Abstract
Spirochetes in the genus Borrelia carry a linear chromosome and numerous linear plasmids that have covalently closed hairpin telomeres. The overall organization of the large chromosome of Borrelia burgdorferi appears to have been quite stable over recent evolutionary time; however, a large fraction of natural isolates carry differing lengths of DNA that extend the right end of the chromosome between about 7 and 20 kbp relative to the shortest chromosomes. We present evidence here that a rather recent nonhomologous recombination event in the B. burgdorferi strain Sh-2-82 lineage has replaced its right chromosomal telomere with a large portion of the linear plasmid lp21, which is present in the strain B31 lineage. At least two successive rounds of addition of linear plasmid genetic material to the chromosomal right end appear to have occurred at the Sh-2-82 right telomere, suggesting that this is an evolutionary mechanism by which plasmid genetic material can become part of the chromosome. The unusual nonhomologous nature of this rearrangement suggests that, barring horizontal transfer, it can be used as a unique genetic marker for this lineage of B. burgdorferi chromosomes.
The Lyme disease spirochetes, Borrelia burgdorferi, Borrelia afzelii, and Borrelia garinii, and most likely all members of the Borrelia genus, have an unusual genome that is made up of an approximately 900-kbp linear chromosome and numerous smaller linear and circular extrachromosomal DNA elements. The nucleotide sequence of the genome from one B. burgdorferi isolate, B31 MI, has been determined (8, 14). Analysis of this strain has shown that the chromosome carries nearly all of its housekeeping genes. Its 21 extrachromosomal elements include 12 linear plasmids 5 to 54 kbp in length and nine circular plasmids 9 to 32 kbp in length that carry mostly genes of unknown function that are unique to the genus Borrelia (8). Examination of numerous independent natural B. burgdorferi isolates has shown that sequences similar to the plasmids in isolate B31 are usually found on plasmids of similar size in other isolates (1, 9, 10, 13, 15, 17-19, 21, 23-25, 27, 31, 32, 34), and a substantial fraction of these plasmids appear to be present in all isolates examined to date (16, 23).
Physical maps of the chromosomes from 25 geographically diverse Borrelia isolates that either cause Lyme disease or are close relatives of ones that do have been constructed (5, 11, 22; R. van Vugt and S. Casjens, unpublished data). These maps are all extremely similar, indicating that the chromosomes carry little gross structural variation across the more than 10 species that make up this cluster of species. This lack of organizational variation in the chromosome is not universal in bacteria (3): for example, the genomes of several gram-negative and -positive bacteria have been found to vary up to tens of percentages in gene content and/or have substantial rearrangements among different isolates (12, 20, 33). On the other hand, there appear to have been numerous and substantial recent rearrangements in the linear plasmid portion of the B. burgdorferi B31 MI genome. Analysis of this genome sequence has shown that 10 of the 12 B31 linear plasmids carry a large amount of DNA that appears to be in a state of mutational decay and not to encode functional proteins. This most likely is a result of the numerous recent duplicative DNA rearrangements (8), since such duplications may release one of the copies from selection, allowing it to mutationally decay. This decay is now observed as a large number of “pseudogenes” that contain many translational frame-disrupting mutations.
Although the bulk of the chromosome appears to be very stable, we have previously noted that in B. burgdorferi sensu stricto the extreme right end of the chromosome (as defined by Fraser et al. [14]) is variable in length, in that additional sequences extend the right telomeric region in some isolates. BB0843 is the rightmost gene in the 903-kbp “constant portion” of the chromosome; it is only a few hundred base pairs from the right telomere in Borrelia isolates with the minimum-size chromosome, such as N40, R-IP3, WI91-23, and HB19 (7, 14). The sequenced chromosome of strain B31 MI has 7.2 kbp of DNA beyond BB0843 at its right telomere. This DNA is almost entirely made up of sequences that have paralogs on the B31 MI plasmids and, except for two apparently intact genes, appears to be largely in a state of severe mutational decay (8). Different B. burgdorferi isolates carry different lengths of DNA to the right of gene BB0843; in 31 B. burgdorferi isolates that we have examined, 21 carry such extensions (7). None have right-end extensions longer than the 19-kbp extension of Sh-2-82. In this study we examine the nature of the right-end chromosomal extension in B. burgdorferi isolate Sh-2-82.
MATERIALS AND METHODS
Bacterial strains.
B. burgdorferi strain Sh-2-82 was isolated from an Ixodes scapularis tick on Shelter Island, N.Y. (26). Passage 6, passage 166, and passage 320 cultures of strain Sh-2-82 were the kind gifts of Tom Schwan, Patti Rosa, and Janis Weis, respectively. Strain 297 passage 5 was the kind gift of Justin Radolf. Strains JD1, 21305, 22921, 29968, 30757, and 28534 are described in the work of Casjens et al. (7); although we do not know exactly how each source laboratory performed these passages, each one typically represents six to nine generations. B. burgdorferi strains were propagated, and whole-cell DNA was prepared in agarose blocks as previously described (6). Contour-clamped homogeneous electric field (CHEF) electrophoresis, DNA transfer to nylon membranes, and Southern hybridizations were performed as previously described (6).
DNA manipulation and nucleotide sequence determination.
The nucleotide sequence of strain Sh-2-82 chromosomal right-end DNA was determined using a primer walking strategy and whole-genome DNA as template. Sequencing reactions with oligonucleotide primers 25 to 28 nucleotides long and Big Dye dideoxy terminator mix (PE Applied Biosystems, Foster City, Calif.) were performed in a thermal cycler as follows: 69 or 99 cycles at 95°C for 0.2 min, 1°C/s to 55°C, 55°C for 0.2 min, 1°C/s to 60°C, and 60°C for 4 min. Automated sequencers (PE Applied Biosystems) were used according to the manufacturer's recommendations. After nearly complete sequences of the two regions described in the text were determined, all ambiguities were resolved by sequencing PCR DNA fragments amplified from Sh-2-82 (passage 320) DNA. All of the sequence reported here was thus determined from both strands, except for the 63-bp repeat regions which could be approached only from one direction on whole-genomic DNA (see elsewhere).
Nucleotide sequence accession numbers.
These sequences have been deposited in GenBank with accession numbers AY309080 and AY309081 for the left and right unique regions, respectively.
RESULTS
Right chromosomal telomere of B. burgdorferi isolate Sh-2-82.
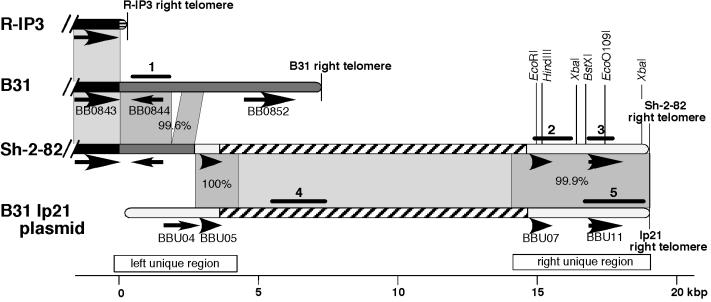
We found by Southern analysis that a DNA probe (probe 1; Table 1 and Fig. 1) from the left part of the 7.2-kbp B31 extension hybridizes with the “extra” DNA at the right telomere of the strain Sh-2-82 chromosome (data not shown), suggesting at least some similarity between the right telomeric regions of these two isolates. To characterize the long right telomeric extension of the Sh-2-82 chromosome in more detail, a primer walking strategy with whole-cell DNA as template was used to determine the nucleotide sequence rightward from the conserved BB0843 gene (see Materials and Methods). Several oligonucleotide sequences were chosen from within the conserved strain B31 gene BB0843 that primed a rightward sequencing reaction on Sh-2-82 template DNA. When a good-quality sequence was obtained with one of them, a second primer was chosen from within the sequence thus determined to sequence further to the right and so on. In this way, nucleotide sequence was determined for 4,173 bp of strain Sh-2-82 DNA rightward from within gene BB0843. The left 2,692 bp of this sequence are nearly identical to similarly located B31 sequence; Sh-2-82 has a 264-bp deletion (between bp 1879 and 1880) relative to B31, but it is otherwise 99.6% identical to the parallel region of the B31 chromosome (Fig. 1). This region in Sh-2-82 contains an apparently intact gene BB0844 homolog that should encode a protein of unknown function that is 99.4% identical to its B31 ortholog. The right 1,481 bp of these 4,173 bp have no similarity to the B31 chromosome, but the sequence is nearly identical to B31 linear plasmid lp21 (similarity extends rightward from bp 2438 on the published lp21 sequence [8]). Between bp 2693 and 3870 the Sh-2-82 sequence is identical to that of the B31 lp21 plasmid. Immediately to the right of bp 3870 the sequence contains about 5.5 tandem copies (there are many more repeats beyond these copies; see below) of 63-bp direct repeats that are very similar to the B31 lp21 63-bp repeat tract which lies at an identical location there. This long repeat tract blocks further rightward sequence determination by this strategy because primers in this region would not have unique binding sites on Sh-2-82 DNA.
TABLE 1.
Southern DNA probes and oligonucleotides used in this study
| Probe or oligonucleotidea | Description (reference) or nucleotide sequence (5′ to 3′) |
|---|---|
| Probes | |
| 1 | Genomic B31 MI DNA clone pMM5 (7) |
| 2 | PCR amplified from Sh-2-82 DNA with primers A and B |
| 3 | PCR amplified from Sh-2-82 DNA with primers C and D |
| 4 | Genomic B31 MI DNA clone CZ32 (23) |
| 5 | Genomic B31 MI DNA clone DF29 (23) |
| Oligonucleotides | |
| A | TATGTGGAGGATATATATATGAGC |
| B | AATATTTTTTTATGTTGTCACCTCG |
| C | TAAAAAATCCCCAATTAATTTA |
| D | ATCTTTCGAGTCAAATTCAATAATAG |
| E | GTATTATAGATCTTGAGTTTGTTGGG |
| F | CAACTTGAACTGGTATTATAATATGG |
Probe locations are indicated in Fig. 1.
FIG. 1.
Sequence relationships among B. burgdorferi B31 MI linear plasmid lp21 and chromosomal right telomeric regions. Shading of the chromosomal and plasmid DNAs represented by rounded bars indicates the following: solid black, the constant portion of the chromosome (which extends from the left chromosomal end through gene BB0843); dark gray, the right-end extension on the B31 chromosome and very similar sequence on the Sh-2-82 chromosome; white, the unique (non-63-bp repeat) regions of B31 plasmid lp21 and the very similar sequence on the Sh-2-82 chromosome; hatched, the region of 63-bp tandem repeats on lp21 and similar sequence on the Sh-2-82 chromosome. Gray areas between DNAs highlight regions of similarity between adjacent DNAs; the darker gray (and percent values there) indicate similarities to sequence determined in this study. Black arrows indicate putative intact genes (pseudogenes are not shown). Numbered black bars show the locations of DNA probes used in this study (also Table 1). The Sh-2-82 nucleotide sequences determined here are indicated by open bars immediately above the kilobase pair scale, whose zero point is the right end of the constant portion of the chromosome.
Since the right portion of the above Sh-2-82 chromosomal sequence is nearly identical to lp21, we attempted to determine whether sequences similar to those on lp21 to the right of its 63-bp repeat tract (the lp21 “right unique region” [Fig. 1]) were also present near the Sh-2-82 right telomere. Opposing oligonucleotides that amplify a 1.2-kbp section of the right unique region of B31 lp21 plasmid (oligonucleotides A and B; Table 1; Fig. 1) were used in a PCR and found to amplify identically sized fragments from whole-cell B31 and Sh-2-82 template DNAs. This Sh-2-82 templated PCR product, called probe 2 (Table 1), was then used in a Southern analysis of Sh-2-82 DNA to determine its location in that strain's genome. It hybridized only with the 920-kbp chromosome in whole Sh-2-82 DNA (Fig. 2) and only with the chromosome's rightmost BssHII and SgrAI fragments (37 and 34 kbp, respectively; data not shown, but see Fig. 4 below), showing that this amplicon lies within 34 kbp of the chromosomal right end. Restriction mapping of the rightmost chromosomal BglII, BsrGI, and NcoI fragments with the same DNA probe (data not shown), in combination with the sequences determined above and below, proved unequivocally that probe 2 hybridizes with lp21-like sequences that are present near the Sh-2-82 right telomere and distal to the 63-bp repeat tract.
FIG. 2.
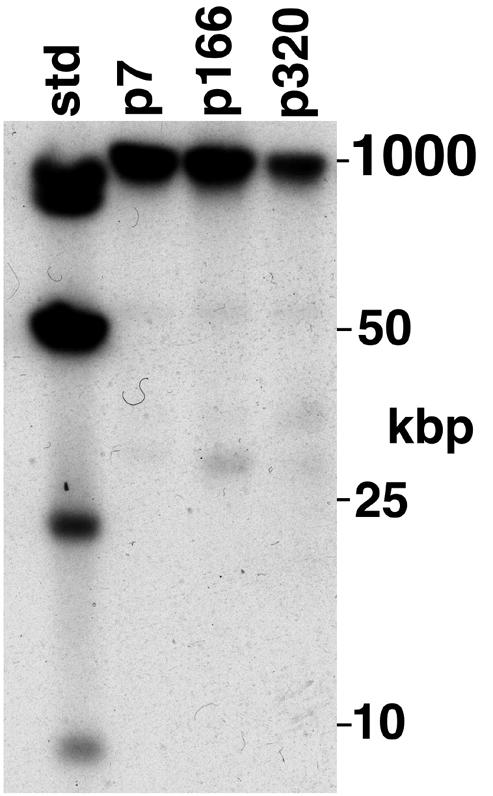
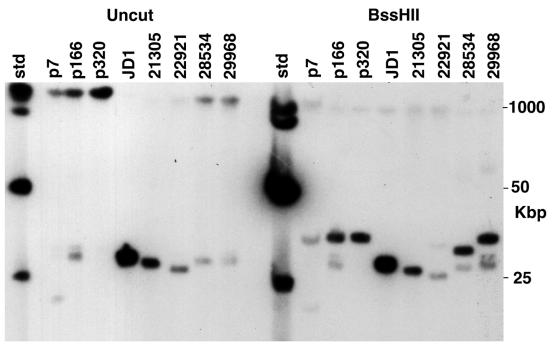
The B. burgdorferi Sh-2-82 chromosome contains strain B31 linear plasmid lp21-like sequences. Whole-genomic Sh-2-82 DNA was prepared in agarose blocks and subjected to CHEF electrophoresis, and Southern analysis was performed with probe 2 (Table 1 and Fig. 1) as described in Materials and Methods. Culture passage number is indicated above the lanes; “std” is HindIII cut plus a ladder of whole bacteriophage λ DNA.
FIG. 4.
Linear plasmid lp21-like sequences lie near the right end of some B. burgdorferi chromosomes. Bacterial DNA was prepared, restricted, separated by CHEF electrophoresis, and subjected to Southern analysis with probe 5 (Table 1 and Fig. 1) DNA from the right unique region of plasmid lp21. Strain names and restriction enzyme cleavage are indicated above the lanes (the p7, p166, and p320 lanes contain Sh-2-82 DNA from cultures with those passage numbers); “std” is HindIII cut plus a ladder of whole bacteriophage λ DNA.
Oligonucleotides A and B were therefore used to prime a second sequencing primer walk on whole-cell Sh-2-82 DNA. The resulting sequence contig had 63-bp repeats at its left end (Fig. 1), and as the walk neared the right telomere, the sequence quality deteriorated, presumably due to competition between primer annealing and snap-back of the telomeric hairpin (see below). A DNA probe derived by PCR amplification near the right end of this sequence contig (probe 3) also hybridized only with the rightmost Sh-2-82 chromosomal BssHII fragment in a Southern analysis (data not shown). In order (i) to determine the location of the right chromosomal telomere more precisely and (ii) to confirm that the sequence determined was actually at this location, probe 3 was used in Southern analyses of singly and doubly restricted whole-genome Sh-2-82 DNA to construct a restriction map of this region of the chromosome that unambiguously located six restriction enzyme cleavage sites (data not shown)—two XbaI sites and one EcoO109I, BstXI, HindIII, and EcoRI site each (Fig. 1). Each of these sites was correctly located in the primer walk sequence, except the rightmost XbaI site, which is very near the telomere and outside the primer walk region. Each of the latter four enzymes' right-end restriction fragments extended about 600 bp beyond the right end of the primer walk contig. Rapid reannealing (snap-back) experiments analogous to those done previously for DNA at the left end of the Sh-2-82 chromosome (7) showed that the two strands of the rightmost EcoO109I, BstXI, EcoRI, and HindIII fragments rapidly anneal (data not shown). We conclude that these fragments are in fact right-terminal chromosomal fragments and that these fragments, like other linear replicon terminal fragments in Borrelia, are tipped by a covalently closed DNA hairpin.
To determine the sequence of the tip of the Sh-2-82 right chromosomal telomere, we gel purified the size fraction 3.8 to 4.1 kbp of HindIII-restricted whole-cell Sh-2-82 DNA, which includes the rightmost HindIII fragment; nicked this DNA's terminal hairpin by S1 nuclease treatment; ligated the resulting DNA to a blunt-ended, double-stranded synthetic oligonucleotide; and used PCR to amplify between a primer that anneals within the primer walk sequence contig and a primer that anneals to the terminal, ligated synthetic sequence as previously described (7). The sequence of this PCR product was determined directly using both the amplification and internal primers, and in addition, the PCR product was cloned into the plasmid vector pCR4-TOPO (Invitrogen, Carlsbad, Calif.) and the sequences of the DNA inserts in several representative plasmids were determined. The resulting sequence overlapped the primer walk right unique region sequence in the manner expected and contains a correctly located telomere-proximal XbaI site (above) 220 bp from the right end; it extends the right unique sequence to the right by about 600 bp as predicted by the restriction map and contains at its extreme right terminus the 23-bp sequence 5′-TTTATACTAAAAAAAACTAATTT-3′, which is similar to the sequences at the tips of other known Borrelia telomeres (7, 14, 15, 35). We also used this methodology to determine the previously unknown telomeric sequence at the right end of strain B31 MI linear plasmid lp21. The reported sequence of plasmid lp21 (GenBank accession no. AE001582 [8]) was found to be lacking the rightmost terminal 25 bp of the plasmid; this 25-bp sequence, 5′-GCTTTATACTAAAAAAAACTAATTT-3′, is identical to the sequence that we determined for the right tip of the Sh-2-82 chromosome. One or a few nucleotides could be missing from the telomeres of these sequences due to possible removal by S1 nuclease. Merging of the right primer walk and the sequence of the terminal PCR amplicon resulted in 4,664 bp that contain the entire right unique region (Fig. 1). The leftmost 501 bp of this sequence contig is composed of approximately eight tandem copies of the same inexact 63-bp repeat present at the right end of the left unique region and is very similar to the parallel portion of lp21. The unique 4,163 bp to the right of bp 501 is 99.9% identical to the parallel region of lp21. In addition to three single nucleotide differences between Sh-2-82 and B31 in this region, there is an inversion of 5 bp (ACTTG centered on bp 2097). These 5 bp precisely separate a perfect 17-bp inverted repeat, which could have mediated the inversion. None of these differences disrupts the reading frame in which it occurs.
We also characterized in more detail the putative 63-bp repeat tract that lies between the two regions that were sequenced above. Southern analysis using a strain B31 probe from the 63-bp repeat region of lp21 (probe 4; Table 1 and Fig. 1) showed that the only Sh-2-82 sequences capable of hybridizing with this probe lie on the chromosome and within the 37-kbp rightmost BssHII chromosomal restriction fragment (Fig. 3). Restriction enzymes MseI (cuts at TTAA) and AseI (ATTAAT) cut the 72% A+T Borrelia DNA extremely frequently, the former giving rise to fragments that are nearly all less than 500 bp in length. There are no MseI or AseI sites in the B31 lp21 repeat tract, so in B31 they give rise to unusually large 63-bp-repeat-containing 11 ± 1.0-kbp and 13 ± 1.0-kbp DNA fragments, respectively (8). The Sh-2-82 passage 320 and B31 MI repeat tract-containing MseI and AseI fragments were indistinguishable in size (data not shown), suggesting that these enzymes also do not cut the Sh-2-82 repeat tract and that the tract length is about the same in B31 linear plasmid lp21 and the Sh-2-82 chromosome. The sequence determinations of the right and left unique regions combined with the length of the 63-bp repeat tract thus show that there is about 19 kbp of extra sequence at the Sh-2-82 right chromosomal telomere, which agrees well with our previous estimate from the length of right terminal restriction fragment sizes (7).
FIG. 3.
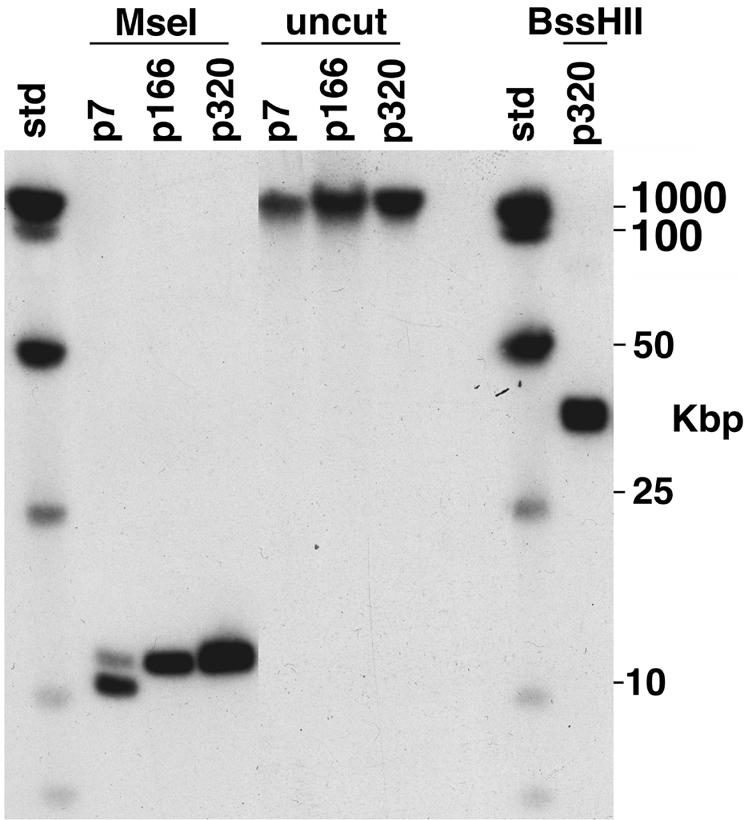
Determination of the length and location of the 63-bp repeat tract present in B. burgdorferi Sh-2-82 DNA. Whole-genomic Sh-2-82 DNA was prepared in agarose blocks, restricted, and subjected to CHEF electrophoresis. Southern analysis was performed with 63-bp repeat probe 4 (Table 1 and Fig. 1). Culture passage number and restriction enzymes used are indicated above the lanes; “std” is HindIII cut plus a ladder of whole bacteriophage λ DNA.
The experiments described above show that DNA probes derived from the right unique region, the 63-bp repeat tract of B31 lp21, and the Sh-2-82 right unique region all hybridize exclusively to the 37-kbp rightmost chromosomal BssHII fragment. Figures 2 and 3 show that this chromosomal hybridization is present at passages 7, 166, and 320 in culture (there are three to five generations per passage), although there is also an approximately 2-kbp-shorter form present (as the majority) in passage 7; it is not present by passage 166. Thus, although a number of linear plasmids have been lost with passage in culture (reference 26 and our unpublished analysis), we have no evidence of any substantial changes in the Sh-2-82 right telomeric region during this period; in particular we note that the length of the 11-kbp form of the chromosomal 63-bp repeat tract appears not to have changed significantly in about 1,500 generations (313 passages). The 13 63-bp repeats in the sequence reported here are all represented exactly among the 34 types of slightly different repeats present in B31 lp21, but the repeat types are not present in the same order, suggesting that gene conversion may have been active in this region since the two sequences diverged.
Other B. burgdorferi isolates with right telomeric chromosomal extensions.
We previously reported that several other B. burgdorferi isolates, 21305, 22921, 29968, and JD1, have approximately 19-kbp right-end chromosomal telomeric extensions (all quite similar in length to Sh-2-82) and 28534 has an approximately 16-kbp extension (7). In addition, strain 297 (28) has a right-end chromosomal extension similar in length to that of Sh-2-82 (J. Aron and S. Casjens, unpublished data). Each of these right-end extensions was found to hybridize to our probe 1 (Table 1). In order to determine whether the extensions in these strains might also contain lp21-like sequences, a Southern analysis was performed using probe 5 (Table 1) from the right unique region of B31 linear plasmid lp21 (this probe is 99.9% identical to, and hybridizes equally well with, the homologous Sh-2-82 right unique region). Figure 4 shows that 28534 and 29968 chromosomes do carry probe 5 sequences near their right telomeres, since the probe hybridizes to the right-end BssHII fragments in these strains (it also hybridizes to this fragment from strain 297; data not shown). MseI and AseI cleavage experiments (as in Fig. 3) estimated the length of the 63-bp repeat tract to be 9 kbp in 28534 and 11 kbp in 29968 (data not shown). Both of these strains also carry 24- to 25-kbp linear plasmids that hybridize (but not as strongly) with this probe (Table 2). Circular DNAs are not resolved into tight bands by the CHEF electrode pulse program used, so the bands observed almost certainly represent linear DNAs. Passage 7 and 166 Sh-2-82 also carry apparently linear plasmids that react very weakly to this probe but which are lost by passage 320 (Table 2). The apparent change in size of these plasmids with passage is difficult to assess since (i) less total DNA was present in the passage 7 lanes; (ii) only a subset of the cells might carry the larger hybridizing plasmids at passage 7, which could have expanded by passage 166; or (iii) there may have been DNA rearrangements among plasmids during growth in culture.
TABLE 2.
Southern hybridization with right-end DNA probes
| Culturec | Repliconb | Probea
|
||||
|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | ||
| Sh-2-82 (p7) | Chromosome | ++ | ++ | ++ | ++ | ++ |
| lp18 | − | − | (+) | − | (+) | |
| Sh-2-82 (p166) | Chromosome | ND | ++ | ++ | ++ | ++ |
| lp26 | − | − | − | (+) | ||
| lp28 | − | − | − | (+) | ||
| Sh-2-82 (p320) | Chromosome | ++ | ++ | ++ | ++ | ++ |
| Linear plasmids | − | − | − | − | − | |
| JD1 (p16) | Chromosome | ++ | − | ND | − | − |
| lp26 | − | ++ | ++ | ++ | ||
| 297 (p6) | Chromosome | ++d | ND | ND | ND | ++ |
| lp29 | (+) | ND | ND | ND | ND | |
| 21305 (p < 20) | Chromosome | ++ | − | ND | − | − |
| lp25 | − | ++ | ++ | ++ | ||
| lp27 | + | − | − | − | ||
| 22921 (p8) | Chromosome | ++ | − | ND | − | − |
| lp24 | − | ++ | ++ | ++ | ||
| lp27 | + | − | − | − | ||
| 28534 (p4) | Chromosome | ++ | ++ | ND | ++ | ++ |
| lp26 | + | (+) | + | + | ||
| 29968 (p < 20) | Chromosome | ++ | ++ | ND | ++ | ++ |
| lp23 | + | − | − | − | ||
| lp26 | − | (+) | + | + | ||
| 30757 (p5) | Chromosome | + | ND | ND | − | − |
| lp24 | − | + | + | |||
Probes are described in Table 1 and Fig. 1. Table symbols are as follows: −, no hybridization; +, moderate hybridization; ++, strong hybridization; ND, not done. Parentheses in the table data indicate very weak hybridization that most likely represents cross-hybridization to nonorthologous, paralogous sequences.
Hybridization with the chromosome includes the demonstration that this hybridization is with the rightmost BssHII and SgrAI restriction fragments. The numbers in the linear plasmid “lp” plasmid names indicate only the apparent size in kilobase pairs and do not indicate any specific relationship to plasmids in other strains with similar sizes or names.
The number of culture passages since the original isolation of the strain is given in parentheses. “p < 20” indicates that cultures were described by the supplier as “low passage.”
A DNA probe that overlaps probe 1 but is not identical to it was used in the case of strain 297 (see text).
Probe 5 does not react well with the chromosome in strains JD1, 21305, and 22921 and hybridizes much better with 23- to 27-kbp linear plasmids in these strains (Fig. 4). These strains also carry 63-bp repeat tracts on similar-size plasmids based on parallel Southern analyses with probe 4, and they carry 9-kbp, 9- and 13-kbp, and 9- and 11-kbp reactive MseI fragments, respectively (data not shown). In a panel of 13 additional isolates with shorter or no right-end extensions, only isolate 30757 (7-kbp right extension) was found to carry the 63-bp-repeat-hybridizing sequences, and these were on an approximately 24-kbp linear plasmid (23).
To test whether the 297, 28534, and 29968 chromosomal right-end extensions are the result of a recombination event that was identical to that of Sh-2-82, we PCR amplified (using oligonucleotides E and F, Table 1) and sequenced a 1,024-bp region that includes the Sh-2-82 recombination event from each strain (from 2515 through 3538 on the Sh-2-82 right unique region sequence). In all three cases the sequence of the amplified product was identical to the parallel Sh-2-82 sequence. Thus, since their recombination joints are identical, the right telomere replacement by lp21 in these three strains almost certainly happened only once in a common ancestor. It is interesting that Stevenson and Miller (30) recently found that Sh-2-82 and 297 also share extensive sequence identity on their cp26 and cp32 circular plasmids, supporting the notion that these are very closely related isolates. Isolates Sh-2-82, 28534, and 29968 are from ticks captured in New York, Maryland, and Connecticut, respectively, and 297 is a human isolate from Connecticut, indicating that geographic movement of the affected chromosome can happen before random mutagenic changes occur in what is thought to be nonfunctional DNA (e.g., in the gene BBU04 pseudogene homologous region). Thus, four of the seven known B. burgdorferi isolates with >15-kbp right-end extensions carry the same extensive homology to B31 linear plasmid lp21 at the right end of their chromosomes. Strains Sh-2-82, 297, 28534, and 29968 have lp21-like extensions at their right chromosomal telomeres, with the 63-bp repeat tract of 28534 being 2 kbp shorter than those of Sh-2-82 and 29968 (strain 297 was not tested). Since they carry probe 1- but not probe 4- or 5-hybridizing DNA near their right chromosomal telomeres, strains JD1, 21305, and 22921 appear to have some other DNA replacing and extending the tip of an ancestral B31-like chromosome; it seems likely that this DNA will be derived from some other Borrelia linear plasmid.
DISCUSSION
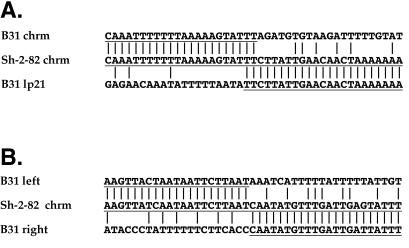
The structure of the right end of the Sh-2-82 chromosome is most easily explained by a simple, single recombination event between the right telomeric region of a B31-like chromosome and a B31 lp21-like linear plasmid, so that the rightmost 16 kbp of the plasmid replaced the distal 4 kbp of that chromosome (Fig. 1). In the Sh-2-82 chromosome right-end extension the proximal 2,692 bp are 99.6% identical to the B31 chromosome, and the distal lp21-like region unique (non-63-bp repeat) sequence is 99.9% identical to linear plasmid lp21. These extremely high similarities allow deduction of the nature of the recombination event in the Sh-2-82 progenitor. Figure 5 shows that there is an abrupt switch in Sh-2-82 from similarity to the B31 chromosome to similarity to the B31 lp21 linear plasmid. There are only 2 bp of sequence identity at the point where this recombination must have taken place, making this an essentially nonhomologous event. There are three additional locations in B. burgdorferi linear replicon sequences in which a similar deduction can be made regarding past rearrangements: (i) the 265-bp deletion in the non-lp21-like Sh-2-82 rightward extension compared to the homologous B31 sequence (Fig. 1 and 5); (ii) linear plasmid lp56 in B31, which appears to have been generated by the integration of a 31-kbp circle (homologous to the cp32 plasmids) into an approximately 24-kbp linear plasmid (8); and (iii) a 900-bp inversion near the left end of B31 lp56 relative to paralogous sequences on B31 plasmids lp28-4 and lp36 (8). In these cases there are 0, 2, and 2 bp of identity (the latter occurred within a 20-bp region of complex imperfect similarity), respectively, at the points where the recombination events must have occurred. In addition, two apparent deletions of circular cp32 plasmids have been reported: an approximately 14-kbp deletion to form cp18 in strain N40, where the putative crossover took place at one side of a 6-of-8-bp match in its closest relative, plasmid cp32-1 (29), and an approximately 10-kbp deletion to form cp18-2 in strain 297, whose crossover point has no base pairs of identity in the two possible parental sequences in plasmid cp32-7 (2). Nonhomologous rearrangements appear to have occurred relatively frequently on these DNAs. However, given the huge number of possible events, it is unlikely that identical nonhomologous recombination events of independent origin will be found, so these unique rearrangements (e.g., the chromosome-lp21 novel joint described here in Sh-2-82, 297, 28534, and 29968) should be useful as genetic markers in the characterization of B. burgdorferi populations.
FIG. 5.
There is little sequence similarity between partners at deduced recombination points. The nucleotide sequence of one strand is shown with the 5′ end at the left; vertical lines indicate identical base pairs in adjacent sequences. The parts of the putative parental sequences that fused to give rise to the Sh-2-82 sequence are underlined. (A) Nucleotide sequences of the two putative parental participants (upper, B31-like chromosome; lower, B31 lp21-like plasmid) in the nonhomologous recombination event that gave rise to the present Sh-2-82 nucleotide sequence (center). (B) Nucleotide sequences of the two putative parental participants (upper, B31-like left location; lower, B31-like right location) in the nonhomologous recombination event that gave rise to the apparent 264-bp deletion in the present Sh-2-82 nucleotide sequence (center).
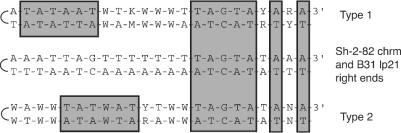
The telomeric sequences at the extreme right ends of the Sh-2-82 chromosome and the B31 lp21 plasmid are identical, but they have interesting differences from the previously characterized Borrelia termini. All previously known Borrelia telomeres have an absolutely conserved TAGTAYANA sequence (5′ to 3′ in the upper strand when the telomere is on the left; Fig. 6) that is 14 bp from the end and a highly conserved TATAAT sequence that is either 1 or 4 bp from the terminus (4). The Sh-2-82 and lp21 right-end terminal sequences determined here have a TAGTAYANA sequence that is 14 bp from the apparent end; however, they have no convincing TATAAT sequence. Compared to the other telomere sequences, AATTAG or TAGTTT occupies the type 1 or type 2 “TATAAT positions,” respectively, and these telomeres do not fit either type (Fig. 6). Tourand et al. (32a) have used mutant target sites to show that the sequence in at least parts of the TATAAT and TAGTA regions of a type 1 telomere is indeed important for telomere formation by protelomerase in vitro. However, the observations made here point out that the target specificity of the Borrelia telomere resolution machinery is not yet fully understood, especially in the “TATAAT portion” of the target sequence, and future new telomere sequences can be expected to shed additional light on terminal sequence constraints and protelomerase recognition.
FIG. 6.
Conservation of telomeric sequences. The 23 terminal bp present at the right ends of the Sh-2-82 chromosome and the B31 lp21 plasmid are shown in the middle, with the covalently closed hairpin telomere on the left for ease of comparison with previous publications (3, 6, 12, 27). Gray boxes highlight the TATAAT and TAGTAYANA conserved regions (see text). Casjens (3) and Tourand et al. (32a) have pointed out that the previously characterized Borrelia telomeres appear to fall into two categories, type 1 and type 2, in which an apparently conserved TATAAT sequence is present 1 or 4 bp, respectively, from the terminus. Among the nine previously sequenced telomeres, five are type 1 and four are type 2; the consensus sequences of the two types are shown at the top and bottom of the figure (R = A or G; Y = C or T; W = A or T; K = G or T; M = A or C; N indicates that three different base pairs are present among the known members of that type).
The data presented here make a particularly clear case for a past recombinational exchange event between a Borrelia linear plasmid and a chromosomal telomere. This, coupled with the finding that the distal, non-Sh-2-82-like portion of the B31 right-end extension is also plasmid related (8) and with the Southern analyses of other strains presented here, suggests that all the observed length variation at the B. burgdorferi chromosomal right end may be due to recombination with linear plasmids. Variations in length at the left end of the Borrelia japonica chromosome (5), coupled with the presence of plasmid-hybridizing sequences there (M. Murphy and S. Casjens, unpublished data), suggest that similar telomeric replacements by linear plasmid DNAs have occurred there as well. It is not known why or how such replacements occur, nor is it known why the phenomenon seems to be restricted to the right end of the B. burgdorferi chromosome and the left end of the B. japonica chromosome among the various Borrelia species examined to date.
The directionality of the postulated right-end telomere exchange event is most likely the replacement of the chromosomal telomere by the lp21 sequences (as opposed to generation of lp21 by “excision” from the end of an Sh-2-82-like parental chromosome), since the recombination event truncated an apparently intact lp21 gene, BBU04, which has paralogs on several other plasmids; it is very unlikely that the intact lp21 gene BBU04 would have been generated by a nonhomologous excision event. Our previous studies suggested that the rightmost 7.2 kbp of the linear chromosome of strain B31 are all derived from plasmids in a similar but more complex manner, so that the Sh-2-82 right end is the result of at least two successive rounds of telomere replacement. B. burgdorferi isolates B31 and Sh-2-82 were both isolated from I. scapularis ticks on Shelter Island, N.Y., but they do not appear to be especially closely related, since the constant portions of their chromosomes have several restriction site polymorphisms among the relatively small number of such sites examined (5); we do note, however, that these two isolates carry an apparently identical cp32 plasmid (30). The lp21 recombination event in the Sh-2-82 chromosome must have been rather recent, since its sequences remain more than 99% identical to the B31 lp21 sequences.
The B31 right-end extension contains two apparently intact genes, BB0844 and BB0852, both of which have paralogs on the B31 linear plasmids. These are surrounded by at least nine severely damaged plasmid-like genes (8), as if plasmid sequences had been joined to the right end of the chromosome, after which most of the plasmid genes were allowed to decay but the two currently intact genes were perhaps selected to remain functional. According to this model, the B31 chromosome had sequences added some time ago and mutational decay processes have partially removed those genes that are of no use there, while the lp21 addition to the Sh-2-82 chromosome (in which three possibly functional genes and six apparent pseudogenes of linear plasmid lp21 replaced one possibly functional gene and several pseudogenes of a B31-like chromosome) happened only rather recently, and further decay has barely begun on the newly added region. This appears to be an evolutionary mechanism which is able to sequentially move genetic material from linear plasmids onto the end of the linear Borrelia chromosome.
Acknowledgments
We thank Tom Schwan, Patti Rosa, Janis Weis, Justin Radolf, and Tom Anderson for Borrelia strains.
This work was supported by NIH grant AI49003 to S.C.
REFERENCES
- 1.Busch, U., C. Hizo-Teufel, R. Bohmer, V. Fingerle, D. Rossler, B. Wilske, and V. Preac-Mursic. 1996. Borrelia burgdorferi sensu lato strains isolated from cutaneous Lyme borreliosis biopsies differentiated by pulsed-field gel electrophoresis. Scand. J. Infect. Dis. 28:583-589. [DOI] [PubMed] [Google Scholar]
- 2.Caimano, M. J., X. Yang, T. G. Popova, M. L. Clawson, D. R. Akins, M. V. Norgard, and J. D. Radolf. 2000. Molecular and evolutionary characterization of the cp32/18 family of supercoiled plasmids in Borrelia burgdorferi 297. Infect. Immun. 68:1574-1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Casjens, S. 1998. The diverse and dynamic structure of bacterial genomes. Annu. Rev. Genet. 32:339-377. [DOI] [PubMed] [Google Scholar]
- 4.Casjens, S. 1999. Evolution of the linear DNA replicons of the Borrelia spirochetes. Curr. Opin. Microbiol. 2:529-534. [DOI] [PubMed] [Google Scholar]
- 5.Casjens, S., M. Delange, H. L. Ley III, P. Rosa, and W. M. Huang. 1995. Linear chromosomes of Lyme disease agent spirochetes: genetic diversity and conservation of gene order. J. Bacteriol. 177:2769-2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Casjens, S., and W. M. Huang. 1993. Linear chromosomal physical and genetic map of Borrelia burgdorferi, the Lyme disease agent. Mol. Microbiol. 8:967-980. [DOI] [PubMed] [Google Scholar]
- 7.Casjens, S., M. Murphy, M. DeLange, L. Sampson, R. van Vugt, and W. M. Huang. 1997. Telomeres of the linear chromosomes of Lyme disease spirochaetes: nucleotide sequence and possible exchange with linear plasmid telomeres. Mol. Microbiol. 26:581-596. [DOI] [PubMed] [Google Scholar]
- 8.Casjens, S., N. Palmer, R. Van Vugt, W. Mun Huang, B. Stevenson, P. Rosa, R. Lathigra, G. Sutton, J. Peterson, R. J. Dodson, D. Haft, E. Hickey, M. Gwinn, O. White, and C. M. Fraser. 2000. A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs in an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol. Microbiol. 35:490-516. [DOI] [PubMed] [Google Scholar]
- 9.Casjens, S., R. van Vugt, K. Tilly, P. A. Rosa, and B. Stevenson. 1997. Homology throughout the multiple 32-kilobase circular plasmids present in Lyme disease spirochetes. J. Bacteriol. 179:217-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Champion, C. I., D. R. Blanco, J. T. Skare, D. A. Haake, M. Giladi, D. Foley, J. N. Miller, and M. A. Lovett. 1994. A 9.0-kilobase-pair circular plasmid of Borrelia burgdorferi encodes an exported protein: evidence for expression only during infection. Infect. Immun. 62:2653-2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davidson, B. E., J. MacDougall, and I. Saint Girons. 1992. Physical map of the linear chromosome of the bacterium Borrelia burgdorferi 212, a causative agent of Lyme disease, and localization of rRNA genes. J. Bacteriol. 174:3766-3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deng, W., S. R. Liou, G. Plunkett III, G. F. Mayhew, D. J. Rose, V. Burland, V. Kodoyianni, D. C. Schwartz, and F. R. Blattner. 2003. Comparative genomics of Salmonella enterica serovar Typhi strains Ty2 and CT18. J. Bacteriol. 185:2330-2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dunn, J. J., S. R. Buchstein, L. L. Butler, S. Fisenne, D. S. Polin, B. N. Lade, and B. J. Luft. 1994. Complete nucleotide sequence of a circular plasmid from the Lyme disease spirochete, Borrelia burgdorferi. J. Bacteriol. 176:2706-2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fraser, C. M., S. Casjens, W. M. Huang, G. G. Sutton, R. Clayton, R. Lathigra, O. White, K. A. Ketchum, R. Dodson, E. K. Hickey, M. Gwinn, B. Dougherty, J. F. Tomb, R. D. Fleischmann, D. Richardson, J. Peterson, A. R. Kerlavage, J. Quackenbush, S. Salzberg, M. Hanson, R. van Vugt, N. Palmer, M. D. Adams, J. Gocayne, and J. C. Venter. 1997. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 390:580-586. [DOI] [PubMed] [Google Scholar]
- 15.Hinnebusch, J., and A. G. Barbour. 1991. Linear plasmids of Borrelia burgdorferi have a telomeric structure and sequence similar to those of a eukaryotic virus. J. Bacteriol. 173:7233-7239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iyer, R., O. Kalu, J. Purser, S. Norris, B. Stevenson, and I. Schwartz. 2003. Linear and circular plasmid content in Borrelia burgdorferi clinical isolates. Infect. Immun. 71:3699-3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kawabata, H., F. Myouga, Y. Inagaki, N. Murai, and H. Watanabe. 1998. Genetic and immunological analyses of Vls (VMP-like sequences) of Borrelia burgdorferi. Microb. Pathog. 24:155-166. [DOI] [PubMed] [Google Scholar]
- 18.Marconi, R. T., D. S. Samuels, R. K. Landry, and C. F. Garon. 1994. Analysis of the distribution and molecular heterogeneity of the ospD gene among the Lyme disease spirochetes: evidence for lateral gene exchange. J. Bacteriol. 176:4572-4582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mathiesen, D. A., J. H. Oliver, Jr., C. P. Kolbert, E. D. Tullson, B. J. Johnson, G. L. Campbell, P. D. Mitchell, K. D. Reed, S. R. Telford III, J. F. Anderson, R. S. Lane, and D. H. Persing. 1997. Genetic heterogeneity of Borrelia burgdorferi in the United States. J. Infect. Dis. 175:98-107. [DOI] [PubMed] [Google Scholar]
- 20.Nakagawa, I., K. Kurokawa, A. Yamashita, M. Nakata, Y. Tomiyasu, N. Okahashi, S. Kawabata, K. Yamazaki, T. Shiba, T. Yasunaga, H. Hayashi, M. Hattori, and S. Hamada. 2003. Genome sequence of an M3 strain of Streptococcus pyogenes reveals a large-scale genomic rearrangement in invasive strains and new insights into phage evolution. Genome Res. 13:1042-1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Norris, S. J., C. J. Carter, J. K. Howell, and A. G. Barbour. 1992. Low-passage-associated proteins of Borrelia burgdorferi B31: characterization and molecular cloning of OspD, a surface-exposed, plasmid-encoded lipoprotein. Infect. Immun. 60:4662-4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ojaimi, C., B. E. Davidson, I. Saint Girons, and I. G. Old. 1994. Conservation of gene arrangement and an unusual organization of rRNA genes in the linear chromosomes of the Lyme disease spirochaetes Borrelia burgdorferi, B. garinii and B. afzelii. Microbiology 140:2931-2940. [DOI] [PubMed] [Google Scholar]
- 23.Palmer, N., C. Fraser, and S. Casjens. 2000. Distribution of twelve linear extrachromosomal DNAs in natural isolates of Lyme disease spirochetes. J. Bacteriol. 182:2476-2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Porcella, S. F., T. G. Popova, D. R. Akins, M. Li, J. D. Radolf, and M. V. Norgard. 1996. Borrelia burgdorferi supercoiled plasmids encode multicopy tandem open reading frames and a lipoprotein gene family. J. Bacteriol. 178:3293-3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Samuels, D. S., R. T. Marconi, and C. F. Garon. 1993. Variation in the size of the ospA-containing linear plasmid, but not the linear chromosome, among the three Borrelia species associated with Lyme disease. J. Gen. Microbiol. 139:2445-2449. [DOI] [PubMed] [Google Scholar]
- 26.Schwan, T. G., W. Burgdorfer, and C. F. Garon. 1988. Changes in infectivity and plasmid profile of the Lyme disease spirochete, Borrelia burgdorferi, as a result of in vitro cultivation. Infect. Immun. 56:1831-1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Simpson, W. J., C. F. Garon, and T. G. Schwan. 1990. Analysis of supercoiled circular plasmids in infectious and non-infectious Borrelia burgdorferi. Microb. Pathog. 8:109-118. [DOI] [PubMed] [Google Scholar]
- 28.Steere, A. C., R. L. Grodzicki, A. N. Kornblatt, J. E. Craft, A. G. Barbour, W. Burgdorfer, G. P. Schmid, E. Johnson, and S. E. Malawista. 1983. The spirochetal etiology of Lyme disease. N. Engl. J. Med. 308:733-740. [DOI] [PubMed] [Google Scholar]
- 29.Stevenson, B., S. Casjens, R. van Vugt, S. F. Porcella, K. Tilly, J. L. Bono, and P. Rosa. 1997. Characterization of cp18, a naturally truncated member of the cp32 family of Borrelia burgdorferi plasmids. J. Bacteriol. 179:4285-4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stevenson, B., and J. C. Miller. 2003. Intra- and interbacterial genetic exchange of Lyme disease spirochete erp genes generates sequence identity amidst diversity. J. Mol. Evol. 57:309-324. [DOI] [PubMed] [Google Scholar]
- 31.Theisen, M. 1996. Molecular cloning and characterization of nlpH, encoding a novel, surface-exposed, polymorphic, plasmid-encoded 33-kilodalton lipoprotein of Borrelia afzelii. J. Bacteriol. 178:6435-6442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tilly, K., S. Casjens, B. Stevenson, J. L. Bono, D. S. Samuels, D. Hogan, and P. Rosa. 1997. The Borrelia burgdorferi circular plasmid cp26: conservation of plasmid structure and targeted inactivation of the ospC gene. Mol. Microbiol. 25:361-373. [DOI] [PubMed] [Google Scholar]
- 32a.Tourand, Y., K. Kobryn, and G. Chaconas. 2003. Sequence-specific recognition but position-dependent cleavage of two distinct telomeres by the Borrelia burgdorferi telomere resolvase, ResT. Mol. Microbiol. 48:901-911. [DOI] [PubMed] [Google Scholar]
- 33.Welch, R. A., V. Burland, C. Plunkett III, P. Redford, P. Roesch, D. Rasko, E. L. Buckles, S. R. Llou, A. Boutin, J. Hackett, D. Stroud, G. F. Mayhew, D. J. Rose, S. Zhou, D. C. Schwartz, N. T. Perna, H. L. Mobley, M. S. Donnenberg, and F. R. Blattner. 2002. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 99:17020-17024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu, Y., and R. C. Johnson. 1995. Analysis and comparison of plasmid profiles of Borrelia burgdorferi sensu lato strains. J. Clin. Microbiol. 33:2679-2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang, J. R., J. M. Hardham, A. G. Barbour, and S. J. Norris. 1997. Antigenic variation in Lyme disease borreliae by promiscuous recombination of VMP-like sequence cassettes. Cell 89:275-285. [DOI] [PubMed] [Google Scholar]