26, 181–194. 10.1007/s00467-010-1585-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf M. T., Lee J., Panther F., Otto E. A., Guan K. L., Hildebrandt F. (2005). Expression and phenotype analysis of the nephrocystin-1 and nephrocystin-4 homologs in Caenorhabditis elegans. J. Am. Soc. Nephrol. 16, 676–687. 10.1681/ASN.2003121025 [DOI] [PubMed] [Google Scholar]
- Won J., Marín de Evsikova C., Smith R. S., Hicks W. L., Edwards M. M., Longo-Guess C., Li T., Naggert J. K., Nishina P. M. (2011). NPHP4 is necessary for normal photoreceptor ribbon synapse maintenance and outer segment formation, and for sperm development. Hum. Mol. Genet. 20, 482–496. 10.1093/hmg/ddq494 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.