Abstract
XCMS Online (xcmsonline.scripps.edu) is a cloud-based informatic platform designed to process and visualize mass-spectrometry-based, untargeted metabolomic data. Initially, the platform was developed for two-group comparisons to match the independent, “control” versus “disease” experimental design. Here, we introduce an enhanced XCMS Online interface that enables users to perform dependent (paired) two-group comparisons, meta-analysis, and multigroup comparisons, with comprehensive statistical output and interactive visualization tools. Newly incorporated statistical tests cover a wide array of univariate analyses. Multigroup comparison allows for the identification of differentially expressed metabolite features across multiple classes of data while higher order meta-analysis facilitates the identification of shared metabolic patterns across multiple two-group comparisons. Given the complexity of these data sets, we have developed an interactive platform where users can monitor the statistical output of univariate (cloud plots) and multivariate (PCA plots) data analysis in real time by adjusting the threshold and range of various parameters. On the interactive cloud plot, metabolite features can be filtered out by their significance level (p-value), fold change, mass-to-charge ratio, retention time, and intensity. The variation pattern of each feature can be visualized on both extracted-ion chromatograms and box plots. The interactive principal component analysis includes scores, loadings, and scree plots that can be adjusted depending on scaling criteria. The utility of XCMS functionalities is demonstrated through the metabolomic analysis of bacterial stress response and the comparison of lymphoblastic leukemia cell lines.
XCMS Online1 is a cloud-based, mass spectrometry data processing platform that was developed in response to the growing need for user-friendly software to process complex untargeted metabolomic results.2−4 Currently, XCMS Online has more than 4500 registered users from 120 different countries. This web-based platform is an extension of the original open-source R package XCMS that was released in 2006.5 Several open-source programs including MetAlign,6 MZmine,7 MAVEN,8 MetaboAnalyst,9 and XCMS,5 as well as commercial programs, such as Mass Profiler Pro (Agilent) and Metabolic Profiler (Bruker) have been successfully developed to facilitate metabolomic data processing. Each of these software tools has its own unique capabilities in supporting metabolomic data storage, analysis, and annotation.10 However, these programs do not allow for complete metabolomic data processing from feature detection and profile alignment to feature annotation and exploratory statistical analyses. For example, the web-based tools MetaboAnalyst and metaP-Server,11 which have been mainly developed for targeted metabolomic data analysis,12 perform comprehensive statistical evaluation of preprocessed metabolomic data, yet rely on external software to process the raw data.
XCMS Online has been designed as a solution for the entire untargeted metabolomic workflow ranging from the computationally expensive raw data processing to direct metabolite assignment through integration with the METLIN database. The platform has been recently upgraded with data streaming capabilities to support high-throughput, cloud-based data processing.13 By continuous improvement of the platform, we aim to facilitate the access and functional interpretation of metabolomic data by scientists with limited background in bioinformatics or statistics.
Our new interactive XCMS Online platform integrates univariate and multivariate data processing, metabolite feature annotation, and metabolite identification. This easy-to-use graphical interface has been significantly improved to support the statistical analysis of more complex experimental designs. It now performs two-group, multiple group, and meta-analysis (Figure 1), providing XCMS Online users with a comprehensive univariate and multivariate statistical report through an interactive display. Novel interactive visualization tools, including two-group and multigroup cloud plots, principal component analysis, and Venn and relational diagrams, have been developed to deconvolve metabolomic data, simplify data analysis, customize data output, and through integration with METLIN, facilitate metabolite identification. The utility of these approaches is demonstrated on several different data sets that fit the experimental design of two-group comparison, of meta-analysis, and of multigroup comparison (Figure 1).
Figure 1.
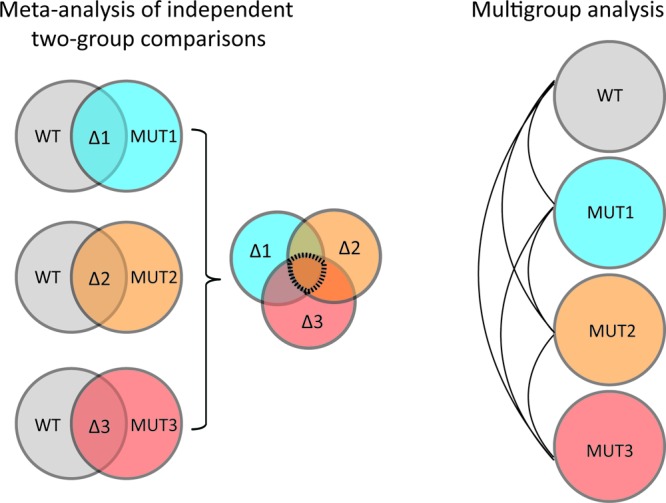
Scheme representing the experimental design of two-group comparison, meta-analysis, and multigroup analysis. Meta-analysis is a higher-order analysis that aims to identify shared metabolic patterns among multiple independent two-group comparisons. Shared dysregulated features are represented by the region at the center of the Venn diagram. In contrast, multigroup analysis aims to identify differences between groups and reveal the diversity of metabolic patterns across different groups: wt, bacterial wild type; mut, bacterial mutant.
Experimental Section
Bacterial Cultures
Wild-type and five different mutants of Desulfovibrio alaskensis G20 were cultured in Wall lactate-sulfate medium.14 The culture medium was supplemented with 0.1% yeast extract. The bacteria were grown at 30 °C in an anaerobic growth chamber (Coy Laboratory Product, Inc., Grass Lake, MI). For exposing bacteria to salt stress, the bacterial cells were washed in minimal media and grown in minimal media to mid log phase (OD = 0.4) at 30 °C. At mid log phase, sodium chloride was added to a final concentration of 800 mM in the culture medium and the cells were cultured for 1 h. The bacterial cells were collected by centrifugation (4000g for 5 min) and the cell pellets were snap frozen in liquid nitrogen and stored at −80 °C until the extraction.
Cell Culture
Cell culture and biological reagents Ramos (CRL-1596), Raji (CCL-86), and SUP-T1 (CRL-1942) cell lines were obtained from American Type Culture Collection (ATCC, Manassas, VA, USA). Cell-culture conditions were as described before.15
Human Blood Sampling
Human blood was sampled from 20 normal, healthy subjects (10 males and 10 females) recruited from the Normal Blood Donor Program and from the general population of The Scripps Research Institute employees. The samples were immediately transferred from the heparin syringe into labeled aliquot tubes and centrifuged at 4 °C at 2000 rpm for 20 min. Blood plasma was removed and frozen at −80 °C.
Metabolite Extraction
The metabolites were extracted from bacterial cell pellets (Desulfovibrio alaskensis G20) using a MeOH/ACN/H2O (2:2:1, v/v) solvent mixture. One milliliter of chilled solvent mixture was added to each bacterial pellet, vortexed for 30 s, and flash frozen in liquid nitrogen for 1 min. The samples were then allowed to thaw at room temperature and sonicated in a water bath for 5 min. This cycle of cell lysis in liquid nitrogen combined with sonication was repeated three times. To precipitate proteins, the samples were incubated for 1 h at −20 °C. The vials were centrifuged at 4 °C for 15 min at 13 000 rpm. The supernatant was collected and evaporated to dryness in a vacuum concentrator. The dry extracts were then reconstituted in 100 μL of ACN/H2O (1:1, v/v), sonicated for 1 min and centrifuged at 4 °C for 15 min at 13 000 rpm. The supernatants were transferred to HPLC vials and stored at −80 °C until LC/MS analysis.
Cell extractions and analyses were performed as described before.16 For normalization, the cells were counted using the Guava Viacount assay Millipore, Billerica, MA, USA) and placed into 5 mL aliquots of 10 million cells per replicate before extraction, to ensure uniform cell numbers in each replicate.
Human plasma samples (200 μL) were extracted using 800 μL of cold MeOH/ACN (1:1, v/v). Protein precipitation and dry extract reconstitution was performed as described above for bacteria pellets.
LC/MS Analysis
Metabolite extracts from bacteria and human blood plasma were analyzed on a 6538 Ultra High Definition QTOF mass spectrometer (Agilent Technologies) interfaced with a 1200 series HPLC system (Agilent Technologies). Metabolite extracts from bacteria were analyzed by HILIC/MS in ESI negative mode. A Phenomenex Luna Aminopropyl column (3 μm, 150 mm × 1.0 mm I.D.) was used for LC separation of metabolites for HILIC/MS analysis. For HILIC, the mobile phase was composed of A = 10 mM ammonium acetate and 10 mM ammonium hydroxide in 95% water and B = 95% acetonitrile (pH-9.7). The samples were loaded onto the column using 100% B (0–5 min) at a flow rate of 50 μL/min and resolved using a linear gradient of 100% B to 100% A over a period of 50 min (5–55 min). A 10 min post-run was applied for HILIC, to ensure column re-equilibration and maintain reproducibility.
Lymphoma metabolomic profiling was performed by RPLC/MS in ESI positive mode. Samples were analyzed by using a XBridge C18, 3.5 μm, 150 mm × 1.0 mm I.D. column (Waters) and the standard mobile phase, A = 0.1% formic acid in water and B = 0.1% formic acid in acetonitrile. A linear gradient elution from 100% A (0–5 min) to 100% B (50–55 min) was applied.
Data Analysis
Data were analyzed by using interactive XCMS Online, which is freely available at https://xcmsonline.scripps.edu. The web interface has been developed for two-group, meta-analysis, and multigroup analysis. It allows users to either upload data sets using a java applet or select preuploaded data sets on XCMS Online. Following the upload of raw data files, users can select preset parameters (or customize them) depending on the instrument platform in which the data were acquired. The parameters are displayed in the web browser using the jQuery-UI framework, with each tab organized by category. Users can define parameters for statistical analysis (parametric/nonparametric, paired/unpaired) based on the type of experiment and data. The raw data files are than processed for peak detection, retention-time correction, chromatogram alignment, metabolite feature annotation, statistical evaluation, and putative identification through METLIN standard database matching. Metabolite features are defined as ions with unique m/z and retention-time values.
Parameter settings for XCMS processing of our demonstration data acquired by HILIC were as follows: centWave for feature detection (Δ m/z = 15 ppm, minimum peak width = 10 s, and maximum peak width = 120 s); obiwarp settings for retention-time correction (profStep = 1); and parameters for chromatogram alignment, including mzwid = 0.015, minfrac = 0.5, and bw = 5. The relative quantification of metabolite features was based on EIC (extracted ion chromatogram) areas. For XCMS processing of reversed-phase LC data, we used the same parameters except for chromatographic peak width settings, which was set as minimum peak width = 10 s and maximum peak width = 60 s. The results output, including EICs, boxplots, cloud plots, Venn/Edwards diagrams and PCA, were exported directly from XCMS Online.
Results and Discussion
The XCMS Online platform was enhanced to implement paired two-group comparisons, higher-order meta-analysis, and multiple group comparisons. Additional statistical tests were introduced, and the interactive visualization tools (Figures 2–7) were improved and developed to help deconvolve complex untargeted metabolomic data sets. The statistical tests are carried out systematically following feature detection and profile alignment, providing users an interface to directly visualize differentially expressed or significantly altered metabolic features. Here we highlight the appropriate usage of different statistical tests and demonstrate the value of interactive, univariate (cloud plot), and multivariate (PCA plots) visualization tools for different experimental designs: two-group comparison, meta-analysis, and multigroup comparison.
Figure 2.
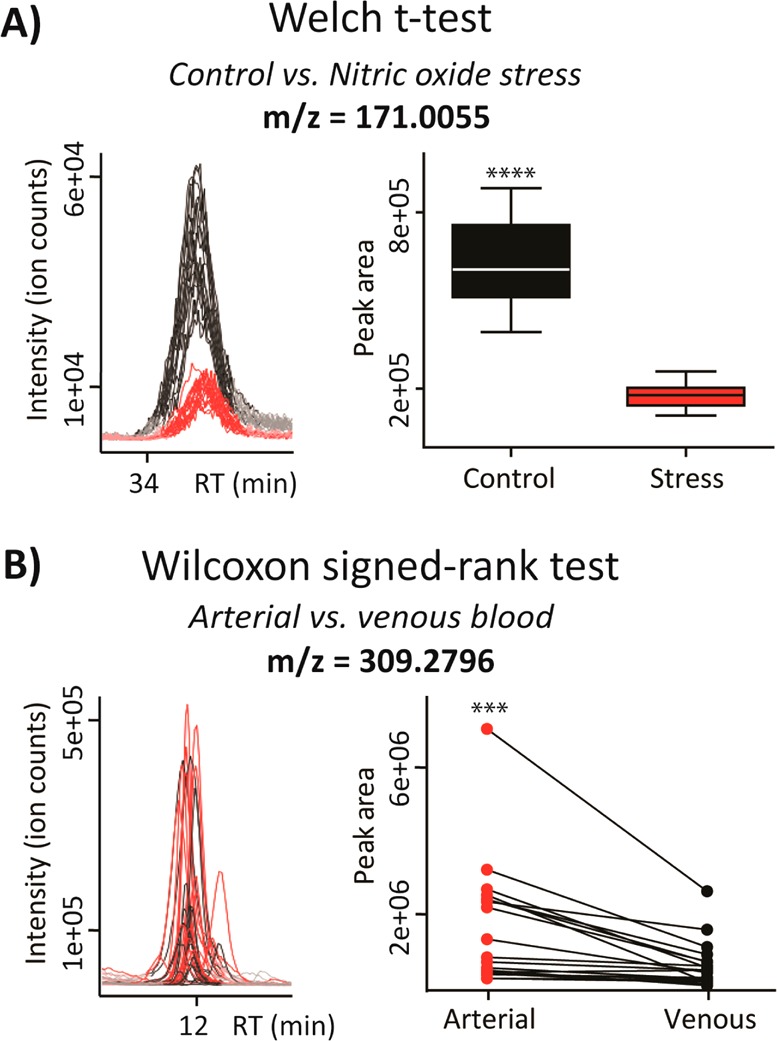
Representative examples of independent and dependent (paired) two-group experimental design. Extracted ion chromatogram and box-plot/paired plot are shown for the features of interest. (A) A significantly down-regulated (p < 0.001) metabolite feature (m/z 171.005; METLIN MS/MS match, glycerol phosphate) in independent group design (control versus stressed bacterial population) was identified by using an independent parametric Welch t test. Welch’s t test is used to compare the means of two independent sample groups with the assumption that two-group variances may differ. (B) A significantly higher level (p < 0.001) of metabolite feature (m/z 309.279; METLIN hit, eicosenoic acid) in arterial blood plasma was determined by a paired nonparametric Wilcoxon test. Wilcoxon signed-rank test is a nonparametric alternative to the paired t test used to compare the related samples.
Figure 7.
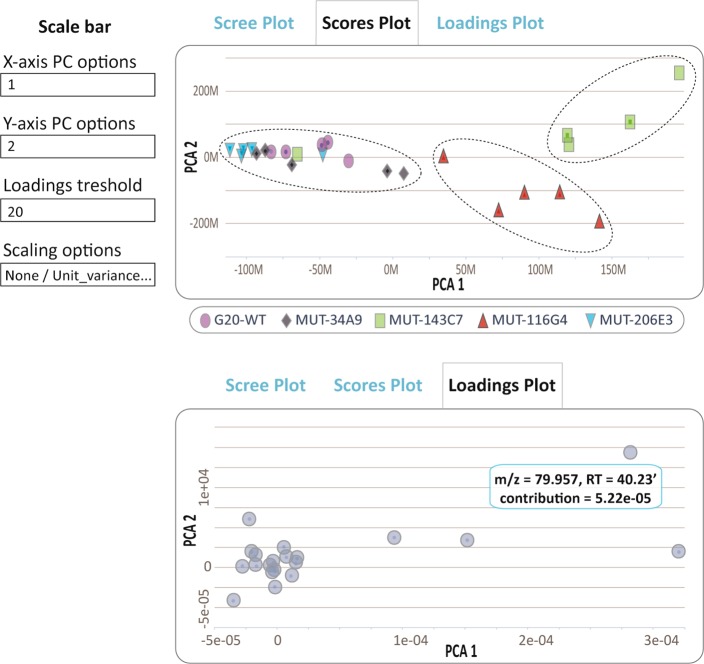
Interactive principal component analysis. A Scores plot showing the correlation between the samples (top panel) and a Loadings plot showing the relationship between the metabolite features that relate to the sample grouping (bottom panel). The clusters represent wild type and different mutant strains of Desulfovibrio alaskensis G20 (WT: wild type; MUT: mutant). The annotations for different mutant strains are given in the legend of Figure 5. The user has the option to set the loadings threshold and to apply different scaling criteria.
Two-Group Comparisons
The most common experimental design in metabolomics is two-group analysis, where “disease” and “control” or “before” and “after” treatment groups are compared. Even in a simple two-group experiment, choosing the right statistical test may be a challenging task for users without a background in the field of biostatistics. Depending on data distribution and experimental design, XCMS Online offers the choice of parametric or nonparametric, independent (unpaired) or dependent (paired) two-group tests (Table 1). Parametric tests are applied when the metabolomic data (peak area or intensity measurements) meet the assumption of normal distribution. When these conditions are not satisfied, the nonparametric alternative tests should be used as they do not rely on assumptions about the parameters of the data distribution.17 Parametric and nonparametric tests can be used to assess the independent (unpaired) and dependent (paired) sample groups. The independent or unpaired group tests are designed to compare means between two groups composed of different subjects who were randomly selected (e.g., metabolic response in “disease” versus “healthy” individuals). In the case of paired tests, the subjects in both groups are the same (or matched pairs) and therefore the mean difference between two repeated observations (peak measurements) is compared (e.g., metabolic response before and after drug treatment). When the choice of paired test is made, the XCMS Online interface provides a separate dialogue-box where users can align the correct pairings by the “drag-n-drop” option.
Table 1. Statistical Tests and Interactive Visualization Tools Implemented within the XCMS Online Platform for Univariate Analysis (One Variable at a Time).
| univariate
statistical analysis | |||
|---|---|---|---|
| experimental design | parametric
test (data follow normal distribution) |
nonparametric alternative
test (no assumption about data distribution) |
visualization tools |
| comparison
of two independent groups |
independent t test (Welch t test) |
Mann–Whitney | interactive two-group cloud plot, box–whisker plots, extracted-ion chromatograms |
| comparison of two dependent (paired or matched) groups |
paired t test | Wilcoxon signed-rank | |
| comparison of ≥ three independent groups |
one-way ANOVA with post-HOC |
Kruskal–Wallis | interactive multigroup cloud plot, box–whisker plots, extracted-ion chromatograms |
| comparison of ≥ three dependent (matched or related) groups |
repeated measures ANOVA | Friedman | |
In general, two-group tests allow users to determine the metabolite features whose levels are significantly different between two defined conditions. The examples from two representative metabolomic experiments are shown in Figure 2. In the first example, Welch’s t test was used to evaluate the metabolic response in bacteria subjected to nitric oxide stress versus control grown in optimal conditions. Each compared group had 5 independent biological replicates. Among the many identified changes, the metabolic feature with m/z 171.005, putatively identified as glycerol-phosphate, was found significantly down-regulated (p < 0.0001) in the stressed bacterial population (Figure 2a). The second example demonstrates the appropriate usage of a paired nonparametric test or a Wilcoxon signed-rank test when comparing metabolic profiles of arterial versus venous blood plasma from 20 human individuals. Each compared group had 20 subjects from whom arterial and venous bloods were sampled simultaneously. Among other subtle differences, the significantly higher (p = 0.0002) abundance of the metabolite feature with m/z 309.280, putatively identified as eicosenoic acid, has been observed in arterial blood, as shown by the paired plot (Figure 2b).
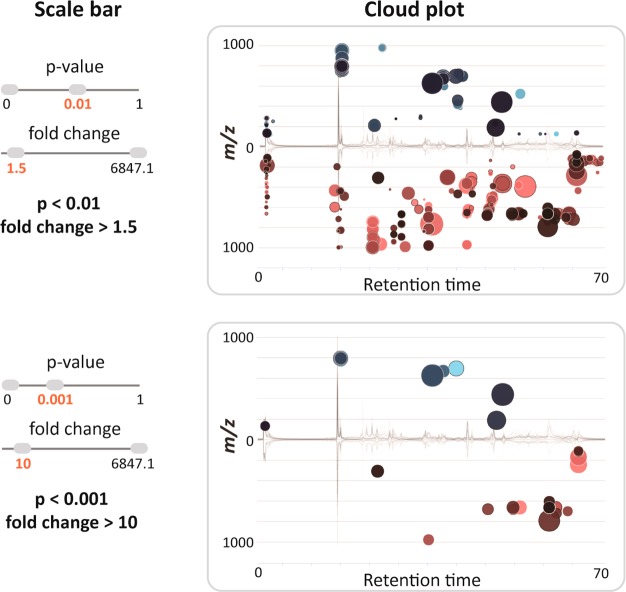
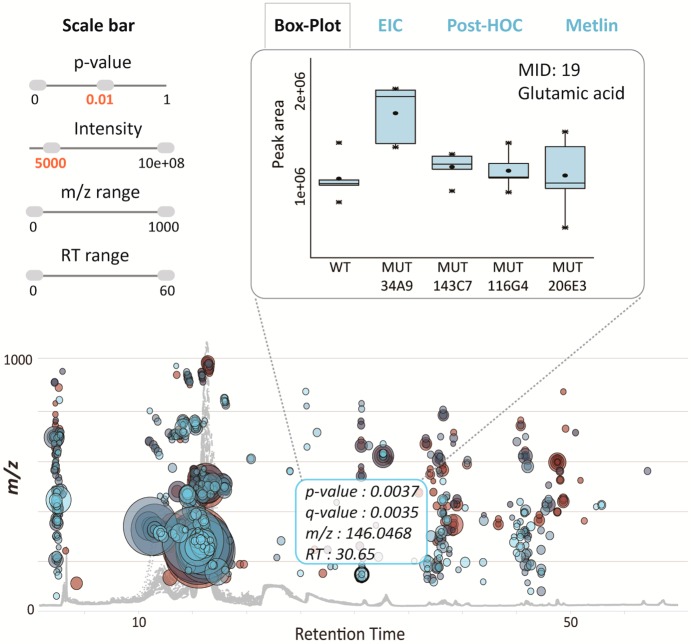
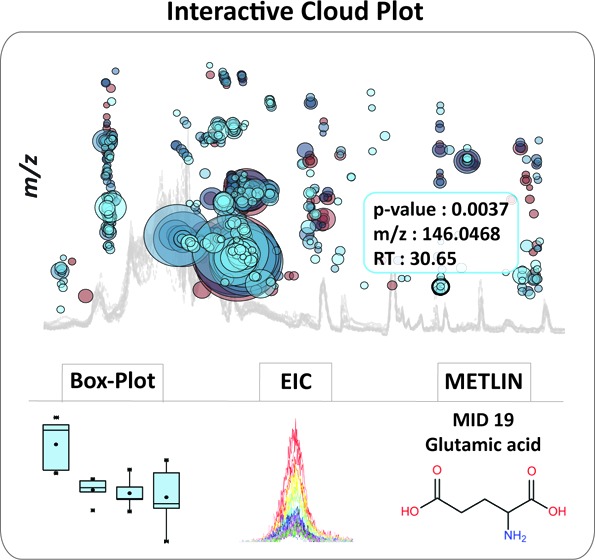
A typical untargeted metabolomic experiment comparing any two conditions (e.g., normal vs disease) yields hundreds of altered features. To visualize the results and filter out significantly altered features, the original cloud plot18 has been expanded from simply providing feature assignments (m/z, retention time, p-value, and directional fold change) to the interactive cloud plot or a dynamic interface enabling users to customize the display (Figure 3 and 4). The default view shows a “main panel” that allows users to modify p-value and fold-change filters (Figure 3), where the thresholds can be changed either by using a slider or the text box below the slider. An “advanced” selection panel allows users to modify m/z range, retention time, and ion-intensity range, depending on the features of interest. Additional choices provided in the advanced selection panel include multiple color options for up-regulated and down-regulated metabolic features, options to enable or disable TICs, colorize TICs, mark features with METLIN hits19 and also to either show or hide isotopic peaks. Furthermore, the “zoom” functionality allows users to focus and magnify a desired area of the plot by dragging the cursor across that area. This functionality is useful in plots with large numbers of data points that cluster together. The plot can be reset to the original view with a “mouse click”.
Figure 3.
Dynamically generated images of the interactive cloud plot based on user-specified thresholds for p-value and fold change. The plot was generated for an untargeted experiment comparing lymphoblastic leukemia cell lines (Raji parental vs SUP-T1 parental line). Each bubble in the plot corresponds to a metabolite feature. Metabolite features are projected depending on their retention time (x-axis) and m/z (y-axis). The color of the bubble denotes directionality of fold change and the size of the bubble denotes the extent of the fold change. Statistical significance (p-value) is represented by the bubble’s color intensity. The features up-regulated in the SUP-T1 line compared to the Raji cell line are displayed in blue.
Figure 4.
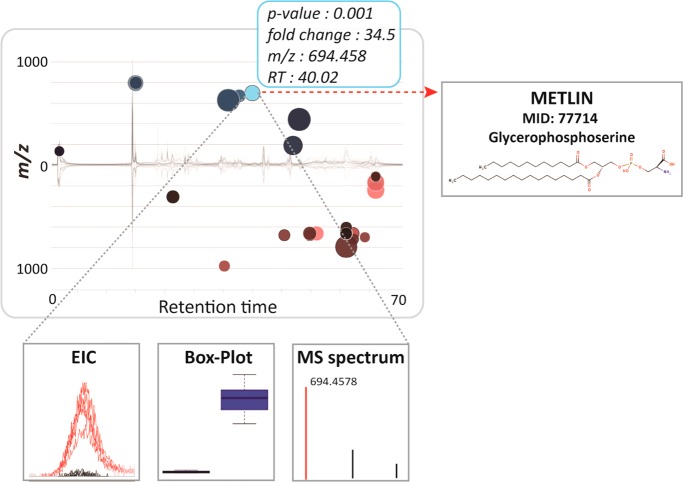
Interactive cloud plot with customized metabolomic data visualization. When a user scrolls the mouse over a bubble, feature assignments are displayed in a pop-up window (m/z, RT, p-value, fold change) with potential METLIN hits. Each bubble is linked to the METLIN database to provide putative identifications based on accurate m/z. When a bubble is selected by a mouse click, its EIC, box–whisker plot, and MS spectrum appear on the bottom of the main panel. The feature with m/z 694.458 and a putative METLIN hit for glycerophosphoserine (PS) seems to be specific to the SUP-T1 parental cell line.
Cloud plots also facilitate characterization and structural identification of metabolite features in untargeted metabolomic experiments. For this purpose, the cloud plot is directly linked to METLIN,20 the in-house developed standard database to allow users to interrogate metabolite matches based on accurate m/z measurements (Figure 4). Dysregulated features with METLIN matches are designated with a dark outline around the bubble, linking the user directly to the METLIN metabolite page with a “mouse click”. METLIN contains 240 000+ metabolic entities of which 11 000+ have experimental MS/MS spectra.
Meta-Analysis: Shared Metabolic Pattern Across Multiple Experiments
Metabolomic studies can reveal hundreds of dysregulated metabolic features, even using stringent statistical criteria. Higher-order meta-analysis across multiple experiments investigating a similar biological question can allow for substantial data reduction.21,22 Meta-analysis can be especially useful for untargeted metabolomic studies, where the identification of metabolite features represents a bottleneck. The integration of data from multiple biologically relevant studies facilitates the prioritization of interesting metabolite features and increases the value and reliability of results.21,23
Meta-analysis has already been extensively used in genome-wide association studies to identify the susceptibility loci for complex disorders24−26 and cancer gene expression signatures when comparing multiple studies carried out by different groups or using different technology platforms.27−29 It has also been applied across different cancer types to identify general signatures associated with cancer metabolism.30
An interface for meta-analysis has been implemented within the XCMS Online platform to enable the identification of shared homologous patterns of metabolic variation across the results of multiple different experiments (Table 2). The main interface is organized as a step-navigation wizard that allows users to select the preprocessed experiments for comparison and define threshold parameters for feature filtering and chromatogram realignment. Metabolite features can be filtered based on fold change, p-value, and ion intensity. Subsequently, the metabolic profiles from multiple experiments are realigned and the results of meta-analysis can be visualized by using two different modalities, the traditional Venn diagram (Figure 5) and the Edwards’s Venn diagram constructed as segments of a sphere. Venn diagrams display the number of shared metabolic features that are hyperlinked to a tabular output providing a list of corresponding m/z and retention-time values along with potential matches to the METLIN database.
Table 2. Interactive Visualization Tools Implemented within XCMS Online Platform for Meta-Analysis.
| meta-analysis | ||
|---|---|---|
| experimental design | method | visualization tools |
| cross comparison of multiple experiments | higher-order analysis | Venn diagram, Edwards diagram |
Figure 5.
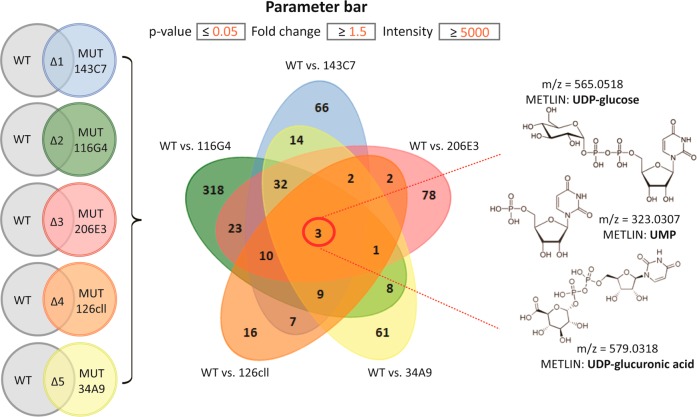
Meta-analysis of the salt-stress response across five different mutant strains of Desulfovibrio alaskensis G20. The results of five two-group comparisons (left). Shared patterns of stress response are characterized by significant up-regulation (p < 0.01) of three metabolites displayed in the center of the Venn diagram (middle). The putative identity of those metabolites, verified by MS/MS matching to standards in METLIN, is shown on the right. Mutant annotations: 143C7, transcriptional regulator (Cro/Cl family); 206E3, potassium uptake protein TrkA; 34A9, lysine 2,3-aminomutase; 126cll, beta-lysine N-acetyltransferase; 116G4, V-type ATPase (subunit J, trk1).
As an example, the common pattern of stress response among five different mutant strains of the sulfate-reducing bacterium, Desulfovibrio alaskensis G20 was identified by meta-analysis (Figure 5). The five different hypersensitive mutants were derived from a fitness profiling experiment to identify the mutants that were susceptible for salt stress as compared to the wild type counterpart. Simple pairwise comparisons of each mutant to their respective wild-type resulted in more than 1000 dysregulated metabolite features of statistical significance. As shown in the Venn diagram, by using meta-analysis we were able to reduce this number to three metabolites, which were significantly altered in all of the different mutant strains versus wild type bacteria culture, when subjected to short-term salt stress. The putatively identified metabolites corresponded to UDP-glucose, UMP, and UDP glucuronic acid, implying that the cell-wall biosynthesis pathways31 may be perturbed as a response to salt stress in all five hypersensitive mutant strains. In a similar manner, meta-analysis has been used to compare different models of pain where histamine has been identified as a commonly altered metabolite.32
Multiple Group Comparisons: Metabolite Variation Patterns Across Multiple Data Groups
Multiple group analysis is an extension of two-group analysis that allows the comparison of means for multiple independent groups (≥3) and enables the identification of metabolite features whose variation pattern is statistically significant. To evaluate the metabolite variation across different experimental groups, XCMS Online provides the univariate analysis of variance (one-way ANOVA) as a parametric test option and the Kruskal–Wallis test as its nonparametric alternative. The Posthoc multiple comparison test is used to determine which groups significantly differ in their metabolite expression pattern.
To visualize the statistically significant results of multigroup analysis and filter out the features of interest, the original two-group cloud plot18 has been adapted for a multigroup output. Multigroup cloud plots display the metabolite features whose level varies significantly across different analyzed groups or data classes. Metabolite features are projected in the same manner as on two-group cloud plots, depending on their m/z ratio and retention time. The new dynamic interface enables users to adjust or determine the statistical significance threshold (ANOVA or Kruskal–Wallis p-value), feature intensity, m/z, and retention-time range for the best representation of targeted features of interest. The box–whisker plot, EIC, post-HOC values, and METLIN hits can be visualized for each metabolic feature with a simple “mouse click” on the specific bubble. The same example of wild type and different mutants of Desulfovibrio alaskensis G20 exposed to salt stress that was processed by the meta-analysis tools was used to demonstrate the multigroup comparison. Relative to meta-analysis where the objective was to identify the shared pattern of metabolic response to stress, the multigroup analysis highlighted the differences in the pattern of stress response across wild type and different, hypersensitive mutants. Among many differentially expressed features, the variation pattern of glutamic acid across defined wild-type and mutant groups is shown in Figure 6. During the exposure to salt stress, the uptake and/or synthesis of glutamic acid was significantly up-regulated in the lysine-aminomutase enzyme mutant (MUT-34A9) when compared to the other mutants and wild type. Multigroup analysis can be essential to discriminate the metabolic response associated with a specific phenotype and therefore to link specific metabolites with distinct functional roles. For example, multigroup comparison could be used to functionally characterize different brain regions or to identify metabolic patterns specific to different types of cancer.
Figure 6.
Interactive multigroup cloud plot with customized metabolomic data visualization. Metabolite features whose level varies significantly (p < 0.01) across wild-type and different mutants are projected on the cloud plot depending on their retention time (x-axis) and m/z (y-axis). Each metabolite feature is represented by a bubble. Statistical significance (p-value) is represented by the bubble’s color intensity. The size of the bubble denotes feature intensity. When the user scrolls the mouse over a bubble, feature assignments are displayed in a pop-up window (m/z, RT, p-value, fold change). When a bubble is selected by a “mouse click”, the EIC, Box-Whisker plot, Posthoc, and METLIN hits appear on the main panel. Each bubble is linked to the METLIN database to provide putative identifications based on accurate m/z. The variation pattern of glutamic acid (m/z 146.0468, MS/MS METLIN match) across different mutants is shown by a box–whisker plot.
In the course of an untargeted metabolomic experiment, many variables or metabolite features are measured simultaneously and the resulting data is multivariate data. Moreover, the metabolites in a biochemical pathway behave in an orchestrated way and tend to covary.33 This relation or interaction between metabolites may be important in the prediction of different groups or classes, and the differences sometimes cannot be revealed by simply analyzing the metabolites separately by univariate tests.33,34 Therefore, in addition to the presented univariate statistical solutions, an interactive multivariate principal component analysis (PCA) has been implemented within the XCMS Online platform for multigroup analysis. PCA is one of the most widely used multivariate analysis tools in metabolomics, especially in the case of untargeted metabolic profiling or fingerprinting. PCA projects the original multidimensional data on a lower dimensional space by capturing as much of the observed variation as possible. It is based on the inherent data structure and makes use of covariances or correlations among the variables or metabolite features.
XCMS Online provides an interactive PCA report sheet, including a scree plot, scores plot, and a loadings plot. The scree plot is used to determine the appropriate number of principal components needed to represent the maximum of observed variation. The scores plot describes the relationship between the samples and the loadings plot is useful for the interpretation of the correlation between metabolites and how they relate to the samples (Table 3). When the data from the multigroup analysis of wild type and mutants of Desulfovibrio alaskensis G20 were projected in two-dimensional space by retaining the metabolite features that explain maximum variation among different groups, three main clusters were observed (Figure 7). Two mutants (for transcriptional regulator, MUT-143C7; for ATP-ase subunit J, MUT-116G4) hypersensitive to salt stress cluster separately, which implies that they have specific metabolic responses to salt stress in comparison to wild type and other mutants (PCA 1), as well as to each other (PCA 2). Metabolite features that drive this sample clustering can be identified on the loadings plot. The metabolite features that show the largest possible variance lie on the first principal component and those that show subsequent largest variance lie on the second principal component. Although the validity of the present PCA model should be explored further, our purpose here was to demonstrate the concept of the interactive PCA as a diagnostic and interpretation tool for untargeted metabolomic experiments. At its current stage of development, the display of scree, scores, and loadings plots can be modified by a user definition of standardization or scaling criteria (none/unit variance/log/pareto), depending on the type of data. Scaling gives the possibility of down-weighting irrelevant or noisy features.
Table 3. Statistical Methods and Interactive Visualization Tools Implemented within the XCMS Online Platform for Multivariate Analysis (Multiple Variables Simultaneously).
| multivariate
statistical analysis | ||
|---|---|---|
| experimental design | method | visualization tools |
| two-group or multigroup comparison |
principal component analysis (PCA) | interactive scree, scores and loadings plot |
| hierarchical cluster analysis (HCA) | heat map, dendogram | |
Conclusion
The untargeted metabolomic workflow relies on statistical data analysis, result visualization, and data presentation, yet these tasks often represent a considerable challenge for scientists with limited bioinformatic expertise. The significant growth of the metabolomic community in the past few years has accelerated the development of user-friendly informatic tools for high-throughput analysis of large metabolomic data sets. In the given context, the interactive XCMS Online platform was developed as an easy-to-use web tool for complete data processing, from raw data upload, peak detection, retention-time correction, profile alignment, comprehensive statistical data evaluation, and visualization to putative metabolite identification. Key to performing more complex data analyses, XCMS Online now incorporates paired two-group analysis, meta-analysis, and multiple group analysis to fit different experimental designs. An interactive visualization scheme has been implemented within these new analysis methods. The interactive, two-group and multigroup cloud plots allow users to navigate across and filter out metabolic features of interest. The variation pattern of each feature can be observed with a box–whisker plot and extracted ion chromatograms. The interactive PCA allows for visualization of the correlations among samples and metabolite features that drive the sample clustering. Finally, a direct link between XCMS Online and the standard METLIN database provides users with putative metabolite identification.
Acknowledgments
This work was supported by the California Institute of Regenerative Medicine no. TR1-01219 (G.S.), the US National Institutes of Health grants R01 CA170737 (G.S.), R24 EY017540 (G.S.), P30 MH062261 (G.S.), RC1 HL101034 (G.S.), P01 DA026146 (G.S.), R01 ES022181 (GJP), and L30 AG0038036 (GJP). Financial support was also received from the U.S. Department of Energy grant nos. FG02-07ER64325 and DE-AC0205CH11231 (G.S.). H. G. is a Wellcome Trust-DBT Early Career Fellow.
Author Contributions
H.G. and J.I. contributed equally
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
References
- Tautenhahn R.; Patti G. J.; Rinehart D.; Siuzdak G. Anal. Chem. 2012, 84, 5035–5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikoff W. R.; Pendyala G.; Siuzdak G.; Fox H. S. J. Clin. Invest. 2008, 118, 2661–2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikoff W. R.; Anfora A. T.; Liu J.; Schultz P. G.; Lesley S. A.; Peters E. C.; Siuzdak G. Proc. Natl. Acad. Sci. U. S. A. 2009, 106, 3698–3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinayavekhin N.; Homan E. A.; Saghatelian A. ACS Chem. Biol. 2010, 5, 91–103. [DOI] [PubMed] [Google Scholar]
- Smith C. A.; Want E. J.; O’Maille G.; Abagyan R.; Siuzdak G. Anal. Chem. 2006, 78, 779–787. [DOI] [PubMed] [Google Scholar]
- Lommen A. Anal. Chem. 2009, 81, 3079–3086. [DOI] [PubMed] [Google Scholar]
- Pluskal T.; Castillo S.; Villar-Briones A.; Oresic M. BMC Bioinf. 2010, 11, 395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melamud E.; Vastag L.; Rabinowitz J. D. Anal. Chem. 2010, 82, 9818–9826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J.; Mandal R.; Sinelnikov I. V.; Broadhurst D.; Wishart D. S. Nucleic Acids Res. 2012, 40, W127–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemela P. S.; Castillo S.; Sysi-Aho M.; Oresic M. J. Chromatogr. B Analyt. Technol. Biomed. Life. Sci. 2009, 877, 2855–2862. [DOI] [PubMed] [Google Scholar]
- Kastenmuller G.; Romisch-Margl W.; Wagele B.; Altmaier E.; Suhre K.. J. Biomed. Biotechnol. 2011, 2011. [DOI] [PMC free article] [PubMed]
- Xia J.; Wishart D. S.. Curr. Protoc Bioinformatics. 2011, Chapter 14, Unit 14 10. [DOI] [PubMed]
- Rinehart D.; Johnson C. H.; Nguyen T.; Ivanisevic J.; Benton H. P.; Lloyd J.; Deutschbauer A.; Arkin A.; Patti G. J.; Siuzdak G.. Nat. Biotechnol. 2014, In Press [DOI] [PMC free article] [PubMed]
- Keller K. L.; Bender K. S.; Wall J. D. Appl. Environ. Microbiol. 2009, 75, 7682–7691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanisevic J.; Zhu Z. J.; Plate L.; Tautenhahn R.; Chen S.; O’Brien P. J.; Johnson C. H.; Marletta M. A.; Patti G. J.; Siuzdak G. Anal. Chem. 2013, 85, 6876–6884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson C. H.; Fisher T. S.; Hoang L. T.; Felding B. H.; Siuzdak G.; O’Brien P. J. Metabolomics 2014, 10.1007/s11306-014-0622-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinaixa M.; Samino S.; Saez I.; Duran J.; Guinovart J. J.; Yanes O. Metabolites 2012, 2, 775–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patti G. J.; Tautenhahn R.; Rinehart D.; Cho K.; Shriver L. P.; Manchester M.; Nikolskiy I.; Johnson C. H.; Mahieu N. G.; Siuzdak G. Anal. Chem. 2013, 85, 798–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautenhahn R.; Cho K.; Uritboonthai W.; Zhu Z.; Patti G. J.; Siuzdak G. Nat. Biotechnol. 2012, 30, 826–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. A.; O’Maille G.; Want E. J.; Qin C.; Trauger S. A.; Brandon T. R.; Custodio D. E.; Abagyan R.; Siuzdak G. Ther. Drug Monit. 2005, 27, 747–751. [DOI] [PubMed] [Google Scholar]
- Patti G. J.; Tautenhahn R.; Johannsen D.; Kalisiak E.; Ravussin E.; Bruning J. C.; Dillin A.; Siuzdak G. Metabolomics 2014, 10.1007/s11306-013-0608-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patti G. J.; Tautenhahn R.; Siuzdak G. Nat. Protoc. 2012, 7, 508–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaever A.; Landesfeind M.; Feussner K.; Morgenstern B.; Feussner I.; Meinicke P. PLoS One 2014, 9, e89297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung C. C.; Kanetsky P. A.; Wang Z.; Hildebrandt M. A.; Koster R.; Skotheim R. I.; Kratz C. P.; Turnbull C.; Cortessis V. K.; Bakken A. C.; Bishop D. T.; Cook M. B.; Erickson R. L.; Fossa S. D.; Jacobs K. B.; Korde L. A.; Kraggerud S. M.; Lothe R. A.; Loud J. T.; Rahman N.; Skinner E. C.; Thomas D. C.; Wu X.; Yeager M.; Schumacher F. R.; Greene M. H.; Schwartz S. M.; McGlynn K. A.; Chanock S. J.; Nathanson K. L. Nat. Genet. 2013, 45, 680–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinds D. A.; McMahon G.; Kiefer A. K.; Do C. B.; Eriksson N.; Evans D. M.; Pourcain B. St; Ring S. M.; Mountain J. L.; Francke U.; Davey-Smith G.; Timpson N. J.; Tung J. Y. Nat. Genet. 2013, 45, 907–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett J. C.; Clayton D. G.; Concannon P.; Akolkar B.; Cooper J. D.; Erlich H. A.; Julier C.; Morahan G.; Nerup J.; Nierras C.; Plagnol V.; Pociot F.; Schuilenburg H.; Smyth D. J.; Stevens H.; Todd J. A.; Walker N. M.; Rich S. S. Nat. Genet. 2009, 41, 703–707.19430480 [Google Scholar]
- Mehra R.; Varambally S.; Ding L.; Shen R.; Sabel M. S.; Ghosh D.; Chinnaiyan A. M.; Kleer C. G. Cancer Res. 2005, 65, 11259–11264. [DOI] [PubMed] [Google Scholar]
- Chen X.; Liang S.; Zheng W.; Liao Z.; Shang T.; Ma W. BMC Genomics 2008, 9, 322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith O. L.; Melck A.; Jones S. J.; Wiseman S. M. J. Clin. Oncol. 2006, 24, 5043–5051. [DOI] [PubMed] [Google Scholar]
- Rhodes D. R.; Yu J.; Shanker K.; Deshpande N.; Varambally R.; Ghosh D.; Barrette T.; Pandey A.; Chinnaiyan A. M. Proc. Natl. Acad. Sci. U. S. A. 2004, 101, 9309–9314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson R. G.; Hussey H.; Baddiley J. Biochem. J. 1972, 127, 11–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautenhahn R.; Patti G. J.; Kalisiak E.; Miyamoto T.; Schmidt M.; Lo F. Y.; McBee J.; Baliga N. S.; Siuzdak G. Anal. Chem. 2011, 83, 696–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worley B.; Powers R. Curr. Metabolomics 2014, 1, 92–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccenti E.; Hoefsloot H. C. J.; Smilde A. K.; Westerhuis J. A.; Hendriks M. M. W. B. Metabolomics 2014, 10, 361–374. [Google Scholar]