Abstract
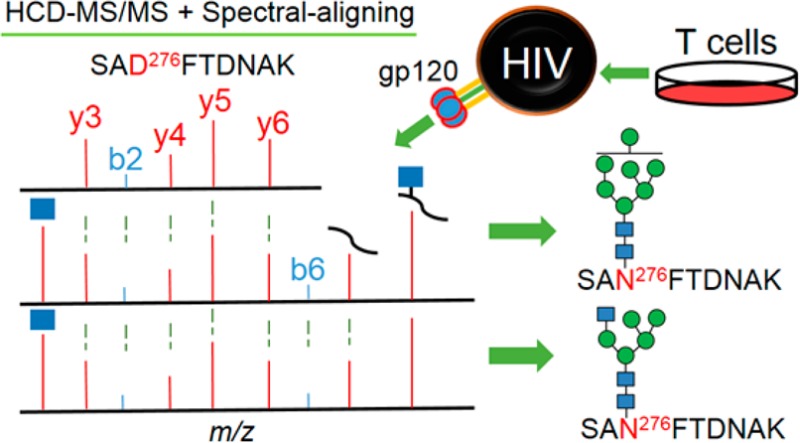
Envelope protein gp120 of human immunodeficiency virus (HIV) is armored with a dense glycan shield, which plays critical roles in envelope folding, immune-evasion, infectivity, and immunogenicity. Site-specific glycosylation profiling of recombinant gp120 is very challenging. Therefore, glycoproteomic analysis of native viral gp120 is still formidable to date. This challenge promoted us to employ a Q-Exactive mass spectrometer to identify low abundant glycopeptides from virion-associated gp120. To search the HCD-MS data for glycopeptides, a novel spectral-aligning strategy was developed. This strategy depends on the observation that glycopeptides and the corresponding deglycosylated peptides share very similar MS/MS pattern in terms of b- and y-ions that do not contain the site of glycosylation. Moreover, glycopeptides with an identical peptide backbone show nearly resembling spectra regardless of the attached glycan structures. For the recombinant gp120, this “copy–paste” spectral pattern of glycopeptides facilitated identification of 2224 spectra using only 18 spectral templates, and after precursor mass correction, 1268 (57%) spectra were assigned to 460 unique glycopeptides accommodating 19 N-linked and one O-linked glycosylation sites (glycosites). Strikingly, we were able to observe five N- and one O-linked glycosites in native gp120. We further revealed that except for Asn276 in the C2 region, glycans were processed to contain both high mannose and hybrid/complex glycans; an additional four N-linked glycosites were decorated with high mannose type. Core 1 O-linked glycan Gal1GalNAc1 was seen for the O-linked glycosite at Thr499. This direct observation of site-specific glycosylation of virion-derived gp120 has implications in HIV glycobiology and vaccine design.
Protein glycosylation functions in protein solubility, stability, intracellular trafficking, cell–cell adhesion, secretion, cell–cell signaling, and notoriously, in the protective shield of viruses.1−5 Protein glycosylation enables attachment of different carbohydrate(s) to proteins resulting in a dramatic increase of protein heterogeneity and diversity.6 The linkage between carbohydrate(s) and proteins defines the type of glycosylation, which can be mainly N- or O-linked.6,7 N-linked glycosylation attaches carbohydrate(s) to asparagine (Asn) residues in the consensus sequon Asn-X-Ser/Thr (X ≠ Pro), whereas any threonine (Thr) or serine (Ser) residues could be a potential site of O-linked glycosylation.6,8 On the basis of the glycan structures, N-linked glycans can be further categorized into high-mannose, hybrid, and complex types.9
HIV envelope (Env) protein gp120, the viral receptor designated for initiation of HIV infection and immune evasion, is protected by a glycan canopy.10 Both N- and O-linked glycosylation are reported.11−13 Site-specific profiling of glycosylation has been reported using recombinant gp120s.12,14 However, there would remain considerable challenges to profile site-specific glycosylation for native gp120 derived from T-cell-expressed virions. One of the main obstacles is the extremely low number of the molecules (i.e., only 21–54 gp120 molecules per virion in contrast to the approximately 1400 Gag molecules per virion).15−17
Mass spectrometry (MS) is the tool of choice to profile protein glycosylation in a site-specific manner.18 MS offers high sensitivity and selectivity, notwithstanding, it is still quite challenging to analyze glycopeptides. In part, the difficulty is introduced by the presence of a different glycan moiety which distributes the otherwise same peptide to different glycopeptide masses (dilution effect) and, even worse, greatly suppresses the signal intensity of glycopeptides on the MS1 level, decreasing the chance of MS/MS to occur for glycopeptide identification.19 Despite the intrinsic pitfalls associated with detection of glycopeptides in the MS analysis, recent studies using high energy C-trap disassociation (HCD)-MS have shown promise to reach subfmol limit of detection.20 Other than HCD fragmentation-type, collision-induced disassociation (CID), infrared multiphotondissociation (IRMPD), electron capture dissociation (ECD), and electron transfer dissociation (ETD) are also applied to study glycopeptides.21−23 The use of multiple fragmentation methods provides comprehensive and confirmatory identification of glycan and peptide moieties of glycopeptides.22,24,25 The advantage of the sole use of the HCD is that it requires minimal time for duty cycle compared to dual-scan (e.g., CID/ETD, HCD/ETD) and favors more MS/MS scans toward extremely low abundant species. The feasibility of using only HCD to study glycopeptides has been established. Oxonium and peptide+HexNAc (Y1) ions have been used to infer confident assignment and the microheterogeneity of each glycosylation site.26 However, unequivocal identification may be compromised using only oxonium and Y1 ions in a complex sample, because peptide backbones of different glycopeptides can generate the same mass even within a mass tolerance of 10 ppm.
Typical HCD MS/MS spectra of glycopeptides consist of three components for a search strategy. First, the oxonium ions are the robust markers at low m/z region.27 Commonly seen oxonium ions are HexNAc at m/z 204.09 and HexHexNAc at m/z 366.14.27 Next comes the peptide fragment b- and y-ions.20 HCD is one of the fragmentation methods that can generate peptide fragment ions.21 Although these ions are less frequently detected according to previous studies,20,22 it is the fingerprint of the peptide backbone that infers the confidence of identification.28 The peptide moiety of a glycopeptide and the peptide without attached glycan should, in theory, produce the same experimental fragment ions because HCD is the fragmentation method applied. The fragmentation pattern of the peptide moiety of the glycopeptide could also be like that of the deglycosylated peptide, whose formerly N-linked glycosylation site (N-glycosite) is deamidated from asparagine to aspartic acid by Peptide-N-Glycosidase F (PNGaseF) treatment.29 The final component is the peptide and peptide + glycan (Y−) ions that appear very frequently in the CID spectra and less frequently in the HCD spectra of glycopeptides.22,30
To improve identification confidence for glycopeptides by HCD exclusively, we evaluated the use of b- and y-ions in MS/MS spectra of deglycosylated peptides and glycopeptides. To do this, characterization of HCD fragmentation of both N- and O-linked glycopeptides was studied and demonstrated that peptide b- and y-ions which do not contain the site of glycosylation are relatively consistent between the HCD glycopeptide fragmentation spectrum and the deglycosylated (via PNGase F) form of the glycopeptide. In addition, MS/MS spectra of glycopeptides with the same peptide moiety are nearly identical regardless of concomitant glycan compositions. This resulted in using a number of 18 spectral templates of glycopeptides to assign 2224 MS/MS spectra to gp120 glycopeptides from which 460 unique glycopeptides were identified within a mass tolerance of 10 ppm. Finally, we observed that the peptide and Y-ions might be absent in the MS/MS spectra of O-linked glycopeptides suggestive of the necessity of using peptide b- and y-ions for identification of O-linked glycopeptides by HCD. These key features were used to profile glycosylation of a recombinant HIV gp120 HxBc2 strain expressed from 293 cells. Strikingly, we were able to detect both N- and O-linked glycopeptides of native gp120 from the T-cell-expressed HIV virion using spectral-aligning strategy. The site-specific glycoform analysis has implications for vaccine design and HIV glycobiology.
Materials and Methods
Materials
HIV envelope protein gp120 (HxBc2 strain, clade B, GenBank No. K03455 and AAB50262.1, amino acid 34–518) was purchased from Immune Technology Corp. (New York, NY). The protein was expressed and purified from 293 cells. The HIV-1 latently infected T cell line, ACH-2 cells, was obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID. NIH: from Dr. Thomas Folks.31−33 Microcon-50 kDa (50 kDa molecular weight cutoff) and Amicon Ultra-0.5 mL-100 kDa (100 kDa molecular weight cutoff) centrifugal filter units were from Millipore (Billerica, MA). Sequencing-grade trypsin was from Promega (Madison, WI). Peptide-N-Glycosidase F (PNGase F) was purchased from New England Biolabs (Ipswich, MA). Sep-Pak C18 1 cm3 Vac Cartridge was from Waters (Milford, MA). Acetonitrile (ACN), ammonium bicarbonate, trifluoroacetic acid (TFA), formic acid, urea, tris (2-carboxyethyl) phosphine (TCEP), iodoacetamide, and phorbol 12-myristate 13-acetate (PMA) were purchased from Sigma-Aldrich (St. Louis, MO).
Expression and Purification of gp120 Derived from T-Cell-Expressed HIV Virion
The ACH-2 cell line is a subclone of the human A3.01 T cell line derived from the acute infection with HIV-1. The HIV DNA (GenBank: K02013.1) has integrated into the genome of the T cell line. The cells were cultured in RPMI 1640 supplemented with 2 mm l-glutamine (Gibco Laboratories, Grand Island, NY), 100 U/ml penicillin, 100 g/mL streptomycin (Invitrogen, Carlsbad, CA), and 10% (v/v) heat-inactivated fetal calf serum (FCS, Hyclone Laboratories, Logan, UT). The cells were pelleted and washed in serum-free medium for five times and resuspended in serum free medium to give a concentration of 1 × 106 cells/ml prior to PMA induction for 72 h. After the incubation, cells were pelleted by low speed centrifugation and medium was harvested and filtered through a 0.22 μm filter unit, and the virus was concentrated by centrifugation through a 20% sucrose cushion at 100 000g for 2 h at 4 °C. The resultant pellets containing HIV virions were resuspended in PBS buffer containing 1% NP40. The supernatant containing native gp120 (GenBank: AAB59751.1) was separated from insoluble particles by centrifugation at 16 100g for 1 h at 4 °C. The sample was then concentrated to 20 μL in 0.4 M ammonium bicarbonate buffer through an Amicon Ultra-0.5 mL-100 kDa filter unit.
Filter-Aided Intact Peptide Preparation
Recombinant and virion-derived gp120 (5 μg) were added to 8 M urea in 0.4 M ammonium bicarbonate buffer and reduced with 50 mM TCEP at room temperature (RT) for 1 h. Proteins were alkylated by iodoacetamide at a final concentration of 10 mM and incubated at RT in the dark with shaking for 30 min. Samples were applied to the Microcon-50 kDa centrifugal filter unit and centrifuged until the solution was minimal in the filter unit. The samples were washed five times with 0.4 M ammonium bicarbonate buffer, and 0.25 μg of trypsin was added in the buffer after the final wash. The digestion was incubated in 30 °C for overnight. The tryptic peptides were harvested by centrifugation for 30 min. The solution containing peptides and glycopeptides was acidified to pH < 3, desalted by C18 cartridge according to manufacturer’s instructions, dried in a speed-vac, and resuspended in 0.1% formic acid.
Deglycosylation of gp120 HxBc2
Glycopeptides of gp120 HxBc2 (2 μg) were dried in a speed-vac and resuspended in PBS buffer. The sample was then treated with 0.1 μg of PNGaseF for 24 h at 37 °C. The resultant deglycosylated peptides were acidified to pH < 3 and desalted by C18 cartridge. Finally deglycosylated peptides were dried in speed-vac and resuspended in 0.1% formic acid.
LC-MS/MS Analysis
The samples (1 μg) were separated through a Dionex Ultimate 3000 RSLC nano system (Thermo Scientific) with a 75 μm × 50 cm C18 PepMapRSLC separating column (Thermo Scientific) protected by a 5 mm guarding column (Thermo Scientific). Mobile phase flow rate was 0.35 μL/min with 0.1% formic acid and 2% acetonitrile in water (A) and 0.1% formic acid 95% acetonitrile (B). The gradient profile was set as following: 4–6% B for 9 min, 6–35% B for 83 min, 35–90% B for 5 min, 90% B for 10 min and equilibrated in 4% B for 12 min. MS analysis was performed using a Thermo Q Exactive mass spectrometer (Thermo Scientific). The spray voltage was set at 1.8 kV. Spectra (AGC target 1 × 106) were collected from 400 to 3000 m/z at a resolution of 140 K followed by data-dependent HCD MS/MS (at a resolution of 17 500, NCE 28%, intensity threshold of 4 × 104 and maximum IT 250 ms) of the 15 most abundant ions using an isolation window of 4 m/z. Charge-state screening was enabled to reject unassigned, singly, eight, and more than eight protonated ions. A dynamic exclusion time of 25 s was used to discriminate against previously selected ions.
Peptide Identification
HIV gp120 peptides were identified by using Proteome Discoverer software (Thermo Fisher Scientific, version 1.4). The database used was recombinant gp120 HxBc2 strain or virion gp120 proteins from LAV strain sequences in a NCBI RefSeq database (downloaded from the NCBI Web site on August, 19, 2013, 53 738 entries). The precursor mass tolerance was set at 20 ppm and the MS/MS tolerance at 0.06 Da. Parameters of the search were modified as follows: oxidized methionines (add Met with 15.995 Da) and a (PNGase F-catalyzed) conversion of Asn to Asp (add Asn with 0.984 Da) set as dynamic modifications and Cys modification (add cysteine with 57 Da) set as a fixed modification. A maximum of two missed tryptic cleavage sites were allowed. MaxQuant version 1.3.0.5 with default setting was also used for database search with default settings.34,35
Analysis of Intact Glycopeptide MS Data
An in-house software tool (manuscript in preparation) was developed to identify the glycopeptides from MS and MS/MS data. The MS raw files were converted to mzXML files by Trans-Proteomic Pipeline (TPP). MatLab version R2013b (MathWorks, Inc.) was then used to extract information from mzXML files and code for the search strategy. The database was gp120 tryptic peptides containing the site of glycosylation. The search strategy is defined in the following steps: (1) identification of deglycosylated peptides—the sequences and glycosylation sites were identified from MS/MS analysis of PNGase F treated tryptic peptide samples using Proteome Discoverer (Thermo Fisher Scientific, version 1.4) of SEQUEST search; (2) detecting the b- and y-ions of glycopeptides—sequences of deglycopeptide were used to pick the b- and y-ions in charge states 1, 2, and 3 within a 20 ppm tolerance; (3) extraction of MS/MS spectra containing glycopeptides—MS/MS spectra containing oxonium ions from Hex, HexNAc, HexHexNAc, and NeuAc at m/z 163, 204, 366, 274, 292 were extracted as putative glycopeptide spectra; (4) assigning glycopeptides—the putative glycopeptide spectra were used to search for b- and y-ions detected from deglycosylated peptides. The ratio of glycopeptide b- and y-ions versus those detected from deglycosylated peptide was calculated. To use the data for each glycopeptide backbone as putative glycopeptides, the ratio above 20% for y+ ion was kept; (5) evaluate the match of glycopeptides—to evaluate glycopeptides matched to deglycosylated peptide sequences containing specific glycosylation sites, the MS1 scan number, MS2 scan number, precursor m/z, precursor charge states, precursor mass, ratio and presence of b- and y-ions between glycopeptides and deglycosylated peptides, other peptide b- and y-ions detected in spectra of glycopeptides but not in deglycosylated peptides, Δglycan mass, potential glycan composition consisting HexNAc, Hexose, Fucose, and NeuAc, Y-ions in charge states 1, 2, and 3 were extracted and computed in the program. These were considered to shortlist the putative spectra for glycopeptides. The best-matched spectrum for individual glycopeptide in different peptide backbones was manually confirmed and used as spectral template to cross-match the putative glycopeptide spectra using b-, y-, peptide, Y-ions, and software Xcalibur version 2.2 (ThermoFisher Scientific). Platelet glycopeptides were enriched by hydrophilic interaction liquid chromatography (HILIC) and analyzed by the same algorithm to evaluate the false discovery rate (FDR). Glycan stuctures reported previously34,35 and ExPASy tools GlycoMod, PeptideMass, and GlycanMass were used to identify glycan compositions and glycan types.36,37 MS analysis infers the glycan compositions rather than actual glycan structures, hence the glycan structures reported here were the representatives of the glycans with the same composition. Xcalibur was used to inspect the correct monoisotopic peak for glycopeptides on the MS1 level.
Results and Discussion
Strategy for Identification of Glycopeptides
We exploited the use of peptide b- and y-ions from deglycosylated peptides and matched them to HCD MS/MS spectra containing oxonium ions to identify glycopeptides. Next, the verified MS/MS spectra of glycopeptides were used to cross-match all the other glycopeptides with the identical peptide backbones but varying in glycoforms using b-, y-, and/or Y-ions. The strategy was developed on the basis of the observation that the presence of peptide b- and y-ions that do not contain a site of glycosylation were relatively consistent between deglycosylated peptides and their glycopeptides, and the MS/MS spectra of glycopeptides with the same peptide backbones but different glycan moieties were nearly identical. Moreover, Y-ions could be absent in the HCD MS/MS spectra of O-linked glycopeptides, suggesting that the use of peptide b- and y-ions was a critical consideration to identify glycopeptides in the HCD-MS. Our search strategy included the following steps: (1) deglycosylated peptides and glycosylation sites were identified from PNGaseF treated sample; (2) b- and y-ions of peptide backbone were determined experimentally using the deglycosylated peptide spectra; (3) MS/MS spectra containing oxonium ions were extracted from the raw file of the sample without PNGaseF treatment; (4) the b- and y-ions of the deglycosylated peptides were used to match and filter the MS/MS spectra of glycopeptides; (5) peptide and Y-ions in the MS/MS spectra of glycopeptides were used to further evaluate the identification; (6) the best-matched MS/MS spectra of glycopeptides were used to cross-match the other MS/MS spectra of glycopeptides with the same peptide backbones; (7) finally, the precursor m/z and the Δmass between the peptide moieties and glycopeptides were used to determine the glycoforms (Figure 1).
Figure 1.
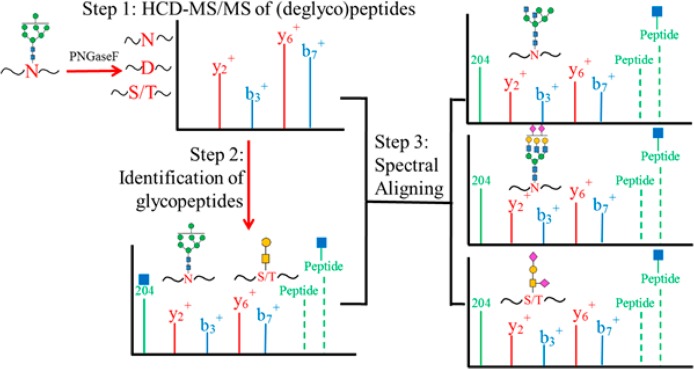
Schematic representation of the HCD spectral-aligning strategy for analysis of glycopeptides. Step 1, experimental b- and y-ions of deglycosylated peptide backbones were recorded from sample treated with PNGaseF. Step 2, these b- and y-ions of the deglycosylated peptides were used to screen the MS/MS spectra containing oxonium ions from the raw file for sample without PNGaseF treatment to identify putative glycopeptides. The putative glycopeptides were further evaluated by precursor ions, mass difference between glycopeptides and deglycosylated peptides, and Y-ions in the MS/MS spectra of the glycopeptides. Step 3, the identified MS/MS spectra of glycopeptides were used as spectral templates to identify other glycopeptides with the same peptide backbone but in different glycoforms.
Determination of Experimental Fragment Ions of Deglycosylated peptides for Identification of Glycopeptides
The strategy described above was applied to the analysis of a recombinant gp120 for identification of glycoforms in a site-specific manner. The protein has 24 known N-linked and one known O-linked glycosites (Figure S1).12 Among these glycosites, 11 N-linked glycosites distributed in the variable regions 1–5 (Figure S1) and the other 13 N-linked glycosites resided in the conserved regions 1–5 (C1–C5) (Figure S1). The single reported O-linked glycosite was located at amino acid position Thr499, which was at the end of the C5 region (Figure S1).12 MS/MS analysis of the gp120 protein with PNGaseF treatment identified 19 of the 24 known N-linked glycosites with 1% FDR. Among the five undetected N-glycosites, one glycosylation site, Asn230, is located in a short peptide backbone CNN230K, and four other N-glycosites, Asn366, Asn392, Asn397, and Asn406, are located in a long peptide. These five N-glycosites are identifiable when a suitable length of deglycosylated peptides is generated using alternative proteases.38 For the proof of principle study of our glycopeptide analysis strategy, we focused on the identification of N-glycopeptides from the 19 N-glycosites identified from deglycosylated peptides using trypsin digestion. The b- and y-ions of the 19 deglycosylated peptides in charge states 1, 2, and 3 were recorded from MS/MS spectra on the basis of their scan number and assigned peptide sequence. Notably, Asn instead of Asp (deamidated asparagine introduced by PNGaseF treatment) in the Asn-X-Ser/Thr sequon (X ≠ Pro) was used for the deglycosylated peptides. This resulted in recording the b- and y-ions from peptide termini to the amino acids prior to the formerly N-glycosylated Asp position, beyond which the b- and y-ions in the MS/MS spectra had an additional mass of 0.984 Da and were not recorded within the mass tolerance of 20 ppm. The rationale for this approach was that the b- and y-ions after the glycosite were less likely to appear in the MS/MS spectra of glycopeptides due to the effect of attached glycans, unless the glycans were cleaved off by HCD fragmentation at the preferred glycosidic bond which could occur in many cases.
Spectral Aligning of MS/MS Spectra of N-Linked Glycopeptides
Next, 12 304 putative glycopeptide spectra containing oxonium ions were extracted from MS/MS spectra from analyzing sample without PNGaseF treatment. The putative glycopeptide spectra were matched to the recorded b- and y-ions from the deglycosylated peptides to identify the peptide moieties of glycopeptides. The ratio of matched b- and y-ions between glycopeptides and deglycosylated peptides was calculated and filtered to generate a list of 9544 putative glycopeptide spectra, as described in Materials and Methods. Each putative spectrum had an average of six putative peptide matches and 921 putative spectra matched to one putative glycopeptide. The spectrum of glycopeptide which scored the highest ratio of match was verified by manual inspection through searching for additional b- and y-ions, intensity of oxonium ions, and appearance of peptide and Y-ions. These verified spectra of glycopeptides served as spectral templates for the subsequent identification of other glycopeptides harboring the same peptide backbone in the list of putative glycopeptide spectra.
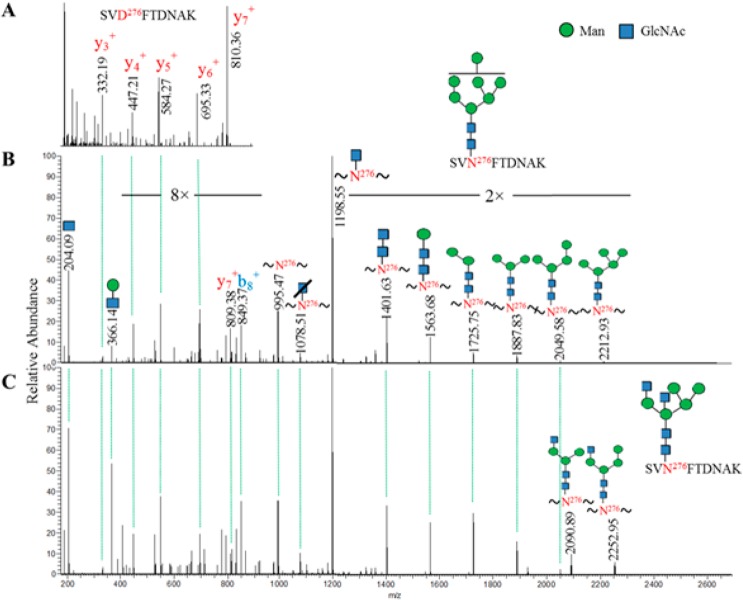
In the HCD MS/MS spectra of glycopeptides, because most glycan structures are fragmented to oxonium ions during MS/MS, the b-, y- and Y-ions appear to be nearly identical among glycopeptides with the same peptide backbone regardless of attached glycan structures. As illustrated, first the peptide b- and y-ions of deglycosylated peptide SVD276FTDNAK were used to identify the glycopeptide having the peptide backbone SVN276FTDNAK and glycan Man7GlcNAc2 (Figure 2A,B). Additional glycopeptide with the same peptide backbone but different glycan, Man5GlcNAc4, was then identified (Figure 2C). Specifically, in the comparison between the MS/MS of the two glycopeptides (Figure 2B,C), the three components, including (i) the oxonium ions at m/z 204, 366, (ii) y-ions at m/z 332 (y3+), 447 (y4+), 548 (y5+), 695 (y6+) from spectrum of deglycosylated peptide and extra b- and y-ions at m/z 849 (b8+), 809 (y7+) from spectrum of glycopeptide, and (iii) peptide ion at m/z 995 and Y-ions from peptide with cross-ring cleavage of GlcNAc at m/z 1079 to peptide with Man4GlcNAc2 at m/z 2049, were matched (Figure 2B,C). One of the differences in these two MS/MS spectra was the detection of peptide with Man5GlcNAc2 at m/z 2213 in the MS/MS spectrum of glycopeptide with Man7GlcNAc2, as well as the peptide with Man3GlcNAc3 at m/z 2091 and Man4GlcNAc3 at m/z 2252 in the MS/MS spectrum of glycopeptide with Man5GlcNAc4 (Figure 2B,C). These differential Y-ions in different MS/MS spectra of glycopeptides could, in part, facilitate the plausible assignment of the glycan composition. Nevertheless, HCD fragmentation of glycopeptides have been shown to improve the number and intensity of peptide b- and y-ions over those observed in CID glycopeptide fragmentation spectra, which shows glycan fragmentation predominantly.30 Therefore, we were able to use b-, y-, and Y-ions to cross-match glycopeptides as illustrated in Figure 2. Using b-, y-, peptide, and/or Y-ions in the verified MS/MS spectral template of glycopeptides to filter against the list of 9544 putative glycopeptide spectra, we obtained 2620 MS/MS spectra for putative glycopeptides. Finally, we manually verified a total number of 2119 MS/MS spectra of N-linked glycopeptides of gp120. We tried to estimate tentative false discovery rate (FDR) using a raw file analyzing platelet glycopeptides that had 18 057 oxonium-ion-containing spectra. Use the same b- and y-ions from gp120 deglycosylated peptides and filtering criteria, 81 spectra were matched to score a tentative FDR of 2.6%. Manual inspection showed that those 81 MS/MS spectra were not similar to spectral templates from recombinant gp120, suggesting that manual inspection was necessary to remove the false-positives. Detail of the tentative FDR estimation and discussion is described in Supporting Information.
Figure 2.
Representative of MS/MS spectra for identification of N-linked glycopeptides using HCD spectral-aligning strategy. (A) MS/MS spectrum of deglycosylated peptide SVD276FTDNAK provided the information on experimentally identified b- and y-ions. (B) MS/MS spectrum of glycopeptide with matched pattern of b- and y-ions to that of deglycosylated peptide was identified. Additional b-, y-, and Y-ions facilitate identification of the glycopeptide SVN276FTDNAK with Man7GlcNAc2, which was used as a spectral template to identify glycopeptides with the same peptide backbone but different glycan Man5GlcNAc4 in (C).
Spectral-Aligning Strategy for Identification of O-Linked Glycopeptides
O-linked glycopeptides contain carbohydrate(s) attaching to Ser or Thr, whose HCD MS/MS spectra could differ from that of its N-linked counterparts. To evaluate the applicability of our strategy for identification of O-linked glycopeptides, experimentally identified peptides IEPLGVAPT499K and IEPLGVAPT499KAK were used to generate theoretical b- and y-ions. Again filtering the presence of peptide b- and y-ions coupled with inspecting for peptide and Y-ions against 341 putative glycopeptide spectra containing at least four y+-ions, we identified O-linked glycopeptides with glycans, Gal1GalNAc1, and other O-linked glycopeptides, one of which was with glycan NeuAc1Gal1GlcNAc1GalNAc1 (Figure 3). In the MS/MS spectra of O-linked glycopeptides, the peptide b- and y-ions were likely to appear because, possibly, the O-linked glycans were easily cleaved off from the peptide moiety at the glycosidic bond resulting in intensive fragmentation of the peptide backbone (Figure 3). In addition, it was noticed that peak intensity of Y1 and other Y-ions was lower to b- and y-ions and could even be absent under the NCE we applied (Figure 3). These observations suggested that oxonium ion HexNAc at m/z 204 and peptide b- and y-ions were highly relevant for identification of O-linked glycopeptides, and the spectral-aligning strategy was applicable to O-linked glycopeptides. Finally, a total number of 105 MS/MS spectra were identified from O-linked glycopeptides bearing peptide backbones of either the IEPLGVAPT499KAK or the IEPLGVAPT499K. Using the same criteria, only one putative O-linked glycopeptide was matched in the platelet raw file scoring approximately 0.6% FDR, and this spectra was dissimilar to its verified glycopeptides from recombinant gp120.
Figure 3.
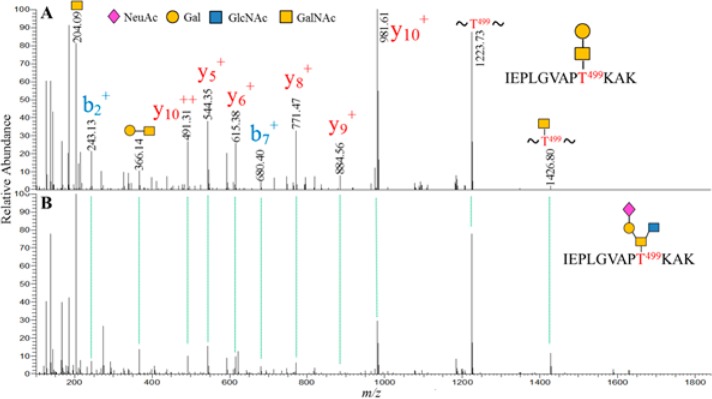
MS/MS spectra of O-linked glycopeptides demonstrating the applicability of spectral-aligning strategy for identification of O-linked glycopeptides. Peptide IEPLGVAPT499KAK in different glycoforms Gal1GalNAc1 in (A) and NeuAc1Gal1GlcNAc1GalNAc1 in (B) are aligned to show a similar pattern of peaks.
Determination of Glycoforms for Site-Specific Profiling of gp120 Glycosylation
After matching glycopeptide spectra to specific glycosylation sites, correct assignment of glycan composition for each glycopeptide was an important step toward site-specific profiling of protein glycosylation. Determination of the correct monoisotopic peak was requisite for glycan assignment but error-prone due to the relatively low abundance of glycopeptides, poor ionization efficiency, and even more prominent, very frequent, intensity of the true 12C monoisotopic peak which was considerably lower than that of 13C peaks (Figure S2). This phenomenon could be used as an additional criterion to select and verify the glycopeptides. However, this also resulted in selection of the 13C peak during data-dependent acquisition of MS/MS and reporting 13C peak as precursor m/z that added at least 1 Da ± 10 ppm to the monoisotopic precursor mass (Figure S2). Without monoisotopic mass correction, it could directly jeopardize the assignment of glycoforms. Indeed, from a total number of 2224 MS/MS spectra with oxomium ions and matched b- or y-ions for N- and O-linked glycopeptides of gp120, only 387 MS/MS spectra (17% of total) were able to match with glycan composition in mass tolerance of 20 ppm. We therefore used MaxQuant to circumvent this issue and correct the precursor m/z for monoisotopic peaks. This step dramatically improved the correct assignment of glycoforms that 1268 MS/MS spectra (57% of total) were assigned within mass tolerance of 10 ppm. These 1268 spectra were corresponding to 460 unique glycopeptides (File S1). These identified spectra were manually verified. Noticeably, glycopeptides with more than one N-linked glycosylation sites could also be assigned to the most likely combined glycoform. For instance, TFN289GTGPCTN295VSTVQCTHGIRPVVSTQLLLN301GSLAEEEVVIR with likely glycan composition Hex17HexNAc4 was observed with five spectral counts and in mass tolerance of 1 ppm (File S1). This was a unique feature for our search strategy that no glycan database was required. Therefore, we were able to identify multiple glycans on the glycopeptides containing sites of glycosylation.
N- and O-Linked Glycopeptides of gp120 from Virion
It is formidable to identify very low abundant glycopeptides in a complex sample. This difficulty was indeed the case for the Env protein gp120 from virions. Here, we employed a Q-Exactive MS instrument, which possesses a high tandem-mass-spectral acquisition speed for an optimized HCD to detect gp120 glycopeptides from T-cell-derived virion. Purification of native gp120 glycopeptides was described in Materials and Methods. MS and MaxQuant identified both gag and gp120 proteins with an ID confidence ≥99% and coverage of 44% and 17%, respectively. The gp120 from ACH-2 cell line was from the LAV strain, which has a 96% identity (amino acid 34–518) with HxBc2 strain determined by NCBI protein–protein BLAST (Figure S3). Using our strategy, five N-linked glycosites were identified from five glycopeptide backbones, that is, GEIKN156CSFN160ISTSIR, SAN276FTDNAK, AKWN339ATLK, WN339ATLK, and CSSSN448ITGLLLTR (Table 1). Of note, the only O-linked glycosite in peptide backbone IEPLGVAPT499K was detected (Table 1 and Figure S4). The ID confidence for native glycopeptides sharing the same peptide backbone with recombinant gp120, first heavily relied on the peptide b- and y-ions, was further supported by matching the MS/MS spectra to that assignment from recombinant gp120 glycopeptides and peptides (Figure S4). The glycoform for the N-linked glycopeptides was assigned to be predominant in high mannose in all four N-linked glycosites and Gal1GalNAc1 for the O-linked glycosite Thr499 (Table 1 and Figure S4). This observation strongly supported previous studies showing relatively abundant oligomannose and presence of O-linked glycosylation in Env protein gp120 from peripheral blood mononuclear cells (PBMC) produced virion.11,12,39 Additional glycoforms of these glycopeptides were investigated at the MS1 level, because glycopeptides with identical peptide backbone but different glycoforms were thought to elute within a small retention time window when C18 reversed-phase liquid chromatography (RPLC) was used. The use of Xcalibur and manual inspection did not seemingly reveal other glycoforms within mass tolerance of 20 ppm. Collectively, we observed that Asn156 and 160 in the V1 V2 region harbored the high mannose structure Man13–15GlcNAc4; Asn276 in C2 region possessed high mannose structure Man6–7GlcNAc2, hybrid/complex structure Hex8HexNAc5, and Hex5HexNAc3. In addition, Asn339 in C3 region had high mannose structures Man8–9GlcNAc2, and Asn448 in C4 region had only high mannose structure Man9GlcNAc2 (Table 1). The Gal1GalNAc1 appeared to be the only glycoform detected for the O-linked glycosite (Table 1). This observation of N-linked glycopeptides from virion suggested that glycans at Asn276 were likely to be hybrid/complex glycans. Broadly neutralizing antibodies (bnAbs) and structural studies has indicated the presence of hybrid/complex glycan at Asn156 and Man5GlcNAc2 glycan at Asn160 for recognition by bnAbs PG9 and PG16 but direct detection of those glycans on native gp120 lacked.9,40−42 Our data directly support that glycans could be processed on native gp120 at these two sites to have the possible glycan structures for bnAbs, but other glycans most likely existed to cause possible reduction in optimal neutralizing potency and complete neutralization. Glycans at Asn276 have been described as providing a protective shield for CD4 bind site (CD4bs) and were targeted by bnAbs HJ16 and 8ANC195.43 The preferential glycan structure for these two bnAbs has not been reported. In our data, the glycans at Asn276 could be relatively more flexible, and bnAbs might need special features to gain adequate affinity to different glycans at this glycosite. Glycans at Asn339 and Asn448 are targeted but not essential by 2G12, which recognizes Manα1–2Man structure at the tip of glycans.44,45 Here, it appeared that glycans at these two sites were mainly Man8–9GlcNAc2, indicating their resistance to glycosidase processing and explaining their supportive role for effective binding by 2G12.39 Moreover, Asn448 has been reported to be critical for efficient MHC class II-restricted presentation of CD4 T cell epitopes and infectivity.46,47 In this regard, the exclusive Man9GlcNAc2 seen at Asn448 implied that the glycan and glycosite might be essential to maintain certain structural feature for infectivity. Overall, it appeared that the glycopeptides from native gp120 presented in extremely low abundance in our sample preparation. This low number precluded the possibility to detect glycopeptides harboring other sites or low abundant glycoforms on identified glycosites. An improved enrichment step for virion gp120 and its glycopeptides would foreseeably improve the detection of low abundant glycoforms.
Table 1. gp120 Glycopeptides Detected from T-Cell-Produced Viriona.
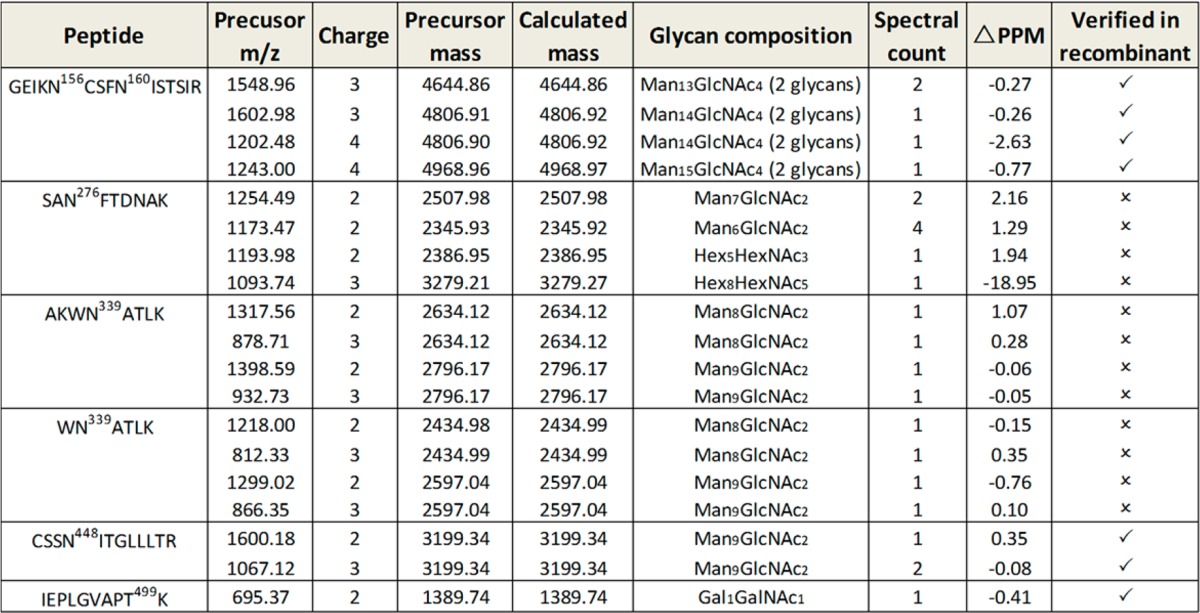
Verified in recombinant indicates that the spectra were compared to that from recombinant gp120 HxBc2 to exclude a false-positive ID (see also Figure S4).
Conclusion
We studied the HCD MS/MS spectra of N- and O-linked glycopeptides and revealed spectral features that can be applied into a data-mining strategy. The central discovery of the features is the “copy–paste” spectral pattern of glycopeptides with identical peptide backbone but different glycoforms. We also re-evaluated the use of peptide b- and y-ions in identification of O-linked glycopeptides in an HCD-MS/MS experiment. The result showed that although the intensity of peptide b- and y-ions was lower than that of Y1-ions for N-linked glycopeptides, they could be higher and became more relevant in identification of O-linked glycopeptides. The b- and y-ions could be obtained by optimizing the collisional energy.48 Using these spectral features, a novel spectral-aligning strategy to search HCD-MS data of both N- and O-linked glycopeptides has been developed. This data-mining strategy used MS2–MS1, which differed from a conventional MS1–MS2 search strategy. In this way, glycopeptides with multiple glycans could be detected for downstream verification using ETD-MS for example.
By applying spectral-aligning strategy, site-specific profile of recombinant gp120 glycosylation was achieved. More importantly, for the first time, we have demonstrated the feasibility to detect both N- and O-linked glycopeptides from virion gp120. Consistent with previous studies, oligomannose structures were present in all N-linked glycosites.39 High mannose glycans at Asn156 and 160, Asn339 and Asn448 might be more resistant to glycosidase processing than that at Asn276, supporting their roles in immune-evasion and infection of HIV. Overall low abundant glycopeptides of gp120 limited our capability to extract more information for site-specific assignment of virion gp120. Our data has primed the technical path to profile glycosylation of HIV native Env that would favor forthcoming studies of HIV glycobiology and rational vaccine design.
Acknowledgments
This research was supported by a National Institutes of Health–National Heart Lung and Blood Institute grant (Program of Excellence in Glycosciences, PEG, P01HL107153) and a National Institutes of Health–National Institute of Allergy and Infectious Diseases grant for the JHU-Guangxi, China Clinical Trials Unit (1U01 AI69482-01) to J.B.J. We thank Dr. Xiao-Fang Yu, who provided the biosafety level-3 lab at the Johns Hopkins Bloomberg School of Public Health.
Supporting Information Available
Additional information as noted in the text. This information is available free of charge via the Internet at http://pubs.acs.org/.
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Patrie S. M.; Roth M. J.; Kohler J. J. Methods Mol. Biol. 2013, 951, 1–17. [DOI] [PubMed] [Google Scholar]
- Scanlan C. N.; Offer J.; Zitzmann N.; Dwek R. A. Nature 2007, 446, 1038–1045. [DOI] [PubMed] [Google Scholar]
- Apweiler R.; Hermjakob H.; Sharon N. Biochim. Biophys. Acta 1999, 1473, 4–8. [DOI] [PubMed] [Google Scholar]
- Sun S.; Wang Q.; Zhao F.; Chen W.; Li Z. PLoS One 2011, 6, e22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S.; Wang Q.; Zhao F.; Chen W.; Li Z. PLoS One 2012, 7, e32119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strum J. S.; Nwosu C. C.; Hua S.; Kronewitter S. R.; Seipert R. R.; Bachelor R. J.; An H. J.; Lebrilla C. B. Anal. Chem. 2013, 85, 5666–5675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoury G. A.; Baliban R. C.; Floudas C. A. Sci. Rep. 2011, 10.1038/srep00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y.; Zhou Y.; Elliott S.; Aebersold R.; Zhang H. Nat. Protoc. 2007, 2, 334–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin M. N.; McLellan J. S.; Huang W.; Orwenyo J.; Burton D. R.; Koff W. C.; Kwong P. D.; Wang L. X. Nat. Chem. Biol. 2013, 9, 521–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crispin M.; Bowden T. A. Nat. Struct. Mol. Biol. 2013, 20, 771–772. [DOI] [PubMed] [Google Scholar]
- Bernstein H. B.; Tucker S. P.; Hunter E.; Schutzbach J. S.; Compans R. W. J. Virol 1994, 68, 463–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Go E. P.; Liao H. X.; Alam S. M.; Hua D.; Haynes B. F.; Desaire H. J. Proteome Res. 2013, 12, 1223–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B.; Gaschen B.; Yusim K.; Thakallapally R.; Kesmir C.; Detours V. Br. Med. Bull. 2001, 58, 19–42. [DOI] [PubMed] [Google Scholar]
- Go E. P.; Hewawasam G.; Liao H. X.; Chen H.; Ping L. H.; Anderson J. A.; Hua D. C.; Haynes B. F.; Desaire H. J. Virol 2011, 85, 8270–8284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muranyi W.; Malkusch S.; Muller B.; Heilemann M.; Krausslich H. G. PLoS Pathog. 2013, 9, e1003198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu P.; Chertova E.; Bess J. Jr.; Lifson J. D.; Arthur L. O.; Liu J.; Taylor K. A.; Roux K. H. Proc. Natl. Acad. Sci. U.S.A. 2003, 100, 15812–15817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chertova E.; Bess J. W. Jr.; Crise B. J.; Sowder I. R.; Schaden T. M.; Hilburn J. M.; Hoxie J. A.; Benveniste R. E.; Lifson J. D.; Henderson L. E.; Arthur L. O. J. Virol. 2002, 76, 5315–5325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alley W. R. Jr.; Mann B. F.; Novotny M. V. Chem. Rev. 2013, 113, 2668–2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavenhagen K.; Hinneburg H.; Thaysen-Andersen M.; Hartmann L.; Varon Silva D.; Fuchser J.; Kaspar S.; Rapp E.; Seeberger P. H.; Kolarich D. J. Mass Spectrom. 2013, 48, i. [DOI] [PubMed] [Google Scholar]
- Hart-Smith G.; Raftery M. J. J. Am. Soc. Mass Spectrom. 2011, 23, 124–140. [DOI] [PubMed] [Google Scholar]
- Zhu Z.; Su X.; Clark D. F.; Go E. P.; Desaire H. Anal. Chem. 2013, 85, 8403–8411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh C.; Zampronio C. G.; Creese A. J.; Cooper H. J. J. Proteome Res. 2012, 11, 4517–4525. [DOI] [PubMed] [Google Scholar]
- Pompach P.; Brnakova Z.; Sanda M.; Wu J.; Edwards N.; Goldman R. Mol. Cell. Proteomics 2013, 12, 1281–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott N. E.; Parker B. L.; Connolly A. M.; Paulech J.; Edwards A. V.; Crossett B.; Falconer L.; Kolarich D.; Djordjevic S. P.; Hojrup P.; Packer N. H.; Larsen M. R.; Cordwell S. J. Mol. Cell. Proteomics 2011, 10.1074/mcp.M000031-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alley W. R. Jr.; Mechref Y.; Novotny M. V. Rapid Commun. Mass Spectrom. 2009, 23, 161–170. [DOI] [PubMed] [Google Scholar]
- Segu Z. M.; Mechref Y. Rapid Commun. Mass Spectrom. 2010, 24, 1217–1225. [DOI] [PubMed] [Google Scholar]
- Nanni P.; Panse C.; Gehrig P.; Mueller S.; Grossmann J.; Schlapbach R. Proteomics 2013, 13, 2251–2255. [DOI] [PubMed] [Google Scholar]
- Michalski A.; Neuhauser N.; Cox J.; Mann M. J. Proteome Res. 2012, 11, 5479–5491. [DOI] [PubMed] [Google Scholar]
- Zhang H.; Li X. J.; Martin D. B.; Aebersold R. Nature Biotechnol. 2003, 21, 660–666. [DOI] [PubMed] [Google Scholar]
- Parker B. L.; Thaysen-Andersen M.; Solis N.; Scott N. E.; Larsen M. R.; Graham M. E.; Packer N. H.; Cordwell S. J. J. Proteome Res. 2013, 12, 5791–5800. [DOI] [PubMed] [Google Scholar]
- Buttke T. M.; Folks T. M. J. Biol. Chem. 1992, 267, 8819–8826. [PubMed] [Google Scholar]
- Clouse K. A.; Powell D.; Washington I.; Poli G.; Strebel K.; Farrar W.; Barstad P.; Kovacs J.; Fauci A. S.; Folks T. M. J. Immunol. 1989, 142, 431–438. [PubMed] [Google Scholar]
- Folks T. M.; Clouse K. A.; Justement J.; Rabson A.; Duh E.; Kehrl J. H.; Fauci A. S. Proc. Natl. Acad. Sci. U.S.A. 1989, 86, 2365–2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S.; Yuan W.; Yang W.; Zhou J.; Harlan R.; Edwards J.; Li S.; Zhang H. Anal. Chem. 2013, 85, 8188–8195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S.; Toghi Eshghi S.; Chiu H.; DeVoe D. L.; Zhang H. Anal. Chem. 2013, 85, 10117–10125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appel R. D.; Bairoch A.; Hochstrasser D. F. Trends Biochem. Sci. 1994, 19, 258–260. [DOI] [PubMed] [Google Scholar]
- Cooper C. A.; Gasteiger E.; Packer N. H. Proteomics 2001, 1, 340–349. [DOI] [PubMed] [Google Scholar]
- Chen R.; Jiang X.; Sun D.; Han G.; Wang F.; Ye M.; Wang L.; Zou H. J. Proteome Res. 2009, 8, 651–661. [DOI] [PubMed] [Google Scholar]
- Doores K. J.; Bonomelli C.; Harvey D. J.; Vasiljevic S.; Dwek R. A.; Burton D. R.; Crispin M.; Scanlan C. N. Proc. Natl. Acad. Sci. U.S.A. 2010, 107, 13800–13805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien J. P.; Cupo A.; Sok D.; Stanfield R. L.; Lyumkis D.; Deller M. C.; Klasse P. J.; Burton D. R.; Sanders R. W.; Moore J. P.; Ward A. B.; Wilson I. A. Science 2013, 342, 1477–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancera M.; Shahzad-Ul-Hussan S.; Doria-Rose N. A.; McLellan J. S.; Bailer R. T.; Dai K.; Loesgen S.; Louder M. K.; Staupe R. P.; Yang Y.; Zhang B.; Parks R.; Eudailey J.; Lloyd K. E.; Blinn J.; Alam S. M.; Haynes B. F.; Amin M. N.; Wang L. X.; Burton D. R.; Koff W. C.; Nabel G. J.; Mascola J. R.; Bewley C. A.; Kwong P. D. Nat. Struct Mol. Biol. 2013, 20, 804–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLellan J. S.; Pancera M.; Carrico C.; Gorman J.; Julien J. P.; Khayat R.; Louder R.; Pejchal R.; Sastry M.; Dai K.; O’Dell S.; Patel N.; Shahzad-ul-Hussan S.; Yang Y.; Zhang B.; Zhou T.; Zhu J.; Boyington J. C.; Chuang G. Y.; Diwanji D.; Georgiev I.; Kwon Y. D.; Lee D.; Louder M. K.; Moquin S.; Schmidt S. D.; Yang Z. Y.; Bonsignori M.; Crump J. A.; Kapiga S. H.; Sam N. E.; Haynes B. F.; Burton D. R.; Koff W. C.; Walker L. M.; Phogat S.; Wyatt R.; Orwenyo J.; Wang L. X.; Arthos J.; Bewley C. A.; Mascola J. R.; Nabel G. J.; Schief W. R.; Ward A. B.; Wilson I. A.; Kwong P. D. Nature 2011, 480, 336–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wibmer C. K.; Bhiman J. N.; Gray E. S.; Tumba N.; Abdool Karim S. S.; Williamson C.; Morris L.; Moore P. L. PLoS Pathog. 2013, 9, e1003738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scanlan C. N.; Pantophlet R.; Wormald M. R.; Ollmann Saphire E.; Stanfield R.; Wilson I. A.; Katinger H.; Dwek R. A.; Rudd P. M.; Burton D. R. J. Virol 2002, 76, 7306–7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duenas-Decamp M. J.; Clapham P. R. J. Virol 2010, 84, 9608–9612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X.; Jin W.; Hu K.; Luo S.; Du T.; Griffin G. E.; Shattock R. J.; Hu Q. Virology 2012, 423, 97–106. [DOI] [PubMed] [Google Scholar]
- Li H.; Xu C. F.; Blais S.; Wan Q.; Zhang H. T.; Landry S. J.; Hioe C. E. J. Immunol. 2009, 182, 6369–6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L.; Tolic N.; Qu Y.; Meng D.; Zhao R.; Zhang Q.; Moore R. J.; Zink E. M.; Lipton M. S.; Pasa-Tolic L.; Wu S. Anal. Biochem. 2014, 452, 96–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.