Abstract
The dynamic glycosylation of serine/threonine residues on nucleocytoplasmic proteins with a single N-acetylglucosamine (O-GlcNAcylation) is critical for many important cellular processes. Cellular O-GlcNAc levels are highly regulated by two enzymes: O-GlcNAc transferase (OGT) is responsible for GlcNAc addition and O-GlcNAcase (OGA) is responsible for removal of the sugar. The lack of a rapid and simple method for monitoring OGT activity has impeded the efficient discovery of potent OGT inhibitors. In this study we describe a novel, single-well OGT enzyme assay that utilizes 6 × His-tagged substrates, a chemoselective chemical reaction, and unpurified OGT. The high-throughput Ni-NTA Plate OGT Assay will facilitate discovery of potent OGT-specific inhibitors on versatile substrates and the characterization of new enzyme variants.
Introduction
Protein glycosylation on serine/threonine residues with N-acetylglucosamine (O-GlcNAcylation) is an abundant post-translational modification that occurs in the nucleocytoplasm. O-GlcNAc is critical for many important cellular processes and has an extensive and dynamic interplay with phosphorylation.1−4 Consequently, dysregulation of the balance between these two modifications has been implicated in cancer,5−7 diabetes,8,9 and neurodegenerative disease.3,10 Control of nuclear and cytoplasmic protein O-GlcNAcylation provides a means to influence numerous cellular events and the potential for management of various human diseases. Our understanding of O-GlcNAc’s biological roles and cellular signaling has rapidly expanded over the past several years due to an increasing number of chemical tools.11−13
Only two enzymes are involved in the regulation of O-GlcNAc cycling in humans: the glycosyltransferase O-GlcNAc transferase (OGT)14−16 and the glycoside hydrolase O-GlcNAcase (OGA).17,18 OGT installs the sugar moiety (GlcNAc) on target proteins using uridine 5′-diphosphate (UDP)-GlcNAc as a substrate and OGA removes O-GlcNAc from the same proteins. Therefore, changes in cellular O-GlcNAc levels can be achieved by manipulating the activities of OGT and OGA. Aided by several high-throughput enzymatic assays, as well as mechanistic and structural insights,19,20 numerous potent and selective OGA inhibitors have been developed.21−25 Indeed, the use of small-molecule OGA inhibitors has greatly assisted in the evolution of our understanding of O-GlcNAc’s cellular roles.
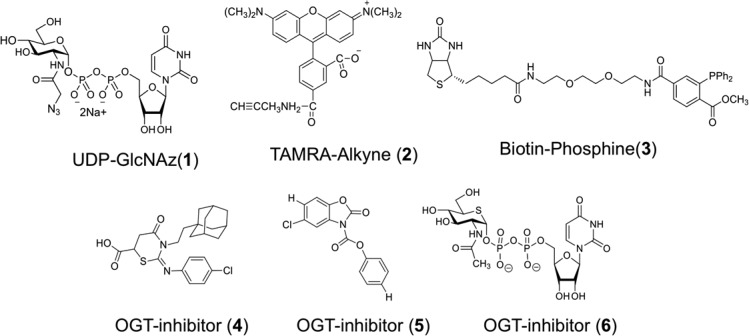
While there are several specific OGA inhibitors, the complexity, high cost, and limited versatility of classical OGT assays, in addition to the enzyme’s instability during purification, have been major obstacles in developing potent and selective OGT inhibitors. The conventional in vitro OGT activity assay26 utilizes a radiolabeled sugar substrate, such as UDP-[14C] GlcNAc or UDP-[3H] GlcNAc. This method measures OGT activity by quantifying radiolabeled GlcNAc incorporation into a protein substrate, but is high in cost and produces radiochemical waste. As an alternative to using radiochemicals, the Walker group introduced a ligand displacement OGT assay27 and a protease-protection assay.28 The former method exploits fluorescence polarization using a fluorophore-containing UDP-GlcNAc analogue. The latter protease-protection assay exploits the fluorescence resonance energy transfer (FRET) phenomenon using specially designed, protease-specific peptide substrates. However, both assays indirectly measure the glycosyltransferase activity of OGT, requiring a second OGT activity assay to validate positive hit compounds. Finally, Leavy et al. introduced an azido-enzyme-linked immunosorbent assay (azido-ELISA) method using the unnatural sugar donor, UDP-N-azidoacetyl glucosamine (UDP-GlcNAz, 1, Figure 1) and biotinylated peptide substrates.29 In this method, a solution-phase enzymatic reaction is transferred into a microplate for subsequent solid-phase reactions, reducing the method’s efficiency.
Figure 1.
Structures of chemical compounds used in Ni-NTA Plate OGT Assays: UDP-GlcNAz (1), TAMRA-Alkyne (2), Biotin-Phosphine (3), and OGT inhibitors (4–6).
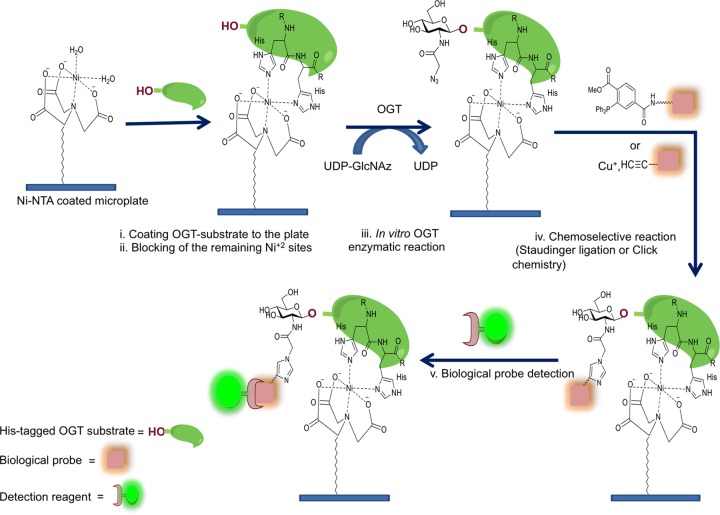
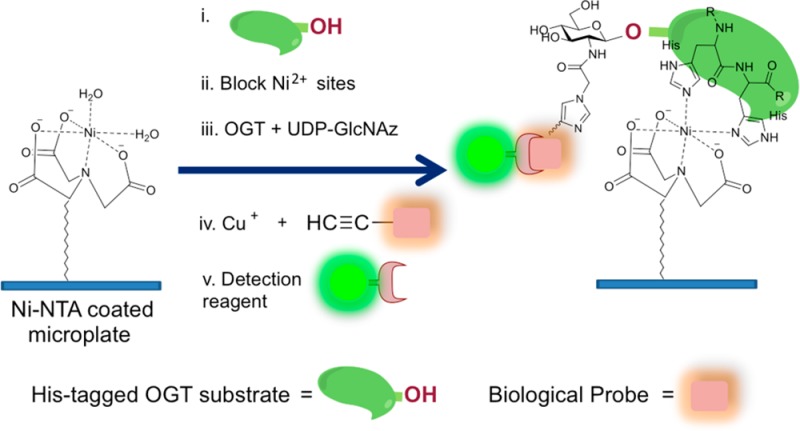
In this Communication, we present a direct and efficient OGT microplate assay in which purification of the enzyme is unnecessary: both bacterial and eukaryotic cell extracts can be readily used as sources of OGT. In addition, because bacterial extracts expressing recombinant OGT isoforms can be employed, the method is applicable to identifying new enzyme variants. This method exploits one of the most widely used affinity tags, a six consecutive histidine (6 × His) tag, and nickel ions immobilized on a chelating microplate. The 6 × His-tag can be genetically engineered at either the amino- or carboxy-terminus of recombinant proteins. The method we developed here uses UDP-GlcNAz (1, Figure 1) as a sugar donor and 6 × His-tagged protein substrates as the sugar acceptors. All reactions are carried out in a microplate coated with Ni2+ ions through coupling with the chelating moiety of nitrilotriacetic acid (NTA). As depicted in Figure 2, the strategy of our OGT assay involves five steps starting with attachment of the 6 × His-tagged OGT substrate to the plate through Ni-NTA chelation, followed by the blocking of unoccupied nickel ion. Protein substrate bound to the plate is then subjected to OGT enzymatic catalysis using UDP-GlcNAz as the sugar donor. Next, a chemoselective reaction, such as the copper-catalyzed “click” reaction, or Staudinger ligation, is performed to label the azido-functionality. Finally, an immunoassay is used to detect the labeled proteins.
Figure 2.
Strategy for Ni-NTA Plate OGT Assay. The high-throughput Ni-NTA OGT assay described herein consists of simple steps as indicated.
All previously published methods for measuring OGT activity, as described above, require high-cost reagents or involve components that necessitate de novo synthesis, increasing both the cost and the time required for these assays. Importantly, the assay described herein is rapid and straightforward, and all components are commercially or readily available. Moreover, our Ni-NTA Plate OGT Assay does not require the purification of the enzyme. This not only is advantageous because OGT is temperamental and does not withstand stringent washes without loss of activity, but also allows for easier characterization of enzyme variants. Beyond directly measuring OGT’s glycosyltransferase activity and rapidly screening chemical libraries to discover OGT inhibitors, our Ni-NTA Plate OGT Assay allows for the study of substrate specificity for each OGT isoform from various organisms.
Results
For our initial studies, we used the 6 × His-tagged, recombinant human nuclear pore protein, Nup62, which is a well-defined OGT substrate that can be heavily modified with GlcNAc.26 UDP-GlcNAz (1, Figure 1) was used as a sugar donor and the TAMRA-conjugated terminal alkyne compound (TAMRA-Alkyne, 2, Figure 1) or biotinylated triarylphosphine compound (Biotin-Phosphine, 3, Figure 1) were used as reagents to selectively react with the azido-functionality. Mammalian OGT has three isoforms: nuclear cytoplasmic OGT (ncOGT), mitochondrial OGT (mOGT), and short OGT (sOGT).15,30 In this study, we used bacterially expressed recombinant human ncOGT31 in both unpurified and partially purified forms.
First, 6 × His-tagged Nup62 was attached to an 8-well strip coated with Ni-NTA by incubating Nup62 dissolved in PBS at room temperature for 1 h. Because the ncOGT construct we used for our assay also has a 6 × His-tag, we wanted to ensure that the ncOGT itself would not bind to the plate during the reaction. As shown in Supporting Information (SI) Figure S1, treatment of each well with blocking reagents including 6 × His oligopeptide (6 × His-Pep), commercially available Odyssey blocking buffer (Ody. Buffer), 5% BSA, and 5% milk abrogated binding of 6 × His-tagged ncOGT to the plate. In our hands, Odyssey blocking buffer is the ideal blocking reagent to consistently minimize background.
Next, the OGT enzymatic reaction was initiated by adding bacterial lysate containing ncOGT or partially purified ncOGT, UDP-GlcNAz, and OGT assay buffer to the bound Nup62. Following the OGT reaction, two azido-selective chemical reactions were carried out to determine the level of OGT glycosyltransferase activity. The copper-catalyzed “click” reaction was carried out using commercially available TAMRA-Alkyne (2, Figure 1) and copper catalyst. Alternatively, the Staudinger ligation reaction was performed by adding Biotin-Phosphine (3, Figure 1) in PBS. Chemoselective reactions were quenched by removing the reaction mixtures and washing the wells several times with PBST (PBS containing 0.1% Tween 20) followed by an immunoassay to probe for bound Biotin- or TAMRA-tags followed by an appropriately conjugated IRDye800CW. After several washes with PBST, each well was filled with PBS, imaged, and the signal quantified. For detailed experimental procedures, see the Supporting Information.
As shown in Figure 3, OGT enzymatic reactions were efficiently detected using an anti-TAMRA antibody, indicating nickel ion precoated on the strip does not interfere with the copper-catalyzed “click” reaction. In addition, the RL2 antibody, which recognizes a number of O-GlcNAc-modified nuclear pore complex proteins, was successfully used to determine OGT activity (see SI Figure S2). These data indicate that, while convenient, a chemoselective reaction to detect O-GlcNAzylated Nup62 is not required when Nup62 is used as the OGT substrate. Importantly, results of OGT assays using partially purified ncOGT (lower panel in Figure 3) were similar to those obtained with bacterial lysate containing ncOGT (upper panel in Figure 3), indicating that OGT purification is not necessary in the Ni-NTA Plate OGT Assay.
Figure 3.
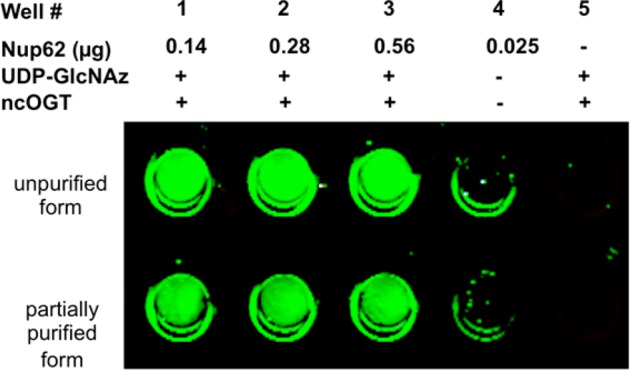
Determination of ncOGT activity. Four concentrations of the 6 × His-tagged Nup62 substrate (as indicated) were used to detect activity of either unpurified or partially purified (p-ncOGT) enzyme as indicated. Glycosyltransferase activity was detected using copper-catalyzed “click” reaction with TAMRA-Alkyne.
Next, we evaluated the minimal amount of Nup62 necessary in order to obtain the maximal IR-signal intensity for this assay. Here, Biotin-Phosphine (3, Figure 1) was chemoselectively reacted with the incorporated azido-moiety and relative IR-signal intensity was determined, which reflects the enzymatic activity of the ncOGT. As shown in Figure 4, IR-signal intensity reached the maximum (wells 4 and 5) when the amount of Nup62 was over 0.14 μg.
Figure 4.
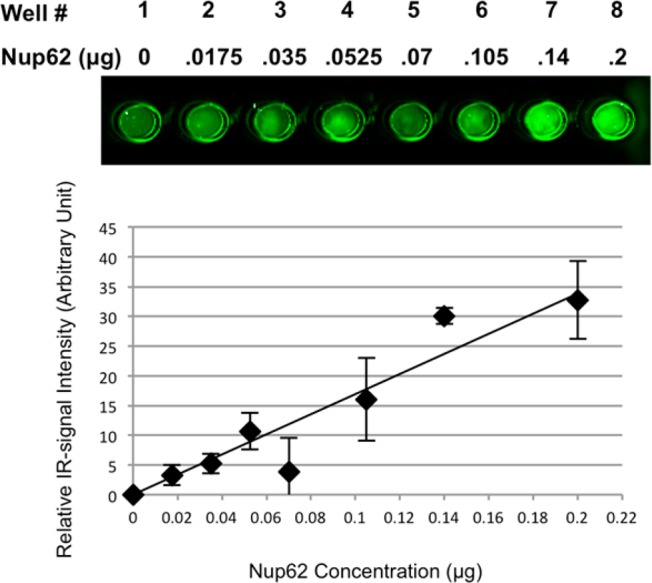
ncOGT activity is linearly dependent on the concentration of Nup62. Increasing amounts of the 6 × His-tagged Nup62 substrate were used as indicated. ncOGT containing lysates were used for these experiments. Glycosyltransferase activity was detected using Staudinger ligation with Biotin-Phosphine reagent. Experiments were performed in duplicate and quantified using the Odyssey Infrared Imaging System at 800 nm. Values were normalized such that no substrate was set to 0. Error bars represent range.
To demonstrate the flexibility of our assay, another well-described OGT substrate, 6 × His-tagged recombinant Casein Kinase 2 alpha protein (CKII-α) was tested. Like Nup62, CKII-α was attached to the well, GlcNAz-labeled by the enzymatic reaction of ncOGT using UDP-GlcNAz, subjected to copper-catalyzed “click” reaction using TAMRA-Alkyne (2, Figure 1) reagent, and immunoassayed with an anti-TAMRA antibody. Resulting IR-signal intensity increased as the amount of incubated CKII-α protein increased until 0.13 μg of the substrate was used (see SI Figure S3). Furthermore, the versatility of our Ni-NTA Plate OGT Assay was demonstrated with OGT from eukaryotic cell lysates as the enzyme source. In the presence of UDP-GlcNAz and Nup62, endogenous OGT glycosyltransferase activity from rabbit reticulocytes was detected (see SI Figure S4), supporting that the activity of OGT enzymes from various organisms can be assayed.
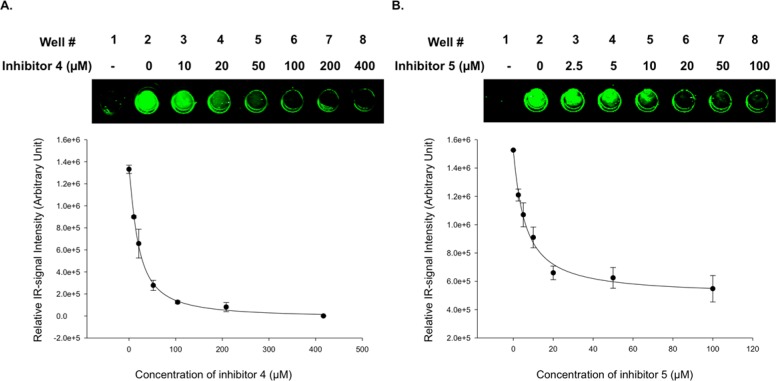
To validate the assay’s ability to identify known inhibitors of OGT, we tested OGT inhibitors (4) and (5), which were reported to have IC50 values of 53 μM (±7 μM) and 10 μM (±1 μM),27 respectively. Prior to the inhibition assays, we examined the amount of OGT-containing lysate and reaction time required for linear enzyme activity [1.4 μL (equivalent to 56 μg of a total protein) of ncOGT-containing lysate and 25 min of reaction time; see SI Figure S5] when 40 μM of UDP-GlcNAz and 0.15 μg of Nup62 were used. 20 mM OGT inhibitors 4 and 5 were prepared in DMSO and serially diluted, and equal volumes were added to each reaction well containing bound Nup62 and UDP-GlcNAz. Enzymatic reactions were initiated by adding 1.4 μL of ncOGT-containing lysate and incubated at 37 °C with gentle shaking for 25 min, followed by quenching by removal of reaction mixtures and PBST washes. Enzyme activity was normalized to a well containing only DMSO but no inhibitor. As shown in Figure 5, compounds 4 and 5 inhibited ncOGT glycosyltransferase activity with IC50 values for 4 and 5 to be 19.7 (±1.4) μM and 6.0 (±0.8) μM, respectively (Figure 5B). Although our results differ from those previously reported,27 these values are well within the expected range. Variations may be caused by differences in the relative sensitivity of methods used for quantification (radiometric assay vs Ni-NTA OGT Assay) and in the choice of OGT substrates (UDP-GlcNAc vs UDP-GlcNAz and peptide vs Nup62).
Figure 5.
Inhibition of ncOGT activity by compounds 4 and 5. Increasing amounts of inhibitors 4 (A) and 5 (B) were added to the Ni-NTA Plate OGT Assay (as indicated). Glycosyltransferase activity was detected using Staudinger ligation with Biotin-Phosphine reagent. Experiments were performed in triplicate (n = 3) and quantified using the Odyssey Infrared Imagining System at 800 nm. IC50 values were calculated using SigmaPlot 12.5 software employing “Four parameter logistic equation in the standard curve analysis”. Error bars represent standard deviations.
Discussion
As OGT is a key regulator of diverse cellular processes whose deregulation has been associated with several diseases, understanding its activity will have major impacts on human health. Indeed, identification of pharmacological inhibitors of OGT has great therapeutic potential. Recent structural and chemical analysis has given us insights into mechanisms of OGT inhibition. OGT consists of two distinct domains: the N-terminal domain containing tetratricopeptide repeat (TPR) motifs, and the C-terminal catalytic domain. Based on a crystal structure of the TPR domain of ncOGT reported in 2004,32 the TPR domain is believed to interact with other proteins and determine substrate specificity. The crystal structure of a human OGT construct containing 4.5 TPR units and the C-terminal domain33 revealed a unique fold of the intervening region between the N-terminal catalytic domain and the C-terminal catalytic domain, and provided clues to the enzyme’s catalytic33,34 and inhibiting35 mechanisms. Additionally, recent studies have uncovered compounds that inhibit OGT. A compound containing a benzoxazolinone (BZX) core structure (5, Figure 1) was reported to inhibit OGT in cells;36 and a nucleotide sugar analogue, UDP-2-acetamido-2-deoxy-5-thio-d-glucopyranoside (UDP-5SGlcNAc, 6, Figure 1) biosynthesized from a synthetic carbohydrate precursor, 2-acetamido-2-deoxy-5-thio-d-glucopyranoside (5SGlcNAc) via the cells’ hexosamine biosynthetic machinery, was also identified as an OGT inhibitor, thus lowering O-GlcNAc levels in cells.37
Because OGT has three isoforms, a layer of complexity is added to understanding mechanisms of inhibition. While all isoforms share the same catalytic domain, they differ in the number of TPR motifs. Interestingly, expression of each OGT isoform varies in different cell types, suggesting that each isoform may have distinct functions that respond differently to cellular signaling depending on its tissue distribution.15,30,31 Therefore, it will be necessary to dissect the functions of each OGT isoform and develop isoform-specific inhibitors.
The high-throughput enzymatic OGT assay described here allows for easy testing of a variety of inhibitors on many substrates. Because this assay does not require OGT purification or the use of radiolabeled substrates, it can be performed more efficiently and cost-effectively than the conventional OGT enzymatic assays. Importantly, our methodology directly measures glycosyltransferase activity, which will decrease the number of false-positive hits as seen with other nonradiometric OGT assays.27,28 Overall, this simple and easy method for continuously monitoring OGT activity will help to discover potent OGT-specific inhibitors, thus advancing our understanding of the functional roles of OGT and O-GlcNAc cycling.
Acknowledgments
This work was supported by NIDDK intramural funds (NIH) and the National Research Foundation of Korea (2011-0027257).
Glossary
Abbreviations
- UDP
uridine diphosphate
- GlcNAc
N-acetylglucosamine
- GlcNAz
N-azidoacetylglucosamine
- O-GlcNAcase
OGA
- O-GlcNAc transferase
OGT
Supporting Information Available
Detailed experimental procedures. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Hu P.; Shimoji S.; Hart G. W. (2010) Site-specific interplay between O-GlcNAcyl-ation and phosphorylation in cellular regulation. FEBS Lett. 584, 2526–2538. [DOI] [PubMed] [Google Scholar]
- Butkinaree C.; Park K.; Hart G. W. (2010) O-linked β-N-acetylglucosamine (O-GlcNAc): Extensive crosstalk with phosphorylation to regulate signaling and transcription in response to nutrients and stress. Biochim. Biophys. Acta 1800, 96–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarus B. D.; Love D. C.; Hanover J. A. (2009) O-GlcNAc cycling: Implications for neurodegenerative disorders. Biochem. Cell Biol. 41, 2134–2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love D. C.; Krause M. W.; Hanover J. A. (2010) O-GlcNAc cycling: Emerging roles in development and epigenetics. Semin. Cell Dev. Biol. 21, 646–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X.; Ongusaha P. P.; Miles P. D.; Havstad J. C.; Zhang F.; So W. V.; Kudlow J. E.; Michell R. H.; Olefsky J. M.; Field S. J.; Evans R. M. (2008) Phosphoinositide signalling links O-GlcNAc transferase to insulin resistance. Nature 451, 964–969. [DOI] [PubMed] [Google Scholar]
- McClain D. A.; Lubas W. A.; Cooksey R. C.; Hazel M.; Parker G. J.; Love D. C.; Hanover J. A. (2002) Altered glycandependent signaling induces insulin resistance and hyperleptinemia. Proc. Natl. Acad. Sci. U.S.A. 99, 10695–10699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vosseller K.; Wells L.; Lane M. D.; Hart G. W. (2002) Elevated nucleocytoplasmic glycosylation by O-GlcNAc results in insulin resistance associated with defects in Akt activation in 3T3-L1 adipocytes. Proc. Natl. Acad. Sci. U.S.A. 99, 5313–5318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W. H.; Kim J. E.; Nam H. W.; Ju J. W.; Kim H. S.; Kim Y. S.; Cho J. W. (2006) Modification of p53 with O-linked N-acetylglucosamine regulates p53 activity and stability. Nat. Cell Biol. 8, 1074–1083. [DOI] [PubMed] [Google Scholar]
- Chou T. Y.; Dang C. V.; Hart G. W. (1995) Glycosylation of the c-Myc transactivation domain. Proc. Natl. Acad. Sci. U.S.A. 92, 4417–4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F.; Iqbal K.; Grundke-Iqbal I.; Hart G. W.; Gong C. X. (2004) O-GlcNAcylation regulates phosphorylation of tau: a mechanism involved in Alzheimer’s disease. Proc. Natl. Acad. Sci. U.S.A. 101, 10804–10809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macauley M. S.; Vocadlo D. J. (2010) Increasing O-GlcNAc levels: An overview of small-molecule inhibitors of O-GlcNAcase. Biochim. Biophys. Acta 1800, 107–121. [DOI] [PubMed] [Google Scholar]
- Gloster T. M.; Vocadlo D. J. (2010) Mechanism, structure, and inhibition of O-GlcNAc processing enzymes. Curr. Signal Transduction Ther. 5, 74–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E. J. (2011) Chemical arsenal for the study of O-GlcNAc. Molecules 16, 1987–2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haltiwanger R. S.; Holt G. D.; Hart G. W. (1990) Enzymatic addition of O-GlcNAc to nuclear and cytoplasmic proteins. Identification of a uridine diphospho-N-acetyl-glucosamine: peptide β-N-acetylglucosaminyltransferase. J. Biol. Chem. 265, 2563–2568. [PubMed] [Google Scholar]
- Kreppel L. K.; Blomberg M. A.; Hart G. W. (1997) Dynamic glycosylation of nuclear and cytosolic proteins. Cloning and characterization of a unique O-GlcNAc transferase with multiple tetratricopeptide repeats. J. Biol. Chem. 272, 9308–9315. [DOI] [PubMed] [Google Scholar]
- Lubas W. A.; Frank D. W.; Krause M.; Hanover J. A. (1997) O-Linked GlcNAc transferase is a conserved nucleocytoplasmic protein containing tetratricopeptide repeats. J. Biol. Chem. 272, 9316–9324. [DOI] [PubMed] [Google Scholar]
- Dong D. L.; Hart G. W. (1994) Purification and characterization of an O-GlcNAc selective N-acetyl-β-D-glucosaminidase from rat spleen cytosol. J. Biol. Chem. 269, 19321–19330. [PubMed] [Google Scholar]
- Gao Y.; Wells L.; Comer F. I.; Parker G. J.; Hart G. W. (2001) Dynamic O-glycosylation of nuclear and cytosolic proteins: cloning and characterization of a neutral, cytosolic β-N-acetylglucosaminidase from human brain. J. Biol. Chem. 276, 9838–9845. [DOI] [PubMed] [Google Scholar]
- Macauley M. S.; Whitworth G. E.; Debowski A. W.; Chin D.; Vocadlo D. J. (2005) O-GlcNAcase uses substrate-assisted catalysis: kinetic analysis and development of highly selective mechanism inspired inhibitors. J. Biol. Chem. 280, 25313–25322. [DOI] [PubMed] [Google Scholar]
- Kim E. J.; Kang D. W.; Love D. C.; Hanover J. A. (2006) Enzymatic characterization of O-GlcNAcase isoforms using a fluorogenic GlcNAc substrate. Carbohydr. Res. 341, 971–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E. J.; Perreira M.; Thomas C. J.; Hanover J. A. (2006) An O-GlcNAcase-specific inhibitor and substrate engineered by the extension of the N-acetyl moiety. J. Am. Chem. Soc. 128, 4234–4235. [DOI] [PubMed] [Google Scholar]
- Cetinbas N.; Macauley M. S.; Stubbs K. A.; Drapala R.; Vocadlo D. J. (2006) Identification of Asp174 and Asp175 as the key catalytic residues of human O-GlcNAcase by functional analysis of site-directed mutants. Biochemistry 45, 3835–3844. [DOI] [PubMed] [Google Scholar]
- Dorfmueller H. C.; Borodkin V. S.; Schimpl M.; Shepherd S. M.; Shpiro N. A.; van Aalten D. M. (2006) GlcNAcstatin: a picomolar, selective O-GlcNAcase inhibitor that modulates intracellular O-GlcNAcylation levels. J. Am. Chem. Soc. 128, 16484–16485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuzwa S. A.; Macauley M. S.; Heinonen J. E.; Shan X.; Dennis R. J.; He Y.; Whitworth G. E.; Stubbs K. A.; McEachern E. J.; Davies G. J.; Vocadlo D. J. (2008) A potent mechanism-inspired O-GlcNAcase inhibitor that blocks phosphorylation of tau in vivo. Nat. Chem. Biol. 4, 483–490. [DOI] [PubMed] [Google Scholar]
- Dorfmueller H. C.; Borodkin V. S.; Schimpl M.; van Aalten D. M. (2009) GlcNAc-statins are nanomolar inhibitors of human O-GlcNAcase inducing cellular hyper-O-GlcNAcylation. Biochem. J. 420, 221–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubas W. A.; Hanover J. A. (2000) Functional expression of O-linked GlcNAc transferase. Domain structure and substrate specificity. J. Biol. Chem. 275, 10983–10988. [DOI] [PubMed] [Google Scholar]
- Gross B. J.; Kraybill B. C.; Walker S. (2005) Discovery of O-GlcNAc transferase inhibitors. J. Am. Chem. Soc. 127, 14588–14589. [DOI] [PubMed] [Google Scholar]
- Gross B. J.; Swoboda J. G.; Walker S. (2008) A strategy to discover inhibition of O-linked glycosylation. J. Am. Chem. Soc. 130, 440–441. [DOI] [PubMed] [Google Scholar]
- Leavy T. M.; Bertozzi C. R. (2007) A high-throughput assay for O-GlcNAc transferase detects primary sequence preferences in peptide substrates. Bioorg. Med. Chem. Lett. 17, 3851–3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanover J. A.; Yu S.; Lubas W. B.; Shin S. H.; Ragano-Caracciola M.; Kochran J.; Love D. C. (2003) Mitochondrial and nucleocytoplasmic isoforms of O-linked GlcNAc transferase encoded by a single mammalian gene. Arch. Biochem. Biophys. 409, 287–97. [DOI] [PubMed] [Google Scholar]
- Lazarus B. D.; Love D. C.; Hanover J. A. (2006) Recombinant O-GlcNAc transferase isoforms: identification of O-GlcNAcase, yes tyrosine kinase, and tau as isoform-specific substrates. Glycobiology 16, 415–421. [DOI] [PubMed] [Google Scholar]
- Jinek M.; Rehwinkel J.; Lazarus B. D.; Izaurralde E.; Hanover J. A.; Conti E. (2004) The superhelical TPR-repeat domain of O-linked GlcNAc transferase exhibits structural similarities to importin alpha. Nat. Struct. Mol. Biol. 11, 1001–1007. [DOI] [PubMed] [Google Scholar]
- Lazarus M. B.; Nam Y.; Jiang J.; Sliz P.; Walker S. (2011) Structure of human O-GlcNAc transferase and its complex with a peptide substrate. Nature 469, 564–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarus M. B.; Jiang J.; Gloster T. M.; Zandberg W. F.; Whitworth G. E.; Vocadlo D. J.; Walker S. (2012) Structural snapshots of the reaction coordinate for O-GlcNAc transferase. Nat. Chem. Biol. 8, 966–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J.; Lazarus M. B.; Pasquina L.; Sliz P.; Walker S. (2012) A neutral diphosphate mimic crosslinks the active site of human O-GlcNAc transferase. Nat. Chem. Biol. 8, 72–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart G. W.; Slawson C.; Ramirez-Correa G.; Lagerlof O. (2011) Cross talk between O-GlcNAcylation and phosphorylation: roles in signaling, transcription, and chronic disease. Annu. Rev. Biochem. 80, 825–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloster T. M.; Zandberg W. F.; Heinonen J. E.; Shen D. L.; Deng L.; Vocadlo D. J. (2011) Hijacking a biosynthetic pathway yields a glycosyltransferase inhibitor within cells. Nat. Chem. Biol. 7, 174–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.