Abstract
Signal transducers and activators of transcription (STATs) were originally discovered as mediators of signal transduction. Persistent aberrant activation of STAT3 is part of the malignant phenotype of hormone-refractory prostate cancer and pancreatic cancer; this is thought to be mediated by homodimers of phosphorylated STAT3, which translocate to the nucleus. One consequence of persistently-activated STAT3 in malignant cells is that they depend upon it for survival. STAT3 is observed to heterodimerize with STAT1 and STAT2; however the contributions of STAT3:STAT1 and STAT3:STAT2 heterodimers to the survival of malignant cells have not been investigated in detail.
Previously we reported that single-stranded oligonucleotides containing consensus STAT3 binding sequences (13410 and 13411) were more effective for inducing apoptosis in prostate cancer cells than antisense STAT3 oligonucleotides. Control oligonucleotides (scrambled sequences) had no effect. STAT3-inhibiting oligonucleotide 13410, but not scrambled-sequence oligonucleotides, induced apoptosis in pancreatic cancer cells as well. Here we report that 13410 and derivative olignucleotides induced apoptosis in STAT1-null and STAT2-null fibrosarcoma cell lines U3A and U6A, as well as in the parental fibrosarcoma cell line 2fTGH. The cell lines expressed constitutively-activated STAT3 and depended on its activity for survival. Forty-eight hr after transfection of 13410 or related oligonucleotides, significant apoptosis was observed in 2fTGH, U3A and U6A cells. Scrambled-sequence oligonucleotides had no effect on survival. These data indicate that neither STAT1 nor STAT2 play significant roles in the maintenance of these cells, and by extension that STAT3:STAT1 and STAT3:STAT2 heterodimers regulate a different set of genes from STAT3:STAT3 homodimers.
Introduction
Transcription factors are latent proteins that bind to the genome upon activation, either inducing or repressing gene expression. After activation, transcription factors bind to specific enhancer sequences on the genome upstream or near the promoter region of the gene regulated by the transcription factor. Signal transducers and activators of transcription (STAT) are part of the signal transduction pathway of many growth factors and cytokines and are activated by phosphorylation of tyrosine and serine residues by upstream kinases (Ihle 1996). For example, signaling by IL-6 generally induces phosphorylation of STAT3 (Ihle 1996). In benign cells, the signaling by STAT3 is under tight regulation, so that the signal is transient. However, aberrant signaling by STAT3 is found in many types of malignancies: multiple myeloma, head and neck cancer, breast cancer, prostate cancer etc. (Barton et al. 2001, 2004a, Buettner et al. 2002, Catlett-Falcone et al. 1999, Epling-Burnette et al. 2001, Grandis et al. 1998, van Bokhoven et al. 2003). Transformed cells often express constitutively-activated STAT3; thereby become dependent on it for survival. Disruption of activation, expression, or activity of STAT3 results in apoptosis when cells express persistently-activated STAT3 (Barton et al. 2004a). STAT3 binds to two known sequences, hSIE and GAS (Seidel et al. 1995, Zhong et al. 1994, 2005), through which its anti-apoptotic and oncogenic effects are directed (Bromberg et al. 1999, Darnell 2005). These sites contain the canonical STAT3 binding motifs TTC(N)2-4GAA or TT(N)4-6AA (Bromberg et al. 1999, Darnell 2005).
Previously, we created a novel strategy for inhibiting STAT3 binding to the genome by employing oligonucleotides containing sequences related to the hSIE binding site (Barton et al. 2004b). We reported that these inhibitors induced apoptosis in prostate and pancreatic cancer cell lines in vitro, and furthermore reduced mean tumor volumes of DU-145 cells in vivo (Barton et al. 2004b, Lewis et al. 2008). We postulated that the inhibitors are specific for STAT3 but still need formal proof. This is especially important, in light of the fact that a) STAT3 and STAT1 share approximately 72% sequence homology (Lui et al. 2007), b) STAT1 is frequently expressed by many tumors expressing STAT3 and c) STAT1 may play a patho-physiological role in certain types of cancer (Buettner et al. 2002, Ernst et al. 2008, Kovacic et al. 2006).
Although STAT3 generally effects gene expression through STAT3:STAT3 homodimers, active heterodimeric forms of STAT3 with STAT1 have been observed (Ichiba et al. 2002, Wegenka et al. 1993). Like STAT3, STAT1, and to a lesser extent, STAT2 are implicated in tumorigenesis (Clifford et al. 2003, Ernst et al. 2008), however, STAT1 activity appears to have both pro- and anti-tumorigenic effects (Khodarev et al. 2004, 2007, Kovacic et al. 2006, Torrero et al. 2006). Inhibition of STAT3 by decoys was observed to have no effect on STAT1 activation in squamous cell carcinoma cell lines (Lui et al. 2007). Under the right circumstances, both STAT1 and STAT2 regulate cytokine-stimulated growth (Gimeno et al. 2005); however STAT1 may be a tumor suppressor in some cells (Lui et al. 2007).
Based on these observations, we decided to see what role, if any STAT1 or STAT2 play in maintaining the survival of STAT3-dependent cells. We determined that the fibrosarcoma cell lines express constitutively-activated STAT3, which had not been examined before. Using STAT1-null and STAT2-null fibrosarcoma cell lines and transfecting them with STAT3-inhibiting oligonucleotides, we observed the induction of apoptosis by STAT3 inhibitors in the absence of STAT1 or STAT2 signaling. Scrambled-sequence oligonucleotides had no effect. We conclude that neither STAT1 nor STAT2 activity contribute to STAT3-mediated survival of transformed cells. Moreover, the activity of our novel anti-STAT3 oligonucleotides did not depend on either STAT1 or STAT2 activation.
Materials and Methods
Oligonucleotides and siRNA
Sequences of oligonucleotides used are as follows: STAT3 inhibitors 13410 5’-AGCTTCATTTCCCGTAAATCCCTA-3’ (14) and 13410a 5’-TCCCGTAAATCCCTA-3’ (15); control oligonucleotides 13778 5’-TATGATCTCCTCCGTAACTCTCAA-3’ (14) and 13778a 5’-TATGATCTCCTCCGT-3’ (15).
Oligonucleotides were synthesized using phosphorothiorate chemistry by the Molecular Resources Facility at University of Medicine and Dentistry, New Jersey Medical School (Newark, NJ). The ribose moie-ties for 5 bp at both 5’ and 3’ ends were modified with 2’-O-methoxyl groups to increase stability of the oligonucleotides and to provide higher hybridization affinity (Goethe et al. 2001, Guvakova et al. 1995). For determinations of transfection efficiencies, a fluorescent oligonucleotide (13778a) was synthesized and included in every transfection experiment. Pooled siRNA oligonucleotides to silence STAT1, STAT2, and STAT3 were obtained from Santa Cruz Biotechnology; specifically the human STAT1 siRNA catalogue #sc-44123, human STAT2 siRNA catalogue #sc-29492, and human STAT3 siRNA catalogue #sc-29493 were used. Transfections were done as directed by the manufacturer, using the transfection reagent supplied by Santa Cruz Biotechnology. A FITC-labeled scrambled sequence siRNA oligonucleotide from Santa Cruz Biotechnology was included in each experiment to determine transfection efficiencies.
Cells and Plasmids
U3A (STAT1-null), U6A (STAT2-null), and 2fTGH (parental) fibrosarcoma cell lines were the kind gift of George Stark, The Cleveland Clinic (Li et al. 1996). They were grown in DMEM plus 10% serum (Hyclone). BPH-1 cells were the gift of Simon Hey-ward, Vanderbilt University School of Medicine, and were grown in RPMI-1640 plus 10% serum. The DU-145 human prostate cancer cell line and the PANC-1 human pancreatic cancer cell line were obtained from the American Type Culture Collection (Manassas, VA); they were grown in DMEM/ Ham's F-12 mixture plus 10% serum. Mycoplasma assays were performed on cell lines by real-time PCR (Mycosensor qPCR Assay Kit; Stratagene, La Jolla, CA); all cell lines were found to be negative.
The dominant-negative STAT3β (DN-STAT3) (Huang et al. 2007) plasmid was the gift of David Tweardy, Baylor College of Medicine. Cell viabilities were determined using fluorescein diacetate (Sigma Chemical Co., St. Louis, MO) and a Universal RIII fluorescence microscope (Zeiss, Jena, Germany).
Creation of STAT3-null BPH-DN-STAT3 cells
LipoFectamine 2000 transfection reagent (Invitrogen) was used to transfect DN-STAT3 into BPH-1 cells as described previously (Barton et al. 2004b). Briefly, cells plated in six-well plates were grown to ~50% confluence. DN-STAT3 and LipoFectamine 2000 were diluted in Opti-MEM I separately as directed by Invitrogen. The diluted LipoFectamine 2000 was allowed to incubate for 5 min at room temperature before mixing it with an equal volume of diluted plasmid for each well; mock transfected wells received LipoFectamine 2000 in the absence of plasmid. The lipo-some-plasmid mixture was incubated at room temperature for 20 min before adding it to each well. Cells were incubated with the mixture for 6 hrs at 37 °C; 1.5 ml per well of cell culture medium containing 30% serum was then added. Selection with G418 began 48 hr post-transfection after cells were sorted for EGFP fluorescence on a FACSVantage SE cell sorter and continued until no surviving cells were observed in the mock-transfected well. The resulting cell line was cloned by limit dilution, which resulted in 1 clone expressing EGFP (BPH-DN-STAT3).
Determination of Activated STAT3 by Intracellular Flow Cytometry
In brief, cells were fixed in Medium A (Invitrogen), then washed in PBS. To enhance intracellular staining, cells were resuspended in ice-cold methanol for 15 minutes on ice. After washing in PBS, cells were re-suspended in Medium B containing 2 mg/ml goat Ig (Invitrogen) for 30 minutes at room temperature, then incubated with phycoerythrin (PE)-labeled rabbit anti-phospho-STAT3 (Santa Cruz) or STAT5 (Cell Signaling) at 1 μg Ab/106 cells in 100 μl buffer for 1 hour on ice, followed by washing in PBS. For analysis, cells were brought to 0.5 ml in buffer. All flow cytometric analyses were performed on a Becton-Dickinson FAC-Scan, using the CellQuest Pro software.
STAT3 Inhibitors
The STAT3 inhibitors 13410 and 13410a, and the scrambled sequence control oligonucleotides have been described elsewhere (Barton et al. 2004b, Lewis et al. 2008). They were transfected into cells using LipoFectamine 2000 as described above. The JAK inhibitor AG490 was obtained from Calbiochem. It was dissolved in DMSO and then diluted in culture medium as needed.
Apoptosis Determinations
Annexin V and propidium iodide (PI) or 7-amino-actonomycin D (7-AAD) staining (Abcam, Malvern PA; Sigma, St. Louis MO) were used to measure the induction of apoptosis. Harvested cells were washed twice in buffer and resuspended, before addition of the forementioned chemicals. Fluorescence was quantified using a FACScan flow cytometer (Becton Dickinson, San Jose, CA) on at least 10,000 events using the CellQuest Pro software (Becton Dickinson).
Statistical Analysis
The graphing program Kaleidagraph 4.2 (Synergy Software, Reading, PA) and the statistical program InStat3 (GraphPad Software, San Diego, CA) were used for data analyses.
Results
2fTGH, U3A, and U6A cells express activated STAT3
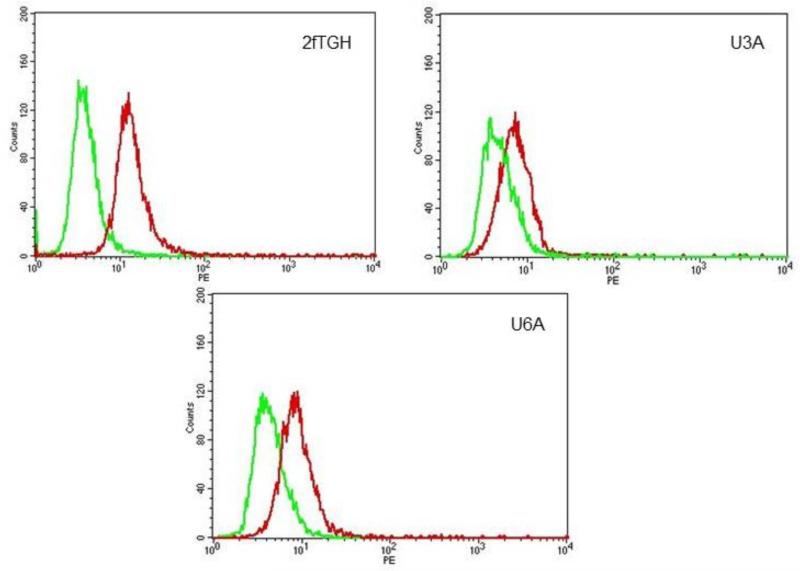
Fibrosarcomas are known to overexpress STAT3 (Lehnhardt et al., 2005) but the question of whether they express persistently-activated STAT3 has not yet been examined. Therefore, cells were stained with anti-phospho-STAT3 and analyzed by flow cytometry. The histograms in Figure 1 show that 2fTGH, U3A, and U6A cells express phospho-STAT3 in the absence of any treatment. Thus our data demonstrate constitutive expression of phosphorylated STAT3 by 2fTGH, U3A, and U6A cells.
Figure 1.
Phospho-STAT3 expression in 2fTGH, U3A and U6A cells. Expression of activated (phospho) STAT3 by fibrosarcoma cell lines. Parental 2fTGH, STAT1-null U3A, and STAT2-null U6A cells were fixed, permeabilized, blocked appropriately and then incubated with PE-anti-phospho-STAT3 (Y-705) Ab. Fluorescence was quantified by flow cytometry. Green = control Ab (anti-P-STAT5 Ab); red = anti-P-STAT3 Ab.
To determine the role of phospho-STAT3 in the survival of the fibrosarcoma cell lines, the cells were treated with the JAK2 inhibitor AG490. Apoptosis was then quantified as described (Barton et al. 2004). Figure 2A shows that 30 to 100 μM treatment with AG490 for 24 hours resulted in nearly 100% apoptosis of U3A and U6A cells, indicating that cells remain sensitive to the STAT3-inhibiting effects of the JAK2 inhibitor AG490, even in the absence of STAT1 or STAT2 expression.
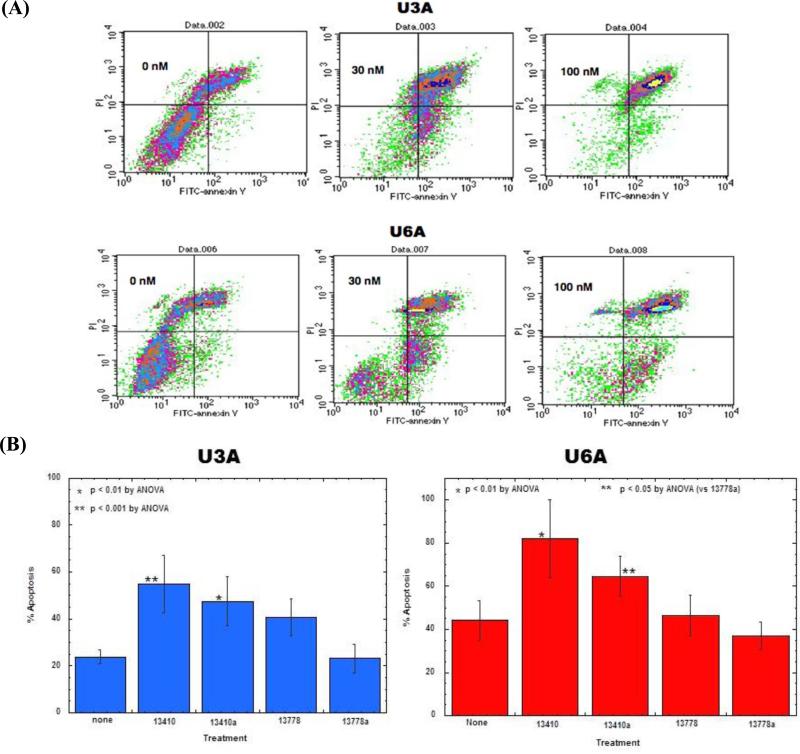
Figure 2.
Effect of STAT3 inhibition on U3A and U6A survival. (A) AG490 treatment x 24 hr; apoptosis. Effect of AG490 treatment on STAT1 or 2 and time they were harvested and stained with FITC-annexin V/PI and analyzed by flow cytometry. (B) Effect of 13410 and 13410a in STAT1- and STAT2-null cells. U3A and U6A cells were treated with STAT3 inhibitors 13410 and 13410a and control oligonucleotides 13778 and 13778a for 48 hr. Cells were harvested and stained with FITC-annexin V/PI to assess apoptosis by flow cytometry. Apoptosis experiments were conducted three times. Data from a representative experiment are given.
Inhibition of STAT3 activity induced apoptosis in U3A and U6A cells
AG490 is a JAK2 inhibitor. Although its effects on U3A and U6A provide evidence that inhibition of STAT3 is deleterious to the survival of these cell lines, this is not proof on its own, as JAK2 also activates STAT5, which has been implicated in tumor cell survival (Harir et al. 2007, Meier et al. 2009, Rocnik et al. 2006). To address the issue of whether direct inhibition of STAT3 results in apoptosis in the absence of expression of either STAT1 or STAT2, oligonucleotide inhibitors of STAT3 activity (Barton et al. 2004b, Lewis et al. 2008) were transfected into 2fTGH, U3A, and U6A cells. Figure 2B shows that STAT3 inhibitors 13410 and 13410a, but not 13778 or 13778a, induced significant apoptosis (60-80%), as measured by FITC-annexin V plus PI staining, in U3A and U6A cells. These results are further proof that STAT3 inhibition induces apoptosis in cancer cells that is independent of STAT1 or STAT2 expression or activity.
Silencing STAT1 or STAT2 expression in STAT3-dependent cancer cell lines did not affect viability
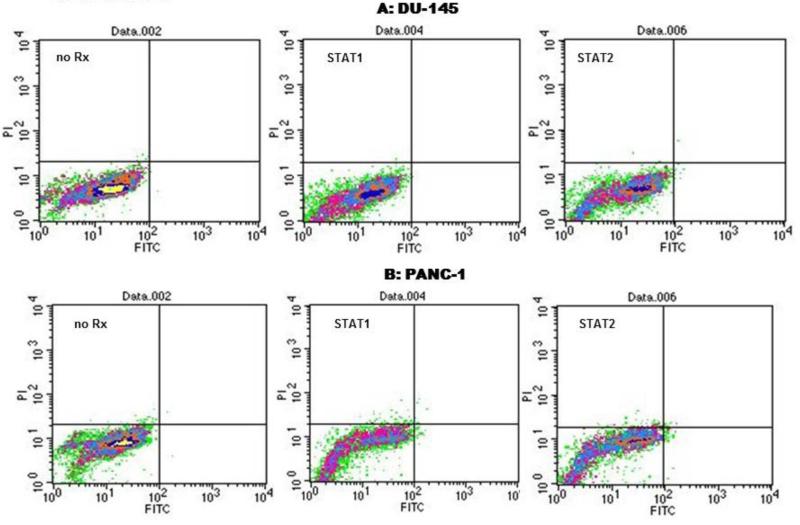
In order to confirm that neither STAT1 nor STAT2 contribute to the survival of malignant cells, siRNA was used to silence their expression in DU-145 and PANC-1 cells. The latter are STAT3-dependent cell lines ie. undergo apoptosis when STAT3 expression or activity is inhibited (Barton et al. 2004a, Lewis et al. 2008). Figure 3 and Table 1 show that 24 to 48 hours after transfection with either STAT1 or STAT2 siRNA, no significant apoptosis was observed in DU-145 or PANC-1 cells. Transfection efficiencies for siRNA were high, averaging >90% (not shown). Transfection of a control siRNA sequence supplied by Santa Cruz had no effect, whereas transfection of STAT3 siRNA induced significant apoptosis (Table 1).
Figure 3.
Effect of STAT1 and STAT2 gene silencing with siRNA in STAT3-dependent cell lines. DU-145 and PANC-1 cells were treated with 500 nM pooled siRNA for 72 hr, at which time cells were harvested and stained with FITC-annexin V/PI and analyzed by flow cytometry. More than 95% of the cells were observed to be viable following siRNA transfection. Transfection efficiencies ranged from 95 to 98% (not shown).
Table 1.
Effect of siRNA-mediation inhibition of STATs 1, 2, and 3 on cell survival. Pooled siRNAs were purchased from Santa Cruz Laboratories. They were transfected into cells using Lipofectamine 2000 according to Invitrogen's protocol. Twenty-four hr after transfection, cells were harvested, washed, stained with FITC-annexin V/PI, and analyzed by flow cytometry. Data from a representative experiment are given.
| Cell Line | siRNA | nM | %Apoptotic |
|---|---|---|---|
| DU-145 | None | - | 0 |
| STAT1 | 250 | 0.04 | |
| 500 | 0.01 | ||
| STAT2 | 250 | 0.02 | |
| 500 | 0.07 | ||
| STAT3 | 250 | 50.62 | |
| 500 | 35 | ||
| Control | 250 | 0.02 | |
| 500 | 0.13 | ||
| PANC-1 | None | - | 0 |
| STAT1 | 250 | 0 | |
| 500 | 0.02 | ||
| STAT2 | 250 | 0.01 | |
| 500 | 0.03 | ||
| STAT3 | 250 | 12.1 | |
| 500 | 46.8 | ||
| Control | 250 | 0.07 | |
| 500 | 0 |
In order to show that neither STAT1 nor STAT2 synergized with STAT3 (thereby contributing to apoptosis), pairs of pooled siRNAs at 250 nM each (i.e., STAT1 plus STAT3 or STAT2 plus STAT3) were transfected into DU-145 and PANC-1 cells. Apoptosis was measured using FITC-annexin V/PI and flow cytometry 24 hours after transfection. The results in Table 2 show that neither STAT1 nor STAT2 siRNA synergized with STAT3 siRNA to induce apoptosis; the amount of apoptosis seen with 250 nM STAT3 siRNA was nearly the same in the presence or absence of either STAT1 or STAT2 siRNA (Table 2). Repeated ANOVA revealed no significant differences in the amount of apoptosis observed whether cells were treated with STAT3 siRNA alone or STAT3 siRNA in combination with STAT1, STAT2, or control siRNA (P value was 0.99 for DU-145 cells and 0.43 for PANC-1 cells; Table 2). We concluded from these experiments that inhibition of STAT3 to induce apoptosis is independent of either STAT1 or STAT2, in DU-145 and PANC-1 cells.
Table 2.
Lack of synergy between STAT1 or STAT2 and STAT3 siRNAs. Pooled siRNAs were purchased from Santa Cruz Laboratories. They were co-transfected into cells using Lipofectamine 2000 according to Invitrogen's protocol. Twenty-four hr after transfection, cells were harvested, washed, stained with FITC-annexin V/PI, and analyzed by flow cytometry. % apoptotic data were normalized to 100% transfection efficiency. P value was determined by repeated ANOVA.
| Cell Line | siRNA | nM (each siRNA) | % Apoptotic | ±SEM | P |
|---|---|---|---|---|---|
| DU-145 | None | - | 0 | 0 | 0.97 |
| STAT3 | 250 | 52.8 | 5.4 | ||
| STAT1+STAT3 | 250 | 49.8 | 10 | ||
| STAT2+STAT3 | 250 | 52.2 | 9.4 | ||
| Control+STAT3 | 250 | 50.8 | 10.7 | ||
| PANC-1 | None | - | 0 | 0 | 0.47 |
| STAT3 | 250 | 38.2 | 10.7 | ||
| STAT1+STAT3 | 250 | 40.2 | 9.9 | ||
| STAT2+STAT3 | 250 | 32.7 | 12 | ||
| Control+STAT3 | 250 | 37.2 | 9.5 |
Absence of STAT3 expression by BPH-DN-STAT3 cells and effect of 13410 and 13410a on them
In order to ensure that the novel STAT3 inhibitors 13410 and 13410a had no pathway for apoptosis induction other than through STAT3 inhibition, we created a STAT3-null cell line from BPH-1 cells. This was generated by forcing the expression of the pseudogene STAT3β in BPH-1 cells by transfecting a plasmid (Huang et al. 2007) and then selecting transfected cells by fluorescence-activated cell sorting. The results in Table 3 demonstrate that BPH-1 and BPH-DNSTAT3 cells showed no apoptosis when either 13410 or 13410a were transfected. These data demonstrate that under conditions where STAT3 activity is suppressed, oligonucleotides 13410 and 13410a exhibited no ancillary pro-apoptotic activity.
Table 3.
Effect of anti-STAT3 oligonucleotides on STAT3-null cells. BPH-1 cells and BPH-DN-STAT3 cells were transfected with olignucleotides as described in Materials and Methods. Forty-eight hrs later, cells were harvested and stained with annexin-V and PI to assess apoptosis by flow cytometry. The means plus standard deviations of three experiments are given.
| Cell Line | OLG | nM | %Apoptotic | ± SD |
|---|---|---|---|---|
| BPH-1 | None | 0 | 0.17 | 0.27 |
| 13410 | 100 | 0 | 0 | |
| 300 | 0 | 0 | ||
| 1000 | 0.04 | 0.07 | ||
| 13410a | 100 | 0 | 0 | |
| 300 | 0.02 | 0.01 | ||
| 1000 | 0.07 | 0.1 | ||
| 13778 | 100 | 0 | 0 | |
| 300 | 0.3 | 0.29 | ||
| 1000 | 0.08 | 0.14 | ||
| 13778a | 100 | 0.04 | 0.03 | |
| 300 | 0.06 | 0.05 | ||
| 1000 | 0.37 | 0.66 | ||
| BPH-DN-STAT3 | None | 0 | 1.94 | 1.77 |
| 13410 | 100 | 0.4 | 0.37 | |
| 300 | 0.52 | 0.46 | ||
| 1000 | 0.87 | 0.12 | ||
| 13410a | 100 | 0.63 | 0.55 | |
| 300 | 1.02 | 0.89 | ||
| 1000 | 2.25 | 1.05 | ||
| 13778 | 100 | 1.29 | 1.13 | |
| 300 | 0.53 | 0.46 | ||
| 1000 | 1.48 | 0.16 | ||
| 13778a | 100 | 0.47 | 0.41 | |
| 300 | 0.56 | 0.49 | ||
| 1000 | 2.60 | 2.55 |
Discussion
Our data demonstrate that neither STAT1 nor STAT2 contributed to the survival of STAT3-addicted tumor cells. Furthermore, our data show that the novel STAT3 inhibiting oligonucleotides 13410 and 13410a have sufficient STAT3 specificity insofar as the oligonucleotides did not induce apoptosis in a STAT3-null BPH cell line, BPH-DN-STAT3. Thus we conclude from our data that STAT3 acts independently of STAT1 or STAT2 in promoting the survival of cancer cells and that our novel STAT3 inhibitors 13410 and 13410 are selective STAT3 inhibitors.
Under the right circumstances, STAT1 and STAT2 can regulate cytokine-stimulated growth, even in opposition to STAT3 effects (Gimeno et al. 2005). However, neither transcription factor is known to act as a proto-oncogene, a trait shared by STAT3 and STAT5. These previous observations did not rule out the possibility that STAT3 interacts with either STAT1 or STAT2 to promote tumor cell survival. Transfection of siRNA to silence expression of either STAT1 or STAT2 had no effect on the survival of DU-145 prostate cancer or PANC-1 pancreatic cells (Figure 3 and Table 1), yet transfection of STAT3 siRNA induced significant apoptosis in these cells lines (Tables 1 and 2). Inhibiting STAT3 in U3A (STAT1-null) or U6A (STAT-2-null) cell lines had no effect on cell survival; however both U3A and U6A cell lines were sensitive to the effects of the JAK2 inhibitor AG490 (which inhibits activation of STAT3) and the effects of the STAT3 inhibitors 13410 and 13410a. No effect on U3A or U6A cell survival was seen when either control oligonucleotides 13778 or 13778a were transfected.
STAT3 usually regulates gene expression through STAT3:STAT3 homodimers; however, active heterodimeric forms of STAT3 with STAT1 have been noted, which bind preferentially to the sequence TTC (N)3GAA, a canonical STAT3 binding motif (Lewis et al. 2008, Wegenka et al. 1993). STAT3:STAT2 heterodimers have not been described, presumably due to the inability of the STAT2 SH2 domain to bind to the major phosphorylated tyrosine site on STAT3 (Wiederkehr-Adam et al. 2003). Independent of interactions with STAT3, STAT1 and STAT2 (in common with STAT3) recruit p300/CBP in the course of trans-activation, albeit to different extents (Paulson et al. 1999). Like STAT3, STAT1, and to a lesser extent, STAT2, are implicated in tumorigenesis (Clifford et al. 2003, Ernst et al. 2008); however STAT1 activity appears to have both pro- and anti-tumorigenic effects depending upon STAT1 activation (transient or constitutive) (Khodarev et al. 2004, Khodarev et al. 2007, Kovacic et al. 2006, Torrero et al. 2006). Inhibition of STAT3 by oligonucleotide decoys was observed to have no effect on STAT1 activation in squamous cell carcinoma cell lines (Lui et al. 2007). In addition, we could find no published data on the effect that STAT3 inhibition has on STAT2 activation. Under the right circumstances, STAT1 and STAT2 can regulate cytokine-stimulated growth, even in opposition to STAT3 effects (Gimeno et al. 2005).
As expected, expression of STAT1 or STAT2 may regulate tumor cell response to interferons. Stable expression of a dominant-negative form of STAT2 in squamous cell carcinoma cell lines blocked interferon-α action as demonstrated by cell growth in its presence. On the other hand, expression of STAT1 or STAT2 in melanoma cell lines did not correlate with interferon-α as adjuvant therapy (Lesinski et al. 2005). More recently, investigators observed that a mutation in the SH2 domain of STAT2 prolonged tyrosine phosphorylation of STAT1, thereby promoting interferon-induced apoptosis (Lesinski et al. 2005). These data may be an indication of interaction between STAT1 and STAT2; more work is needed to determine if this is true in other cancer cell lines. In addition, very recent data demonstrated that the STAT1 gene is a target of microRNA-145 in colon cancer cells, indicating that inhibiting STAT1 may be therapeutic for cancer treatment (Gregersen et al. 2010). Clearly, more work delineating the roles of STAT1, STAT2, and STAT3 in cancer is in order for understanding oncogenesis and for developing better treatment options.
Acknowledgements
The authors acknowledge the Molecular Resources Facility, New Jersey Medical School, for synthesizing the modified oligonucleotides used. This work was supported by NIH grant CA 121782, an award from the Elsa U. Pardee Foundation, and a Research & Development Merit Award from the Department of Veterans Affairs (BEB).
Footnotes
Author Contributions
A.L.S. contributed significantly to this manuscript. K.G. and H.D.L. contributed equally to this manuscript. S.R. contributed to this manuscript. B.E.B. is the corresponding author.
Competing interests
The authors declare no conflicts of interest.
References
- Barton BE, Karras JG, Murphy TF, Barton AB, Huang HF. STAT3 Activation in Prostate Cancer: Direct STAT3 Inhibition Induces Apoptosis in Prostate Cancer Lines. Mol Cancer Ther. 2004a;3:11–20. [PubMed] [Google Scholar]
- Barton BE, Murphy TF, Adem P, Watson RA, Irwin RJ, Huang HS. IL-6 signaling by STAT3 participates in the change from hyperplasia to neoplasia in NRP-152 and NRP-154 rat prostatic epithelial cells. BMC Cancer. 2001;1:19. doi: 10.1186/1471-2407-1-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton BE, Murphy TF, Shu P, Huang HF, Meyenhofen M, Barton AB. Novel Single-Stranded Oligonucleotides that Inhibit STAT3 Induce Apoptosis In Vitro and In Vivo in Prostate Cancer Cell Lines. Mol Cancer Ther. 2004b;3:1183–1191. [PubMed] [Google Scholar]
- Bromberg JF, Wrzeszczynska MH, Devgan G, Zhao Y, Pestell RG, Albanese C, Darnell JE. Stat3 as an oncogene. Cell. 1999;98:295–303. doi: 10.1016/s0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- Buettner R, Mora LB, Jove R. Activated STAT signaling in human tumors provides novel molecular targets for therapeutic intervention. Clin Cancer Res. 2002;8:945–954. [PubMed] [Google Scholar]
- Catlett-Falcone R, Landowski TH, Oshiro MM, Turk son J, Levitzki A, Savino R, Ciliberto G, Moscinski L, Fernández-Luna JL, Nuñez G, Dalton WS, Jove R. Constitutive activation of Stat3 signaling confers resistance to apoptosis in human U266 myeloma cells. Immunity. 1999;10:105–115. doi: 10.1016/s1074-7613(00)80011-4. [DOI] [PubMed] [Google Scholar]
- Clifford JL, Yang X, Walch E, Wang M, Lippman SM. Dominant negative signal transducer and activator of transcription 2 (STAT2) protein: stable expression blocks interferon alpha action in skin squamous cell carcinoma cells. Molecular Cancer Therapeutics. 2003;2:453–459. [PubMed] [Google Scholar]
- Darnell JE., Jr Validating Stat3 in cancer therapy. Nat Med. 2005;11:595–596. doi: 10.1038/nm0605-595. [DOI] [PubMed] [Google Scholar]
- Epling-Burnette PK, Liu JH, Catlett-Falcone R, Turkson J, Oshiro M, Kothapalli R, Li Y, Wang JM, Yang-Yen HF, Karras J, Jove R, Loughran TP., Jr Inhibition of STAT3 signaling leads to apoptosis of leukemic large granular lymphocytes and decreased Mcl-1 expression. J Clin Invest. 2001;107:351–362. doi: 10.1172/JCI9940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst M, Najdovska M, Grail D, Lundgren-May T, Buchert M, Tye H, Matthews VB, Armes J, Bhathal PS, Hughes NR, Marcusson EG, Karras JG, Na S, Sedgwick JD, Hertzog PJ, Jenkins BJ. STAT3 and STAT1 mediate IL-11-dependent and inflammation-associated gastric tumorigenesis in gp130 receptor mutant mice. J Clin Invest. 2008;118:1727–1738. doi: 10.1172/JCI34944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimeno R, Lee CK, Schindler C, Levy DE. Stat1 and Stat2 but not Stat3 arbitrate contradictory growth signals elicited by alpha/beta interferon in T lymphocytes. Mol Cell Biol. 2005;25:5456–5465. doi: 10.1128/MCB.25.13.5456-5465.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goethe JW, Bittova M, Vogel JU, Kotchekov R, Doerr HW, Cinati J., Jr. Antisense oligonucleotide ISIS 2922 targets IE-expression and prevents HCMVIE-induced suppression of TSP-1 and TSP-2 expression. Nucleosides Nucleotides Nucleic Acids. 2001;20:1425–1428. doi: 10.1081/NCN-100002569. [DOI] [PubMed] [Google Scholar]
- Grandis JR, Drenning SD, Chakraborty A, Zhou M-Y, Zang Q, Pitt AS, Tweardy DJ. Requirement of Stat3 but not Stat1 activation for epidermal growth factor receptor-mediated cell growth in vitro. J Clin Invest. 1998;102:1385–1392. doi: 10.1172/JCI3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregersen LH, Jacobsen AB, Frankel LB, Wen J, Krogh A, Lund AH. MicroRNA-145 targets YES and STAT1 in colon cancer cells. PLoS One. 2010;5:e8836. doi: 10.1371/journal.pone.0008836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guvakova MA, Yakubov LA, Vlodavesky I, Tonkinson JL, Stein CA. Phosphothiorate oligodeoxynucleotides bind to basic fibroblast growth factor, inhibit its binding to cell surface receptors, and remove it from low affinity binding sites on extracellular matrix. J Biol Chem. 1995;270:2620–2627. doi: 10.1074/jbc.270.6.2620. [DOI] [PubMed] [Google Scholar]
- Harir N, Pecquet C, Kerenyi M, Sonneck K, Kovacic B, Nyga R, Brevet M, Dhennin I, Gouilleux-Gruart V, Beug H, Valent P, Lassoued K, Moriggl R, Gouil leux F. Constitutive activation of Stat5 promotes its cytoplasmic localization and association with PI3-kinase in myeloid leukemias. Blood. 2007;109:1678–1686. doi: 10.1182/blood-2006-01-029918. [DOI] [PubMed] [Google Scholar]
- Huang Y, Qiu J, Dong S, Redell MS, Poli V, Mancini MA, Tweardy DJ. Stat3 isoforms, alpha and beta, demonstrate distinct intracellular dynamics with prolonged nuclear retention of Stat3beta mapping to its unique C-terminal end. J Biol Chem. 2007;282:34958–34967. doi: 10.1074/jbc.M704548200. [DOI] [PubMed] [Google Scholar]
- Ichiba M, Miyazaki Y, Kitamura S, Kiyohara T, Shinomura Y, Matsuzawa Y. Epidermal growth factor inhibits the growth of TE8 esophageal cancer cells through the activation of STAT1. J Gastroenterol. 2002;37:497–505. doi: 10.1007/s005350200077. [DOI] [PubMed] [Google Scholar]
- Ihle JN. STATs and MAPKs: obligate or opportunistic partners in signaling. BioEssays. 1996;18:95–98. doi: 10.1002/bies.950180204. [DOI] [PubMed] [Google Scholar]
- Khodarev NN, Beckett M, Lebay E, Darga T, Roizman B, Weichselbaum RR. STAT1 is overexpressed in tumors selected for radioresistance and confers protection from radiation in transduced sensitive cells. Proc Natl Acad Sci USA. 2004;101:1714–1719. doi: 10.1073/pnas.0308102100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodarev NN, Minn AJ, Efimova EV, Darga TE, Labay E, Beckett M, Mauceri HJ, Roizman B, Weichselbaum RR. Signal Transducer and Activator of Transcription 1 Regulates Both Cytotoxic and Prosurvival Functions in Tumor Cells. Cancer Res. 2007;67:9214–9220. doi: 10.1158/0008-5472.CAN-07-1019. [DOI] [PubMed] [Google Scholar]
- Kovacic B, Stoiber D, Moriggl R, Weisz E, Ott RG, Kreibich R, Levy DE, Beug H, Freissmuth M, Sexl V. STAT1 acts as a tumor promoter for leukemia development. Cancer Cell. 2006;10:77–87. doi: 10.1016/j.ccr.2006.05.025. [DOI] [PubMed] [Google Scholar]
- Lehnhardt M, Klein-Hitpass L, Kuhnen C, Homann HH, Daigeler A, Steinau HU, Roehrs S, Schnoor L, Steinstraesser L, Mueller O. Response rate of fibrosarcoma cells to cytotoxic drugs on the expression level correlates to the therapeutic response rate of fibrosarcomas and is mediated by regulation of apoptotic pathways. BMC Cancer. 2005;5:74. doi: 10.1186/1471-2407-5-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesinski GB, Valentino D, Hade EM, Jones S, Magro C, Chaudhury AR, Walker MJ, Carson WE., 3rd Expression of STAT1 and STAT2 in malignant melanoma does not correlate with response to interferon-alpha adjuvant therapy. Cancer Immunol Immunother. 2005;54:815–825. doi: 10.1007/s00262-004-0649-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis HD, Winter A, Murphy TF, Tripathi S, Pandey VN, Barton BE. STAT3 inhibition in prostate and pancreatic cancer lines by STAT3 binding sequence oligonucleotides: differential activity between 5′ and 3′ ends. Mol Cancer Ther. 2008;7:1543–1550. doi: 10.1158/1535-7163.MCT-08-0154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Leung S, Qureshi S, Darnell JE, Jr, Stark GR. Formation of STAT1-STAT2 heterodimers and their role in the activation of IRF-1 gene transcription by interferon-alpha. J Biol Chem. 1996;271:5790–5794. doi: 10.1074/jbc.271.10.5790. [DOI] [PubMed] [Google Scholar]
- Lui VW, Boehm AL, Koppikar P, Leeman RJ, Johnson D, Ogagan M, Childs E, Freilino M, Grandis JR. Antiproliferative Mechanisms of a Transcription Factor Decoy Targeting Signal Transducer and Activator of Transcription (STAT) 3: The Role of STAT1. Mol Pharmacol. 2007;71:1435–1443. doi: 10.1124/mol.106.032284. [DOI] [PubMed] [Google Scholar]
- Meier C, Hoeller S, Bourgau C, Hirschmann P, Schwaller J, Went P, Pileri SA, Reiter A, Dirnhofer S, Tzankov A. Recurrent numerical aberrations of JAK2 and deregulation of the JAK2-STAT cascade in lymphomas. Mod Pathol. 2009;22:476–487. doi: 10.1038/modpathol.2008.207. [DOI] [PubMed] [Google Scholar]
- Paulson M, Pisharody S, Pan L, Guadagno S, Mui AL, Levy DE. Stat protein transactivation domains recruit p300/CBP through widely divergent sequences. J Biol Chem. 1999;274:25343–25349. doi: 10.1074/jbc.274.36.25343. [DOI] [PubMed] [Google Scholar]
- Rocnik JL, Okabe R, Yu JC, Lee BH, Giese N, Schenkein DP, Gilliland DG. Roles of tyrosine 589 and 591 in STAT5 activation and transformation mediated by FLT3-ITD. Blood. 2006;108:1339–1345. doi: 10.1182/blood-2005-11-011429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidel H, Milocco L, Lamb P, Darnell JJ, Stein R, Rosen J. Spacing of palindromic half sites as a determinant of selective STAT (signal transducers and activators of transcription) DNA binding and transcriptional activity. Proc Natl Acad Sci USA. 1995;92:3041–3045. doi: 10.1073/pnas.92.7.3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrero MN, Xia X, Henk W, Yu S, Li S. Stat1 deficiency in the host enhances interleukin-12-mediated tumor regression. Cancer Res. 2006;66:4461–4467. doi: 10.1158/0008-5472.CAN-05-3554. [DOI] [PubMed] [Google Scholar]
- van Bokhoven A, Varella-Garcia M, Korch C, Johannes WU, Smith EE, Miller HL, Nordeen SK, Miller GJ, Lucia MS. Molecular characterization of human prostate carcinoma cell lines. Prostate. 2003;57:205–225. doi: 10.1002/pros.10290. [DOI] [PubMed] [Google Scholar]
- Wegenka UM, Buschmann J, Lutticken C, Heinrich PC, Horn F. Acute-phase response factor, a nuclear factor binding to acute-phase response elements, is rapidly activated by interleukin-6 at the posttranslational level. Mol Cell Biol. 1993;13:276–288. doi: 10.1128/mcb.13.1.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiederkehr-Adam M, Ernst P, Müller K, Bieck E, Gombert FO, Ottl J, Graff P, Grossmüller F, Heim MH. Characterization of Phosphopeptide Motifs Specific for the Src Homology 2 Domains of Signal Transducer and Activator of Transcription 1 (STAT1) and STAT3. J Biol Chem. 2003;278:16117–16128. doi: 10.1074/jbc.M300261200. [DOI] [PubMed] [Google Scholar]
- Zhong M, Henriksen MA, Takeuchi K, Schaefer O, Liu B, ten Hoeve J, Ren Z, Mao X, Chen X, Shuai K, Darnell JE., Jr Implications of an antiparallel dimeric structure of nonphosphorylated STAT1 for the activation-inactivation cycle. Proc Natl Acad Sci U S A. 2005;102:3966–3971. doi: 10.1073/pnas.0501063102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Z, Wen Z, Darnell JE., Jr Stat3: a STAT family member activated by tyrosine phosphorylation in response to epidermal growth factor and interleukin-6. Science. 1994;264:95–98. doi: 10.1126/science.8140422. [DOI] [PubMed] [Google Scholar]