Abstract
The motile bacterium Vibrio fischeri is the specific bacterial symbiont of the Hawaiian squid Euprymna scolopes. Because motility is essential for initiating colonization, we have begun to identify stage-specific motility requirements by creating flagellar mutants that have symbiotic defects. V. fischeri has six flagellin genes that are uniquely arranged in two chromosomal loci, flaABCDE and flaF. With the exception of the flaA product, the predicted gene products are more similar to each other than to flagellins of other Vibrio species. Immunoblot analysis indicated that only five of the six predicted proteins were present in purified flagella, suggesting that one protein, FlaF, is unique with respect to either its regulation or its function. We created mutations in two genes, flaA and flaC. Compared to a flaC mutant, which has wild-type flagellation, a strain having a mutation in the flaA gene has fewer flagella per cell and exhibits a 60% decrease in its rate of migration in soft agar. During induction of light organ symbiosis, colonization by the flaA mutant is impaired, and this mutant is severely outcompeted when it is presented to the animal as a mixed inoculum with the wild-type strain. Furthermore, flaA mutant cells are preferentially expelled from the animal, suggesting either that FlaA plays a role in adhesion or that normal motility is an advantage for retention within the host. Taken together, these results show that the flagellum of V. fischeri is a complex structure consisting of multiple flagellin subunits, including FlaA, which is essential both for normal flagellation and for motility, as well as for effective symbiotic colonization.
All animals and plants associate with bacteria at some stage during their lifetimes, and in many cases this native microbiota must be acquired directly from the host's environment. The process of establishing a relationship between environmental bacteria and a host requires adaptation by both partners, which is mediated by changes in behavioral, metabolic, and developmental characteristics. One bacterial function that is often required for establishment of such an association is flagellation and motility. In addition to a requirement for flagella so that an organism can reach the site of colonization, flagella and virulence traits have been linked through the following mechanisms: (i) motility and virulence genes can be coordinately regulated (1, 3), (ii) the flagellar apparatus can secrete virulence proteins in addition to flagellar subunits (50), and (iii) the flagellum itself can act as a mediator of adhesion or contact signaling to host cells (4, 17). Thus, the role of motility in host-bacterium interactions can be a complex role in which some or all of these mechanisms contribute to the motility phenotype. Because of these multiple effects and the limitations for study in many pathogenic associations, defining the role of motility in host-bacterium associations would benefit from the use of a natural model system that is easy to manipulate.
The association between the Hawaiian bobtail squid, Euprymna scolopes, and the luminous bacterium Vibrio fischeri is a useful model for studying the role of bacterial motility for several reasons. First, only motile cells of V. fischeri in the environment can colonize newly hatched juvenile animals, a process that can be closely mimicked in the laboratory. Second, the colonization process can be monitored continuously and indirectly by measuring the luminescence of intact animals. Third, the genetics of V. fischeri are well established and have proven to be useful for mutant studies (43, 48). Finally, the intimate nature of the association can be visualized by confocal laser scanning microscopy (CLSM) of green fluorescent protein (GFP)-labeled bacteria during their colonization of live juvenile squid (30, 34).
Within minutes to hours after the initial contact, both partners in this symbiosis respond to the others' presence with developmental and behavioral changes. One of the first known processes to occur is the secretion by the animal of an external mucus web that is used to trap environmental bacteria (32) and that shows selectivity for V. fischeri (33). Interestingly, while motile and nonmotile cells of V. fischeri are recruited similarly at this step, cells that are hyperflagellated are not recruited (29). After attaching to the mucus, the bacteria migrate along its strands and enter one of six pores, a process that appears to be dependent on flagellum-mediated motility (13, 34). Once the cells enter the pores, they traverse one of several ciliated ducts in order to colonize the internal nascent light organ, where the bacteria grow, divide, and, when a critical cell density is reached, produce luminescence. Each morning, a time when light is no longer needed by the animal, approximately 95% of the symbiotic cells are expelled, and the remaining 5% repopulate the light organ by the next evening. The squid light organ changes dramatically upon colonization by symbiotic V. fischeri as a result of changes at both the molecular (22) and protein (11) levels. Similarly, the bacteria respond to the host environment through altered gene expression that results in changes in at least two known colonization factors, motility (38) and luminescence (6).
V. fischeri cells are motile by means of a tuft of polar sheathed flagella that typically contains between one and five flagellar filaments (29). Although in most bacteria the flagellum is comprised of a single flagellin protein, some organisms can assemble flagella that contain multiple flagellin subunits (27, 40). In Vibrio species, for example, the flagella can consist of up to six different kinds of subunits (27); however, the functions of multiple flagellins in these species are not known. Interestingly, at least one of the flagellin subunits from each of the three Vibrio species in which such subunits have been described has distinct regulation and/or a distinct function (23; for a review see reference 27). In Vibrio cholerae, transcription of the gene encoding this subunit, flaA, is directly dependent on the motility master regulator, FlrA, and the alternative sigma factor, σ54, while transcription of the remaining four flagellin genes is dependent on the motility sigma factor, σ28 (36). Furthermore, the FlaA protein is essential for motility in V. cholerae (23) and for the production of a full-length flagellum in Vibrio anguillarum (31), while deletion of the gene encoding any one of the other four flagellins has no apparent effect on motility. Interestingly, in V. anguillarum FlaA, as well as FlaD and FlaE, are essential for virulence in fish when the fish are infected by both the immersion and intraperitoneal routes. In V. fischeri, flaA is subject to regulation by σ54 (49), as it is in V. cholerae. Thus, flaA was deemed a likely candidate for our studies.
To begin to characterize the flagellar apparatus in V. fischeri, we identified the genes encoding six flagellin subunits. Our results revealed that the flagellum of V. fischeri, like the flagella of other Vibrio species, is complex and is comprised of multiple flagellin subunits. We found that at least one of the V. fischeri flagellins, FlaA, is both required for full motility and essential for normal symbiotic colonization.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and reagents.
Wild-type V. fischeri strain ES114, which was isolated from E. scolopes (6), was used as the parent strain. The mobilizable plasmid pKV111, containing a red-shifted GFP derivative (34, 42), was used in CLSM studies. V. fischeri cells were generally grown at 28°C either in SWT medium (6), which contained 0.5% Bacto Tryptone (Difco Co., St. Louis, Mo.), 0.3% yeast extract, and 0.3% glycerol in 70% seawater, or in LBS medium, which contained 1% Bacto Tryptone, 0.5% yeast extract, 2% NaCl, and 20 mM Tris-HCl (pH 7.4). Chemotaxis plates were prepared with tryptone media (TM) containing 1% Bacto Tryptone, 0.88% NaCl, 0.62% MgSO4, 0.072% CaCl2, and 0.038% KCl, and motility media (MM) were prepared with SWT medium. Agar was added at a concentration of 1.5% for solid media, at a concentration of 0.25% for TM, and at a concentration of 0.3 to 0.7% for MM. Growth of mutant strains in cultures was determined by using SWT media, and luminescence was determined as described previously (29). When chloramphenicol was necessary to maintain V. fischeri plasmids, it was added to growth media (5 μg ml−1) or to seawater in squid colonization experiments (2 μg ml−1). Kanamycin was added to growth media at a concentration of 50 μg ml−1 for Escherichia coli strains and at a concentration of 100 μg ml−1 for V. fischeri strains. All chemicals were obtained from Sigma Chemical Co. (St. Louis, Mo.). Restriction enzymes and DNA ligase were obtained from New England Biolabs (Beverly, Mass.). AmpliTaq DNA polymerase was obtained from Perkin-Elmer (Branchburg, N.J.). Oligonucleotides were synthesized by Operon Technologies, Inc. (Alameda, Calif.).
Molecular genetic techniques.
Chromosomal and plasmid DNA were isolated and purified by using QIAGEN spin columns as suggested by the manufacturer (QIAGEN, Valencia, Calif.). DNA sequencing was conducted with an ABI automated DNA sequencer at the University of Hawaii Biotechnology/Molecular Biology Instrumentation Training Facility. Overlapping contiguous sequences were aligned by using Sequencer (Gene Codes Corp., Ann Arbor, Mich.). A sequence analysis was performed by using the BLAST program for database searches, and multiple-sequence alignments were constructed by using the ClustalWprogram (46). Consensus binding sequences for σ54 and σ28 were obtained from previous reports (5, 16). Phylogenetic analyses were performed with PAUP (45).
Cloning, sequence analysis, and disruption of flagellin genes.
Degenerate oligonucleotide primers for PCR were designed by using alignments of the predicted amino acid sequences for V. cholerae and Vibrio parahaemolyticus flagellins (GenBank accession numbers AF007121, AF007122, and AF069392). The PCR was performed as follows: 35 cycles of 95°C for 1 min, 48°C for 1 min, and 72°C for 1 min, followed by a 10-min extension at 72°C. The oligonucleotides used were either DM1 and DM3 or DM2 and DM4 (Table 1). The resulting PCR products were cloned by using the pCR2.1 vector and a TA cloning kit (Invitrogen) and were sequenced. Analysis of the resulting cloned products revealed the presence of at least three unique amplicons. One of the 568-bp PCR products (later identified as a portion of flaA) was used as a probe to screen a library of XbaI-digested V. fischeri ES114 chromosomal fragments. Consecutive screening of smaller and smaller pools of clones led to identification of a single clone, inserted into pDM13 (Table 1), that contained 9,275 bp of the V. fischeri sequence, including flaA. To create insertions in flagellin genes, pDM13 was digested with HindIII and ligated with pEVS79 that was similarly digested. One of the resulting plasmids, pDM88, contained a 3.4-kb HindIII fragment containing the entire flaA gene and a portion of the upstream sequence corresponding to the V. cholerae HAP2 gene. Another plasmid, pDM89, contained a 2.9-kb subclone that included the complete flaB and flaC open reading frames (ORFs). Both pDM88 and pDM89 were subjected to in vitro mutagenesis (Epicentre Technologies, Madison, Wis.), and the resulting plasmid pool was transformed into competent E. coli DH5α cells. Plasmids were isolated from kanamycin-resistant colonies, screened by restriction digestion analysis, and sequenced to confirm the location and orientation of the transposon insertion. The flaA::Knr allele from pDM88-16, which was found to contain an insertion in the flaA gene at codon position 141, was crossed into the chromosome of V. fischeri ES114 by marker exchange as previously described (44), which generated strain DM143 (Table 1). The flaC::Knr allele from one clone, pDM89-33, which contained an insertion at codon position 72 of the flaC gene, was similarly crossed into the chromosome of strain ES114, which generated strain DM138.
TABLE 1.
Strains, plasmids, and oligonucleotides used in this study
| Strain, plasmid, or oligonucleotide | Relevant characteristicsa | Reference or source |
|---|---|---|
| V. fischeri strains | ||
| Wild-type ES114 | Wild-type E. scolopes isolate | 6 |
| flrA mutant DM127 | flrA inactivated by insertion of a kanamycin resistance marker (kanR) | 30 |
| flaC mutant DM138 | flaC inactivated by insertion of a kanamycin resistance marker (kanR) | This study |
| flaA mutant DM143 | flaA inactivated by insertion of a kanamycin resistance marker (kanR) | This study |
| Plasmids | ||
| pCR2.1 | E. coli cloning vector; Knr | Invitrogen, Inc. |
| pEVS79 | Allelic exchange vector | 44 |
| pVO8 | V. fischeri cloning vector; Cmr/Err | 47 |
| pKV111 | gfp-containing derivative of pVO8 | 42 |
| pDM13 | 9.3-kb XbaI fragment containing flagellin sequences | This study |
| pDM88 | pEVS79 with 3.4-kb flaA-containing HindIII fragment from pDM13 | This study |
| pDM89 | pEVS79 with 2.9-kb flaC-containing HindIII fragment from pDM13 | This study |
| pDM88-16 | flaA::Knr at nucleotide position 424 | This study |
| pDM89-33 | flaC::Knr at nucleotide position 216 | This study |
| pDM104 | pVO8 with 1.2-kb flaA-containing PstI fragment from pDM13 | This study |
| Oligonucleotides | ||
| DM1 | TCNATGGARCGBYTNKCDTCNGG | This study |
| DM2 | ATGRCVRTTAANGTWARYAC | This study |
| DM3 | TTGVCCRTTRATGTAVGTNGC | This study |
| DM4 | CCRTTRATGTAVGTNGCHAVCTC | This study |
Knr, kanamycin resistance; Cmr, chloramphenicol resistance; Err, erythromycin resistance.
Motility assays.
The motilities of different strains of V. fischeri were measured by determining the movement of bacterial cells through MM containing different agar concentrations. Equal numbers of exponentially growing cells suspended in 2 μl of TM or SWT medium were spotted in the centers of MM or TM plates; the rate of movement away from the inoculation point in each plate was measured over the course of several hours, and the results were expressed as a rate relative to the wild-type movement. Digital images were obtained 8 h after inoculation. Comparable strains (strains ES114 and DM143, strains ES114/pDM104 and DM143/pDM104, or strains ES114/pVO8 and DM143/pVO8) were inoculated into the same plate to account for differences in hydration or medium composition among plates. Strains carrying plasmids (either the complementing plasmid, pDM104, or the parent plasmid, pVO8) were inoculated into soft agar plates containing chloramphenicol.
Preparation of flagellin proteins.
Flagella were isolated from cells grown to the mid-exponential phase as described previously (29). Briefly, cells were collected by centrifugation at 8,000 × g for 10 min at 4°C, resuspended in 1 ml of chilled 70% artificial seawater (37), and transferred to 25-ml polycarbonate tubes containing 10 ml of chilled 70% artificial seawater. Flagella were sheared by vigorous vortexing for 2 min, and the resulting cell suspension was examined by phase-contrast microscopy to ensure that cells were neither motile nor lysed after this treatment. The cells were removed by two rounds of centrifugation at 6,000 × g for 10 min at 4°C. Flagella were concentrated in a pellet by centrifuging the cell-free supernatant at 38,000 × g for 40 min at 4°C. This pellet was resuspended in 80 μl of a standard loading buffer before the protein was separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) on a 12.5% polyacrylamide gel (39). Separated proteins representing approximately 5 × 108 cells were detected by Coomassie brilliant blue staining.
2D polyacrylamide gel analysis.
Samples were prepared for two-dimensional (2D) analysis as described above, except that purified flagella were resuspended in sample buffer for 2D gel electrophoresis according to the manufacturer's suggestions (Amersham Biosciences, Uppsala, Sweden). Preparations of flagellar proteins, corresponding to the proteins from approximately 2 × 109 bacteria, were loaded onto a first-dimension gel strip (Immobiline Dry Strip; pH 4 to 7; 18 cm; Amersham Biosciences) and were separated by using a Multiphor II flatbed system (Pharmacia Biotech) as recommended by the manufacturer. The second dimension consisted of an Excel Gel XL SDS 12-14 (Pharmacia Biotech) gel, and after separation, the proteins were visualized by silver staining.
Immunoblot analysis.
Following SDS-PAGE and following second-dimension electrophoresis, proteins were transferred to and immobilized on Trans-Blot 0.45-μm-pore-size nitrocellulose membranes (Bio-Rad, Hercules, Calif.) by using 120 and 200 mA, respectively, for 1 h. The membranes were blocked by incubation in 4% dry milk in TTBS (0.5 M NaCl, 0.5% Tween 20, 10 mM Tris-HCl [pH 7.5]) for 16 h. Then the membranes were incubated for 2 h with primary polyclonal antibody 129, which was directed against the six diverse V. parahaemolyticus polar flagellins (26), that had been added to 2% dry milk in TTBS. The membranes were then washed three times for 10 min in TTBS, incubated for 45 min with secondary antibody (goat anti-rabbit immunoglobulin G conjugated to horseradish peroxidase) and avidin-horseradish peroxidase for detection of biotinylated standards (Amersham Biosciences, Little Chalfont, England) in TTBS, and washed three times for 10 min in TTBS. Antibody-reactive proteins were detected by using the ECL+plus Western blotting detection system (Amersham Biosciences, Little Chalfont, England).
Electron microscopy.
V. fischeri cells were prepared for transmission electron microscopy as described previously (29). Briefly, Formvar-coated copper grids (Ted Pella Co., Tustin, Calif.) were floated on suspensions of cells grown to the mid-exponential phase (optical density at 600 nm, 0.4) in SWT medium and then transferred to a drop of fixative solution (2.5% glutaraldehyde and 2.5% paraformaldehyde in 0.1 M sodium cacodylate buffer, pH 7.4) for 10 min. The grids were washed twice with Nanopure (Millipore Corp., New Bedford, Mass.) water for 30 s and negatively stained for 1 min with freshly prepared filtered 1% uranyl acetate. Sample grids were examined with a LEO 912 EF electron microscope at an accelerating voltage of 100 kV or with a Philips transmission electron microscope at an accelerating voltage of 60 kV. The presence and number of flagella for a minimum of 50 cells were determined for each strain by using at least two separate preparations.
Squid colonization.
Within 3 h of hatching, E. scolopes juveniles were exposed to V. fischeri cells as described previously (6), with several modifications. Animals were exposed to one of the following six inocula: wild-type strain ES114, ES114 carrying pVO8 (vector control), ES114 carrying pDM104 (containing a wild-type copy of flaA), flaA mutant strain DM143, DM143 carrying pVO8, or DM143 carrying pDM104. The final inoculum concentration was between 1 × 103 and 4 × 103 cells ml of seawater−1. The levels of colonization were determined either indirectly by measuring luminescence with a modified scintillation counter or directly by homogenizing light organs and plating the homogenate on SWT agar (29). To visualize bacterial cells during the early stages of colonization, newly hatched squid were exposed to an inoculum consisting of either the wild-type V. fischeri parent or the flaA mutant strain (each carrying the GFP-encoding plasmid pKV111) at a concentration of 1 × 105 cells ml of seawater−1. Beginning at 3 h postinoculation, animals were anesthetized and examined by CLSM as described previously (29).
Competition assays were performed as previously described (29). Briefly, different ratios of wild-type and flaA mutant strains were combined in seawater, which was then used as the inoculum for animal experiments. The exact ratio of the wild type to the mutant was determined by plating a dilution of the inoculum and testing at least 100 individual colonies for the Knr phenotype of the flaA mutant. At 24 h after inoculation animals were sacrificed, and the total numbers of symbiotic bacteria in light organ homogenates were determined. The relative numbers of wild-type and flaA mutant cells in the symbiotic population were estimated by testing at least 100 colonies from each animal for Knr. The relative competitive index (RCI) was determined by dividing the ratio of the mutant to the wild type for the colonizing population by the same ratio for the inoculum.
The competitiveness of the flaA mutant during the first 24 h of colonization was determined as follows. A group of animals was exposed to a single inoculum containing a mixture of ES114 and DM143 cells and was maintained in the dark for 12 h. At 12 h postinoculation, a subset of the animals was collected and homogenized for plating by using red light illumination to avoid light-induced expulsion. Another subset of animals was exposed to artificial light to induce the expulsion behavior. At 12.5 h (0.5 h after exposure to light), the seawater containing the expelled bacteria was diluted and plated, the animals were rinsed extensively in autoclaved seawater, and the bacterial population still present in the light organs was isolated after homogenization. The remaining animals were maintained until 24 h postcolonization, at which time the colonizing bacteria were isolated. The ratio of flaA mutant cells to wild-type cells in each of the four populations was determined as described above.
Nucleotide sequence accession number.
The nucleotide sequences of the fla loci reported here have been deposited in the GenBank database under accession number AY514454. The complete nucleotide sequence of V. fischeri flaE and the sequence of flaF can be obtained from the Vibrio fischeri Genome Project website (http://ergo.integratedgenomics.com/Genomes/VFI).
RESULTS
V. fischeri has six flagellin genes arranged in two distinct chromosomal loci.
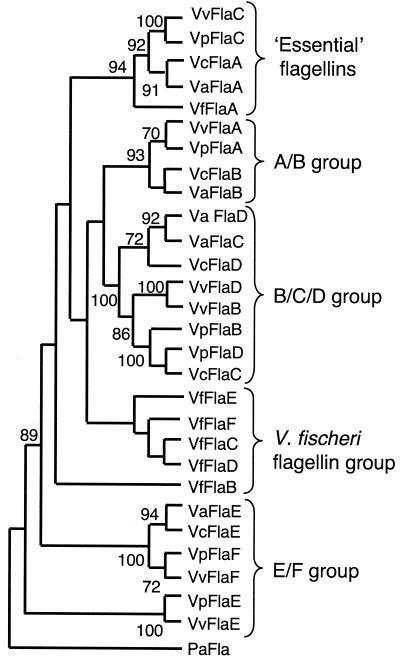
To identify flagellin genes in V. fischeri, we used degenerate oligonucleotide primers (Table 1) to PCR amplify chromosomal fragments corresponding to three unique ORFs with similarity to sequences encoding flagellins. Southern hybridization with the PCR-amplified products revealed a single XbaI-digested fragment that exhibited similarity to sequences encoding flagellins. The complete sequence of this 9.3-kb cloned fragment in pDM13 was determined, which revealed one partial and four complete flagellin ORFs whose products were subsequently designated FlaA through FlaE (Fig. 1A). The tandem arrangement of five flagellin genes (Fig. 1A) is unique among the Vibrio species whose flagellin sequences have been determined (Fig. 1B). Completion of the genome sequence of V. fischeri ES114 subsequently revealed the presence of a sixth gene located at a site that is distinct from the other five flagellin-encoding sites (Fig. 1A) and does not contain additional flagellar genes. Interestingly, there are corresponding loci in V. cholerae, V. parahaemolyticus, and Vibrio vulnificus that contain the flanking genes but lack a homolog of flaF or any other gene at the position of flaF.
FIG. 1.
Chromosomal arrangement of flagellin genes of V. fischeri (Vf) (A) and other Vibrio species, including V. anguillarum (Va), V. cholerae (Vc), V. parahaemolyticus (Vp), and V. vulnificus (Vv) (B) (21, 23, 28). The arrows indicate the direction of transcription, and, when known, the presence of promoter consensus sequences is indicated by arrowheads. Empirical evidence that transcription requires the alternative sigma factors σ54 and σ28 is indicated by solid arrowheads, while the presence of putative promoter consensus sequences upstream of the corresponding genes is indicated by open arrowheads.
The flaABCDE locus appears to be monocistronic, and all of these ORFs except flaE exhibit similarity to known promoter sequences (Fig. 1A). The promoter region upstream of flaA exhibits similarity to σ54 consensus binding sequences, while the sequences upstream of flaB, flaC, flaD, and flaF exhibit similarity to σ28 binding sequences (i.e., at least 9 of 12 residues were identified). σ54, encoded by the rpoN gene, is an alternative sigma factor for RNA polymerase that controls the expression of flagellar genes in V. cholerae (36) and V. fischeri (30, 49). In a previous study the workers demonstrated that the flaA gene is expressed from a σ54 promoter and that it requires both RpoN and the transcriptional activator FlrA for expression (49). σ28, encoded by the fliA gene, is a known alternative sigma factor in enteric bacteria that is required for the expression of certain motility genes. In V. cholerae, σ28 is necessary for transcription of flaB, flaC, flaD, and flaE but not for transcription of flaA (Fig. 1B).
The six flagellins of V. fischeri are homologous to each other.
The predicted products of the six flagellin genes have calculated molecular masses and predicted isoelectric points (Table 2) that are similar. The flagellins are homologous to each other, and the levels of identity range from 61 to 79% for the entire predicted protein sequences; the highest levels of homology are at the amino and carboxy termini of the proteins.
TABLE 2.
Immunoreactive proteins in the V. fischeri flagellar filament
| Predicted flagellin species
|
Reactive speciesa | Protein characteristics estimated from 2D gel
|
|||
|---|---|---|---|---|---|
| Protein | Mass (kDa) | pI | Mass (kDa) | pI | |
| FlaA | 40.2 | 4.46 | a | 45.4 | 4.5 |
| FlaB | 40.2 | 4.39 | b | 45.6 | 4.42 |
| FlaC | 42.3 | 4.53 | c | 46.6 | 4.54 |
| FlaD | 39.7 | 4.27 | d | 49.3 | 4.25 |
| FlaE | 40.6 | 4.45 | e | 46.1 | 4.48 |
| FlaF | 39.9 | 4.15 | NDb | ND | ND |
See Fig. 3.
ND, not detected.
The predicted amino acid sequences of the six V. fischeri flagellins also exhibit similarity to the amino acid sequences of flagellins of other Vibrio species, and the levels of identity range from 44 to 73%. In a previous review the workers performed a phylogenetic analysis of the flagellins of V. parahaemolyticus, V. anguillarum, and V. cholerae, and the results indicated that each flagellin is more closely related to the predicted product of the spatially equivalent ORF in the other organisms than to the other flagellins in the same organism (27). We were interested in determining whether the same relationship was true for the V. fischeri flagellins and performed a maximum-parsimony and distance-based analysis to assess the phylogenetic relationship among the known flagellin sequences from five Vibrio species (a total of 29 sequences). The representative cladogram in Fig. 2 shows that while the predicted FlaA protein of V. fischeri grouped with one flagellin from each of the other four species, the remaining flagellins did not cluster with sequences from the other organisms. In contrast, each of the flagellins from the other Vibrio species used in this analysis contained sequences that were more similar to the sequences of the product encoded by the corresponding gene in another organism than to the sequences of the product encoded by any other gene in that organism. Interestingly, four of the six V. fischeri flagellins formed a unique clade, although in some cases (as in the tree shown) the node could not be supported. Thus, we concluded that these four flagellins either are unique to V. fischeri or are evolutionarily further derived than the flagellins present in other species. Two of the flagellins in V. fischeri, FlaC and FlaD, are more closely related to each other (79% identity) than to any other protein, suggesting that there is a possible gene duplication. One of the flagellins, FlaB, was unique since it did not group with any other flagellin (Fig. 2).
FIG. 2.
Phylogenetic analysis of flagellin proteins in the genus Vibrio. Predicted amino acid sequences were obtained either from sequencing (V. fischeri [Vf]) (this study) or from the GenBank database (V. anguillarum [Va], V. cholerae [Vc], V. parahaemolyticus [Vp], and V. vulnificus [Vv]). Both maximum-parsimony (data shown) and distance-based (data not shown) analyses were performed by using PAUP (45), and similar results were obtained. Bootstrap values greater than 70 are indicated at nodes. The Pseudomonas aeruginosa (Pa) flagellin sequence of fliC served as an outgroup.
Detection of V. fischeri flagellins.
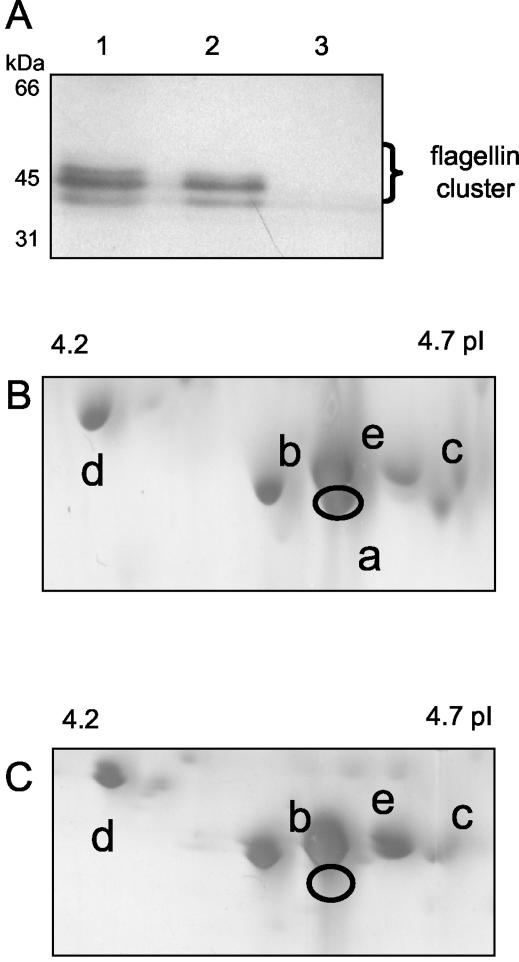
Because all six flagellin genes are apparently transcribed during exponential growth of V. fischeri (A. Schaefer, personal communication), we used PAGE to determine whether the predicted protein products were present in preparations of flagellar filaments. Separation of the flagellar proteins by SDS-PAGE resulted in only three or four bands, which were clustered around 45 kDa and were the only bands that showed immunological reactivity to a polyclonal flagellin antibody (Fig. 3A). 2D gel electrophoresis provided greater resolution of the five proteins in this region (Fig. 3B), and immunoblot analysis confirmed that all five proteins were reactive with the flagellin antiserum (data not shown). The estimated molecular masses and pIs of these proteins roughly correspond to the estimated molecular masses and pIs of five of the predicted flagellin gene products, FlaA, FlaB, FlaC, FlaD, and FlaE (Table 2). As expected, the protein corresponding to FlaA was missing in flagellum preparations from a strain with an insertional mutation in flaA (Fig. 3C). In all cases the proteins occurred at a higher molecular mass than predicted, suggesting that V. fischeri flagellins may be secondarily modified, as reported for flagellins of other species (7, 20, 24). Interestingly, there was no evidence that the flagellar filament preparations of culture-grown cells contained a sixth protein species at the expected pI that represented the FlaF protein (pI 4.15) (data not shown).
FIG. 3.
Electrophoretic separation of V. fischeri flagellar filament proteins. (A) Flagellar proteins from cells of strain ES114 (lane 1), strain DM143 (lane 2), and nonmotile strain DM127 (lane 3) immobilized on nitrocellulose and immunoblotted with a polyclonal flagellin antibody (see Materials and Methods). (B and C) Flagellar proteins were prepared from either strain ES114 (B) or strain DM143 (C), separated by 2D gel electrophoresis, and detected by silver staining. Immunoblotting with the polyclonal flagellin antibody detected the five protein species indicated; a to e indicate the assignments shown in Table 2.
flaA, but not flaC, is essential for normal motility in V. fischeri.
To better understand the roles of the different flagellins of V. fischeri in motility, we examined mutants in which two of the six flagellins were defective (Table 1). An insertional mutation in flaC had no effect on the rate of cell migration through soft agar, nor was the mutant's behavior visibly different from the behavior of the wild type as determined by phase-contrast microscopy. In contrast, the flaA mutation resulted in a 50 to 60% decrease in the rate of migration through motility agar (Fig. 4). In addition, microscopic observation of cells grown in liquid medium revealed that the percentage of the mutant cells that were nonmotile was higher than the percentage of wild-type cells that were nonmotile. Motile FlaA mutant cells appeared to exhibit wild-type behavior, including the ability to reverse direction (data not shown). Both the flaA and flaC mutant cells produced the two chemotaxis rings typically displayed by wild-type V. fischeri growing on motility agar (Fig. 4A and B) (10), and the mutants responded normally to the presence of the chemoattractant serine (data not shown). Thus, neither a loss of flaA or flaC nor a reduction in motility per se had a detectable effect on the ability of V. fischeri to sense and respond to its chemical environment.
FIG. 4.
Motility agar plates showing the decreased rate of migration of flaA mutant strain DM143 (B) compared to the rate of migration of the wild type (A). Mid-exponential cells were inoculated into motility agar plates containing 0.25% agar and incubated for 8 h, at which time the plates were photographed. Although not clearly visible, the flaA mutant (B) exhibited rings of chemotaxis similar to those of the parent (A). (C) Rates of migration of strains DM143 (gray bars), DM143 carrying flaA in trans (solid bars), and DM143 carrying a control plasmid (open bars) relative to the rates of migration of the wild type through media containing different concentrations of agar. Measurements were made in triplicate, and pairs of strains (strains ES114 and DM143, strains ES114/pDM104 and DM143/pDM104, and strains ES114/pVO8 and DM143/pVO8) were tested on the same plates. The error bars indicate standard errors; not all error bars are visible. The experiment was repeated three times, and similar results were obtained in all experiments.
Flagellar structure of the FlaA and FlaC mutants.
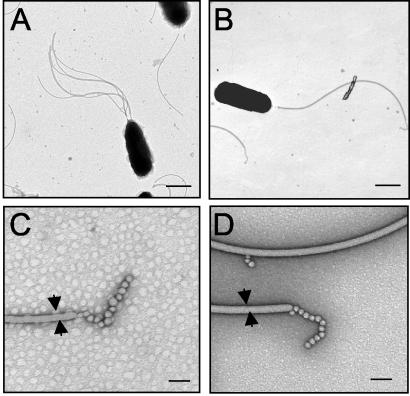
In previous studies workers have demonstrated the presence of a tuft of polar, sheathed flagella on cells of V. fischeri (2). Transmission electron microscopy revealed that the percentage of cells of the FlaA mutant that elaborated flagella (66%) was significantly lower than the percentage of wild-type cells that elaborated flagella (89%). In addition, the mutant had slightly, although significantly (P < 0.005%), fewer flagella per flagellated cell (1.7 ± 0.1 flagella per cell) than the wild type had (3.4 ± 0.3 flagella per cell) (Fig. 5). In contrast, the number of flagella per cell for the FlaC mutant was similar to the number of flagella for the wild type (data not shown). In a separate experiment, we found that the percentage of cells of the FlaA mutant carrying a wild-type copy of flaA in trans (pDM104) that had flagella (92%) was comparable to the percentage of wild-type cells carrying the same plasmid that had flagella (95%). In addition, the number of flagella per cell for the complemented mutant was equivalent to the number of flagella per cell for the wild type carrying the complementing plasmid. No other structural differences, such as the length or width of the filaments, were apparent when the two mutants and the wild type were compared, and all three strains had the bead-like structures typically observed at the distal end of the flagellar filament (Fig. 5C and D). The images also indicated that a sheath structure is present on flagellar filaments of both the wild-type and mutant cells.
FIG. 5.
Transmission electron micrographs of V. fischeri cells in the mid-exponential growth phase. (A and B) Cells of wild-type strain ES114 (A) have more sheathed flagella than cells of the FlaA mutant DM143 (B). Bars = 500 nm. (C and D) At a higher magnification, the presence of a sheath, the diameter of the filament (arrows), and the presence of unknown structures at the distal end of the filament were similar for wild-type flagella (C) and FlaA mutant flagella (D). Bars = 50 nm.
Colonization by the FlaA mutant is delayed, and this mutant colonizes less than the wild type colonizes.
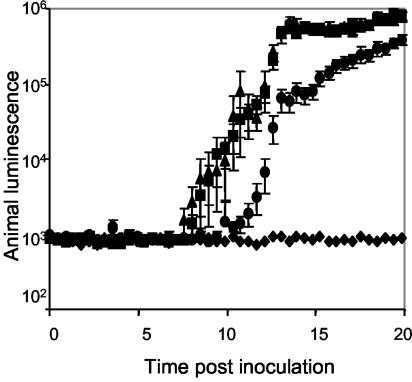
To determine whether the flagellin proteins play a role in colonization, we exposed newly hatched juvenile squid to seawater containing either mutant or wild-type V. fischeri cells. Because colonized animals produce luminescence, the onset and levels of colonization could be monitored indirectly by using an automated photometer. We found that when animals were inoculated with FlaA mutant cells, but not when they were inoculated with the FlaC mutant, the initial detection of luminescence was delayed by about 3 h (Fig. 6). Furthermore, the level of luminescence did not reach the level of luminescence in animals colonized by the wild type until after 24 h (data not shown). In addition, even after 24 h the level of colonization by the FlaA mutant was only 20 to 25% of the wild-type level of colonization (Fig. 7), and the difference was independent of the length of time that the animals were exposed to the inoculum. In contrast, the luminescence and growth of the FlaA mutant strain in culture were indistinguishable from the luminescence and growth of the wild type, suggesting that this mutant had a specific defect in initiating the association and that this defect was likely independent of growth and was not overcome by increasing the length of exposure.
FIG. 6.
Colonization of squid by different V. fischeri strains, as indicated by the development of symbiotic luminescence. Newly hatched E. scolopes juveniles were exposed to seawater containing either no V. fischeri (♦), strain ES114 (▪), flaC strain DM138 (▴), or flaA strain DM143 (•). The bioluminescence emission values are averages for 10 animals for each treatment. The error bars indicate the standard errors of the means. Similar results were obtained in two separate experiments.
FIG. 7.
Relative levels of colonization by wild-type strain ES114 (solid bars) and FlaA mutant strain DM143 (open bars) 24 h after juvenile squid were exposed to an inoculum of each strain for either 3 or 14 h. The error bars indicate the standard errors of the means. Similar results were obtained in two additional experiments.
The FlaA mutant is outcompeted by the wild type.
To further understand the colonization defect of the FlaA mutant, we exposed juvenile squid to a mixed inoculum containing both wild-type and FlaA mutant cells. Even when the inoculum contained twofold more mutant cells, in each of five experiments the resulting symbiotic population was dominated by the wild type in all 50 animals tested (data not shown). The RCI ranged from 0.02 to 0.06. When the mutant was coinoculated into seawater, it survived as well as wild-type cells for at least 14 h (data not shown), suggesting that a decreased ability to remain viable in seawater during the inoculation process did not underlie the FlaA mutant's colonization defects. Furthermore, in three separate experiments, the presence of the complementing pDM104 plasmid eliminated the competitive defect of the FlaA mutant (data not shown), suggesting that this defect was due solely to the loss of FlaA.
To determine whether an increased ratio of mutant cells to wild-type cells could overcome the competitive defect of FlaA, animals were exposed to a mixed inoculum containing between 2- and 100-fold more mutant cells (Table 3). We found that at ratios of mutant cells to wild-type cells between 24:1 and 100:1, the resulting colonizing populations were comprised of approximately equal numbers of the two strains. However, the RCI remained essentially the same regardless of the initial inoculum ratio, suggesting that the competitive defect of the FlaA mutant could not be overcome by increasing the absolute number of mutant cells in the colonizing population.
TABLE 3.
Effect of inoculum ratio on the competition between the FlaA mutant and its wild-type parent during symbiotic colonization
| Expt | Inoculum conditions
|
No. of animals | Symbiotic colonization
|
||||
|---|---|---|---|---|---|---|---|
| Total no. of CFU/ml | No. of ES114 CFU/ml | No. of DM143 CFU/ml | Ratio of DM143 to ES114 | CI Competitive indexa | RCIb | ||
| I | 5,720 | 1,887 | 3,832 | 2.0 | 5 | 0.038 | 0.019 |
| II | 7,960 | 1,432 | 6,527 | 4.6 | 5 | 0.21 | 0.045 |
| III | 8,340 | 1,334 | 7,005 | 5.3 | 5 | 0.55 | 0.10 |
| IV | 6,980 | 279 | 6,700 | 24 | 5 | 0.76 | 0.031 |
| V | 6,900 | 69 | 6,831 | 100 | 5 | 2.6 | 0.026 |
The competitive index was the ratio of DM143 to ES114 in a symbiotic colonization at 48 h. A competitive index of <1 indicated that the colonizing population was dominated by ES114.
The RCI was the competitive index divided by the ratio of DM143 to ES114 in the inoculum.
FlaA mutant cells are preferentially expelled from the host light organ.
To further characterize the nature of the competitive defect of the FlaA mutant during the first 24 h, we monitored the competitiveness of the mutant during each of the following three stages of the colonization process: (i) initiation and growth (during the first 12 h after inoculation), (ii) expulsion of 95% of the colonizing population (at 12 h postinoculation), and (iii) subsequent regrowth in the light organ (during the 11.5 h following expulsion). At each of these three stages, in one experiment a subset of animals was removed, and the ratio of the colonizing populations was determined (Table 4). The composition of the expelled cells was also determined. In three experiments the RCI were lower for the animals examined after expulsion (after venting, 12.5 h) than for the animals examined before expulsion (before venting, 12 h); the data for two experiments are shown in Table 4. A third experiment revealed a similar trend; however, the data for this experiment are not shown because the ratio of mutant cells to wild-type cells in the inoculum was four- to fivefold higher the ratio in the first two experiments. The P values calculated for the two data sets were between 4 and 26% for the three experiments. Thus, while the difference between the two data sets is only slightly significant, the finding that there was a decreased RCI in postexpulsion animals was repeatable. In contrast, there was no significant difference between the RCI values for symbiont populations immediately after expulsion (after venting, 12.5 h) and the RCI values 11.5 h later (after venting plus growth, 24 h), suggesting that while the FlaA mutant is between 6 and 18 times more likely to be expelled during the host's normal venting behavior, its competitive defect is not expressed during the postexpulsion regrowth of the light organ population. This suggestion was supported by the results of one experiment in which we determined that the proportions of expelled cells were significantly different for the population before venting and the population after venting (Table 4). Interestingly, the RCI values for symbiont populations at 48 and 72 h were not significantly different from the RCI value obtained at 24 h, suggesting either that the wild type can complement the FlaA mutant defect at later stages of colonization or that FlaA is essential for only two of the initial stages of colonization, growth and expulsion. Taken together, these results suggest that at the first two stages (i.e., initiation and expulsion) the ability of the FlaA mutant to compete with the wild type is limited.
TABLE 4.
Competition between the FlaA mutant and its wild-type parent during the first 24 h of symbiosis
| Expt | Inoculuma
|
RCIb
|
||||
|---|---|---|---|---|---|---|
| Total no. of CFU/ml | Ratio of DM143 to ES114 | Before venting (12 h) | Expelled cells (12 h) | After venting (12.5 h) | After venting growth (24 h) | |
| 1 | 3,280 | 1.5 | 0.36 (0.18-.60) | 0.5 | 0.06 (0.0-.14) | 0.06 |
| 2 | 7,440 | 1.2 | 0.18 (0.08-.31) | NDc | 0.01 (0.0-.02) | 0.04 |
| Avgd | 6,730 | 1.3 | 0.35 | 0.5 | 0.037 | 0.038 |
Animals (33 animals in experiment 1 and 30 animals in experiment 2) were exposed to the inoculum for 12 h.
Average RCI (confidence range).
ND, not determined.
The values are averages for all 63 animals for a condition, not the average for each of the two experiments weighted equally.
Colonization of the deep crypt regions by the FlaA mutant is delayed.
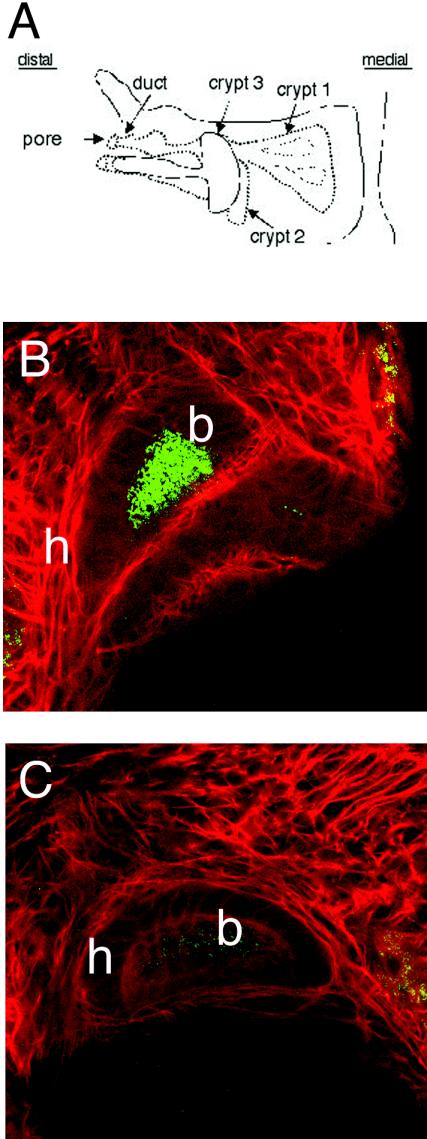
One hypothesis to explain the observed competitive defect of the FlaA mutant during the expulsion stage is that certain subpopulations that colonize discrete regions of the light organ crypts are maintained during this stage; FlaA mutant cells either may be unable to colonize such sites or are outcompeted by the wild type for occupation of this privileged niche. To determine whether the FlaA mutant is able to colonize all of the normal sites in the three crypts (Fig. 8A), we used confocal microscopy to investigate the behavior during the first 39 h of colonization. When animals were exposed to an inoculum consisting of approximately 1.4 ×105 cells of either strain, aggregations of cells whose sizes were similar were attached to external mucus at similar times postinoculation (data not shown). Furthermore, between 5 and 7 h postinoculation, in three separate experiments, similar percentages of animals exposed to the wild-type strain (68%; n = 22) and animals exposed to the FlaA mutant (65%; n = 17) had GFP-labeled cells in the light organ pores, suggesting that the initial entry of the FlaA mutant into the light organ is not delayed (data not shown). However, while subsequent observations revealed that the wild type densely colonized the deep regions of the crypts prior to 16 h (Fig, 8B), mutant cells were not observed at these sites even at 20 h postinoculation. Only after 24 h were a few mutant cells observed in these regions (Fig. 8C).
FIG. 8.
Confocal microscopy of GFP-labeled V. fischeri cells during the early stages of light organ colonization. (A) Schematic drawing of one half of a juvenile light organ, showing the pore and duct through which V. fischeri must enter to colonize the crypts. Animals colonized with GFP-labeled wild-type strain ES114 for 16 h contained fluorescent cells in the deep regions of crypts 1 and 2 (data not shown), as well as of crypt 3 (B). In contrast, colonization of the same deep crypt regions by the FlaA mutant DM143 was delayed for an additional 8 h, and low numbers of the mutant were present in crypt 3 24 h after inoculation (C). b, GFP-labeled bacteria; h, host cells stained with Cell-Tracker Red.
DISCUSSION
In this study, we identified six flagellin subunit genes in V. fischeri and found that in cultured cells only five of the six genes encode proteins localized to the flagellum. Furthermore, we found that FlaA, but not FlaC, is required for normal motility and plays an essential role in colonizing the juvenile squid host. Specifically, a mutation in flaA severely compromises the ability of the bacterium to compete with the wild type in a mixed infection, both during initiation and during the first daily expulsion event that occurs after 12 h. Comparative studies in which CLSM analysis was used indicated that the presence of the flaA mutant in the deep crypts is delayed, suggesting that colonization of these regions might be important for resisting the expulsion process.
Genomic analysis of V. fischeri flagellins.
We identified six flagellin genes in V. fischeri that are located in two chromosomal loci. The first locus contains genes encoding five flagellins and is located in a region that contains the majority of the flagellar and chemotaxis genes. In contrast, other Vibrio species whose flagellar gene arrangements are known (V. cholerae, V. anguillarum, V. parahaemolyticus, and V. vulnificus) contain two major chromosomal loci for flagellar genes, which in V. cholerae are separated by approximately 33 kb of sequence. The intervening sequence does not contain motility-related genes but does show synteny to a region located 40 kb from the V. fischeri flagellar locus. Interestingly, the presence of the insertion element IS1004 downstream of the flaC gene, as well as a number of tRNA genes in this region in V. cholerae, suggests a possible mechanism for chromosomal rearrangement at this locus. The presence and location of the sixth flagellin gene in V. fischeri, flaF, are also unique in Vibrio spp. Surprisingly, each of the V. fischeri subunits except FlaA is more similar to the other flagellins of V. fischeri than to the flagellins of any of the five Vibrio species examined, suggesting that the V. fischeri flagellins are evolutionarily further derived. This relationship is in contrast to the relationship of flagellins in other Vibrio species, in which there appears to be a gene whose location and predicted protein sequence are equivalent for each of the subunits. The one exception is the clustering of the FlaA sequence of V. fischeri with other, paralogous FlaA sequences, which may indicate that there are special functional constraints for the flaA gene.
Although the flaF gene has a possible σ28-binding sequence upstream and appears to be transcribed (Schaefer, personal communication), the V. fischeri flagellum did not appear to contain the FlaF protein under the growth conditions used in this study. Perhaps FlaF is subjected to posttranscriptional regulation, or alternatively, the FlaF protein is a component of the V. fischeri flagellum only under specific conditions.
In vitro phenotypes of the FlaA mutant.
In each of the three Vibrio species in which it has been described, one flagellin gene appears different from the other flagellin genes with respect to both transcriptional regulation and cellular function (for a review see reference 27). In V. cholerae and V. anguillarum (31) this gene is flaA, and in V. parahaemolyticus it is the ortholog flaC. A mutation in flaA in V. cholerae results in a loss of motility (23), and in V. anguillarum a loss of flaA results in cells with truncated flagella that exhibit decreases in both motility and virulence (31). In V. fischeri, flaA is also subject to regulation by σ54, similar to the regulation in V. cholerae (49). Phenotypes of the V. fischeri flaA mutant revealed that the FlaA protein is essential for production of a normal number of flagella, but when flagella were present, they did not appear to be different from wild-type flagella in terms of length or helical nature (Fig. 5). Mutants that elaborate a greater number of flagella per cell have also been shown to have greater swimming speeds than the wild type (29). Thus, it is likely that the smaller percentage of flagellated cells and the fewer flagella per cell expressed by the flaA mutant contributed to the observed decrease in motility. Interestingly, the level of expression of four additional flagellin subunits remains high in a flaA mutant (Fig. 3), indicating that although these subunits are homologous, they cannot replace some essential function of the FlaA protein in assembling a flagellum. In contrast, a flaC mutant strain was as motile as the wild type, and it elaborated a similar number of flagella, suggesting that the FlaC protein is dispensable. Thus, although homologous, these proteins apparently do not have identical functions in all Vibrio species.
In most bacteria, the flagellum contains a single type of flagellin. However, flagellar filaments containing multiple subunits have been found in a variety of bacterial genera, including Helicobacter (18), Yersinia (19), Caulobacter (12), Campylobacter (15), Agrobacterium (9), and Rhizobium (35, 40). While the function underlying the expression of multiple subunits is not known, the subunits may play different and unique roles in coordinating the assembly of a complex filament (12) or in producing flagella that are particularly suited for motility in a given environment. In this regard, it is interesting that V. fischeri flaA transcription was reduced threefold in the flaA mutant compared to transcription in the wild type (data not shown), suggesting that there is feedback control of flagellin biosynthesis when a nonfunctional combination of subunits is expressed. Such transcriptional feedback control has been found in Sinorhizobium meliloti (40), in which a combination of FlaA and at least one accessory flagellin is required to assemble a functional filament. In the absence of such a combination, flaA transcription is reduced (40). Thus, bacteria may have evolved mechanisms to respond to the expression of inappropriate combinations of flagellin subunits, perhaps to adjust their motility for a particular environment.
An alternative function for multiple flagellins could be to provide antigenic variation for the flagellum as a means of evading host defenses (41). In Salmonella enterica serovar Typhimurium, antigenic variation involves the expression of one flagellin gene at a time, while the other gene remains silent. In contrast, the multiple subunits of Vibrio species appear to be coexpressed under laboratory conditions (23, 26, 28). Future studies might reveal an exclusive role for one or more flagellins, in particular the FlaF protein. DNA microarray analysis of colonizing V. fischeri cells should help define the transcriptional regulation of flagellins in the host environment. Interestingly, sequencing of the E. scolopes transcriptome has revealed several Toll-like receptors (M. Goodson, unpublished observations), one of which (Toll-like receptor 5) is known in other organisms to recognize bacterial flagellins that, like the flagellin of V. fischeri, have a potential binding site for Toll-like receptor 5 (17).
Symbiotic colonization.
The initiation of symbiotic colonization of E. scolopes by V. fischeri can be defined by several temporally and spatially separated events. By creating mutations in genes of the flagellar regulon we are beginning to define the requirements for motility at each of the early colonization stages. In this study, a FlaA mutant (but not a FlaC mutant) was compromised in terms of the ability to colonize juvenile squid; in animals colonized with the FlaA mutant there was a delay in the onset of luminescence and there were fewer colonizing bacteria at 24 h. Motility of V. fischeri has been shown to be required for colonization (13) and is probably influenced by chemotaxis (10). The decreased motility of the FlaA mutant strain, and thus the decreased chemotaxis, could by itself account for the observed delay in colonization. Interestingly, colonization by strains that are hyperflagellated is also delayed, but presumably this is due to a mechanism different than the mechanism underlying the delay exhibited by the FlaA mutant (29); that is, unlike hyperflagellated cells, the FlaA mutant was able to form normal aggregates. In addition, hyperflagellated cells were found to colonize nonspecific sites of the host, a behavior not observed for the FlaA mutant. Therefore, FlaA plays a critical role in colonization of a discrete site(s) within the light organ. Interestingly, compared to the wild-type cells, 23% fewer FlaA mutant cells grown in culture are flagellated; thus, the actual number of cells of the FlaA mutant that are able to initiate colonization is less than the number of wild-type cells. Could this difference be responsible for the observed delay in initiation observed with the FlaA mutant? We determined the estimated number of flagellated cells present in the inoculum of FlaA mutant cells (between 660 and 2,640 cells) and found that it compared well with the estimated number of flagellated cells present in the wild-type inoculum (between 890 and 3,560 cells) and was within the range expected to show the same kinetics of colonization (25). Furthermore, the FlaA mutant reaches the light organ pores as quickly as wild type reaches these pores. Taken together, these data demonstrate that the observed delay in FlaA mutant colonization is not due to a defect in the ability of the mutant to attach to external mucus or to initiate entry into the light organ but is likely due to a delay or to an inability of the cells to colonize discrete sites within the crypts.
To understand better the initial colonization events, we exposed animals to a mixed inoculum containing both wild-type and mutant cells and monitored the ratio of the strains in the light organ. After 24 h, the colonizing population was always dominated by wild-type cells, except when the FlaA mutant was given an initial advantage of greater than 24:1 (Table 4). When we more closely examined the appearance of competitiveness over the first 24 h of colonization, we found that during two events (the initial growth period and the expulsion event that occurs at 12 h) the wild type outcompeted the FlaA mutant for colonization. To explain the defect during the first event, we hypothesized that the FlaA mutant is unable to compete due to the delay in colonization observed for the bacteria (see above).
The second event, expulsion of 95% of the symbiont population, involves muscular contraction of the light organ of the animal (14). Because the FlaA mutant is selected against during expulsion, this strain may be defective in forming specific bacterium-host interactions or may be physically separated from the wild-type symbionts. Analysis by confocal microscopy showed that colonization of the deep crypts by the FlaA mutant cells is delayed (Fig. 8), suggesting that colonization of these sites might be a mechanism for preventing expulsion. Coinoculation studies performed with strains carrying different fluorescence labels would contribute to our understanding of the mechanisms that mediate bacterial selection during expulsion and to our overall understanding of crypt complexity.
It is possible that FlaA and/or the presence of an appropriate number of flagella mediates attachment to host cells and that the attached cells are not expelled. Interestingly, FlaA apparently is not required for attachment to glass surfaces (unpublished observations) or to host mucus (this study), suggesting that the FlaA mutant is not generally impaired in terms of surface attachment. Regardless, as in some pathogens (3, 4, 8), some aspect of flagellar regulation or the flagellum itself may contribute to specific attachment to host epithelial cells. While the details of the mechanism(s) remain to be described, the results presented here show that a functional FlaA is required for normal symbiotic colonization by V. fischeri.
Acknowledgments
We thank L. McCarter for generous donation of flagellin antibody, B. Holland for his expertise in phylogenetic analyses, and J. Stewart and J. Kimbell for assistance with 2D gel electrophoresis. K. Visick and J. Graber provided helpful comments on the manuscript.
In its later stages, this work was aided by data provided by the Vibrio fischeri Genome Project (http://ergo.integratedgenomics.com/Genomes/VFI), which was made possible by a grant from the W. M. Keck Foundation. This work was supported by an NSF postdoctoral fellowship in microbial biology to D. S. Millikan, by NSF grant IBN9904601 to M. McFall-Ngai and E. G. Ruby, and by NIH grant RR12294 to E. G. Ruby and M. McFall-Ngai.
REFERENCES
- 1.Akerley, B. J., D. M. Monack, S. Falkow, and J. F. Miller. 1992. The bvgAS locus negatively controls motility and synthesis of flagella in Bordetella bronchiseptica. J. Bacteriol. 174:980-990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allen, R. D., and P. Baumann. 1971. Structure and arrangement of flagella in species of the genus Beneckea and Photobacterium fischeri. J. Bacteriol. 107:295-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arora, S. K., B. W. Ritchings, E. C. Almira, S. Lory, and R. Ramphal. 1997. A transcriptional activator, FleQ, regulates mucin adhesion and flagellar gene expression in Pseudomonas aeruginosa in a cascade manner. J. Bacteriol. 179:5574-5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arora, S. K., B. W. Ritchings, E. C. Almira, S. Lory, and R. Ramphal. 1998. The Pseudomonas aeruginosa flagellar cap protein, FliD, is responsible for mucin adhesion. Infect. Immun. 66:1000-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barrios, H., B. Valderrama, and E. Morett. 1999. Compilation and analysis of sigma (54)-dependent promoter sequences. Nucleic Acids Res. 27:4305-4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boettcher, K. J., and E. G. Ruby. 1990. Depressed light emission by symbiotic Vibrio fischeri of the sepiolid squid Euprymna scolopes. J. Bacteriol. 172:3701-3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brimer, C. D., and T. C. Montie. 1998. Cloning and comparison of fliC genes and identification of glycosylation in the flagellin of Pseudomonas aeruginosa a-type strains. J. Bacteriol. 180:3209-3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clyne, M., T. Ocroinin, S. Suerbaum, C. Josenhans, and B. Drumm. 2000. Adherence of isogenic flagellum-negative mutants of Helicobacter pylori and Helicobacter mustelae to human and ferret gastric epithelial cells. Infect. Immun. 68:4335-4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deakin, W. J., V. E. Parker, E. L. Wright, K. J. Ashcroft, G. J. Loake, and C. H. Shaw. 1999. Agrobacterium tumefaciens possesses a fourth flagellin gene located in a large gene cluster concerned with flagellar structure, assembly and motility. Microbiology 145:1397-1407. [DOI] [PubMed] [Google Scholar]
- 10.DeLoney-Marino, C. R., A. J. Wolfe, and K. L. Visick. 2003. Chemoattraction of Vibrio fischeri to serine, nucleosides and N-acetylneuraminic acid, a component of squid light organ mucus. Appl. Environ. Microbiol. 69:7527-7530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doino Lemus, J., and M. J. McFall-Ngai. 2000. Alterations in the proteome of the Euprymna scolopes light organ in response to symbiotic Vibrio fischeri. Appl. Environ. Microbiol. 66:4091-4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Driks, A., R. Bryan, L. Shapiro, and D. J. DeRosier. 1989. The organization of the Caulobacter crescentus flagellar filament. J. Mol. Biol. 206:627-636. [DOI] [PubMed] [Google Scholar]
- 13.Graf, J., P. V. Dunlap, and E. G. Ruby. 1994. Effect of transposon-induced motility mutations on colonization of the host light organ by Vibrio fischeri. J. Bacteriol. 176:6986-6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Graf, J., and E. G. Ruby. 1998. Host-derived amino acids support the proliferation of symbiotic bacteria. Proc. Natl. Acad. Sci. USA 95:1818-1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guerry, P., S. M. Logan, S. Thornton, and T. J. Trust. 1990. Genomic organization and expression of Campylobacter flagellin genes. J. Bacteriol. 172:1853-1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Helmann, J. D., and M. J. Chamberlin. 1987. DNA sequence analysis suggests that expression of flagellar and chemotaxis genes in Escherichia coli and Salmonella typhimurium is controlled by an alternative sigma factor. Proc. Natl. Acad. Sci. USA 84:6422-6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jacchieri, S. G., R. Torquato, and R. R. Brentani. 2003. Structural study of binding of flagellin by Toll-like receptor 5. J. Bacteriol. 185:4243-4247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Josenhans, C., R. L. Ferrero, A. Labigne, and S. Suerbaum. 1999. Cloning and allelic exchange mutagenesis of two flagellin genes of Helicobacter felis. Mol. Microbiol. 33:350-362. [DOI] [PubMed] [Google Scholar]
- 19.Kapatral, V., and S. A. Minnich. 1995. Coordinate temperature-sensitive regulation of transcription of three Yersinia enterocolitica flagellin genes. Mol. Microbiol. 17:49-56. [DOI] [PubMed] [Google Scholar]
- 20.Kelly-Wintenberg, K., T. Anderson, and T. C. Montie. 1990. Phosphorylated tyrosines in the flagellum filament protein of Pseudomonas aeruginosa. J. Bacteriol. 172:5135-5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim, Y. K., and L. L. McCarter. 2000. Analysis of the polar flagellar gene system of Vibrio parahaemolyticus. J. Bacteriol. 182:3693-3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kimbell, J., and M. McFall-Ngai. 2003. The squid-vibrio symbioses: from demes to genes. Integ. Comp. Biol. 43:254-260. [DOI] [PubMed] [Google Scholar]
- 23.Klose, K. E., and J. J. Mekalanos. 1998. Differential regulation of multiple flagellins in Vibrio cholerae. J. Bacteriol. 180:303-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Logan, S. M., T. J. Trust, and P. Guerry. 1989. Evidence for posttranslational modification and gene duplication of Campylobacter flagellin. J. Bacteriol. 171:3031-3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCann, J. R., E. V. Stabb, D. S. Millikan, and E. G. Ruby. 2003. Population dynamics of Vibrio fischeri during infection of Euprymna scolopes. Appl. Environ. Microbiol. 69:5928-5934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCarter, L. L. 1995. Genetic and molecular characterization of the polar flagellum of Vibrio parahaemolyticus. J. Bacteriol. 177:1595-1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCarter, L. L. 2001. Polar flagellar motility of the Vibrionaceae. Microbiol. Mol. Biol. Rev. 65:445-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McGee, K., P. Horstedt, and D. L. Milton. 1996. Identification and characterization of additional flagellin genes from Vibrio anguillarum. J. Bacteriol. 178:5188-5198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Millikan, D. S., and E. G. Ruby. 2002. Alterations in Vibrio fischeri motility correlate with a delay in symbiosis initiation and are associated with additional symbiotic colonization defects. Appl. Environ. Microbiol. 68:2519-2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Millikan, D. S., and E. G. Ruby. 2003. FlrA, a σ54 transcriptional activator in Vibrio fischeri, is required for motility and symbiotic light-organ colonization. J. Bacteriol. 185:3547-3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Milton, D. L., R. O'Toole, P. Hoerstedt, and H. Wolf-Watz. 1996. Flagellin A is essential for the virulence of Vibrio anguillarum. J. Bacteriol. 178:1310-1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nyholm, S. V., B. Deplancke, H. R. Gaskins, M. A. Apicella, and M. J. McFall-Ngai. 2002. Roles of Vibrio fischeri and nonsymbiotic bacteria in the dynamics of mucus secretion during symbiont colonization of the Euprymna scolopes light organ. Appl. Environ. Microbiol. 68:5113-5122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nyholm, S. V., and M. J. McFall-Ngai. 2003. Dominance of Vibrio fischeri in secreted mucus outside the light organ of Euprymna scolopes: the first site of symbiont specificity. Appl. Environ. Microbiol. 69:3932-3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nyholm, S. V., E. V. Stabb, E. G. Ruby, and M. J. McFall-Ngai. 2000. Establishment of an animal-bacterial association: recruiting symbiotic vibrios from the environment. Proc. Natl. Acad. Sci. USA 97:10231-10235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pleier, E., and R. Schmitt. 1989. Identification and sequence analysis of two related flagellin genes in Rhizobium meliloti. J. Bacteriol. 171:1467-1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prouty, M. G., N. E. Correa, and K. E. Klose. 2001. The novel σ54- and σ28-dependent flagellar gene transcription hierarchy of Vibrio cholerae. Mol. Microbiol. 39:1595-1609. [DOI] [PubMed] [Google Scholar]
- 37.Reichelt, J. L., and P. Baumann. 1973. Taxonomy of the marine, luminous bacteria. Arch. Mikrobiol. 94:283-330. [DOI] [PubMed] [Google Scholar]
- 38.Ruby, E. G., and L. M. Asato. 1993. Growth and flagellation of Vibrio fischeri during initiation of the sepiolid squid light organ symbiosis. Arch. Microbiol. 159:160-167. [DOI] [PubMed] [Google Scholar]
- 39.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 40.Scharf, B., H. Schuster-Wolff-Buhring, R. Rachel, and R. Schmitt. 2001. Mutation analysis of the Rhizobium lupini H13-3 and Sinorhizobium meliloti flagellin genes: importance of flagellin A for flagellar filament structure and transcriptional regulation. J. Bacteriol. 183:5334-5342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Silverman, M., J. Zieg, M. Hilmen, and M. Simon. 1979. Phase variation in Salmonella: genetic analysis of a recombinational switch. Proc. Natl. Acad. Sci. USA 76:391-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stabb, E., K. Visick, D. S. Millikan, A. A. Corcoran, L. Gilson, S. V. Nyholm, M. McFall-Ngai, and E. Ruby. 2001. The Vibrio fischeri-Euprymna scolopes symbiosis: a model marine animal-bacteria interaction, p. 269-277. In Recent advances in marine science and technology. Pacon International, Honolulu, Hawaii.
- 43.Stabb, E. V., K. A. Reich, and E. G. Ruby. 2001. Vibrio fischeri genes hvnA and hvnB encode secreted NAD+-glycohydrolases. J. Bacteriol. 183:309-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stabb, E. V., and E. G. Ruby. 2003. New RP4-based plasmids for conjugation between Escherichia coli and members of the Vibrionaceae. Methods Enzymol. 358:413-426. [DOI] [PubMed] [Google Scholar]
- 45.Swofford, D. L. 2002. PAUP*. Phylogenetic analysis using parsimony (* and other methods). Sinauer Associates, Sunderland, Mass.
- 46.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Visick, K. L., and E. G. Ruby. 1997. New genetic tools for use in the marine bioluminescent bacterium Vibro fischeri, p. 119-122. In Bioluminescence and chemiluminescence. John Wiley and Sons, Chichester, United Kingdom.
- 48.Visick, K. L., and L. M. Skoufos. 2001. Two-component sensor required for normal symbiotic colonization of Euprymna scolopes by Vibrio fischeri. J. Bacteriol. 183:835-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wolfe, A. J., D. S. Millikan, J. M. Campbell, and K. L. Visick. 2004. Vibrio fischeri σ54 controls motility, biofilm formation, luminescence, and colonization. Appl. Environ. Microbiol. 70:2520-2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Young, G. M., M. J. Smith, S. A. Minnich, and V. L. Miller. 1999. The Yersinia enterocolitica motility master regulatory operon, flhDC, is required for flagellin production, swimming motility, and swarming motility. J. Bacteriol. 181:2823-2833.10217774 [Google Scholar]