Abstract
We have characterized a transferable tetracycline resistance (Tcr) element from a Streptococcus intermedius isolate. The gene responsible for this resistance was identified by PCR and Southern hybridization as tet(S). Furthermore, the genetic support for this determinant was shown to be a conjugative transposon closely related to Tn916. This element has been designated Tn916S.
Tetracycline-resistant streptococci are frequently isolated from the oral cavity of humans (13), and resistance is most commonly conferred by Tet(M), a ribosomal protection protein often associated with the conjugative transposon (cTn) Tn916 (4). Tn916 belongs to a family of cTns that are composed of functional modules (10, 12) involved in conjugation, antibiotic resistance, regulation, and integration and excision. Different members of this family of cTns are comprised of different modules.
The Tcr determinant tet(S) encodes a ribosomal protection protein showing 79% amino acid identity with Tet(M). It was initially identified in a multiresistant Listeria monocytogenes strain on a 37-kb conjugative plasmid, pIP811 (2). Subsequently tet(S) has been found on plasmid pK214 from Lactococcus lactis (8) and in the chromosome of Enterococcus faecalis (3). The tet(S) gene in L. lactis and L. monocytogenes is linked to homologues of the Tn916 orf6, orf9, and orf7. In this work, we show that the tet(S) gene from a Streptococcus intermedius isolate, originally isolated from a 5-year-old human child, is contained within a functional Tn916-like element.
All chemicals were purchased from BDH (Poole, United Kingdom). Antibiotics were purchased from Sigma-Aldrich (Poole, United Kingdom) and used at concentrations of 8 μg/ml for tetracycline and 25 μg/ml for rifampin. All enzymes were purchased from Promega (Southampton, United Kingdom), and all growth media were purchased from Oxoid (Basingstoke, United Kingdom). All bacterial strains and plasmids used are shown in Table 1. The primers used are shown in Table 2. The S. intermedius strain 15.3T.2 was grown on Iso-Sensitest agar containing 5% defibrinated horse blood (E&O Laboratories, Bonneybridge, United Kingdom) and tetracycline at 37°C in an anaerobic cabinet (Don Whitley Scientific Ltd., Shipley, United Kingdom) containing a mixture of 80% nitrogen, 10% hydrogen, and 10% carbon dioxide. All other strains were grown aerobically at 37°C. The filter-mating recipients E. faecalis and Streptococcus spp. were grown in brain heart infusion broth.
TABLE 1.
Bacterial strains and plasmids used throughout this study
| Strain or plasmid | Commentsa | Source or reference |
|---|---|---|
| Strains | ||
| S. intermedius 15.3T.2 | Tcr donor strain | This study |
| B. subtilis BS34A | TcrB subtilis::Tn916 | 11 |
| E. faecalis JH2-2 | Rifr recipient strain | 14 |
| E. coli pAT451 | TcrE. coli::pAT451b | 1 |
| S. sobrinus NCTC 12279 (type strain) | Rifr recipient strain | Health Protection Agencyc |
| S. sanguinis NCTC 7863 (type strain) | Rifr recipient strain | Health Protection Agency |
| Plasmids | ||
| pAM120 | pUC18::Tn916 | 5 |
| pAT451 | pUC18::tet(S) | 1 |
Tcr; tetracycline resistant, Rifr; rifampin resistant.
pUC18 carrying a 4.5-kb ClaI fragment of pIP811 with the tet(S) gene.
TABLE 2.
Oligonucleotide primers used in this study
| Primer name | Primer sequence or reference | Expected PCR product |
|---|---|---|
| 1-26 | 15a | orf24 to orf14 of Tn916 |
| RT1 | 5′-CTCTATCCTACAGCGACAGC-3′ | PCR product containing orf13, orf12, tet(M), and orf6 |
| RT4 | 5′-TCTTTGCGTCTGGCTCTGTA-3′ | |
| RT6 | 5′-TATGATTTTAGAGCCCTTTGG-3′ | PCR product containing orf6 and orf9 |
| RT11 | 5′-ACAGAGCCAGACGCAAAGAC-3′ | |
| RT7 | 5′-CTTGTATGCTGGGGTGTTGC-3′ | PCR product containing orf9 and orf7 |
| RT14 | 5′-TAATTCTTCCGCTCGTCGTC-3′ | |
| RT13 | 5′-AAAGAAAGGGGGTGAAC-3′ | PCR product containing orf7 and orf8 |
| RT15 | 5′-GGTTAATCGCTTCTGTATCG-3′ | |
| RT12 | 5′-ATTTCACGTTTCTTGTCTGG-3′ | Primer reading upstream from within orf7 |
| RT18 | 5′-AAGTATGGTCGTTGATGAAG-3′ | Primer reading downstream from within orf8 |
| SRV | 7 | 667-bp product of the tet(S) gene |
| SFW | 7 | |
| intxis1 | 6 | xis and int fragment of Tn916 |
| intxis2 | 6 | |
| REO | 14 | Joint of the circular form of Tn916 |
| LEO | 14 | |
| SFW(2) | 5′-GTGTCCAGGAGTATCTAC-3′ | Sequencing between RT1 and RT4 |
| RT1(2) | 5′-CTGTCAATTAGATAGCGGG-3′ | Sequencing between RT1 and RT4 |
| RT4rev | 5′-GATTTGAATTAAAGTGTAAAGGAGG-3′ | Primer reading downstream from within orf6 |
| RT7rev | 5′-ATGTATAGAGTGGTCTACTATGCG-3′ | Primer reading upstream from within orf9 |
Thirteen pairs of sequences.
Filter-mating experiments were carried out as previously described (14). Transconjugants were selected on brain heart infusion agar, containing 5% horse blood, rifampin, and tetracycline at 4 μg/ml. Spontaneous mutations to rifampin resistance in the donor were not detected. The streptococcal transconjugants were also subcultured onto esculin agar to confirm that they were negative for hydrolysis, in order to distinguish them from the donor, which was positive.
S. intermedius 15.3T.2 was grown overnight, and genomic DNA was extracted with the Yeast and Gram Positive Bacteria Genomic DNA kit (Genetra, Minneapolis, Minn., supplied through Flowgen) according to the manufacturer's instructions. PCR for the detection of a variety of Tcr genes was carried out as described by Ng et al. (7) and Villedieu et al. (13). Positive PCR products were sequenced with the Big Dye Terminator ready reaction mixture (PE Biosystems, Warrington, United Kingdom) and an ABI310 genetic analyzer (PE Biosystems) or sent to Oswel Sequencing (Romsey, United Kingdom).
Southern blotting and hybridization were carried out with an ECL Direct Nucleic Acid Labeling and Detection system (Amersham Biosciences, Little Chalfont, United Kingdom). Southern blots were probed with pAM120 (Table 1) and PCR products derived from tet(S), tet(M), and the int and xis genes of Tn916 (Table 1). PCR assays were also carried out as previously described by Wang et al. (15) to detect all regions of Tn916. The region between orf13 and orf6 (RT1 to RT4 on Fig. 1) was sequenced in triplicate.
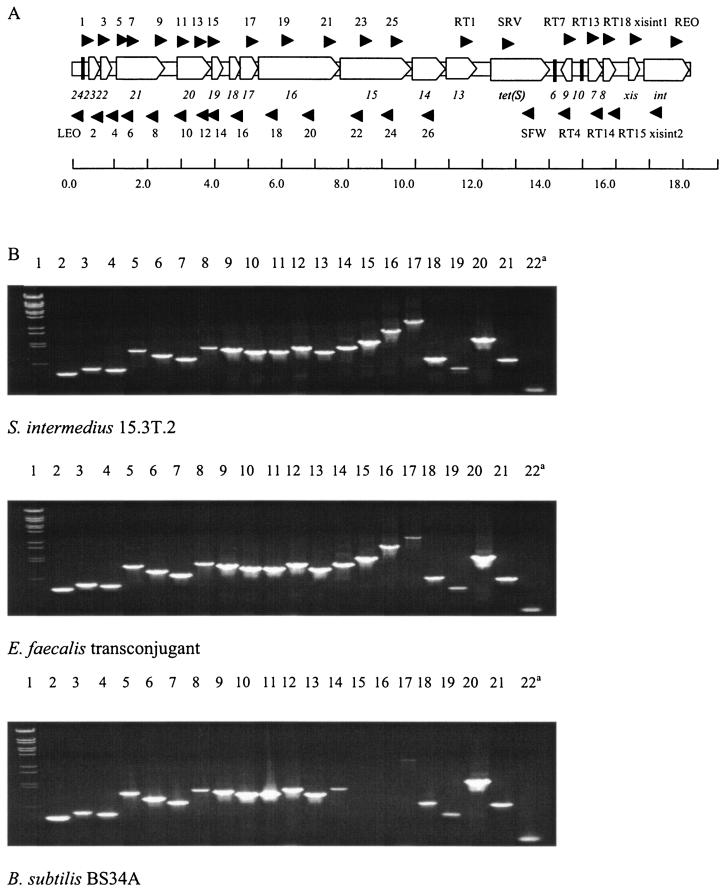
FIG. 1.
(A) Predicted schematic of Tn916S based on the structure of Tn916. Shown are the positions and names of primers (solid triangles) used to amplify the amplicons shown in panel B below. The origins and sequences of the primers are given in Table 1. The last line represents the scale in kilobases. (B) Amplicons from S. intermedius 15.3T.2, the E. faecalis transconjugant, and BS34A (contains Tn916). Lanes: 1, lambda BstEII molecular marker; 2, primers 1 and 2; 3, primers 3 and 4; 4, primers 5 and 6; 5, primers 7 and 8; 6, primers 9 and 10; 7, primers 11 and 12; 8, primers 13 and 14; 9, primers 15 and 16; 10, primers 17 and 18; 11, primers 19 and 20; 12, primers 21 and 22; 13, primers 23 and 24; 14, primers 25 and 26; 15, primers RT1 and SFW; 16, primers RT4 and SRV [reactions 15 and 16 should be negative for BS34A, as this strain does not contain tet(S)]; 17, primers RT1 and RT4; 18, primers RT7 and RT14; 19, primers RT13 and RT15; 20, primers RT18 and intxis2; 21, primers intxis1 and intxis2; and 22, primers REO and LEO (a, this primer pair amplifies the circular form of the element).
Tcr from the S. intermedius 15.3T.2 donor was transferable to E. faecalis JH2-2, S. sobrinus, and S. sanguinis at frequencies of 4.5 × 10−7, 2.5 × 10−7, and 1.0 × 10−5 per donor, respectively. PCR amplifications specific for tet(S) on S. intermedius 15.3T.2 and transconjugant genomic DNA demonstrated that tet(S) was present. No other Tcr genes could be amplified. No tet genes could be detected in the recipients. PCRs for the entire length of Tn916 were carried out on S. intermedius 15.3T.2, the E. faecalis transconjugant, and the E. faecalis JH2-2 recipient. S. intermedius 15.3T.2 and the transconjugant had PCR amplicons [with the exception of the tet(M)-containing amplicon] the same size as the positive control of Bacillus subtilis BS34A (which contains a single copy of Tn916), indicating that Tn916S has the same genetic organization as Tn916. The PCR assay on the recipient (E. faecalis JH2-2) yielded no amplicons. The sequence data of the region between RT1 and RT4 (Fig. 1A) showed that tet(S) has effectively replaced tet(M) with the upstream region lacking repeat regions that are involved in transcriptional control in Tn916.
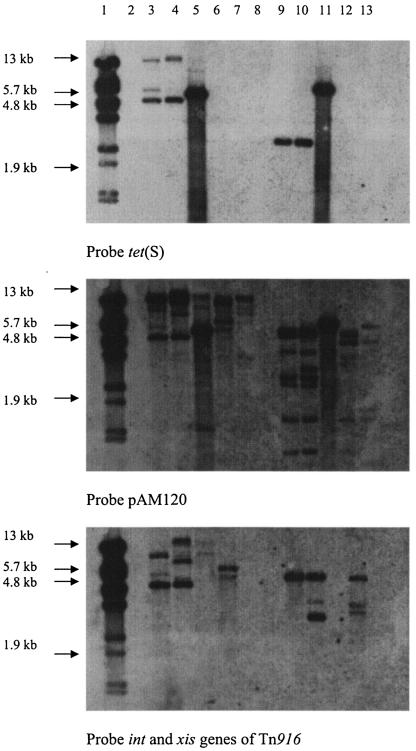
DNA from the parents and one of the transconjugants was subjected to Southern blot analysis (Fig. 2). HindIII digests were probed with tet(S), and two hybridizing fragments were observed. As there is one HindIII site in the tet(S) gene, the two hybridizing fragments are likely to be Tn916S-genome junction regions. A single HincII fragment is seen in both the donor and transconjugant, which corresponds to an internal fragment (9).
FIG. 2.
Southern blot analysis of DNA from parents and transconjugants containing Tn916S. Lanes 3 to 7 contained HindIII-digested genomic DNA, and lanes 9 to 13 contained HincII-digested genomic DNA. Lanes: 1, lambda BstEII molecular marker; 2, blank; 3, S. intermedius 15.3T.2; 4, E. faecalis transconjugant; 5, Escherichia coli pAT451; 6, B. subtilis BS34A; 7, E. faecalis JH2-2; 8, blank; 9, S. intermedius 15.3T.2; 10, E. faecalis transconjugant; 11, tet(S)-positive strain; 12, B. subtilis BS34A; and 13, E. faecalis JH2-2.
When probed with int/xis, two HindIII fragments are observed in the donor and three are observed in the transconjugant. As this probe should hybridize to one junction fragment within Tn916S, this means either that there are two copies of the element in the donor and three in the transconjugant or that there is another int/xis-containing genetic element in the donor that may have transferred to the recipient. We prefer the latter explanation, as the 4.8-kb hybridizing HindIII fragment with the int/xis probe is the same size as one of the fragments when tet(S) is used as the probe, indicating that the tet(S) gene and one of the int/xis regions are linked. When pAM120 is used to probe the blots, HincII digestion shows similar hybridizing fragments, as would be expected from DNA containing an integrated copy of Tn916 but with the extra HincII fragment (see above).
When the blots were probed with tet(M), there was no hybridization (data not shown). Taken together, these data indicate the donor strain contains at least two mobile elements containing xis and int, one of which contains tet(S). Both are capable of transfer to the recipient.
The finding of tet(S) in the same relative position as tet(M) in a broad-host-range Tn916-related element supports the view that conjugative transposons are composed of modules that are able to exchange with modules from other elements (10, 12), possibly by homologous recombination. It now seems apparent that not only is Tn916 involved in the dissemination of tet(M), it is also involved in the dissemination of tet(S).
Nucleotide sequence accession number.
The sequence of the region between orf13 and orf6 has been deposited in GenBank under accession no. AY534326.
Acknowledgments
This work was supported by the Wolfson Trust.
REFERENCES
- 1.Aminov, R. I., N. Garrigues-Jeanjean, and R. I. Mackie. 2001. Molecular ecology of tetracycline resistance: development and validation of primers for detection of tetracycline resistance genes encoding ribosomal protection proteins. Appl. Environ. Microbiol. 67:22-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Charpentier, E., G. Gerbaud, and P. Courvalin. 1993. Characterization of a new class of tetracycline-resistance gene tet(S) in Listeria monocytogenes BM4210. Gene 131:27-34. [DOI] [PubMed] [Google Scholar]
- 3.Charpentier, E., G. Gerbaud, and P. Courvalin. 1994. Presence of the Listeria tetracycline resistance gene tet(S) in Enterococcus faecalis. Antimicrob. Agents Chemother. 38:2330-2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Flannagan, S. E., L. A. Zitzow, Y. A. Su, and D. B. Clewell. 1994. Nucleotide sequence of the 18-kb conjugative transposon Tn916 from Enterococcus faecalis. Plasmid 32:350-354. [DOI] [PubMed] [Google Scholar]
- 5.Gawron-Burke, C., and D. B. Clewell. 1984. Regeneration of insertionally inactivated streptococcal DNA fragments after excision of transposon Tn916 in Escherichia coli: strategy for targeting and cloning of genes from gram-positive bacteria. J. Bacteriol. 159:214-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marra, D., and J. R. Scott. 1999. Regulation of excision of the conjugative transposon Tn916. Mol. Microbiol. 31:609-621. [DOI] [PubMed] [Google Scholar]
- 7.Ng, L. K., I. Martin, M. Alfa, and M. Mulvey. 2001. Multiplex PCR for the detection of tetracycline resistant genes. Mol. Cell Probes 15:209-215. [DOI] [PubMed] [Google Scholar]
- 8.Perreten, V., F. Schwarz, L. Cresta, M. Boeglin, G. Dasen, and M. Teuber. 1997. Antibiotic resistance spread in food. Nature 389:801-802. [DOI] [PubMed] [Google Scholar]
- 9.Roberts, A. P., G. Cheah, D. Ready, J. Pratten, M. Wilson, and P. Mullany. 2001. Transfer of Tn916-like elements in microcosm dental plaques. Antimicrob. Agents Chemother. 45:2943-2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roberts, A. P., P. A. Johanesen, D. Lyras, P. Mullany, and J. I. Rood. 2001. Comparison of Tn5397 from Clostridium difficile, Tn916 from Enterococcus faecalis and the CW459tet(M) element from Clostridium perfringens shows that they have similar conjugation regions but different insertion and excision modules. Microbiology 147:1243-1251. [DOI] [PubMed] [Google Scholar]
- 11.Roberts, A. P., C. Hennequin, M. Elmore, A. Collignon, T. Karjalainen, N. Minton, and P. Mullany. 2003. Development of an integrative vector for the expression of antisense RNA in Clostridium difficile. J. Microbiol. Methods 55:617-624. [DOI] [PubMed] [Google Scholar]
- 12.Toussaint, A., and C. Merlin. 2002. Mobile elements as a combination of functional modules. Plasmid 47:26-35. [DOI] [PubMed] [Google Scholar]
- 13.Villedieu, A., M. L. Diaz-Torres, N. Hunt, R. McNab, D. A. Spratt, M. Wilson, and P. Mullany. 2003. Prevalence of tetracycline resistance genes in oral bacteria. Antimicrob. Agents Chemother. 47:878-882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang, H., A. P. Roberts, D. Lyras, J. I. Rood, M. Wilks, and P. Mullany. 2000. Characterization of the ends and target sites of the novel conjugative transposon Tn5397 from Clostridium difficile: excision and circularization is mediated by the large resolvase, TndX. J. Bacteriol. 182:3775-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang, H., A. P. Roberts, and P. Mullany. 2000. DNA sequence of the insertional hot spot of Tn916 in the Clostridium difficile genome and discovery of a Tn916-like element in an environmental isolate integrated in the same hot spot. FEMS Microbiol. Lett. 192:15-20. [DOI] [PubMed] [Google Scholar]