Abstract
Since the physical process of fluorescence resonance energy transfer (FRET) was elucidated more than six decades ago, this peculiar fluorescence phenomenon has turned into a powerful tool for biomedical research due to its compatibility in scale with biological molecules as well as rapid developments in novel fluorophores and optical detection techniques. A wide variety of FRET approaches have been devised, each with its own advantages and drawbacks. Especially in the last decade or so, we are witnessing a flourish of FRET applications in biological investigations, many of which exemplify clever experimental design and rigorous analysis. Here we review the current stage of FRET methods development with the main focus on its applications in protein studies in biological systems, by summarizing the basic components of FRET techniques, most established quantification methods, as well as potential pitfalls, illustrated by example applications.
1. Introduction
1.1 The concept of FRET
Fluorescence resonance energy transfer (FRET) is an electromagnetic phenomenon in which quantum energy is transferred non-radiatively from an excited donor fluorophore to an acceptor molecule within close proximity [1, 2]. The term “resonance energy transfer” refers to the fact that energy transfer is by means of intermolecular dipole-dipole coupling, that is, the process does not involve emission and reabsorption of photons. The donor fluorophore typically emits at shorter wavelengths that overlap with the absorption spectrum of the acceptor molecule (which may be a fluorophore or a non-fluorescent molecule). The rate of energy transfer, kT (r), falls off with the sixth power of the distance between the donor and acceptor molecules:
| Equation 1 |
where τD is the decay time of the donor in the absence of acceptor, r is the actual distance between the chromophores (the symbol “r” used here and in Equations 3a and 3b should not be confused with the same symbol used later for anisotropy), and R0 is the Förster distance or the distance at which the energy transfer efficiency is 50% [2]. R0 depends on the quantum yield of the donor, the extinction coefficient of the acceptor, the overlap of the donor emission and acceptor excitation spectra, and the relative orientation of the chromophores:
| Equation 2 |
where κ2 is the orientation factor between donor and acceptor; n is the refractive index of the medium; QD is the quantum yield of the donor in the absence of the acceptor; J(λ) is the degree of spectral overlap between the donor emission spectrum and the acceptor absorbance spectrum [3].
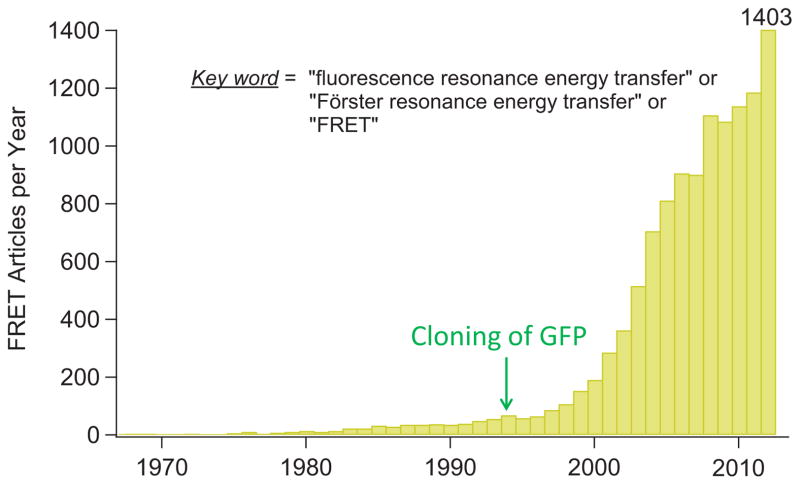
The phenomenon of FRET is extremely useful for biomedical research for three major reasons. First, due to its exquisite sensitivity to molecular distance, FRET makes a great reporter for close proximity as well as very small perturbation in proximity. For this reason, FRET has been referred as a “spectroscopic ruler” [4]. FRET is most sensitive to distance changes when the donor and acceptor are separated by a distance near R0 (typically 20–60 Å). The second reason FRET is particularly suitable for biomedical research is that the distance range at which FRET occurs matches well with the dimension of many biological molecules, such as the size of proteins and polynucleotides, the distance between sites on multi-subunit proteins and interacting molecules, the thickness of the cell membrane, etc. Furthermore, while the distance resolution of FRET is much lower than that of X-ray crystallography, FRET has the advantage of being applicable in living biological systems under physiological conditions and, in many cases, in real time. Indeed, since the physical process of FRET was elucidated more than six decades ago [1, 2], its application in biomedical research has exploded (Fig. 1).
Figure 1.
Explosive increase of FRET applications in biomedical research. The number of FRET papers published each year is determined by a Pubmed search with keywords combination “fluorescence resonance energy transfer” or “Förster resonance energy transfer” or “FRET”.
1.2 FRET efficiency
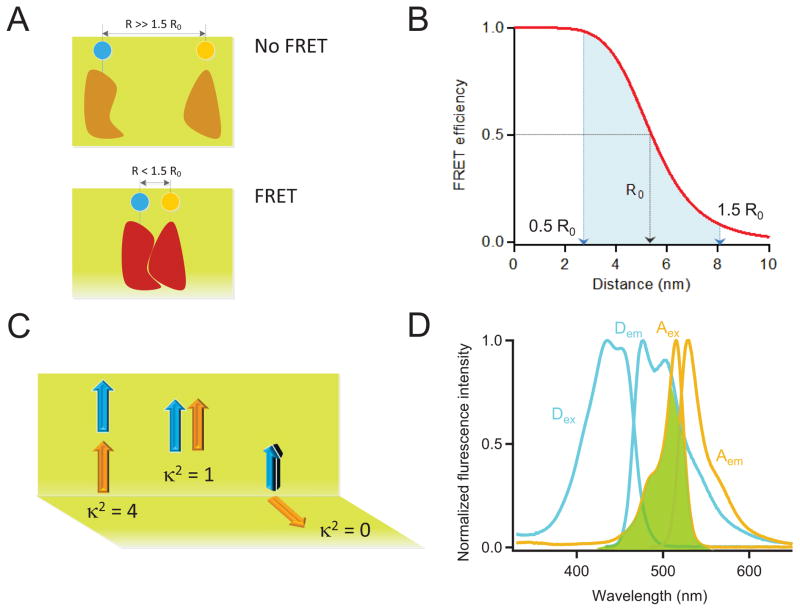
The efficiency of FRET (EFRET), which is the fraction of the energy absorbed by the donor that is transferred to the acceptor, is given by the Förster equation (Equation 3). Similar to the rate of energy transfer, EFRET falls off with sixth power of the distance between the donor and acceptor molecules, leading to a large change in its value over the range of 0.5 R0 to 1.5 R0. Below 0.5 R0, energy transfer efficiency remains 98–100%, and there would be not much change to see. Beyond 1.5 R0, the efficiency of energy transfer is very low (< 8.1%) (Fig. 2A–B).
Figure 2.
Basic concepts in FRET. (A) Schematic diagram illustrating the proximity requirement of FRET to occur. Two target proteins are labelled with a donor FP and an acceptor FP respectively. Energy transfer between the two FPs occurs when the two target proteins interact, which shortens the distance between FPs from beyond 1.5 R0 (top) to within 1.5 R0 (bottom). (B) FRET efficiency as a function of the distance between the donor and the acceptor fluorophores with R0 = 54 Å (an R0 value similar to that for the Cerulean/Venus pair or the EGFP/Alexa Fluor 568 pair). At R = R0, FRET efficiency = 0.5. The range between 0.5 R0 to 1.5 R0, indicated by the blue shade, is where FRET changes can be reliably measured. Notice the linearity of the FRET efficiency values at distances near R0. (C) Schematic representation of the orientation factor κ2 between the transition dipole moments of the two fluorophores. There is no FRET when κ2 = 0. (D)Absorption and emission spectra of Cerulean (donor, cyan) and Citrine (acceptor, green). The overlap between Cerulean emission spectrum and Citrine absorption spectrum is highlighted by green shade.
| Equation 3a |
| Equation 3b |
EFRET is affected by all the factors in Equation 2. While most factors determining EFRET are straightforward and well-discussed in the literature, an important parameter for FRET that often causes confusion is the orientation factor κ2, which describes the relative orientation of the donor and acceptor transition dipoles and ranges from 0 to 4. When the donor and acceptor transition dipoles are aligned to each other, κ2 = 4, the orientation is ideal for energy transfer; whereas in a perpendicular orientation, κ2 = 0, preventing any energy transfer disregard of the distance (Fig. 2C). It is important to realize that κ2 matters only during the lifetime of the donor excited state when energy coupling is possible. For most fluorophores, this time is 1-to-10 ns. It means that if the fluorophores, or their chromophore part, or the molecules the fluorophores are attached to, move at frequencies significantly higher than 108–109 Hz, the fluorophore pair will experience varying orientation while FRET occurs. In biological systems, proteins labelled with fluorophores generally can adopt a variety of conformations, and the covalent linkage between the fluorophore and the host protein can rotate. All these lead to randomization of the orientation both spatially and temporally. Thus for most macromolecular interactions in solutions, κ2 value of 2/3 is often assumed.
Minor polarizations of donor and acceptor molecules will not lead to major variations in the determination of R0. But if one assumes that a range of static donor-acceptor orientations are present and do not change during the lifetime of the excited state, κ2 = 0.476 should be used instead [3, 5]. Nevertheless, under certain experimental conditions such assumption does not hold and more rigorous treatments of κ2 are called for [6, 7]. More detailed basic concepts of FRET could be found in classical reviews and textbooks [3, 8, 9].
1.3 The first key step for a FRET experiment: choose the right fluorophore pair
When designing a FRET experiment, the first key step of performing a successful measurement is to choose a suitable pair of donor and acceptor molecules based on their spectroscopic characteristics and experimental needs. Several fundamental conditions should be met: 1) sufficient separation in excitation spectra so that the donor could be selectively activated by the light source (perfect selectivity is almost never achievable, see below); 2) enough overlap between the emission spectrum of the donor and the excitation spectrum of the acceptor to ensure efficient energy transfer (Fig. 2D); 3) reasonable separation in emission spectra to allow selective measurement of the emitted fluorescence from the acceptor (again, exclusive measurement is hard to achieve in practice) [10]; 4) The choice of the donor and the acceptor fluorophores should also allow the distance between them, when attached to biomolecules, to be within 10–100 Å.
There are plenty of choices when it comes to fluorophores, such as organic dyes (e.g., fluorescein, FITC (fluorescein isothiocyanate), rhodamine, Cyanine-dyes, Alexa-dyes and Atto-dyes), inorganic ions (e.g., lanthanide ions Eu3+, Tb3+, Yb3+, etc.), and fluorescent proteins (FP). The application of FRET technique in biomedical research really started to grow exponentially following the development of green fluorescent protein (GFP) from the jellyfish Aequoria victoria and its many derivatives (Fig. 1) [11]. Wild-type GFP (26.9 kDa) consists of a β-barrel structure in which the essential chromophoric moiety lies at the center and could form automatically under physiological conditions due to an autocatalyzed biosynthesis of imidazolinone from residues Ser65-Tyr66-Gly67 [12]. The same process occurs when the fluorescent protein is expressed in jellyfish, as an exogenous protein, or as part of a fusion protein [13, 14]. The β-barrel structure surrounding the tripeptide influences its fluorescent properties and protects the chromophore from environmental influences.
Following the discovery of GFP, many FPs from other species, such as the Anthozoan button polyp Zoanthus (ZsYellow), sea anemone Discosoma (DsRed) [15] and Anemonia majano (AmCyan1), were identified and isolated. Also, researchers have achieved great success in modifying FPs by mutagenesis to expand the color spectrum, narrow the emission peak, improve the photostability, or enhance the quantum yield (Table 1). As a result, FPs spanning the full visible spectrum have been available, and sparked a revolution in the FRET study in living cells (Fig. 1; Table 1).
Table 1.
| FP | λex (nm)a | λem (nm)b | ε (M−1cm−1)c | Φ (%)d | Brightnesse | pKaf | Photostability (s)g |
|---|---|---|---|---|---|---|---|
| EBFP | 383 | 445 | 31,000 | 25 | 23 | 5.8 | 3.5 |
| ECFP | 433 | 476 | 29,000 | 37 | 32 | 4.7 | 98 |
| Cerulean | 433 | 475 | 43,000 | 63 | 81 | 4.7 | 126 |
| mEmerald | 487 | 509 | 57,000 | 68 | 116 | 6.0 | 101 |
| EGFP | 488 | 507 | 56,000 | 60 | 100 | 6.0 | 150 |
| UnaG | 498 | 527 | 77,300 | 51 | 117 | <4.0 | ND |
| mNeonGreen | 506 | 517 | 116,000 | 80 | 276 | 158 | 5.7 |
| LanYFP | 513 | 524 | 150,000 | 95 | 424 | ND | 3.5 |
| EYFP | 514 | 527 | 83,000 | 61 | 151 | 6.9 | 60 |
| mVenus | 515 | 528 | 92,000 | 57 | 156 | 6.0 | 15 |
| mBanana | 540 | 553 | 6,000 | 70 | 13 | 6.7 | 1.4 |
| mOrange | 548 | 562 | 71,000 | 69 | 146 | 6.5 | 6.4 |
| tdTomato | 554 | 581 | 138,000 | 69 | 283 | 4.7 | 70 |
| dsRED | 558 | 583 | 75,000 | 79 | 176 | 4.7 | 167 |
| mStrawberry | 574 | 596 | 90,000 | 29 | 78 | <4.5 | 11 |
| mRFP1 | 584 | 607 | 50,000 | 25 | 37 | 4.5 | 6 |
| mCherry | 587 | 610 | 72,000 | 22 | 47 | <4.5 | 68 |
λex: Excitation maximum.
λem: Emission maximum.
Extinction coefficient (ε): an intrinsic property of the species that quantifies how strongly a chemical species absorbs light at a given wavelength, which is usually determined by alkali-denaturation method.
Quantum yield (Φ): the ratio of the number of photons emitted to the number of photons absorbed, which can be determined by the rate constants of emission (Γ) and the sum of all non-radiative decay processes (knr) that depopulate the excited state. Φ = Γ/(Γ+ knr).
Product of ε and Φ, expressed as a percentage of mEGFP brightness.
pH at which fluorescence intensity is 50% of its maximum value.
Time to bleach to half-maximal emission intensity.
The choice of FRET pair depends on the purpose of the FRET study, the microscopy setup and the quantification method to use. For example, EGFP is extensively used due to its high quantum yield and resistance to photobleaching. The nice separation of the ECFP emission spectrum and the EYFP excitation spectrum, as well as the high absorption and quantum yield of the latter have made them one of the most commonly used FRET pairs [14, 16]. Very recently, Shaner et al. reported a monomeric yellow-green fluorescent protein, mNeonGreen, the brightest monomeric green or yellow fluorescent protein available so far, which is much brighter than cyanine dyes or Alexa dyes and is of similar brightness as ATTO 550 (brightness calculated as the product of extinction coefficient and quantum yield), thus can serve as an excellent FRET acceptor for cyan fluorescent proteins [17]. Another exciting progress is that the Miyawaki group cloned a brand new fluorescent protein, UnaG, from a type of Japanese eel. When triggered by an endogenous chromogenic ligand, bilirubin, UnaG can produce bright oxygen-independent green fluorescence [18]. As the first FP from the vertebrate subphylum, UnaG is not only a great clinical tool for quantifying human bilirubin level [18], but also will be a brilliant FRET donor fluorophore candidate since it has much smaller size than GFP (139 aa vs 238 aa) and one can achieve conditional switch of its fluorescence emission.
1.4. Quantum-dots
Over the past decades, rapid evolution in bionanotechnology has led to the development of luminescent nanoparticles with outstanding physical and chemical properties that are unmatched by other fluorophores. Quantum dots (QDs) have emerged as exceptional representatives among them. QDs are inorganic fluorescent semiconductor nanocrystals, generally 2–10 nm in diameter, that are composed of elements in the periodic groups of II–VI, III–V or IV–VI [19]. The emission color of QDs, which depends on their size, chemical composition, and surface chemistry, can be tuned from ultraviolet to the visible and near-infrared wavelengths. In general, the smaller the QDs are, the bluer the emission light is [20, 21].
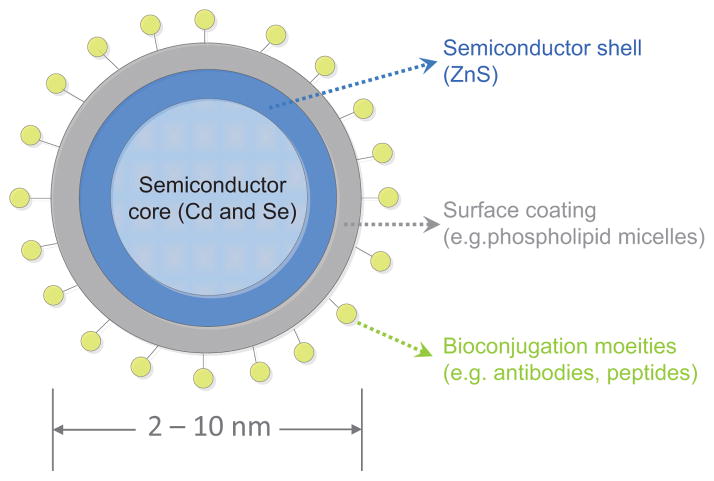
The general structure of a QD (Fig. 3) comprises an inorganic core semiconductor material (e.g. CdTe or CdSe), an inorganic shell of a higher band gap semiconductor material (e.g. ZnS), and an aqueous organic coating layer to which biomolecules of interest can be conjugated (e.g. antibodies, peptides, nucleic acids, etc.) [22]. The shell and coatings not only confer different photophysical properties to the QDs, but also form physical barriers that separate the nanocrystal core from the surrounding environment, thus making QDs resistant to photobleaching.
Figure 3.
Schematic structure of a quantum dot. These inorganic fluorescent semiconductor nanocrystals often have a diameter of 2–10 nm.
QDs have many advantageous features over traditional fluorophores, which make them attractive in imaging applications like FRET [20, 22–24]. First, as expected, QDs are extremely photostable. They can maintain high brightness even after repeated cycles of excitation and fluorescence for hours. Therefore, background signals from short-lived fluorescent species can be corrected by time-gating techniques, allowing QDs be used for long-term monitoring and cell-tracking studies [25, 26]. Second, each QD has a quite broad absorption spectrum and a narrow and symmetrical emission spectrum. This property makes extensive multiplexed imaging much easier, because a single light source can be used to excite multiple QDs simultaneously; and due to the narrow emission spectrum of each QD, there won’t be much overlap between different emission spectra [24]. On the practical end, broad absorption of QDs also leaves plenty of choices for the excitation source from simple voltaic arc lamps to most commercially available lasers (e.g. argon-ion, helium-cadmium and krypton-argon). For FRET applications, the broad absorption spectrum permits easy pairing with a donor fluorophore. However, due to cross-talk contamination, this also makes it problematic to engineer QDs as acceptors in QD-FRET. Third, as mentioned above, different QDs cover broad emission spectra from ultraviolet to near-infrared, which contributes to a better separation of donor and acceptor emission peaks, thereby avoiding donor bleed-through contamination effectively. Fourth, due to their particularly large extinction coefficients (> 106 M−1cm−1 vs < 105 M−1cm−1 for typical organic dyes and FPs), large absorbance cross-section and saturation intensity, QDs have exceptional brightness (about 10–100 times brighter than organic fluorogenic dyes). Fifth, QDs have slow excited-state decay rates (lifetime 30-to-100 ns). This is valuable in overcoming the background autofluorescence, therefore improving signal-to-noise ratio.
Nevertheless, QDs are not perfect. The toxicity of QDs has limited their applications in biological studies [27, 28]. Their “blinking” behavior, i.e. rapidly alternating between an emitting and non-emitting state, may cause problems in single-molecule imaging or tracking [29]. In addition, QD “photobrightening” (increase in fluorescence intensity upon extended excitation) could be troublesome in fluorescence quantitation studies [20]. In terms of QD-FRET, the proximity requirement for FRET determines that the major limitation for QDs is their large size. Practically, large (red/infrared emitting) QDs cannot be used [30]. Furthermore, unlike FPs, QD-protein fusions cannot be genetically coded and thus cannot be produced in living cells.
Despite these restrictions, numerous QD-based FRET applications have been reported in the literature, the majority of which are in vitro studies using QDs as the donor and organic fluorophores as the acceptor [24, 31–36]. The continuous efforts in generating smaller, brighter, and non-blinking QDs may allow novel applications of QD-FRET techniques and significantly expand our ability to deal with challenging questions, such as dynamics of protein-protein interactions at a long time scale.
2. Potential pitfalls and practical considerations in FRET experiments
In order to minimize artifacts and maximize signal-to-noise ratio in FRET studies, it is critical to be aware of potential contamination sources, design effective controls as well as understand that FRET measurements are usually based on specific assumptions. In this section, we will go through some common problems and discuss useful strategies to deal with them.
2.1 Artifacts caused by orientation issue
As discussed above, FRET efficiency is dependent on both the distance and the relative orientation of the fluorophore pair. If the fluorophores do not have any preferential orientation, or are wobbling at rates comparable to fluorescence emission, the issue of orientation dependence should be negligible for the majority of FRET studies [3]. Many chemical fluorophores appear to fall into this category. Even covalently attached FP molecules are generally found to be highly mobile under physiological conditions [37]. Nevertheless, it is reassuring to confirm mobility of the fluorophores by measuring their anisotropy in the FRET experimental environment. For example, binding of Ca2+-calmodulin was observed to decrease FRET efficiency between FPs attached to cyclic nucleotide-gated (CNG) channels, which was attributed to disruption of an inter-subunit interaction. An alternative possibility in the case was that Ca2+-calmodulin might immobilize the fluorescent proteins in unfavorable orientations. To rule out this possibility, anisotropy from eYFP attached to different channel sites was measured before and after Ca2+-calmodulin modulation, which showed no detectable difference. These results confirmed that binding of Ca2+-calmodulin per se did not affect the properties of the fluorescence [38].
2.2 Cross-talk
Ideally, there should be extensive overlap between the donor emission spectrum and the acceptor excitation spectrum, but no overlap in the excitation spectra or emission spectra between the FRET pair. Unfortunately, this is rarely true in reality as the Stoke’s shift (the wavelength difference between excitation and emission spectra) for most fluorophores is comparable or smaller than the width of the spectra. As a result of overlap in the excitation spectra, the acceptor can be excited directly by the donor excitation light, which would lead to contaminating fluorescence signals in FRET experiments. This type of error is often called “cross-talk”. The extent of cross-talk is determined by the extinction coefficient of the acceptor fluorophore at the donor excitation wavelength and the relative excitation light intensities for the donor and the acceptor. One way to estimate the extent of cross-talk is to measure the fluorescence intensity ratio of an acceptor-only sample excited with the donor and the acceptor excitation light. Using this approach, the CFP-YFP pair measured under typical physiological conditions was found to exhibit a cross-talk contamination accounting for 15–25% of the YFP peak emission [39].
There have been several approaches developed to minimize or correct for cross-talk in FRET experiments. One approach is to separate the cross-talk emission spectrum of the acceptor from the total emission spectrum using spectral imaging techniques, such as linear unmixing and Spectra FRET (as discussed below) [40–42]. Other options, when possible, are to use an optimized excitation wavelength to minimize direct acceptor excitation [43] or to apply fluorescence lifetime measurement for FRET quantification (FLIM, see below). A different FRET-type approach, bioluminescence resonance energy transfer (BRET), uses an enzyme (typically luciferase, which emits light in the presence of its substrate) to generate donor light and a fluorophore (typically a fluorescent protein) as the acceptor for resonance energy transfer [44, 45]. Since in BRET the donor is excited chemically instead of optically, the problem of excitation cross-talk is eliminated; this is one of the advantageous features of this method.
2.3 Bleed-through
Another source of contamination from spectra overlap is often called “bleed-through”, which refers to fluorescence emission from the donor fluorophore that is detected within the range of acceptor fluorescence emission. The extent of bleed-through is determined by the relative fluorescence intensities of donor and acceptor within the detection wavelength range. Bleed-through could be a serious problem for some FRET pairs if not properly corrected. For example, the emission of CFP at the YFP peak emission wavelength (530 nm) could be as high as 50% of the CFP peak emission measured at 480 nm. Similar to cross-talk, bleed-through can also be eliminated by subtracting it from the total emission spectrum using spectrum-based FRET techniques [46, 47].
2.4 Non-specific FRET
It is almost always desirable to have stronger fluorescence signals in FRET studies. With a fixed FRET pair, one way to achieve high fluorescence intensity is to increase the power of the excitation light, which has an upper limit and is always penalized by increased photobleaching. An alternative is to introduce more fluorophores into the FRET system by having higher densities of fluorophore-tagged molecules. However, this could cause problems of non-specific FRET. As the fluorophore density increases, the average distance between non-associated donor and acceptor fluorophores drops. As a result, the probability of finding a donor and an acceptor within the FRET distance increases. Fluorophore aggregation may also occur when clusters of fluorescently labelled molecules exist, for example, at the synapse or caveolae. A simple and often effective method to detect such non-specific FRET is to plot the measured FRET values against the fluorescence intensity of either the donor or the acceptor (assuming that association of the FRET pair is not affected by concentration). Since the fluorescence intensity represents roughly the fluorophore density, a detectable dependence of FRET efficiency on the fluorescence intensity may suggest that the fluorophore density is too high and non-specific FRET has occurred.
An additional issue arising from high density of fluorophores is that the donor fluorescence emission could also be directly reabsorbed by the acceptor (as opposed to the non-radiative energy transfer of FRET). This non-FRET, re-absorption mechanism is normally assumed to be negligible at low fluorophore densities, but could be an issue when the fluorophore density is high or the sample volume is large.
2.5 Mixture of fluorophore populations
In FRET experiments, a general assumption is that there is a single donor and a single acceptor present as a FRET couple. This simple case scenario, unfortunately, is often not true in biological experiments. In case of one donor coupled to multiple acceptors, the most commonly used model is a simple kinetic model assuming that the donor interacts separately with each acceptor and the collective EFRET is calculated from the sum of all FRET transfer rates divided by the sum of all radiative and non-radiative energy release rates [3]. Used properly, this strategy could be applied to analyze rather complex questions quantitatively [46, 48]. Interestingly, Koushik et al observed anomalous surplus energy transfer with multiple acceptors [49]. The mechanism behind this phenomenon is not known yet, but the authors speculate that “either an additional energy transfer pathway exists when multiple acceptors are present, or that a theoretical assumption on which the kinetic model prediction is based is incorrect” [49]. It is important to realize that for the kinetic approach to work, the assumption of independent coupling between fluorophores has to be met, that is, at any moment, one donor can only be coupled to one acceptor independent of other fluorophores nearby. This assumption is only true at low excitation condition when the probability of a fluorophore being in the excited state is low.
In biological preparations, uncoupled fluorophores and pairs of like fluorophores are very likely to co-exist with paired donor-acceptors. Emission from these fluorophores contributes to the total fluorescence intensity but not to the FRET signal, thus reducing the apparent FRET efficiency. To illustrate this issue, one can consider a simple scenario in which the biological sample contains the donor-acceptor pair as well as the free donor and the free acceptor at concentrations of [DA], [D], and [A], respectively. This is the situation when donor and acceptor labelled molecules interact but do not form obligated complexes. Without FRET, the donor and acceptor fluorescence intensities can be calculated as:
| Equation 4 |
| Equation 5 |
in which SD and SA are constants reflecting the properties of the recording system and fluorophores, such as excitation light intensity, the transfer function of the fluorescence detector, extinction coefficient and quantum yield of fluorophores, etc. FRET causes a decrease in the donor intensity and an increase in the acceptor intensity. As will be discussed below, FRET efficiency E can be measured by either the fractional decrease in the donor intensity (Equation 6) or the fractional increase in the acceptor intensity (Equation 7). The apparent FRET efficiency, Eapp, of the system would be:
| Equation 6 |
| Equation 7 |
As we can see from Equation 6 and 7, no matter how E is quantified, if singular fluorophores exist, the observed Eapp will be lower than the true efficiency E. If only half of the donors/acceptors can form FRET pairs, for example, the apparent FRET efficiency will be only one half of the true efficiency, Eapp = 0.5E.
Obviously, a straightforward strategy to minimize the error caused by mixed population in calculating Eapp is to make sure [DA]≫[D] in donor-based calculation or [DA]≫[A] in acceptor-based calculation [50]. If the system involves affinity binding between D and A as mentioned above, one may achieve the goal by ensuring [A] ≫ [D] or [D] ≫ [A]. This useful practice would yield much more reliable FRET measurements than a surprisingly common practice of picking cells with equal donor and acceptor intensities. However, the amount of labelled molecules is not always easily controllable. In a biological system involving co-transfection of donor and acceptor, for example, the expression levels of the donor and the acceptor fluorophores would vary greatly from one cell to another. If Eapp is calculated from the enhanced acceptor emission and is plotted against the fluorescence intensity ratio between the donor and acceptor, the figure can be divided into three regions (Figure 4I). In region I, there is a substantial amount of uncoupled acceptor molecules that do not contribute to FRET ([DA] ≪ [A]). As a result, Eapp would be much lower than the true efficiency if it is estimated from the acceptor enhanced emission. Region III, on the other hand, is ideal for acceptor-based estimation in that most of the acceptor fluorophores are coupled to a donor ([DA] ≫ [A]) and Eapp approaches the true value. Region II is where the donor intensity and the acceptor intensity are comparable and easy to measure, but Eapp depends strongly on the donor-to-acceptor ratio. Ideally, we want all the cells to be in region III. However, the high donor-to-acceptor ratio often translates into low acceptor intensities, which would be hard to quantify accurately and more prone to contaminations from autofluorescence and background light. Particularly in the presence of the high donor intensity, the calculated FRET efficiency tends to be inaccurate. Therefore, more accurate estimation of the intrinsic FRET efficiency is likely to be achieved by taking into account the variable donor-to-acceptor ratio and getting the real EFRET by fitting to data from all three regions [46, 51].
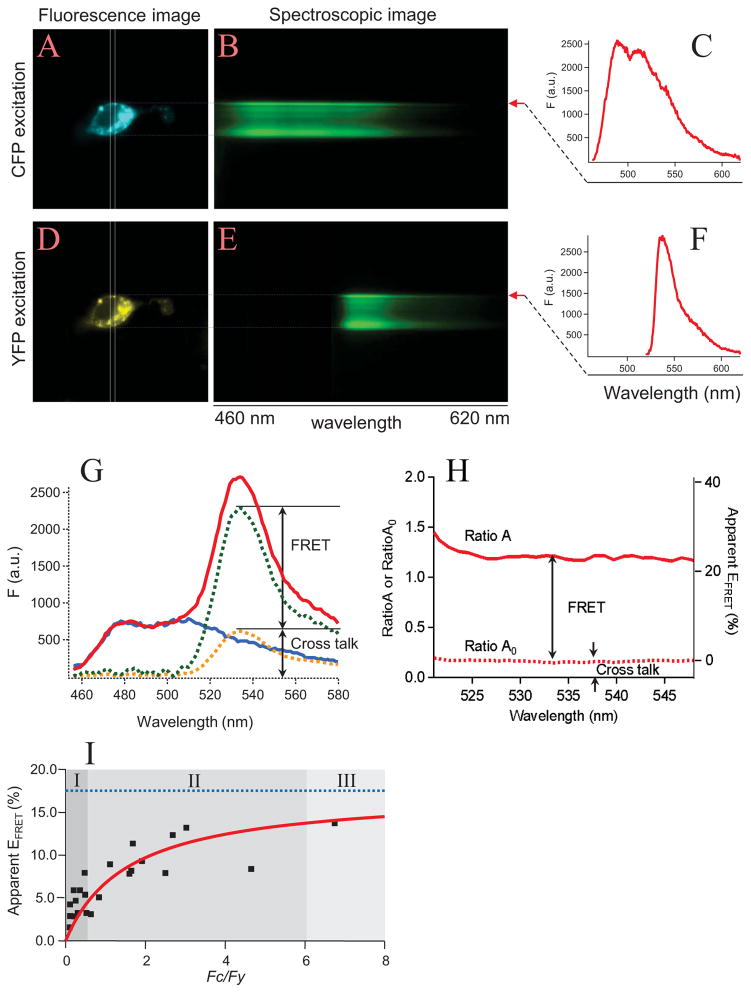
Figure 4.
Spectra FRET. (A–F) The collection of spectral information. A single HEK 293 cell co-transfected with chloride channel subunits CLC-0-CFP and CLC-0-YFP is shown with CFP (A) or YFP (D) excitation light. The formation of CLC-0 dimeric channels is expected to bring CFP and YFP together. Spectroscopic images from the cellular region under the slit (indicated by rectangles) are shown in B and E, in which the y axis represents the position of the cell, the x axis represents the wavelength. Using line-scanning measurement of the fluorescence intensity values along the upper membrane region (red arrows) of the spectroscopic images, the emission spectra of CFP (C) and YFP (F) are constructed. (G&H) FRET quantification. To remove the bleed-through and cross-talk contaminations, a CFY emission spectrum (blue curve in G) is normalized to the CFP peak of an emission spectrum from a cell expressing CFP-YFP tandem dimers (red curve). The difference between the red curve and the blue curve (green curve) contains both the FRET component and the cross-talk component (yellow curve, measured from CFP excitation light). The difference between RatioA (solid line in H) and RatioA0 (dotted line in H) indicates the existence of a FRET signal. Note that RatioA0 corresponds to the zero FRET efficiency level (right axis). (I) An example of the Spectra FRET measurement showing the dependence of apparent FRET efficiency on the donor-to-acceptor fluorescence ratio. Measurements from a number of cells (square symbols) were fitted to a model for the dependence of the apparent FRET efficiency on the donor-to-acceptor fluorescence intensity ratio (red curve). The dotted blue line represents the expected FRET efficiency. The figure was divided into three regions depending on the donor-to-acceptor ratio (I to III). Only at high donor-to-acceptor ratios (Region III), the apparent FRET efficiency approaches the true FRET efficiency. This figure is modified from Fig. 2 in [58] with permission from Elsevier.
2.6 Change of fluorescence property upon the binding of labelled molecules
In an ideal experiment, the emission properties of the fluorophores should not be affected by interaction between the fluorescently labelled molecules per se. However, in practice, this is sometimes not true. For example, hybridization of an unlabelled strand of polynucleic acids was found to quench the fluorophore on a labelled complementary strand [52]. Interestingly, many factors could affect the extent of this type of quenching, including the number of nucleotide pairs in the hybrid, the length of the hydrocarbon spacer between the terminal nucleotide and the fluorophore, the temperature, and the ionic strength of the solution [53]. The study suggests that unintended changes in fluorescence property may be more common than they were assumed. It is therefore helpful to determine the fluorescence property of the donor in the presence of the unlabeled complementary interacting molecule. This can be easily implemented for example in experiments using “donor-dequenching after acceptor photobleaching” quantification method (as discussed below), in which the donor fluorescence intensity in the absence of the acceptor is measured after the acceptor fluorophore is photobleached [54].
In addition to the potential pitfalls mentioned above, there are also other sources of contaminations that should draw our attention, such as autofluorescence and the “background” scattered light, which should always be minimized if possible, or to be quantified and subtracted from FRET measurements [3]. When fluorophores are tethered to biological molecules of interests, one should test whether the spectral properties of these fluorophores are unchanged [13].
3. FRET quantification techniques
Many FRET quantification techniques have been developed, and novel approaches are continuously being invented. On the basis of practical considerations, four general approaches are widely applied: (i) change in the fluorescence emission intensity of the donor; (ii) sensitized steady-state fluorescence emission intensity of the acceptor; (iii) decrease in the donor lifetime; and (iv) change of fluorescence polarization.
3.1 Change in donor fluorescence intensity – donor quenching upon FRET or donor de-quenching after acceptor photobleaching
Quenching is a phenomenon by which interaction of a molecule (the quencher) with the fluorophore reduces the quantum yield or the lifetime of the fluorophore. In this sense, the acceptor in FRET works as a quencher for the donor, so the FRET efficiency can be quantified as the decrease of the donor emission fluorescence upon coupling with the acceptor. An interesting example of FRET measurement using this quantification method is “transition metal FRET” [55]. The idea behind this method is that transition metal ions such as Ni2+, Co2+ and Cu2+ are colored, and their absorbance spectra overlap with the emission spectra of fluorescent dyes such as fluorescein. Hence transition metals can serve as the acceptor to “quench” the donor fluorescence. Transition metals are not sensitive to the orientation problems usually associated with FRET and do not require long flexible linker as FPs do. Also, because the R0 value for Ni2+ or Cu2+ and fluorescein (12 Å and 16 Å respectively), for example, is much lower than standard FRET pairs such as CFP and YFP (50 Å), the fluorescein /transition metal ion FRET pair could be used to monitor short-range FRET changes. In this regard, it helps that the transition metals are much smaller than most fluorophores. The transition metals can form a metal bridge between two or more cysteines or histidines. To direct transition metal ions to a target site of protein, one could introduce two histidines spaced one turn away on an α-helix to minimize the backbone perturbation and ensure the spacing between the two sites. A donor fluorophore such as fluorescein can bind to a cysteine residue specifically introduced to the site of interest in the protein. FRET efficiency can then be easily calculated by measuring fluorescence before (F) and after (Fmetal) the addition of metals [55–57]:
| Equation 8 |
While transition metal FRET measures the decrease in donor emission upon addition of the acceptor, a more widely used approach for donor-based FRET measurement is to measure the increase in donor emission upon photobleaching the acceptor. When the acceptor is destroyed optically, its quenching effect on the donor is eliminated, which results in an increase in the fluorescence intensity of the donor. In a typical experiment using this method, often called “donor de-quenching” [58] or “acceptor photobleaching FRET” [23], the fluorescence intensities of the donor are measured before (FDA) and after photobleaching of the acceptor (FD) in a limited area. The FRET efficiency is calculated as [59]:
| Equation 9 |
This is a relatively straightforward method and can be well adapted to any fluorescence microscope provided with a powerful light source to selectively bleach the acceptor. As donor de-quenching measures changes in donor fluorescence, it is less affected by either cross-talk (the excitation light used to bleach the acceptor usually bleach the donor much less efficiently) or bleed-through (donor emission can generally be well separated from the acceptor emission). It is also attractive because unlike the photobleaching process intrinsic to photoexcitation, an increase in fluorescence is generally hard to achieve unless FRET is present [60]. As expected, incomplete acceptor photobleaching, accidental donor bleaching, or mixtures of fluorophore populations can lead to underestimation of FRET [61].
Disadvantages of the donor dequenching method include: (i) The photobleaching process can cause photo-damage to the sample; (ii) For many live-cell studies where freely diffusible molecules are under investigation, this method is inappropriate because it is possible that the FRET measurement will be invalidated by recovery of the acceptor fluorophore [62]; (iii) There have been studies reporting the photoconversion artifacts (shift of the acceptor emission spectrum) in this method [63, 64]. A nice modification of this method demonstrated that the obscured artifacts in donor de-quenching analysis may be circumvented by measuring the donor intensity as a function of time during gradual acceptor photobleaching. In this way, both potential donor photobleaching and the presence of any background intensity can be detected without additional measurements [65].
3.2. An increase in the steady-state fluorescence emission intensity of the acceptor - donor sensitized acceptor emission
Instead of measuring donor fluorescence intensity change, one can quantify FRET by measuring the enhancement of acceptor emission, known as sensitized emission. In this method, the FRET efficiency is quantified as
| Equation 10 |
in which FA and F′A are the acceptor intensity in the absence and in the presence of the donor, respectively, and εD and εA are the molar extinction coefficient for the donor and the acceptor at the excitation wavelength range.
This is one of the simplest ways to measure FRET, especially when the donor and acceptor emissions can be cleanly separated. As discussed above, particular precautions must be taken to deal with potential problems like cross-talk and bleed-through. Several different methods have been designed to measure FRET in this way, such as “netFRET” [66, 67], “three-cube FRET” [68], and “Spectra FRET”[39]. These approaches share similar designs and handle potential contaminations with independent controls. A major difference in these approaches is the way of quantifying fluorescence intensities (point measurement versus range measurement). Here we will use the spectroscopy-based “Spectra FRET” as an example to explain in detail how to measure FRET in living cells and calculate EFRET from the enhanced acceptor emission.
The recording system for Spectra FRET could be built on an epifluorescence microscope equipped with a shutter-controlled excitation light source (e.g. mercury lamp, laser, or LED) to minimize photobleaching, as well as a spectrograph in conjunction with a cooled CCD camera for spectroscopic imaging. Two filter cubes (instead of three for three-cube FRET) are needed to excite the donor and the acceptor respectively. For example, when cerulean/EYFP are used as the FRET pair, the excitation filter D436/20 and dichroic mirror 455dclp can be used for donor excitation, and the excitation filter HQ500/20 and dichroic mirror Q515lp would be appropriate for acceptor excitation. Unlike three-cube FRET, no emission filter is required as the fluorescence emission will be collected and separated spectroscopically. The spectrograph normally has an input slit with adjustable width and a grating allowing easy selection of the recording wavelength range. When the input slit is wide open and the grating set at a small angle, the cell image can be projected to the camera (Fig. 4A and 4D); whereas when a narrow slit is in the light path and the grating is rotated to the desired angle to record at a selected wavelength range, one can get the spectroscopic images from the cellular region visible through the slit (Fig. 4B and 4E). A fluorescence emission spectrum can then be constructed from each spectroscopic image using the fluorescence intensity values along a horizontal line whose position corresponds to the part of the cell to be measured (Fig. 4C and 4F) [42]. Like three-cube FRET and other methods based on enhanced acceptor emission, three samples are required in order to obtain FRET estimates: (i) cells expressing only the donor; (ii) cells expressing only the acceptor; and (iii) cells co-expressing both the donor and the acceptor - the actual FRET experiment sample.
To quantify FRET, first, an emission spectrum from the sample group (iii) is acquired after excitation by donor excitation light (Ftotal, Fig. 4G red curve). This is the total emission spectrum that contains three components: the donor bleed-through, the acceptor cross-talk and the real FRET signal. Second, get the emission spectrum from the sample group (i) with donor excitation light, and normalize it to the peak fluorescence in the donor emission region of Ftotal (Fdonor, Fig. 4G blue curve). When Fdonor is subtracted from Ftotal, the contamination from donor bleed-through is eliminated (Ftotal − Fdonor, Fig. 4G green curve). Third, collect two emission spectra from the sample group (ii) using the donor and the acceptor excitation light respectively. The ratio between these two spectra, which is termed RatioA0, represents the percentage of cross-talk that is specific to this FRET pair and the recording system – hence the same as in Ftotal (Fig. 4H dotted red line). Multiplying RatioA0 to (Ftotal−Fdonor) yields the amount of cross-talk signal (Fcross-talk, Fig. 4G, dotted yellow curve). The real FRET signal can be calculated as FFRET = Ftotal − Fdonor − Fcross-talk (Fig. 4G the difference between the green curve and the yellow curve). Finally, record another emission spectrum from the sample group (iii) by acceptor excitation light (Facceptor, not shown). The ratio between (Ftotal − Fdonor) and Facceptor is defined as Ratio A (Fig. 4H, red line), and the apparent FRET efficiency Eapp can be calculated as:
| Equation 11 |
Having the potential contamination from bleed-through and cross-talk taken care of through the processes described above, there are still potential errors due to mixed fluorophore populations and variable expression levels, which cannot be corrected in individual cell measurement. In the Spectra FRET method, Eapp measured from each cell is plotted against the ratio between the peak intensity of Fdonor (Fc) and the peak intensity of Facceptor (Fy) (defined as Fc/Fy, reflecting the relative expression level of the donor and the acceptor) of the same cell (Fig. 4I). As expected, as Fc/Fy increases, so does the Eapp, which eventually plateaus off at high Fc/Fy values. While the shape of the distribution depends on the biological system that yields the measurements, the plot should reach a plateau at high Fc/Fy ratios.
One advantage of the Spectra FRET method is that fluorescence signals from different parts of the cell can be conveniently identified and selected. This is particularly useful in studies of plasma membrane proteins, for which collecting signals from the surface membrane is a key for reliable FRET measurement [38, 39, 46, 47, 51]. Moreover, this method is more tolerant to low signal-to-noise ratios thus more reliable, because it provides built-in controls for system linearity (both RatioA and RatioA0 are expected to be wavelength-independent) and accurate subtraction of potential light contaminants (not only from bleed-through and cross-talk, but also from autofluorescence and background light) [58].
3.3 Change in donor fluorescence lifetime – FLIM FRET
After a fluorophore is excited, the fluorescence emission decays exponentially. The fluorescence lifetime (τ), the time needed for the fluorescence intensity to decrease to 1/e (= 0.37) of its initial value, reflects the time that a molecule spends in the excited state. Fluorescence lifetime varies between 1 ns to 10 ns for most probes used in biological studies [69]. Kinetically, the lifetime equals the inverse of the sum of rate constants of all the mechanisms leaving the excited state, including the rate constant of emitting a photon (f), non-radiative decay (kNR), quenching (q), and FRET (kET):
| Equation 12 |
Since τ is shortened by kET, its change can be used to reflect the occurrence of FRET. Many potential artefacts intrinsic to intensity-based FRET quantification methods, such as signal cross-contamination, concentration dependence of the probes, variations in local excitation intensity and exposure duration, as well as moderate level of photobleaching, could be overcome by measuring the reduction in the donor fluorescence lifetime that results from the quenching effect of the acceptor using fluorescence lifetime imaging microscopy (FLIM). The lifetime of the donor fluorophore with (τDA) and without (τD) an acceptor can be directly measured from the fluorescence decay time course and the FRET efficiency can be calculated as
| Equation 13 |
It is noteworthy that although the fluorescence lifetime is largely insensitive to the aforementioned potential problems, it is very sensitive to the local chemical environment such as changes in the ion concentration, oxygen, pH, polarity, refractive index of the medium and temperature [23]. Therefore, it is pivotal to control, as much as possible, these external factors during FLIM-FRET measurements to minimize non-FRET donor fluorescence lifetime relaxation. In practice, besides the experimental group, it helps to include good controls such as a positive control with donor fluorophore fused to acceptor fluorophore, a negative control with free donor fluorophore alone to calibrate the system, negative controls with co-existence of free donor fluorophore and free acceptor fluorophore, and a negative control with co-transfection of donor fluorophore-labeled target protein and free acceptor fluorophore.
FLIM requires specialized and expensive instrumentation, but besides the analysis of protein-protein interactions with high temporal specificity and protein conformational changes [70, 71], FLIM-FRET was also used to map the spatial distribution of probe lifetimes in vivo [72–74]. In addition, it has been used in ion concentration imaging, in measuring oxygen concentration as well as in various medical applications [75–77]. More information about time-resolved fluorescence technologies will be described in detail by Dr. Alarcon in this volume (REF).
3.4 Change of polarization of the fluorophore - fluorescence anisotropy
In section 1.2 the orientation of fluorophore dipole movement and its time-dependent changes are discussed. A unique FRET quantification method can be devised based on this phenomenon. When a pool of randomly oriented fluorophores are excited with polarized light, only those that have their transition dipole moments nearly aligned to the plane of applied polarization will be excited (a process called photoselection). This is because excitation involves the redistribution of electrons about the fluorophore. As a result, only when the electromagnetic field of a photon is oriented to a particular axis about the fluorophore could its energy be captured. Release of the captured energy by fluorescence emission or by resonance coupling to another fluorophore is similarly polarized, that is, the fluorescence can be considered “anisotropic”. The intrinsic anisotropy of the fluorophore (denoted r0) can be measured by embedding the fluorophore in a frozen polyol. Hence photoselection of donors produces a pool of photoselected acceptors that would emit polarized fluorescence light. However, rotation of both donor and acceptor fluorophores during their lifetimes would reduce the polarization. The extent of polarization remaining in the fluorescence is called anisotropy (r, measured with respect to excitation polarization; the symbol “r” used here should not be confused with the same symbol used earlier for the distance between donor and acceptor), which depends on the ratio between the rotational lifetime constant (Φ) and the fluorescence lifetime (τ) of the fluorophore [78–80].
| Equation 14 |
The slower the rotation of the fluorescent species, the higher the value of r would be. Since the depolarization of the acceptor originated from the compounding rotations of the donor and the acceptor, acceptor emission is expected to be more depolarized compared to the donor emission.
Measurement of fluorescence anisotropies can be performed using a vertically polarized excitation source and collect vertically (parallel to excitation) and horizontally (perpendicular to excitation) polarized emission respectively. Polarization (P) and anisotropy (r) are simply two different measures of the same phenomenon, defined as:
| Equation 15 |
| Equation 16 |
where Fparallel and Fperpendicular are the intensities of the emitted light parallel and perpendicular to the excitation source, respectively [3, 81, 82].
Since the most commonly used fluorophores in FRET, fluorescence proteins (FPs), are large in size and their fluorescence lifetime is much shorter compared to its rotational lifetime constant (2.9 ns vs 20 ns), they produce highly polarized emission when excited by polarized light [83]. In a FRET experiment, such as one using mCerulean as donor and mVenus as acceptor, the collected emission would contain both highly polarized cyan fluorescence and less polarized yellow fluorescence. The magnitude of decreased anisotropy is correlated to the amount of FRET collected in the acceptor channel. Even a small amount of FRET could generate strongly depolarized fluorescence.
A unique application of the anisotropy-based FRET is the homoFRET method, in which only one type of fluorophore is used. Since for most fluorophores the Stoke’s shift is not very large, the excitation and emission spectra overlap appreciably. Therefore when two fluorophores of the same type are close to each other, FRET occurs just as when two different fluorophores are in close proximity. The depolarization process occurs in homoFRET as well. However, because there is only one type of fluorophore, there is no bleed-through or cross-talk to worry about. The stoichiometry/mixed population issue discussed above is also irrelevant [84, 85].
It has been reported that anisotropy-FRET can resolve FRET with 10-fold greater contrast than FLIM [86]. Moreover, with limited requirement for the microscopy setup, this method can be easily adapted to any imaging modality. Furthermore, accurate fluorescence anisotropy can be collected quickly thanks to the minimal image processing. So this approach is well suited for applications in high-throughput screening. Using more sophisticated equipment, picosecond time-resolved fluorescence anisotropy decay can be used to measure the molecular rotation time, dynamic oligomeric state and the relative orientation of two dimer subunits quantitatively [87].
On the other hand, this method also has its weaknesses. First, the very key facility for the measurement, the polarizer, would cause a significant reduction in the emission fluorescence intensity. Second, this approach is susceptible to polarization artifacts from elements in the optical path such as beam-splitters, filters, mirrors. Also, high numerical aperture objectives could reduce the polarization, so objectives with numerical apertures more than 1.0 should be avoided. Finally, this is an excellent method in qualitative study (FRET or no FRET), but does not have a high enough resolution for quantitative FRET measurement [23].
4. Biological applications of FRET
The advance in fluorescence microscopy techniques combined with the development of new FRET pairs has resulted in fast adoption of the FRET technique to a wide spectrum of biological studies, such as protein trafficking and co-localizations, protein-protein interactions, conformational rearrangements, nucleic acid studies, characterization of gene expression, real-time PCR assays and FRET-based bio-sensors [88–91]. Here we discuss several key applications of FRET using mostly examples from ion channel studies.
4.1 Intermolecular FRET to visualize protein-protein interactions
Protein-protein interactions govern a variety of biological processes. Numerous biochemical and molecular methods, e.g. co-immunoprecipitation, pull-down assays, yeast two-hybrid analysis, isothermal titration calorimetry (ITC), and surface plasmon resonance (SPR), have been developed to detect such interactions. Nevertheless, FRET has been an irreplaceable approach and gained its remarkable popularity due to its ability to detect interactions with very high temporal and spatial resolutions in a non-invasive way in living cells. It is also one of the most sensitive measurements because of its very small working distance (1–10 nm).
Good examples in this application are countless. In ion channel field, for example, FRET has witnessed the progress of knowledge in calcium store depletion-dependent dynamic interactions between store-operated calcium channel Orai1 and its calcium-sensing partner STIM1 [92–97]; how ATP dose-dependently inhibits membrane-bound SNARE protein syntaxin-1A (Syn-1A) binding to sulfonylurea receptors, which releases the inhibition of K(ATP) channel activity [98]; the close spatial proximity between IP3 receptor 1 and TRPC3 in arterial myocytes, leading to nonselective cation currents and vasoconstriction [99]; the interaction between TRPV4 and F-actin, which might play a role in sensing hypotonicity and the onset of regulatory volume decrease [100]; the association between actin-binding protein cortactin and potassium channel Kv1.2, thus providing a direct link between actin dynamics and membrane excitability [101]. Very recently, FRET also helped to set up the link between the tumor suppressor protein Numb and TRPV6, shedding light on another potential mechanism on the involvement of ion channels in tumorgenesis [102]. The relationship between two known players in Long QT syndrome-9, caveolin3 and Kir2.1 channel, was established with the help of FRET as well [103].
4.2 Intramolecular FRET to decode the stoichiometry of protein complexes
Most proteins function as a multi-subunit complex in vivo. Ion channels, for example, are dimers (CLC Cl− channels, K2P K+ channels), trimers (P2X, ASIC), tetramers (Kv channels, TRP channels, iGluR), pentamers (cys-loop ion channel receptors). They often exist as heteromeric complexes (such as native CNG channels, NMDA receptors, GABA receptors). FRET is a convenient way to detect subunit composition of protein complexes. This is because FRET only occurs within 100 Å. When fluorophore-labeled proteins are found to position so closely, they are most likely to interact with each other. In the case when the total number of subunits is known, it may also be used to determine the subunit stoichiometry. For example, CNG channels in the rod photoreceptor neurons have been known to exist as tetramers composed of CNGA1 and CNGB1 subunits. For the FRET approach, it was predicted that if the subunit stoichiometry between CNGA1 and CNGB1 is 2:2, then co-expression of CNGA1-CFP, CNGA1-YFP and CNGB1 should yield FRET [39]. Similarly, co-expression of CNGB1-CFP, CNGB1-YFP and CNGA1 should also yield FRET. However, if the subunit stoichiometry is 3:1 instead, only the first combination but not the second will yield FRET. Conversely, if the subunit stoichiometry is 1:3, only the second combination but not the first will yield FRET. Using this simple logic, it was determined that rod CNG channels are composed of three CNGA1 and one CNGB1 subunit [39].
The subunit stoichiometry of several other channels has been successfully determined using a similar method. For example, it has been confirmed that olfactory CNG channels are composed of two CNGA2, one CNGA4, and one CNGB1b [47]; NMDG receptors have two NR1 and two NR2 subunits in a dimer-of-dimers arrangement [104]. In multimeric channels with arbitrary subunit assembly, the total apparent FRET efficiency can be calculated from a binomial distribution, from which the real FRET efficiency can be derived. For example, this strategy has been used to investigate the relative arrangement of SUR1 and Kir 6.2 in KATP channels [105] and the assembly of heteromeric TRP channels [48].
While FRET reports the existence of multiple subunits in a protein complex, it is in general not an effective approach to determine the number of subunits. In this case, other fluorescence-based methods may be used, such as single-molecule photobleaching. In this method, target subunits labeled with FPs are expressed at low density in either mammalian cells or Xenopus oocytes, which are then imaged with total internal reflection fluorescence (TIRF) microscopy to selectively detect fluorescence signals from the plasma membrane and effectively reduce the background fluorescence to increase the signal-to-noise ratio. The idea is that fluorophore photobleaching is a discrete process, so the number of fluorescently labelled subunits in a complex can be deduced by counting fluorophore photobleaching steps as long as only one complex exist in a single pixal (hence the need of a low expression level). This method is pretty straightforward and easy to adapt to different systems, but one should be aware that many versions of FPs tend to dimerize [106] and transiently switch to a nonfluorescent state (“blinking”). Depending on the derivative of FP, the expression system and the temperature, the rate of maturation can also vary. Generally speaking, the maturation rate decreases with increased incubation temperature [106–111]. Oocyte system is therefore preferable in this regard as the eggs are normally incubated at 18°C. Whereas for expression in mammalian cells, an optimized version of GFP for expression at higher temperatures called “superfolder” GFP may be used [109]. Due to partial labelling and photobleaching during sample preparation, even a complex with a fixed number of subunits usually has a binomial distribution of the ensemble photobleaching steps [112]. The probability of observing k photobleaching steps P(k) could be calculated as:
| Equation 17 |
where N is the number of subunits, k is the number of steps observed and Pf is the fraction of mature FP.
When alternatively labeling different subunits in the protein complex, it is possible to deduce the subunit stoichiometry with single-molecule photobleaching. Using this method, Ulbrich and Isacoff showed that the subunit composition of the NMDA receptors composed of NR1 and NR3 subunits is 2:2 [113]; Demuro et al. found that both Orai1 and Orai3 channels undergo a dimer-to-tetramer transition to be activated as a store-operated Ca2+ channel and Orai3 forms a dimeric nonselective cation pore upon activation by 2-APB [114], which confirmed similar observations previously made by Cahalan group [115]; and Coste et al. recently demonstrated that the newly identified mechanically activated ion channel family Piezo proteins homo-multimerize with four subunits [116].
4.3 Probing protein conformational changes with FRET
Due to its remarkable distance sensitivity and the capacity to be used in living cells, FRET has been extensively applied in probing real-time protein conformational changes during protein function under physiological condition. Examples can be found in elegant applications of FRET to understand how the voltage-sensor of voltage-gated cation channels move during the channel activation process. Bezanilla and colleagues applied lanthanide-based resonance energy transfer (LRET), where lanthanide was used as a donor and organic fluorescent dye fluorescein was the acceptor, and recorded angstrom-scale relative “tilting” movements between S4 segments upon depolarization [117]. In the same year, Isacoff group used FM/TMRM pair (Fuorescein maleimide / tetramethylrhodamine maleimide) to stoichiometrically label (i.e. with different concentrations of donor and acceptor resulting in a biased binomial donor-acceptor distribution) the S3–S4 linker, and observed that S4 twists during the activation [118].
A similar approach was applied to other voltage-gated channels such as Na+ channel NaChBac and K+ channel KvAP [119, 120]. Besides this small “tilting” movement, S4 segments also make vertical movement relative to the plasma membrane, which was also detected by FRET studies. In this case, the FRET pair was formed by sulphorhodamine or ABD (7-fluorobenz-2-oxa-1,3-diazole-4-sulphonamide, donor) with DPA (dipicrylamine, acceptor) that can be dissolved in the plasma membrane [121]. The picture of S4 segments movements during voltage-sensing set up by these FRET studies was supported by later studies using other methods such as crystallization and molecular dynamics simulation [122–125].
Gating of ion channels always involves complex conformational changes, and FRET has been a very useful tool in monitoring these changes thus providing mechanistic insights into gating mechanisms. Thoughtfully designed FRET measurements have been applied in studies on voltage-gated Kv2.1 channels [126], voltage-gated Cav1.2 channels [127], CLC chloride channels [46, 51], ryanodine receptors [128], L-type Ca2+ channels [129], P2X channels [130], gramicidin channels [131, 132], Large-conductance voltage- and calcium-dependent potassium channels (BK channels) [133], and store-operated calcium channels [134].
Study of another branch of protein conformational changes, protein folding, has also benefited greatly from FRET technique, particularly single-molecule FRET. This specific application of FRET is beyond the interest of this review, but details could be found in many interesting reviews [135–139].
4.4 FRET based biosensors
Biosensors based on the FRET principle have been rapidly developed in the past decade and have become powerful tools in monitoring and identifying cellular molecular dynamics, cellular physiology and cell-cell interactions. A FRET biosensor generally comprises an acceptor fluorophore, a donor fluorophore, a ligand domain, a sensor domain, and linkers in between. The sensor domain responds to a specific signal, such as phosphorylation, GTP loading, or phospholipid binding, by changing its conformation. The ligand domain would bind to the conformation-changed sensor domain, thereby changing the distance (perhaps also orientation) between the acceptor and donor fluorophores, which generates FRET changes.
There are generally two types of FRET biosensors: the intermolecular (bimolecular) and intramolecular (unimolecular) FRET biosensors. The former consists of two separate molecules, one comprised of a donor fluorophore and the sensor (or ligand) domain and the other comprised of an acceptor fluorophore and the ligand (or sensor) domain. This format is particularly useful for detecting protein-protein interaction within the cells, but the potential contaminations from cross-talk and bleed-through need to be well controlled. In intramolecular FRET biosensors, all the components are combined into a single molecule, therefore the order of these components needs be optimized [140]. Fundamentally three approaches are commonly applied to the design of FRET-based biosensors:
Interaction between the sensor domain and the ligand domain upon a cellular stimulation results in an increased FRET signal due to shortening of the distance between donor and acceptor. In this design, the distance-dependence of FRET development should ideally not be disturbed by change of orientations between the two fluorophores. Note that GFP-based fluorophores tend to dimerize which causes high FRET efficiency to start with, so the binding between the sensor and the ligand might cause a relative rotation of the FRET pair and reduce the FRET efficiency instead of increasing it. One reported approach to overcome this problem is to introduce a very long linker between the ligand domain and the sensor domain to eliminate the FRET pair dimerization, thus rendering the FRET efficiency mostly dependent on distance [141].
When the FRET pair are simultaneously attached to one molecule at close positions and generate positive FRET continually, proteolysis of this intramolecularly labeled biomolecule triggered by a cellular process would lead to separation of the donor and the acceptor and loss of FRET signal [142].
Alternatively, an intramolecularly labeled biomolecule, which has two fluorophores labeled at a distance beyond FRET sensitivity, could undergo a conformational change upon stimulation, resulting in the initiation of FRET [143].
An example of a very widely used FRET biosensor is the intracellular Ca2+ indicator “cameleon”, which is composed of a calmodulin as the sensor domain, a M13 (containing a calmodulin-binding sequence) as the ligand domain, and CFP/YFP as the FRET fluorophore pair. With an increase in the Ca2+ concentration, calmodulin is prompted to bind to M13, bringing CFP and YFP closer to yield an increased FRET signal. The Ca2+ sensitivity range of cameleons can be tuned by mutations in the Ca2+-binding sites of calmodulin [144].
FRET biosensors are great tools for kinase inhibitors screening in live cells, or as biomarkers in early cancer detection, cancer prognosis, and therapeutic efficacy monitor. In one successful application of FRET biosensor, Mizutani et al. engineered an intramolecular FRET biosensor with CrkL, a major substrate of the BCR-ABL kinase (a kinase that plays a critical role in the pathogenesis of chronic myeloid leukemia, CML), sandwiched between Venus and ECFP, so that when the BCR-ABL kinase is active, the phosphorylated tyrosine site on CrkL can bind to its intramolecular SH2 domain to cause a conformational change and subsequently increase FRET efficiency. The final version of this FRET biosensor displayed an impressive 80% increase of FRET ratio in vitro upon stimulation by BCR-ABL. This FRET biosensor showed remarkable sensitivity not only in screening the efficacy of CML specific drugs, but also in detecting rare drug-resistant cancer cells from a large cell population. This is particularly interesting, because these drug-resistant cells likely constitute the main reason for CML therapeutic failure and relapse; this biosensor, therefore, can be used as a powerful tool to assess the biopsy samples from patients and to predict the probability of drug resistance and prognosis [145]. Success has also been gained in FRET biosensors developed to detect other oncogene-related kinase activities including Src, FAK, PKA, EGFR, and ABL [146, 147] and of other molecules important for migration and cancer invasion [148].
There is still plenty of room to improve FRET biosensors. For example, there are needs for more efficient methods to deliver biosensors to live cells, particularly primary cells, since currently available methods, including liposome-based delivery, electroporation, virus infection and “cell-loading” techniques, only have ~20–30% efficiency [149]. FRET biosensors with the capacity to simultaneously visualize multiple molecular events in the same cell would also be very valuable. Indeed, the future of FRET biosensors is quite promising, particularly in clinical research with applications to visualize subcellular molecular signaling events in real time and to identify novel targeting molecules and pathways.
4.5 Single-molecule FRET for nucleic acid and protein dynamics study
FRET measurement from a population of molecules has two major drawbacks: first, it generally reports an ensemble average of the behavior of individual molecules due to a lack of synchronization; second, it is often impractical to detect transitional conformations. These drawbacks can be remedied by single-molecule FRET (smFRET), which detects structural changes in individual biological molecules in real time and discriminate the distribution of several conformations, including those short-lived and low-populated ones [150–152].
smFRET requires a high signal to noise ratio, so background fluorescence needs be minimized and the signal maximized. To achieve this, on the one hand, objectives with high numerical aperture and detectors with high quantum efficiency and low noise are required. Though some studies used confocal microscopy [153, 154], smFRET time trajectories are most commonly acquired by imaging surface immobilized molecules with TIRF microscopy combined with a highly sensitive electron-multiplying charge-coupled device (EM-CCD) camera [155, 156]. On the other hand, it is pivotal to choose the right fluorophores. Ideally, the fluorophore for single molecule studies should be bright (extinction coefficient, ε > 50,000 M−1 cm−1; quantum yield, QY > 0.1), small, photostable, and water-soluble with different forms of bio-conjugation chemistries available [157]. Additionally, to form a powerful FRET pair for smFRET, besides the general requirements for FRET pair as mentioned earlier, comparable emission quantum yield and detection efficiencies for donor and acceptor are preferred, because this facilitates anti-correlated intensity changes in donor and acceptor. Among various dyes being used in smFRET studies, Cy3 and Cy5 have been the most popular donor and acceptor pair due to their superior photophysical properties, such as the large spectral separation (~100 nm), good photostability, comparable quantum yields (~0.25) and commercial availability of various reactive forms (amino-, thiol-, etc) [112, 158].
The first key step in a smFRET experiment is to label the target molecule with fluorescent dyes. Nucleic acids are best labeled during synthesis or by introducing amine-modified bases to a specific site and labeling with the NHS-ester form of a dye that react with the amine group, which can then be separated from unlabeled molecules by polyacrylamide gel electrophoresis. For proteins, a popular approach is to use the maleimide form of a dye which would specifically react with a thiol group (-SH) on cysteines [158]. No matter whether smFRET will be conducted in a sealed sample chamber or in a flow cell, the next critical step is to clean up the binding surface and immobilize the fluorophore-labeled samples to the surface for imaging. Detailed protocols can be found in [159].
Similar as in single-channel recordings, single-molecule fluorescence data also follow the stochastic fluctuations as single proteins switch between different conformations. Accordingly, similar quantification/analysis methods can be used, such as amplitude and dwell-time histograms analysis and Hidden Markov modellings [155, 158, 160–165].
As a strong tool for detecting the conformational diversity and dynamics of nucleic acids/protein molecules and their interactions, smFRET has been widely applied to various biological systems [155, 166–170]. It is a useful complementary strategy to other atomic-level structural techniques, such as X-ray crystallography, NMR or cryo-electron microscopy, which cannot provide dynamic structural information in real time. Moreover, interesting developments of traditional smFRET are rapidly springing up, such as detailed structural determination of DNA and RNA molecules [171] and smFRET with multi-fluorophores [172–176].
8. Conclusion
Being a highly sensitive reporter for intermolecular/intramolecular distances in living cells, FRET has gained tremendous popularity in life sciences, drug development, and clinical medicine. It is promising that some of the current problems associated with FRET measurement may be solved in the near future; meanwhile, we must keep in mind that FRET imaging, like any experimental measurement, requires both a sound understanding of the theory and careful attentions to practical details, such as the target system used for imaging, choice of fluorophore pair and microscopy instruments, quantification method to be used, controls for appropriate corrections, etc. Progress in this field has never stopped. Research into new fluorescent molecules continues to generate improved donor-acceptor pairs. Enhancements in the optical instrumentation, such as new light sources, faster shutters, more sensitive CCD cameras, will undoubtedly continue to enable novel FRET applications. Developments in quantitative high-throughput FRET screening arrays, and improvements in algorithms to better manage variables in FRET experiment will make this technique a more and more useful “molecular ruler”.
Highlights.
FRET is a highly distance-sensitive measurement that is compatible with protein studies
Choosing the right fluorophores is a key for the designing of successful FRET experiment
It is important to be aware of potential pitfalls and practical considerations in FRET experiments
There are a wide variety of FRET quantification techniques that are suitable for specific experimental designs.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Förster T. Naturwissenschaften. 1946;6:166–175. [Google Scholar]
- 2.Förster T. Annalen der Physik. 1948;2:55–75. [Google Scholar]
- 3.Lakowicz JR. Principles of fluorescence spectroscopy. 3. Springer; Berlin, Heidelberg, New York: 2006. [Google Scholar]
- 4.Stryer L, Haugland RP. Proc Natl Acad Sci U S A. 1967;58:719–726. doi: 10.1073/pnas.58.2.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Steinberg IZ. Annu Rev Biochem. 1971;40:83–114. doi: 10.1146/annurev.bi.40.070171.000503. [DOI] [PubMed] [Google Scholar]
- 6.Loura LM. Int J Mol Sci. 2012;13:15252–15270. doi: 10.3390/ijms131115252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Medintz I, Hildebrandt N. FRET - Förster Resonance Energy Transfer: From Theory to Applications. Wiley-VCH; Weinheim, Germany: 2013. [Google Scholar]
- 8.Clegg RM. Methods Enzymol. 1992;211:353–388. doi: 10.1016/0076-6879(92)11020-j. [DOI] [PubMed] [Google Scholar]
- 9.Selvin PR. Methods Enzymol. 1995;246:300–334. doi: 10.1016/0076-6879(95)46015-2. [DOI] [PubMed] [Google Scholar]
- 10.Pollok BA, Heim R. Trends Cell Biol. 1999;9:57–60. doi: 10.1016/s0962-8924(98)01434-2. [DOI] [PubMed] [Google Scholar]
- 11.Chalfie M, Tu Y, Euskirchen G, Ward WW, Prasher DC. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 12.Ormo M, Cubitt AB, Kallio K, Gross LA, Tsien RY, Remington SJ. Science. 1996;273:1392–1395. doi: 10.1126/science.273.5280.1392. [DOI] [PubMed] [Google Scholar]
- 13.Heim R, Tsien RY. Curr Biol. 1996;6:178–182. doi: 10.1016/s0960-9822(02)00450-5. [DOI] [PubMed] [Google Scholar]
- 14.Tsien RY. Annu Rev Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 15.Matz MV, Fradkov AF, Labas YA, Savitsky AP, Zaraisky AG, Markelov ML, Lukyanov SA. Nat Biotechnol. 1999;17:969–973. doi: 10.1038/13657. [DOI] [PubMed] [Google Scholar]
- 16.Patterson G, Day RN, Piston D. J Cell Sci. 2001;114:837–838. doi: 10.1242/jcs.114.5.837. [DOI] [PubMed] [Google Scholar]
- 17.Shaner NC, Lambert GG, Chammas A, Ni Y, Cranfill PJ, Baird MA, Sell BR, Allen JR, Day RN, Israelsson M, Davidson MW, Wang J. Nature methods. 2013;10:407–409. doi: 10.1038/nmeth.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumagai A, Ando R, Miyatake H, Greimel P, Kobayashi T, Hirabayashi Y, Shimogori T, Miyawaki A. Cell. 2013;153:1602–1611. doi: 10.1016/j.cell.2013.05.038. [DOI] [PubMed] [Google Scholar]
- 19.Smith AM, Duan H, Mohs AM, Nie S. Advanced drug delivery reviews. 2008;60:1226–1240. doi: 10.1016/j.addr.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alivisatos AP, Gu W, Larabell C. Annual review of biomedical engineering. 2005;7:55–76. doi: 10.1146/annurev.bioeng.7.060804.100432. [DOI] [PubMed] [Google Scholar]
- 21.Drummen GP. Int J Mol Sci. 2010;11:154–163. doi: 10.3390/ijms11010154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rizvi SB, Ghaderi S, Keshtgar M, Seifalian AM. Nano reviews. 2010;1 doi: 10.3402/nano.v1i0.5161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ishikawa-Ankerhold HC, Ankerhold R, Drummen GP. Molecules. 2012;17:4047–4132. doi: 10.3390/molecules17044047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Medintz IL, Mattoussi H. Phys Chem Chem Phys. 2009;11:17–45. doi: 10.1039/b813919a. [DOI] [PubMed] [Google Scholar]
- 25.Dahan M, Laurence T, Pinaud F, Chemla DS, Alivisatos AP, Sauer M, Weiss S. Optics letters. 2001;26:825–827. doi: 10.1364/ol.26.000825. [DOI] [PubMed] [Google Scholar]
- 26.Mitchell AC, Dad S, Morgan CG. J Microsc. 2008;230:172–176. doi: 10.1111/j.1365-2818.2008.01973.x. [DOI] [PubMed] [Google Scholar]
- 27.Kirchner C, Liedl T, Kudera S, Pellegrino T, Munoz Javier A, Gaub HE, Stolzle S, Fertig N, Parak WJ. Nano letters. 2005;5:331–338. doi: 10.1021/nl047996m. [DOI] [PubMed] [Google Scholar]
- 28.Shiohara A, Hoshino A, Hanaki K, Suzuki K, Yamamoto K. Microbiology and immunology. 2004;48:669–675. doi: 10.1111/j.1348-0421.2004.tb03478.x. [DOI] [PubMed] [Google Scholar]
- 29.Yao J, Larson DR, Vishwasrao HD, Zipfel WR, Webb WW. Proc Natl Acad Sci U S A. 2005;102:14284–14289. doi: 10.1073/pnas.0506523102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clapp AR, Medintz IL, Mauro JM, Fisher BR, Bawendi MG, Mattoussi H. Journal of the American Chemical Society. 2004;126:301–310. doi: 10.1021/ja037088b. [DOI] [PubMed] [Google Scholar]
- 31.Clapp AR, Medintz IL, Mattoussi H. Chemphyschem. 2006;7:47–57. doi: 10.1002/cphc.200500217. [DOI] [PubMed] [Google Scholar]
- 32.Barroso MM. The journal of histochemistry and cytochemistry : official journal of the Histochemistry Society. 2011;59:237–251. doi: 10.1369/0022155411398487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alam R, Zylstra J, Fontaine DM, Branchini BR, Maye MM. Nanoscale. 2013;5:5303–5306. doi: 10.1039/c3nr01842c. [DOI] [PubMed] [Google Scholar]
- 34.Algar WR, Ancona MG, Malanoski AP, Susumu K, Medintz IL. ACS nano. 2012;6:11044–11058. doi: 10.1021/nn304736j. [DOI] [PubMed] [Google Scholar]
- 35.Dennis AM, Rhee WJ, Sotto D, Dublin SN, Bao G. ACS nano. 2012;6:2917–2924. doi: 10.1021/nn2038077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sapsford KE, Granek J, Deschamps JR, Boeneman K, Blanco-Canosa JB, Dawson PE, Susumu K, Stewart MH, Medintz IL. ACS nano. 2011;5:2687–2699. doi: 10.1021/nn102997b. [DOI] [PubMed] [Google Scholar]
- 37.Hink MA, Griep RA, Borst JW, van Hoek A, Eppink MH, Schots A, Visser AJ. J Biol Chem. 2000;275:17556–17560. doi: 10.1074/jbc.M001348200. [DOI] [PubMed] [Google Scholar]
- 38.Zheng J, Varnum MD, Zagotta WN. J Neurosci. 2003;23:8167–8175. doi: 10.1523/JNEUROSCI.23-22-08167.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zheng J, Trudeau MC, Zagotta WN. Neuron. 2002;36:891–896. doi: 10.1016/s0896-6273(02)01099-1. [DOI] [PubMed] [Google Scholar]
- 40.Thaler C, Koushik SV, Blank PS, Vogel SS. Biophys J. 2005;89:2736–2749. doi: 10.1529/biophysj.105.061853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zimmermann T, Rietdorf J, Girod A, Georget V, Pepperkok R. FEBS Lett. 2002;531:245–249. doi: 10.1016/s0014-5793(02)03508-1. [DOI] [PubMed] [Google Scholar]
- 42.Zheng J. Methods Mol Biol. 2006;337:65–77. doi: 10.1385/1-59745-095-2:65. [DOI] [PubMed] [Google Scholar]
- 43.van Rheenen J, Langeslag M, Jalink K. Biophys J. 2004;86:2517–2529. doi: 10.1016/S0006-3495(04)74307-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bacart J, Corbel C, Jockers R, Bach S, Couturier C. Biotechnol J. 2008;3:311–324. doi: 10.1002/biot.200700222. [DOI] [PubMed] [Google Scholar]
- 45.Gandia J, Lluis C, Ferre S, Franco R, Ciruela F. Bioessays. 2008;30:82–89. doi: 10.1002/bies.20682. [DOI] [PubMed] [Google Scholar]
- 46.Bykova EA, Zhang XD, Chen TY, Zheng J. Nat Struct Mol Biol. 2006;13:1115–1119. doi: 10.1038/nsmb1176. [DOI] [PubMed] [Google Scholar]
- 47.Zheng J, Zagotta WN. Neuron. 2004;42:411–421. doi: 10.1016/s0896-6273(04)00253-3. [DOI] [PubMed] [Google Scholar]
- 48.Cheng W, Yang F, Takanishi CL, Zheng J. J Gen Physiol. 2007;129:191–207. doi: 10.1085/jgp.200709731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Koushik SV, Blank PS, Vogel SS. PloS one. 2009;4:e8031. doi: 10.1371/journal.pone.0008031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zheng J. J Gen Physiol. 2013;142:347–350. doi: 10.1085/jgp.201311090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ma L, Rychkov GY, Bykova EA, Zheng J, Bretag AH. Biochem J. 2011;436:415–428. doi: 10.1042/BJ20102153. [DOI] [PubMed] [Google Scholar]
- 52.Cardullo RA, Agrawal S, Flores C, Zamecnik PC, Wolf DE. Proc Natl Acad Sci U S A. 1988;85:8790–8794. doi: 10.1073/pnas.85.23.8790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cardullo RA. Methods Cell Biol. 2007;81:479–494. doi: 10.1016/S0091-679X(06)81023-X. [DOI] [PubMed] [Google Scholar]
- 54.Kenworthy AK. Methods. 2001;24:289–296. doi: 10.1006/meth.2001.1189. [DOI] [PubMed] [Google Scholar]
- 55.Taraska JW, Puljung MC, Olivier NB, Flynn GE, Zagotta WN. Nature methods. 2009;6:532–537. doi: 10.1038/nmeth.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Puljung MC, Zagotta WN. J Biol Chem. 2013;288:12944–12956. doi: 10.1074/jbc.M113.464123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Taraska JW, Puljung MC, Zagotta WN. Proc Natl Acad Sci U S A. 2009;106:16227–16232. doi: 10.1073/pnas.0905207106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Takanishi CL, Bykova EA, Cheng W, Zheng J. Brain Res. 2006;1091:132–139. doi: 10.1016/j.brainres.2006.01.119. [DOI] [PubMed] [Google Scholar]
- 59.Wouters FS, Bastiaens PI, Wirtz KW, Jovin TM. The EMBO journal. 1998;17:7179–7189. doi: 10.1093/emboj/17.24.7179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Miyawaki A, Tsien RY. Methods Enzymol. 2000;327:472–500. doi: 10.1016/s0076-6879(00)27297-2. [DOI] [PubMed] [Google Scholar]
- 61.Berney C, Danuser G. Biophys J. 2003;84:3992–4010. doi: 10.1016/S0006-3495(03)75126-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bacskai BJ, Skoch J, Hickey GA, Allen R, Hyman BT. J Biomed Opt. 2003;8:368–375. doi: 10.1117/1.1584442. [DOI] [PubMed] [Google Scholar]
- 63.Kremers GJ, Hazelwood KL, Murphy CS, Davidson MW, Piston DW. Nature methods. 2009;6:355–358. doi: 10.1038/nmeth.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Valentin G, Verheggen C, Piolot T, Neel H, Coppey-Moisan M, Bertrand E. Nature methods. 2005;2:801. doi: 10.1038/nmeth1105-801. [DOI] [PubMed] [Google Scholar]
- 65.Van Munster EB, Kremers GJ, Adjobo-Hermans MJ, Gadella TW., Jr J Microsc. 2005;218:253–262. doi: 10.1111/j.1365-2818.2005.01483.x. [DOI] [PubMed] [Google Scholar]
- 66.Gordon GW, Berry G, Liang XH, Levine B, Herman B. Biophys J. 1998;74:2702–2713. doi: 10.1016/S0006-3495(98)77976-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xia Z, Liu Y. Biophys J. 2001;81:2395–2402. doi: 10.1016/S0006-3495(01)75886-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Erickson MG, Alseikhan BA, Peterson BZ, Yue DT. Neuron. 2001;31:973–985. doi: 10.1016/s0896-6273(01)00438-x. [DOI] [PubMed] [Google Scholar]
- 69.Sun Y, Hays NM, Periasamy A, Davidson MW, Day RN. Methods Enzymol. 2012;504:371–391. doi: 10.1016/B978-0-12-391857-4.00019-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Parsons M, Messent AJ, Humphries JD, Deakin NO, Humphries MJ. J Cell Sci. 2008;121:265–271. doi: 10.1242/jcs.018440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kinoshita K, Goryo K, Takada M, Tomokuni Y, Aso T, Okuda H, Shuin T, Fukumura H, Sogawa K. The FEBS journal. 2007;274:5567–5575. doi: 10.1111/j.1742-4658.2007.06075.x. [DOI] [PubMed] [Google Scholar]
- 72.Sun Y, Wallrabe H, Booker CF, Day RN, Periasamy A. Biophys J. 2010;99:1274–1283. doi: 10.1016/j.bpj.2010.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yasuda R. Curr Opin Neurobiol. 2006;16:551–561. doi: 10.1016/j.conb.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 74.Wouters FS, Bastiaens PI. Curr Biol. 1999;9:1127–1130. doi: 10.1016/s0960-9822(99)80484-9. [DOI] [PubMed] [Google Scholar]
- 75.Bastiaens PI, Squire A. Trends Cell Biol. 1999;9:48–52. doi: 10.1016/s0962-8924(98)01410-x. [DOI] [PubMed] [Google Scholar]
- 76.van Munster EB, Gadella TW., Jr Cytometry Part A : the journal of the International Society for Analytical Cytology. 2004;58:185–194. doi: 10.1002/cyto.a.20013. [DOI] [PubMed] [Google Scholar]
- 77.Jose M, Nair DK, Reissner C, Hartig R, Zuschratter W. Biophys J. 2007;92:2237–2254. doi: 10.1529/biophysj.106.092841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Perrin F. Conférences D’Actualités Scientifiques et Industrielles. 1931;22:2–41. [Google Scholar]
- 79.Prerrin F. Photochimie Ann Phys, Ser. 1929;10:169–275. [Google Scholar]
- 80.Perrin F. J Phys Radium V, Ser. 1926;6:390–401. [Google Scholar]
- 81.Jabłoński A. Acta Physica Polonica. 1957;16:471–479. [Google Scholar]
- 82.Jabłoński A. Bull Acad Polon Sci, Ser sci math astr phys. 1960;8:259–264. [Google Scholar]
- 83.Swaminathan R, Hoang CP, Verkman AS. Biophys J. 1997;72:1900–1907. doi: 10.1016/S0006-3495(97)78835-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ghosh S, Saha S, Goswami D, Bilgrami S, Mayor S. Methods Enzymol. 2012;505:291–327. doi: 10.1016/B978-0-12-388448-0.00024-3. [DOI] [PubMed] [Google Scholar]
- 85.Bader AN, Hoetzl S, Hofman EG, Voortman J, van Bergen en Henegouwen PM, van Meer G, Gerritsen HC. Chemphyschem. 2011;12:475–483. doi: 10.1002/cphc.201000801. [DOI] [PubMed] [Google Scholar]
- 86.Rizzo MA, Piston DW. Biophys J. 2005;88:L14–16. doi: 10.1529/biophysj.104.055442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gautier I, Tramier M, Durieux C, Coppey J, Pansu RB, Nicolas JC, Kemnitz K, Coppey-Moisan M. Biophys J. 2001;80:3000–3008. doi: 10.1016/S0006-3495(01)76265-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Miyawaki A. Dev Cell. 2003;4:295–305. doi: 10.1016/s1534-5807(03)00060-1. [DOI] [PubMed] [Google Scholar]
- 89.Greeson JN, Raphael RM. J Biomed Opt. 2007;12:021002. doi: 10.1117/1.2717499. [DOI] [PubMed] [Google Scholar]
- 90.Organ LE, Raphael RM. J Biomed Opt. 2007;12:021003. doi: 10.1117/1.2715202. [DOI] [PubMed] [Google Scholar]
- 91.Spector AA, Deo N, Grosh K, Ratnanather JT, Raphael RM. J Membr Biol. 2006;209:135–152. doi: 10.1007/s00232-005-0843-7. [DOI] [PubMed] [Google Scholar]
- 92.Barr VA, Bernot KM, Srikanth S, Gwack Y, Balagopalan L, Regan CK, Helman DJ, Sommers CL, Oh-Hora M, Rao A, Samelson LE. Molecular biology of the cell. 2008;19:2802–2817. doi: 10.1091/mbc.E08-02-0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Navarro-Borelly L, Somasundaram A, Yamashita M, Ren D, Miller RJ, Prakriya M. The Journal of physiology. 2008;586:5383–5401. doi: 10.1113/jphysiol.2008.162503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Calloway N, Vig M, Kinet JP, Holowka D, Baird B. Molecular biology of the cell. 2009;20:389–399. doi: 10.1091/mbc.E07-11-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sampieri A, Zepeda A, Asanov A, Vaca L. Cell calcium. 2009;45:439–446. doi: 10.1016/j.ceca.2009.02.010. [DOI] [PubMed] [Google Scholar]
- 96.Wang Y, Deng X, Zhou Y, Hendron E, Mancarella S, Ritchie MF, Tang XD, Baba Y, Kurosaki T, Mori Y, Soboloff J, Gill DL. Proc Natl Acad Sci U S A. 2009;106:7391–7396. doi: 10.1073/pnas.0900293106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Huang PC, Chiu TY, Wang LC, Teng HC, Kao FJ, Yang DM. Microscopy and microanalysis : the official journal of Microscopy Society of America, Microbeam Analysis Society. Microscopical Society of Canada. 2010;16:313–326. doi: 10.1017/S1431927610000188. [DOI] [PubMed] [Google Scholar]
- 98.Kang Y, Zhang Y, Liang T, Leung YM, Ng B, Xie H, Chang N, Chan J, Shyng SL, Tsushima RG, Gaisano HY. J Biol Chem. 2011;286:5876–5883. doi: 10.1074/jbc.M109.089607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Adebiyi A, Zhao G, Narayanan D, Thomas-Gatewood CM, Bannister JP, Jaggar JH. Circulation research. 2010;106:1603–1612. doi: 10.1161/CIRCRESAHA.110.216804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Becker D, Bereiter-Hahn J, Jendrach M. European journal of cell biology. 2009;88:141–152. doi: 10.1016/j.ejcb.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 101.Williams MR, Markey JC, Doczi MA, Morielli AD. Proc Natl Acad Sci U S A. 2007;104:17412–17417. doi: 10.1073/pnas.0703865104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kim SY, Yang D, Myeong J, Ha K, Kim SH, Park EJ, Kim IG, Cho NH, Lee KP, Jeon JH, So I. Cell calcium. 2013;53:102–111. doi: 10.1016/j.ceca.2012.10.005. [DOI] [PubMed] [Google Scholar]
- 103.Vaidyanathan R, Vega AL, Song C, Zhou Q, Tan BH, Berger S, Makielski JC, Eckhardt LL. J Biol Chem. 2013 doi: 10.1074/jbc.M112.435370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Qiu S, Hua YL, Yang F, Chen YZ, Luo JH. J Biol Chem. 2005;280:24923–24930. doi: 10.1074/jbc.M413915200. [DOI] [PubMed] [Google Scholar]
- 105.Wang S, Makhina EN, Masia R, Hyrc KL, Formanack ML, Nichols CG. J Biol Chem. 2013;288:4378–4388. doi: 10.1074/jbc.M112.388629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.McGuire H, Aurousseau MR, Bowie D, Blunck R. J Biol Chem. 2012;287:35912–35921. doi: 10.1074/jbc.M112.402057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Garcia-Parajo MF, Koopman M, van Dijk EM, Subramaniam V, van Hulst NF. Proc Natl Acad Sci U S A. 2001;98:14392–14397. doi: 10.1073/pnas.251525598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ulbrich MH, Isacoff EY. Nat Methods. 2007;4:319–321. doi: 10.1038/NMETH1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Pedelacq JD, Cabantous S, Tran T, Terwilliger TC, Waldo GS. Nat Biotechnol. 2006;24:79–88. doi: 10.1038/nbt1172. [DOI] [PubMed] [Google Scholar]
- 110.Zacharias DA, Violin JD, Newton AC, Tsien RY. Science. 2002;296:913–916. doi: 10.1126/science.1068539. [DOI] [PubMed] [Google Scholar]
- 111.Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY. Nat Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- 112.Ha T, Tinnefeld P. Annual review of physical chemistry. 2012;63:595–617. doi: 10.1146/annurev-physchem-032210-103340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ulbrich MH, Isacoff EY. Nature methods. 2007;4:319–321. doi: 10.1038/NMETH1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Demuro A, Penna A, Safrina O, Yeromin AV, Amcheslavsky A, Cahalan MD, Parker I. Proc Natl Acad Sci U S A. 2011;108:17832–17837. doi: 10.1073/pnas.1114814108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Penna A, Demuro A, Yeromin AV, Zhang SL, Safrina O, Parker I, Cahalan MD. Nature. 2008;456:116–120. doi: 10.1038/nature07338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Coste B, Xiao B, Santos JS, Syeda R, Grandl J, Spencer KS, Kim SE, Schmidt M, Mathur J, Dubin AE, Montal M, Patapoutian A. Nature. 2012;483:176–181. doi: 10.1038/nature10812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Cha A, Snyder GE, Selvin PR, Bezanilla F. Nature. 1999;402:809–813. doi: 10.1038/45552. [DOI] [PubMed] [Google Scholar]
- 118.Glauner KS, Mannuzzu LM, Gandhi CS, Isacoff EY. Nature. 1999;402:813–817. doi: 10.1038/45561. [DOI] [PubMed] [Google Scholar]
- 119.Richardson J, Blunck R, Ge P, Selvin PR, Bezanilla F, Papazian DM, Correa AM. Proc Natl Acad Sci U S A. 2006;103:15865–15870. doi: 10.1073/pnas.0607532103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sandtner W, Bezanilla F, Correa AM. Biophys J. 2007;93:L45–47. doi: 10.1529/biophysj.107.119073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Chanda B, Asamoah OK, Blunck R, Roux B, Bezanilla F. Nature. 2005;436:852–856. doi: 10.1038/nature03888. [DOI] [PubMed] [Google Scholar]
- 122.Long SB, Tao X, Campbell EB, MacKinnon R. Nature. 2007;450:376–382. doi: 10.1038/nature06265. [DOI] [PubMed] [Google Scholar]
- 123.Long SB, Campbell EB, Mackinnon R. Science. 2005;309:897–903. doi: 10.1126/science.1116269. [DOI] [PubMed] [Google Scholar]
- 124.Jensen MO, Jogini V, Borhani DW, Leffler AE, Dror RO, Shaw DE. Science. 2012;336:229–233. doi: 10.1126/science.1216533. [DOI] [PubMed] [Google Scholar]
- 125.Yarov-Yarovoy V, DeCaen PG, Westenbroek RE, Pan CY, Scheuer T, Baker D, Catterall WA. Proc Natl Acad Sci U S A. 2012;109:E93–102. doi: 10.1073/pnas.1118434109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kobrinsky E, Stevens L, Kazmi Y, Wray D, Soldatov NM. J Biol Chem. 2006;281:19233–19240. doi: 10.1074/jbc.M601231200. [DOI] [PubMed] [Google Scholar]
- 127.Kobrinsky E, Schwartz E, Abernethy DR, Soldatov NM. J Biol Chem. 2003;278:5021–5028. doi: 10.1074/jbc.M211254200. [DOI] [PubMed] [Google Scholar]
- 128.George CH, Jundi H, Walters N, Thomas NL, West RR, Lai FA. Circulation research. 2006;98:88–97. doi: 10.1161/01.RES.0000199296.70534.7c. [DOI] [PubMed] [Google Scholar]
- 129.Kobrinsky E, Kepplinger KJ, Yu A, Harry JB, Kahr H, Romanin C, Abernethy DR, Soldatov NM. Biophys J. 2004;87:844–857. doi: 10.1529/biophysj.104.041152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Fisher JA, Girdler G, Khakh BS. J Neurosci. 2004;24:10475–10487. doi: 10.1523/JNEUROSCI.3250-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Harms GS, Orr G, Montal M, Thrall BD, Colson SD, Lu HP. Biophys J. 2003;85:1826–1838. doi: 10.1016/S0006-3495(03)74611-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Borisenko V, Lougheed T, Hesse J, Fureder-Kitzmuller E, Fertig N, Behrends JC, Woolley GA, Schutz GJ. Biophys J. 2003;84:612–622. doi: 10.1016/S0006-3495(03)74881-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Miranda P, Cadaveira-Mosquera A, Gonzalez-Montelongo R, Villarroel A, Gonzalez-Hernandez T, Lamas JA, Alvarez de la Rosa D, Giraldez T. J Neurosci. 2013;33:2684–2696. doi: 10.1523/JNEUROSCI.3442-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Yu F, Sun L, Hubrack S, Selvaraj S, Machaca K. J Cell Sci. 2013 doi: 10.1242/jcs.117200. [DOI] [PubMed] [Google Scholar]
- 135.Haas E. Chemphyschem. 2005;6:858–870. doi: 10.1002/cphc.200400617. [DOI] [PubMed] [Google Scholar]
- 136.Schuler B, Eaton WA. Curr Opin Struct Biol. 2008;18:16–26. doi: 10.1016/j.sbi.2007.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Gambin Y, Deniz AA. Molecular bioSystems. 2010;6:1540–1547. doi: 10.1039/c003024d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ferreon AC, Deniz AA. Biochimica et biophysica acta. 2011;1814:1021–1029. doi: 10.1016/j.bbapap.2011.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Schuler B, Hofmann H. Curr Opin Struct Biol. 2013;23:36–47. doi: 10.1016/j.sbi.2012.10.008. [DOI] [PubMed] [Google Scholar]
- 140.Aoki K, Komatsu N, Hirata E, Kamioka Y, Matsuda M. Cancer science. 2012;103:614–619. doi: 10.1111/j.1349-7006.2011.02196.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Komatsu N, Aoki K, Yamada M, Yukinaga H, Fujita Y, Kamioka Y, Matsuda M. Molecular biology of the cell. 2011;22:4647–4656. doi: 10.1091/mbc.E11-01-0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kohl T, Heinze KG, Kuhlemann R, Koltermann A, Schwille P. Proc Natl Acad Sci U S A. 2002;99:12161–12166. doi: 10.1073/pnas.192433499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Kajihara D, Abe R, Iijima I, Komiyama C, Sisido M, Hohsaka T. Nature methods. 2006;3:923–929. doi: 10.1038/nmeth945. [DOI] [PubMed] [Google Scholar]
- 144.Miyawaki A, Llopis J, Heim R, McCaffery JM, Adams JA, Ikura M, Tsien RY. Nature. 1997;388:882–887. doi: 10.1038/42264. [DOI] [PubMed] [Google Scholar]
- 145.Mizutani T, Kondo T, Darmanin S, Tsuda M, Tanaka S, Tobiume M, Asaka M, Ohba Y. Clin Cancer Res. 2010;16:3964–3975. doi: 10.1158/1078-0432.CCR-10-0548. [DOI] [PubMed] [Google Scholar]
- 146.Wang Y, Botvinick EL, Zhao Y, Berns MW, Usami S, Tsien RY, Chien S. Nature. 2005;434:1040–1045. doi: 10.1038/nature03469. [DOI] [PubMed] [Google Scholar]
- 147.Zhang J, Allen MD. Molecular bioSystems. 2007;3:759–765. doi: 10.1039/b706628g. [DOI] [PubMed] [Google Scholar]
- 148.Wang Y, Shyy JY, Chien S. Annual review of biomedical engineering. 2008;10:1–38. doi: 10.1146/annurev.bioeng.010308.161731. [DOI] [PubMed] [Google Scholar]
- 149.Morris MC. Cell biochemistry and biophysics. 2010;56:19–37. doi: 10.1007/s12013-009-9070-7. [DOI] [PubMed] [Google Scholar]
- 150.McKinney SA, Declais AC, Lilley DM, Ha T. Nature structural biology. 2003;10:93–97. doi: 10.1038/nsb883. [DOI] [PubMed] [Google Scholar]
- 151.Lee JY, Okumus B, Kim DS, Ha T. Proc Natl Acad Sci U S A. 2005;102:18938–18943. doi: 10.1073/pnas.0506144102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Cornish PV, Ha T. ACS chemical biology. 2007;2:53–61. doi: 10.1021/cb600342a. [DOI] [PubMed] [Google Scholar]
- 153.Chung HS, Gopich IV, McHale K, Cellmer T, Louis JM, Eaton WA. The journal of physical chemistry A. 2011;115:3642–3656. doi: 10.1021/jp1009669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Huang F, Ying L, Fersht AR. Proc Natl Acad Sci U S A. 2009;106:16239–16244. doi: 10.1073/pnas.0909126106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Ha T. Methods. 2001;25:78–86. doi: 10.1006/meth.2001.1217. [DOI] [PubMed] [Google Scholar]
- 156.Zhuang X, Bartley LE, Babcock HP, Russell R, Ha T, Herschlag D, Chu S. Science. 2000;288:2048–2051. doi: 10.1126/science.288.5473.2048. [DOI] [PubMed] [Google Scholar]
- 157.Roy R, Hohng S, Ha T. Nature methods. 2008;5:507–516. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Joo CaHT. Cold Spring Harb Protoc. 2012 doi: 10.1101/pdb.top072058. [DOI] [PubMed] [Google Scholar]
- 159.Kruger AC, Hildebrandt LL, Kragh SL, Birkedal V. Methods Cell Biol. 2013;113:1–37. doi: 10.1016/B978-0-12-407239-8.00001-X. [DOI] [PubMed] [Google Scholar]
- 160.Blunck R, McGuire H, Hyde HC, Bezanilla F. Proc Natl Acad Sci U S A. 2008;105:20263–20268. doi: 10.1073/pnas.0807056106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Sonnleitner A, Mannuzzu LM, Terakawa S, Isacoff EY. Proc Natl Acad Sci U S A. 2002;99:12759–12764. doi: 10.1073/pnas.192261499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Zhao Y, Terry D, Shi L, Weinstein H, Blanchard SC, Javitch JA. Nature. 2010;465:188–193. doi: 10.1038/nature09057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Zhao Y, Terry DS, Shi L, Quick M, Weinstein H, Blanchard SC, Javitch JA. Nature. 2011;474:109–113. doi: 10.1038/nature09971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Borsch M, Wrachtrup J. Chemphyschem. 2011;12:542–553. doi: 10.1002/cphc.201000702. [DOI] [PubMed] [Google Scholar]
- 165.Holden SJ, Uphoff S, Hohlbein J, Yadin D, Le Reste L, Britton OJ, Kapanidis AN. Biophys J. 2010;99:3102–3111. doi: 10.1016/j.bpj.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Deindl S, Hwang WL, Hota SK, Blosser TR, Prasad P, Bartholomew B, Zhuang X. Cell. 2013;152:442–452. doi: 10.1016/j.cell.2012.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Clerte C, Declerck N, Margeat E. Nucleic Acids Res. 2013;41:2632–2643. doi: 10.1093/nar/gks1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Liu S, Harada BT, Miller JT, Le Grice SF, Zhuang X. Nat Struct Mol Biol. 2010;17:1453–1460. doi: 10.1038/nsmb.1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Gansen A, Valeri A, Hauger F, Felekyan S, Kalinin S, Toth K, Langowski J, Seidel CA. Proc Natl Acad Sci U S A. 2009;106:15308–15313. doi: 10.1073/pnas.0903005106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Myong S, Stevens BC, Ha T. Structure. 2006;14:633–643. doi: 10.1016/j.str.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 171.Sindbert S, Kalinin S, Nguyen H, Kienzler A, Clima L, Bannwarth W, Appel B, Muller S, Seidel CA. Journal of the American Chemical Society. 2011;133:2463–2480. doi: 10.1021/ja105725e. [DOI] [PubMed] [Google Scholar]
- 172.Hohng S, Joo C, Ha T. Biophys J. 2004;87:1328–1337. doi: 10.1529/biophysj.104.043935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Kapanidis AN, Lee NK, Laurence TA, Doose S, Margeat E, Weiss S. Proc Natl Acad Sci U S A. 2004;101:8936–8941. doi: 10.1073/pnas.0401690101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Muller BK, Zaychikov E, Brauchle C, Lamb DC. Biophys J. 2005;89:3508–3522. doi: 10.1529/biophysj.105.064766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Uphoff S, Holden SJ, Le Reste L, Periz J, van de Linde S, Heilemann M, Kapanidis AN. Nature methods. 2010;7:831–836. doi: 10.1038/nmeth.1502. [DOI] [PubMed] [Google Scholar]
- 176.Lee J, Lee S, Ragunathan K, Joo C, Ha T, Hohng S. Angew Chem Int Ed Engl. 2010;49:9922–9925. doi: 10.1002/anie.201005402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Day RN, Davidson MW. Bioessays. 2012;34:341–350. doi: 10.1002/bies.201100098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Kremers GJ, Goedhart J, van Munster EB, Gadella TW., Jr Biochemistry. 2006;45:6570–6580. doi: 10.1021/bi0516273. [DOI] [PubMed] [Google Scholar]
- 179.Rizzo MA, Springer GH, Granada B, Piston DW. Nat Biotechnol. 2004;22:445–449. doi: 10.1038/nbt945. [DOI] [PubMed] [Google Scholar]
- 180.Shaner NC, Patterson GH, Davidson MW. J Cell Sci. 2007;120:4247–4260. doi: 10.1242/jcs.005801. [DOI] [PubMed] [Google Scholar]
- 181.Ward WW. Biochemical and physical properties of GFP. In: Chalfie M, Kain S, editors. Green Fluorescent Protein: Properties, Applications, and Protocols. Wiley; New York: 1998. [Google Scholar]