Abstract
Understanding the phylogenetic placement of crown turtles (Testudines) among amniotes has been a source of particular contention. Recent morphological analyses suggest that turtles are sister to all other reptiles, whereas virtually all analyses of gene sequences support turtles as being inside Diapsida, and usually as sister to crown Archosauria (birds and crocodylians). Previously, a study using microRNAs (miRNAs) placed turtles inside diapsids, but as sister to lepidosaurs (lizards and Sphenodon) rather than archosaurs. Here, we test this result with an expanded dataset, and employ proper criteria for miRNA annotation. Significantly, we find no support for a turtle + lepidosaur sister-relationship; intstead, we recover strong support for turtles sharing a more recent common ancestor with archosaurs as the living sister group to birds + crocodylians. These results are in accordance with most gene sequence studies, providing strong, consilient evidence from diverse independent datasets for the phylogenetic position of turtles.
Keywords: Testudines, Amniota, Archosauria, Reptilia, microRNA, Turtles
Introduction
The phylogenetic position of turtles represents one of the most recalcitrant problems in vertebrate biology, with contrasting hypotheses arising from different datasets. In recent years, three contending hypotheses have been put forth for the phylogenetic placement of turtles: i) turtles represent the sister group to all diapsid reptiles (mainly supported by morphological datasets and developmental data, e.g. Gauthier et al. 1988, Lee 1997, Werneburg and Sanchez-Villagra 2009, Lyson et al. 2010, Lyson et al. 2013); ii) turtles are the sister group to Lepidosauria (Sphenodon and lizards, including snakes; supported mainly by expressed microRNAs as well as some morphological analyses, e.g. Rieppel and deBraga 1996, deBraga and Rieppel 1997, Rieppel and Reisz 1999, Li et al. 2008, Lyson et al. 2012); and iii), turtles are the sister taxon to, or are nested within, Archosauria (birds and crocodylians; supported mainly by gene-sequence datasets e.g. Zardoya and Meyer 1998, Hedges and Poling 1999, Kumazawa and Nishida 1999, Iwabe et al. 2005, Shen et al. 2011, Tzika et al. 2011, Chiari et al. 2012, Crawford et al. 2012, Fong et al. 2012, Shaffer et al. 2013, Wang et al. 2013, Lu et al. 2013). In the absence of a well-resolved phylogenetic hypothesis for Amniota, outstanding macroevolutionary questions, including those regarding the acquisition of the unique turtle body plan, cannot be adequately addressed.
Although in contradiction to most molecular studies, the miRNA data supporting a turtle + lepidosaur clade (Lyson et al. 2012) was not entirely unexpected (Rieppel and deBraga 1996, Becker et al. 2010). miRNAs are ~22-nucleotide noncoding RNA molecules that have been heralded as especially useful phylogenetic characters due to their continuous addition to animal genomes through time, comparatively low rates of secondary loss, and the largely conservative nature of the mature gene product’s primary sequence, and have been used to reconstruct the phylogenetic interrelationships of numerous animal clades at all levels in metazoan phylogeny (Sperling and Peterson 2009, Tarver et al. 2013). Lyson et al. (2012) showed that the lizard Anolis carolinensis and the turtle Chrysemys picta shared four putative microRNAs, and that these nucleotide sequences were not recovered in a small RNA library derived from a total RNA preparation of an alligator, nor present in any sequenced bird genome. On the basis of these apparent synapomorphic miRNAs, these authors concluded that turtles were likely the extant sister group of the lepidosaurs.
As with any dataset though, the robustness of the characters used directly dictates the robustness of the analysis. With miRNAs, care must be taken in distinguishing them from other types of RNA molecules including other small RNAs (e.g., piRNAs, tRNAs), and fragments of larger RNA molecules (in particular, fragments of rRNAs and mRNAs). Recent clarifications of the criteria for miRNA annotation have challenged the diagnosis of many sequences previously identified as miRNAs (Tarver et al. 2012), and the four miRNA sequences identified as turtle + lepidosaur synapomorphies by Lyson et al. (2012) do not meet the minimal criteria established for miRNA annotation (Tarver et al. 2012, Kozomara and Griffiths-Jones 2011). Thus, the discordance between the miRNA dataset of Lyson et al. (2012) and most sequence-based datasets to this point, including the recent phylogenomic analysis of Chiari et al. (2012), could be due to mistaken miRNA homologies in the Lyson et al. (2012) study. To address this issue, we characterized the near-complete miRNA repertoire of the turtle Chrysemys picta using both small RNA library reads and genomic sequences, and compared this repertoire to the near-complete repertoires of the snake Python bivittatus, the crocodylian Alligator mississippiensis, and the avian Columba livia, in addition to previously published lizard (Lyson et al. 2012) and bird data (miRBase v.19; Kozomara and Griffiths-Jones 2011). Further, we sequenced small RNA libraries from three additional species from across the lizard tree - the gecko Coleonyx variegatus, the xantusiid Xantusia wigginsi, and the snake Chionactis occipitalis - and queried the genomes of one additional crocodylian (A. sinensis), and three other turtles (the cheloniid Chelonia mydas, and the trionychids Pelodiscus sinensis and Apalone spinifera). Our analyses fully support an archosaur affinity for turtles, as the original ‘miRNAs’ identified by Lyson et al. (2012) appear to be spurious, and turtles share several bona fide miRNAs with archosaurs not found or expressed in lepidosaurs, mammals, or any other metazoans. Further, a Bayesian phylogenetic analysis of 238 precursor miRNA sequences fully supports a close relationship between turtles and archosaurs rather than an affinity between turtles and lepidosaurs. Therefore, according to these analyses, turtles are strongly supported as diapsid reptiles sharing a more recent common ancestor with archosaurs than with lepidosaurs. These results alleviate a major discordance between miRNA and gene sequence datasets regarding the phylogenetic position of Testudines within Amniota.
Materials and Methods
Total RNA (Wheeler et al. 2009) was extracted from single late-stage embryos of the pigeon Columba livia, an adult gecko Coleonyx variegatus, an adult xantusiid Xantusia wigginsi, and a juvenile snake Chionactis occipitalis, following standard animal care protocols (IACUC number 2009-11302). Small RNA libraries were prepared at the Yale W. M. Keck Facility according to manufacturer’s instructions, and sequenced using their Illumina Genome Analyzer II platform. The number of reads sequenced per library is detailed in Table 1.
Table 1.
Read Count and Genome Assembly Information.
| species | total reads | collapsed reads* | genome assembly accesion number |
|---|---|---|---|
| Alligator mississippiensis | 21,731,314 | 97,477 | AKHW00000000.1 |
| Columba livia | 45,635,579 | 86,204 | AKCR00000000.1 |
| Chrysemys picta | 23,765,521 | 104,168 | AHGY00000000.1 |
| Chionactis occipitalis | 44,178,821 | 29,885 | NA |
| Coleonyx variegatus | 110,883,152 | 102,251 | NA |
| Python bivittatus | NA | NA | AEQU010000000.1 |
| Xantusia wigginsi | 63,299,421 | 93,742 | NA |
This number represents the number of non-redundant sequences 20–25 nucleotides in length that were expressed two or more times in the respective small RNA library and annotated to miRBase (v. 19) using miRMiner.
An updated version of miRMiner (Wheeler et al. 2009) was used to identify both orthologues of previously identified miRNAs (miRBase v. 19; Kozomara and Griffiths-Jones 2011) and novel miRNA families from these four taxa, in addition to reanalyzing the raw data from Lyson et al. (2012) for the turtle C. picta, the alligator A. mississippensis, and the lizard A. carolinensis. Because published genomes are now available for C. picta (Shaffer et al. 2013) and A. mississippensis (St John et al. 2012), the near-complete complements of miRNAs from these two taxa were assembled (ESM 1–2, respectively). In addition, the reads from C. livia were used to query the recently released pigeon genome (Shapiro et al. 2013), and its near-complete miRNA repertoire was assembled (ESM 3). Finally, the near-complete ancestral miRNA complement of macrostomate snakes (Lee et al. 2007) was assembled (ESM 4) using the reads from the snake C. occipitalis and the genome of the python Python bivittatus (Castoe et al. 2011).
Next, the miRNAs constituting each of these complements (ESM 1–4) were used as queries to search the genomes of three other turtles (the cheloniid Chelonia mydas, and the two trionychids Pelodiscus sinensis and Apalone spinifera), the chinese alligator Alligator sinensis, and the coelacanth Latimeria chalumnae, using the default blastn parameters. A data matrix of 57 miRNAs (ESM 5), including 32 new miRNA families specific to either the snake, alligator, or turtle lineages, was assembled that included all known miRNAs to have evolved in the reptile lineage since their split with the mammalian stem-group ~310 Ma ago (ESM 6), excluding autapomorphies. Each putative miRNA was aligned with its known orthologues using Macvector v. 10.02 (Symatec Co., Mountain View, CA; alignments available upon request), and a dataset of presences/absences (ESM 5) was assembled using MacClade v. 4.08 (Maddison and Maddison 2005). This character matrix was analyzed using both Dollo parsimony (PAUP* 4.0b10; Swofford 2002) with all characters given equal weight and using the branch and bound search algorithm, and Bayesian analysis (BEAST 1.8; Drummond et al. 2012) using the stochastic Dollo model. Two runs of 10,000,000 generations were performed with the starting tree for each of these analyses randomly generated. Because BEAST requires the use of a molecular clock in conjunction with standard phylogenetic analyses, we incorporated an uncorrelated exponential clock using four calibration points (all of which were modeled as normal distributions centered on the midpoint value between the minimum and the maximum – see ESM 6). The root node was also modeled using a normal distribution centered on the age of the split between Actynopterygii and Sarcopterygii (416 to 421 Ma with average distribution = 418.5 and SD = 3; Benton et al. 2009). Clade support was estimated using Bremer support values (Bremer 1994) for the parsimony analysis, or posterior probabilities for the Bayesian analysis.
Finally, a concatenated dataset of 238 pre-miRNA sequences was assembled for 17 tetrapod taxa: the frog Xenopus tropicalis; Homo sapiens; the mouse Mus musculus; the marsupials Monodelphis domestica and Macropus eugenii; the platypus Ornithorhynchus anatinus; the lepidosaurs Anolis carolinensis and Python bivittatus, the birds Gallus gallus, Taenopygia guttata and Columba livia; the alligators Alligator mississippensis and A. sinensis; and the turtles Chrysemys picta, Chelonia mydas, Pelodiscus sinensis and Apalone spinifera; as well as two outgroup species, the coelacanth Latimeria chalumnae and the zebrafish Danio rerio. This dataset (ESM 7) was assembled in two stages. First, orthology of each miRNA reconstructed as present in the last common ancestor (LCA) of Tetrapoda was determined for six taxa (H. sapiens, M. musculus, G. gallus, A. carolinensis, X. tropicalis, and D. rerio) using both phylogenetic (Macvector v. 10.02) and syntenic (Ensembl release 72) analysis (ESM 8). Because the current miRNA annotation system (Kozomara and Griffiths-Jones 2011) is not amenable to orthology analysis, a new nomenclature system was erected to make orthology recognition readily apparent among multi-gene families (ESM 8). Once orthology was determined for all multi-gene miRNA family members for these six taxa, all members of each of these families from the remaining taxa were aligned and phylogenetically analyzed by distance analysis (Macvector v. 10.02). Subsequently, each miRNA gene was assigned to a particular paralogy group, giving a total of 238 miRNA genes reconstructed to have been present in the tetrapod LCA (ESM 9). These 238 genes were then concatenated for each taxon, and finally analyzed using Bayesian phylogenetics. Phylogenetic analyses were performed under the GTR + G, the CAT–GTR + G, and the QMM + G models using Phylobayes (Lartillot et al. 2009). The difference between these models is that while a single GTR matrix is applied to an unpartitioned superalignment under GTR + G, under CAT–GTR +G and QMM + G the data are automatically partitioned (during tree search) in an optimal number of compositionally defined partitions. In addition, in the CAT–GTR substitution rates are modeled using one GTR matrix (common to all the partitions), while in the QMM model each partition is assigned its own partition–specific GTR matrix. For each considered model, two independent analyses were run until convergence (for the two analyses the burnin was different, but the chains were always subsampled every 100 generations). From the concatenation of the miRNA dataset we also performed a molecular clock analysis aimed at identifying lineage specific rates of microRNA evolution. Molecular clock analyses were run in Phylobayes (under both the autocorrelated lognormal model (Thorne et al. 1998) and the uncorrelated gamma multiplier model (Drummond et al. 2006). Substitutions were modeled using the CAT-GTR model; calibration points are those in ESM 6. Clock analyses were run to completion and ratograms were derived in which branch lengths represent lineage specific rates of evolution.
Results
A total of 267 miRNA loci were found in the genome of the turtle Chrysemys picta (Table 2), with 251 supported with reads from at least one arm of the miRNA hairpin (ESM 1). Of these 267 miRNAs, 20 are novel miRNA families, acquired hierarchically in the turtle lineage (Fig. 1) as expected (Sperling and Peterson 2009, Tarver et al. 2013). A further three loci are likely present in the C. picta genome: miR-15-P4 (= Hsa-miR-497), miR-138-P1 (= Hsa-miR-138-1), and miR-150, as reads were detected for these miRNAs in the C. picta small RNA library, and these loci are present in other turtle genomes, but a corresponding locus was not present in the deposited trace archives at Genbank. The number of loci annotated in C. picta is similar to the three unambiguous diapsids described herein (Table 2). The completeness of the C. picta genome appears to be slightly higher than that of the crocodylian Alligator mississippensis and the avian Columba livia (at least by assessing the number of missing miRNA loci), but seems comparable to that of the python P. bivittatus. A. mississippiensis appears to be missing 10–11 loci, although six of these missing loci are linked in a single cluster in all other amniotes (the miR-18b/miR-106a/miR-363 cluster), and were sequenced in the close relative A. sinensis, whereas C. livia is missing 15–18 loci based on the appearance of reads in its small RNA library.
Table 2.
MicroRNA Loci.
| species | miRNA loci | read support | novel loci | inferred missing loci* |
|---|---|---|---|---|
| Alligator mississippiensis | 244 | 235 | 18 | 10–11$ |
| Columba livia | 250 | 241 | 39 | 15–18& |
| Chrysemys picta | 267 | 251 | 20 | 3 |
| Python bivittatus | 215 | 203 | 1# | 2 |
Ascertained by the presence of reads in the small RNA library, most of which are also present in the genomic sequence of a near relative.
This represents the shared complement between P. molurus and C. occipitalis, the latter the source of the RNA used to query the genome of the former.
The presence vs. absence of miR-103-P2 cannot be confirmed because of the sequence identity of both the 5p and 3p arms with other paralogues as ascertained by the sequence of A. sinensis.
The presence vs. absence of three loci cannot be determined because of the identity of read sequences (both 5p and 3p) with paralogues (mir-9-P3, mir-124-P1, and mir-196-P1) in other Neoaves.
Figure 1.
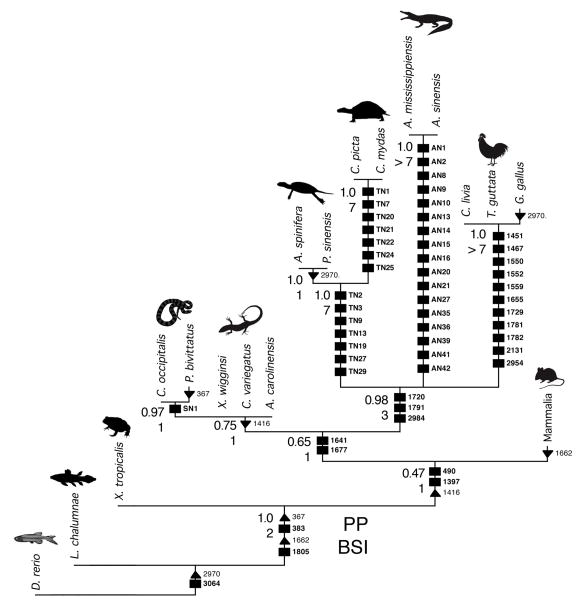
microRNAs support a turtle-archosaur relationship. Sixteen tetrapod taxa were scored for the presence/absence of 57 miRNA families using the coelacanth L. chalumnae and the zebrafish D. rerio as outgroups. A single shortest tree (tree length = 62) was found using Dollo parsimony (PAUP* 4.0b10, ref. 41) with all characters given equal weight and using the branch and bound search algorithm. Bremer support indexes (BSI) were calculated using PAUP* and the values are indicated at the nodes. Posterior probabilities (PP) were calculated using Bayesian analysis (BEAST 1.8, ref. 48) using the stochastic Dollo model. These data support an archosaur affinity of turtles (PP = 0.98, BSI = 3) as turtles share three miRNAs with archosaurs not found or expressed in any other tetrapod taxon. microRNAs that are not secondarily lost are shown as boxes; those that are secondarily lost are shown as triangles, with the loss denoted by an upside-down triangle.
Of particular interest to us was confirming the presence of the four synapomorphic miRNAs used by Lyson et al. (2012) to demonstrate a phylogenetic affinity between the turtle Chrysemys picta and the lizard Anolis carolinensis: miR-5390, -91, -92, and miR-5393. Lyson et al. (2012) showed that these reads were present in both the C. picta and A. carolinensis small RNA libraries, and absent in the Alligator mississippensis small RNA library; additionally, a locus for each of these reads was present in the genomic sequence of A. carolinensis. However, star reads were not recorded for any of these four putative miRNAs, which is problematic given that the relative position of the enzymatic cuts between the two arms of the putative hairpin is essential for recognizing bona fide miRNAs (Tarver et al. 2012). Using these four putative miRNA sequences as queries against all diapsid genome databases in Genbank reveals that there are no corresponding loci in any other genome including C. picta, the supposed source of the shared reads with A. carolinensis (Lyson et al. 2012), for three of the four loci – only miR-5391 has a corresponding locus in other reptiles (including A. mississippiensis, contra Lyson et al. 2012). However, closer examination of this sequence reveals that the supposed mature miRNA read is actually the terminal portion of an exon, and a consensus splice site sits immediately 3′ of the putative mature read. Further, reads for none of these four miRNAs were found in any of our new libraries, including the lizards Coleonyx variegatus, Xantusia wigginsi, and the snake Chionactis occipitalis. Therefore, it appears that none of these four sequences can be confirmed as miRNAs, and none of the four support any sort of phylogenetic argument for the placement of turtles.
Instead, three miRNAs – miR-1720, miR-1791 and miR-2984 –are present in archosaur genomes and in all four turtle genomes (ESM 10–12) that are absent in the lizards Anolis carolinensis and Python bivittatus. These miRNAs have not been reported in any other animal genome, although the squamate sister clade, Sphenodon punctatus, has yet to be assayed for them. Curiously, none of these miRNAs were detected in our single-ontogenetic-stage library of Chrysemys picta (which is why they were missed by Lyson et al. 2012, as publically available genomes for turtles were not available to the authors at that time), suggesting that these miRNAs are expressed either at very low levels in turtles, or (and more likely) at different ontogenetic stages. Indeed, reads for most of these miRNAs were found in the late-stage pigeon embryo, whereas neonatal turtle and alligator individuals were used for the C. picta and Alligator mississippensis libraries in Lyson et al. (2012). Therefore, it is possible that profiling miRNAs from late-stage embryos would reveal transcripts of these lowly expressed miRNAs in turtles.
Both our maximum parsimony (BSI = 3) and Bayesian analysis (PP = 0.98) strongly support an archosaur affinity for turtles, with no support for a lepidosaur affinity based on shared miRNA sequences (Fig. 1). Nonetheless, despite both archosaurs and turtles evolving a suite of novel miRNAs (ESM 1–3), no synapomorphic miRNAs appear to exist that enable resolution of the interrelationships among turtles and archosaurs (whether turtles are the extant sister group of archosaurs, or alternatively nested within Archosauria as the living sister to either birds or crocodylians).
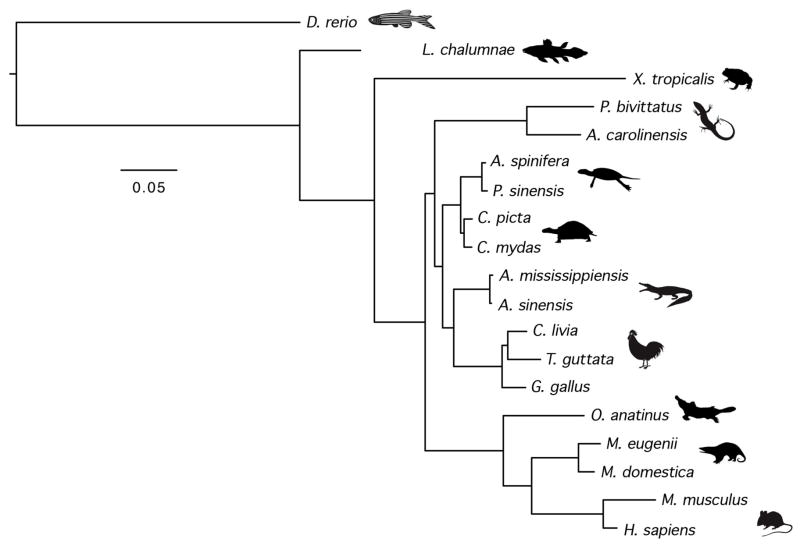
To test this result, and to see if a more precise position of turtles relative to archosaurs could be inferred, we analyzed the primary nucleotide sequences of the precursor miRNAs for every miRNA sequence reconstructed as present in the last common ancestor of Tetrapoda (= Amphibia + Amniota; ESM 7), including many new miRNA sequences not currently deposited in miRBase v. 20 (ESM 13–15), using standard Bayesian phylogenetics. When the superalignment of the considered microRNAs was analyzed, we found that turtles resolve as sister to archosaurs under all considered models (PP = 1 under all models investigated: GTR, CAT–GTR, and QMM; Fig. 2).
Figure 2.
Bayesian phylogenetic analysis of 238 concatenated pre-miRNA sequences in sixteen tetrapod taxa using the coelacanth L. chalumnae and the zebrafish D. rerio as the outgroups. These data strongly support the hypothesis that turtles are the extant sister group of archosaurs (PP = 1.0 in all considered models, see text).
Discussion
Contrary to the results reported by Lyson et al. (2012), both the pattern of acquisition of post-tetrapod miRNAs (Fig. 1), and a phylogenetic analysis of the primary sequences of pre-tetrapod pre-miRNAs (Fig. 2), robustly support an archosaur, rather than a lepidosaur, affinity for turtles. Indeed, the latter analysis strongly supports a sister group relationship between crown turtles and crown archosaurs, as do most recent studies addressing amniote interrelationships using gene sequence data (Zardoya and Meyer 1998, Hedges and Poling 1999, Kumazawa and Nishida 1999, Iwabe et al. 2005, Shen et al. 2011, Tzika et al. 2011, Chiari et al. 2012, Crawford et al. 2012, Fong et al. 2012, Shaffer et al. 2013, Wang et al. 2013, Lu et al. 2013).
The reasons for the different results obtained in our analysis and Lyson et al. (2012) are not due to problems with miRNAs per se (as suggested, e.g. Chiari et al. 2012), as turtles show both slow rates of miRNA evolution (ESM 16) and minimal secondary miRNA gene loss (ESM 17). Instead, the reason for the apparent incongruence is simply due to misrecognition of primary homologies by Lyson et al. (2012). The four miRNAs purported to be shared between the lizard Anolis carolinensis and the turtle Chrysemys picta did not express both arms of the hairpin in A. carolinensis (the “mature” and the “star”; Ambros et al. 2003). Normally, this is not a problem; deep phylogenetic conservation can substitute for absence of star reads when annotating miRNAs (Ambros et al. 2008), as star sequences are often expressed at much lower levels than their corresponding mature reads. However, in this case, deep phylogenetic conservation was the issue at hand, and thus Lyson et al. (2012) essentially made a circular argument – they used phylogenetic conservation to justify the robustness of the new miRNAs discovered in A. carolinensis, and then used these miRNAs to propose a close affinity between turtles and lepidosaurs. More recent work on miRNA annotation strongly indicates that obtaining reads from both arms of the hairpin is essential for the recognition of new miRNAs (e.g., Tarver et al. 2012) and, indeed, each of the three miRNAs shared between turtles and archosaurs presented here express both arms of the hairpin in at least one species (ESM 10–12).
One final contrast between our study and that of Lyson et al. (2012) is that none of the miRNAs supporting a turtle + archosaur grouping were expressed in our single-ontogenetic-stage turtle library, and thus, as suggested (Crawford et al.), sampling biases – in this case the absence of sequenced genomes in key areas of the tree – resulted not only in the misrecognition of putative miRNAs, but also in the non-recognition of bona fide miRNAs. Nonetheless, given the concordance between our two independent analyses using miRNAs (one a presence/absence analysis, and the other a primary sequence analysis), and virtually every other study of gene sequences focused on amniote phylogeny, we conclude that molecular data in general strongly support an exclusive turtle + archosaur clade (but see Lu et al. 2013).
Despite concordance among studies using molecules to address turtle affinities, the turtle + archosaur sister-group hypothesis has yet to find much support in morphological and fossil datasets, which argue instead for turtles as sister to total group pan-diapsids (Gauthier et al. 1988, Lee 1997, Werneburg and Sanchez-Villagra 2009, Lyson et al. 2010, Lyson et al. 2013), or as being closely related to marine lepidosauromorph sauropterygians (Rieppel and deBraga 1996, deBraga and Rieppel 1997, Rieppel and Reisz 1999; although this latter hypothesis has not enjoyed much recent support). Although the putative ‘parareptilian’ affinities of turtles inferred from recent morphological datasets appear to stand in stark contrast to molecular results, recent work applying molecular scaffolds to turtle morphological datasets underscores the lability of reptile interrelationships in morphological analyses. In particular, Lee (2013) demonstrated that the interrelationships of turtles, parareptiles and diapsids exhibit little consistency among phylogenetic analyses employing different optimality criteria, ingroup compositions and character sets. Significantly, this study demonstrated that morphological and genomic analyses may be more congruent than generally espoused, with only minor decreases in fit incurred when constraining morphological data to molecular topologies. Such analyses indicate the intriguing possibility that turtles may simultaneously share a recent common ancestor with ‘parareptiles’ such as Eunotosaurus (as frequently supported by paleontological data (Lyson et al. 2010, Lyson et al. 2013, Lyson et al. 2013, Carroll 2013), and be most closely related to archosaurs amongst extant taxa (Lee et al. 2013). Much progress in our understanding of morphological evolution stands to be made from the simultaneous phylogenetic analysis of parareptiles, basal stem diapsids, and crown reptiles; however, no relevant matrices have so far been constructed (Lee 2013).
If the morphological hypothesis that turtles represent the extant sister group of living reptiles accurately reflects turtle origins, it would indicate that virtually the entire genome (Matsuda et al. 2005, Shedlock et al. 2007), including mitochondrial genes (Zardoya and Meyer 1998, Kumazawa and Nishida 1999), ribosomal RNA genes (Hedges and Poling 1999), protein coding genes (Hedges and Poling 1999, Iwabe et al. 2005, Shen et al. 2011, Tzika et al. 2011, Chiari et al. 2012, Fong et al. 2012, Shaffer et al. 2013, Wang et al. 2013, Lu et al. 2013), ultraconserved elements (Crawford et al. 2012), and miRNAs (Figs. 1 and 2), exhibit astonishing levels of homoplasy in a surprisingly congruent pattern.
Consilience amongst independent datasets remains the most reliable way to adjudicate phylogenetic hypotheses, and the strongly-supported miRNA results presented herein add to the considerable (and ever-growing) body of evidence that turtles represent the extant sister-group to archosaurs. This work is necessary to provide an accurate evolutionary framework from which patterns of trait evolution, such as the origin of the unique turtle body plan, and their fascinating physiology (Gilbert and Corfe 2013), can be inferred. The accurate interpretation of relevant fossils may await the development of a comprehensive morphological phylogenetic matrix incorporating all relevant taxa, and such work may be necessary for basal stem turtles with diapsid features to come to light. Although this pursuit will be a major undertaking, it is only fitting that a ‘slow and steady’ approach to reptile systematics will be necessary to confidently reconstruct the evolutionary history of this fascinating clade.
Supplementary Material
Acknowledgments
We thank K. Kuester for providing an egg of C. livia, J. Musser and G. Watkins-Colwell for assistance processing the pigeon, A. Heimberg for comments, J. Tarver for laboratory assistance, and G. Wagner for providing lab space. Funding for this project was provided by a NSERC CGS and Lougheed Award of Distinction to DJF, and Yale Peabody Museum of Natural History to JAG. BLK and KJP were supported by NASA-Ames.
Footnotes
Author contributions:
D.J.F. and K.J.P. conceived of the project. D.J.F., K.J.P., and T.R.L. collected data. K.J.P, B.L.K., D.P., and D.J.F. analyzed data. All authors contributed in preparing the manuscript.
References
- Ambros V, Bartel B, Bartel DP, Burge CB, Carrington JC, Chen X, Dreyfuss G, Eddy SR, Griffiths-Jones S, Marshall M, Matzke M, Ruvkun G, Tuschl T. A uniform system for microRNA annotation. RNA. 2003;9:277–279. doi: 10.1261/rna.2183803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker RE, Valverde RA, Crother BI. Proopiomelanocortin (POMC) and testing the phylogenetic position of turtles (Testudines) J Zool Syst Evol Res. 2010;49:148–159. [Google Scholar]
- Benton MJ, Donoghue PCJ, Asher RJ. Calibrating and constraining molecular clocks. In: Hedges SB, Kumar S, editors. The Timetree of Life. Oxford University Press; Oxford: 2009. pp. 35–86. [Google Scholar]
- Bremer K. Branch support and tree stability. Cladistics. 1994;10:295–304. [Google Scholar]
- Carroll RL. Problems of the ancestry of turtles. In: Brinkman DB, Holroy PA, Gardner JD, editors. Morphology and Evolution of Turtles. Springer; Netherlands: 2013. pp. 19–36. [Google Scholar]
- Castoe TA, de Koning APJ, Hall KT, Yokoyama KD, Gu W, Smith EN, Feschotte C, Uetz P, Ray DA, Dobry J, Bogden R, Mackessy SP, Bronikowski AM, Warren WC, Secor SM, Pollock DD. Sequencing the genome of the Burmese python (Python molurus bivittatus) as a model for studying extreme adaptations in snakes. Genome Biol. 2011;12:406. doi: 10.1186/gb-2011-12-7-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiari Y, Cahais V, Galtier N, Delsuc F. Phylogenomic analyses support the position of turtles as the sister group of birds and crocodiles (Archosauria) BMC Biol. 2012;10:65. doi: 10.1186/1741-7007-10-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford NG, Faircloth BC, McCormack JE, Brumfield RT, Winker K, Glenn TC. More than 100 ultraconserved elements provide evidence that turtles are the sister group of archosaurs. Biol Letters. 2012;8:783–786. doi: 10.1098/rsbl.2012.0331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- deBraga M, Rieppel O. Reptile phylogeny and the interrelationships of turtles. Zool J Linn Soc. 1997;120:281–354. [Google Scholar]
- Drummond AJ, Ho SYW, Phillips MJ, Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006;4:e88. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 2012;29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong JJ, Brown JM, Fujita MK, Boussau BA. Phylogenomic approach to vertebrate phylogeny supports a turtle-archosaur affinity and a possible paraphyletic Lissamphibia. PLoS ONE. 2012;7:e48990. doi: 10.1371/journal.pone.0048990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier J, Kluge Arnold G, Rowe T. Amniote phylogeny and the importance of fossils. Cladistics. 1988;4:105–209. doi: 10.1111/j.1096-0031.1988.tb00514.x. [DOI] [PubMed] [Google Scholar]
- Gilbert SF, Corfe I. Turtle origins: picking up speed. Dev Cell. 2013;25:326–328. doi: 10.1016/j.devcel.2013.05.011. [DOI] [PubMed] [Google Scholar]
- Hedges SB, Poling LL. A molecular phylogeny of reptiles. Science. 1999;283:998–1000. doi: 10.1126/science.283.5404.998. [DOI] [PubMed] [Google Scholar]
- Iwabe N, Hara Y, Kumazawa Y, Shibamoto K, Saito Y, Miyata T, Katoh K. Sister group relationship of turtles to the bird-crocodilian clade revealed by nuclear DNA-coded proteins. Mol Biol Evol. 2005;22:810–813. doi: 10.1093/molbev/msi075. [DOI] [PubMed] [Google Scholar]
- Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nuc Acids Res. 2011;39:D152–D157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumazawa Y, Nishida M. Complete mitochondrial DNA sequences of the green turtle and blue-tailed mole skink: statistical evidence for archosaurian affinity of turtles. Mol Biol Evol. 1999;16:784–792. doi: 10.1093/oxfordjournals.molbev.a026163. [DOI] [PubMed] [Google Scholar]
- Lartillot N, Lepage T, Blanquart S. PhyloBayes 3: a Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics. 2009;25:2286–2288. doi: 10.1093/bioinformatics/btp368. [DOI] [PubMed] [Google Scholar]
- Lee MSY. Reptile relationships turn turtle. Nature. 1997;389:246–246. [Google Scholar]
- Lee MSY, Hugall AF, Lawson R, Scanlon JD. Phylogeny of snakes (Serpentes): combining morphological and molecular data in likelihood, Bayesian and parsimony analyses. System Biodiv. 2007;5:371–389. [Google Scholar]
- Lee MSY. Turtle origins: insights from phylogenetic retrofitting and molecular scaffolds. J Evol Biol. 2013;26:2729–2738. doi: 10.1111/jeb.12268. [DOI] [PubMed] [Google Scholar]
- Li C, Wu XC, Rieppel O, Wang LT. An ancestral turtle from the Late Triassic of southwestern China. Nature. 2008;456:497–501. doi: 10.1038/nature07533. [DOI] [PubMed] [Google Scholar]
- Lu B, Yang W, Dai Q, Fu J. Using genes as characters and a parsimony analysis to explore the phylogenetic position of turtles. PLoS ONE. 2013;8:e79348. doi: 10.1371/journal.pone.0079348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyson TR, Bever GS, Bhullar BAS, Joyce WG, Gauthier JA. Transition fossils and the origin of turtles. Biol Letters. 2010;6:453–455. doi: 10.1098/rsbl.2010.0371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyson TR, Sperling EA, Heimberg AM, Gauthier JA, King BL, Peterson KJ. MicroRNAs support a turtle + lizard clade. Biol Letters. 2012;8:104–107. doi: 10.1098/rsbl.2011.0477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyson TR, Bever GS, Scheyer TM, Hsiang AY, Gauthier JA. Evolutionary origin of the turtle shell. Curr Biol. 2013;23:1113–1119. doi: 10.1016/j.cub.2013.05.003. [DOI] [PubMed] [Google Scholar]
- Lyson TR, Bhullar BAS, Bever GS, Joyce WG, de Quieroz K, Abzhanov A, Gauthier JA. Homology of the enigmatic nuchal bone reveals novel reorganization of the shoulder girdle in the evolution of the turtle shell. Evolution & Development. 2013;15:317–325. doi: 10.1111/ede.12041. [DOI] [PubMed] [Google Scholar]
- Maddison DR, Maddison WP. MacClade. Vol. 4. Sinauer Associates; Sunderland: 2005. [Google Scholar]
- Matsuda Y, Nishida-Umehara C, Tarui H, Kuroiwa A, Yamada K, Isobe T, Ando J, Fujiwara A, Hirao Y, Nishimura O, Ishijima J, Hayashi A, Saito T, Murakami T, Murakami Y, Kuratani S, Agata K. Highly conserved linkage homology between birds and turtles: Bird and turtle chromosomes are precise counterparts of each other. Chromosome Res. 2005;13:601–615. doi: 10.1007/s10577-005-0986-5. [DOI] [PubMed] [Google Scholar]
- Rieppel O, deBraga M. Turtles as diapsid reptiles. Nature. 1996;384:453–455. [Google Scholar]
- Rieppel O, Reisz RR. The origin and early evolution of turtles. Ann Rev Ecol Syst. 1999;30:1–22. [Google Scholar]
- Shaffer HB, Minx P, Warren DE, Shedlock AM, Rhomson RC, Valenzuela N, Abramyan J, Amemiya CT, Badenhorst D, Biggar KK, Borchert GM, Borka CW, Bowden RM, Braun EL, Bronikowski AM, Bruneau BG, Buck LT, Capel B, Castoe TA, Czerwinski M, Delehaunty KD, Edwards SV, Fronick CC, Fujita MK, Fulton L, Graves TA, Green RE, Haerty W, Hariharan R, Hernandez O, Hillier LW, Holloway AK, Janes D, Janzen FJ, Kandoth C, Kong L, de Koning APJ, Li Y, Literman R, McGaugh SE, Mork L, O’Laughlin M, Paitz RT, Pollock DD, Ponting CP, Radhakrishnan S, Raney BJ, Richman JM, St John J, Schwartz T, Sethuraman A, Spinks PQ, Storey KB, Thane N, Vinar T, Zimmerman LM, Warren WC, Mardis ER, Wilson RK. The western painted turtle genome, a model for the evolution of extreme physiological adaptations in a slowly evolving lineage. Genome Biol. 2013;1:R:28. doi: 10.1186/gb-2013-14-3-r28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro MD, Kronenberg Z, Li C, Domyan ET, Pan H, Campbell M, Tan H, Huff CD, Hu H, Vickrey AI, Nielsen SCA, Stringham SA, Hu H, Willerslev E, Gilbert MTP, Yandell M, Zhang G, Wang J. Genomic Diversity and Evolution of the Head Crest in the Rock Pigeon. Science. 2013;339:1063–1067. doi: 10.1126/science.1230422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shedlock AM, Borka CW, Zhao S, Shetty J, Zhang T, Liu JS, Deschavanne PJ, Edwards SV. Phylogenomics of nonavian reptiles and the structure of the ancestral amniote genome. Proc Nat Acad Sci USA. 2007;104:2767–72. doi: 10.1073/pnas.0606204104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen XX, Liang D, Wen JZ, Zhang P. Multiple genome alignments facilitate development of NPCL markers: a case study of tetrapod phylogeny focusing on the position of turtles. Mol Biol Evol. 2011;28:3237–3252. doi: 10.1093/molbev/msr148. [DOI] [PubMed] [Google Scholar]
- Sperling EA, Peterson KJ. microRNAs and metazoan phylogeny: big trees from little genes. In: Telford MJ, Littlewood DTJ, editors. Animal Evolution - Genomes, Trees and Fossils. Oxford University Press; Oxford: 2009. pp. 157–170. [Google Scholar]
- St John JA, Braun EL, Isberg SR, Miles LG, Chong AY, Gongora J, Dalzell P, Moran C, Bed’Hom B, Abzhanov A, Burgess SC, Cooksey AM, Castoe TA, Crawford NG, Densmore LD, Drew JC, Edwards SV, Faircloth BC, Fujita MK, Greenwold MJ, Hoffmann FG, Howard JM, Iguchi T, Janes DE, Khan SY, Kohno S, de Koning APJ, Lance SL, McCarthy FM, McMormack JE, Merchant ME, Peterson DG, Pollock DD, Pourmand N, Raney BJ, Roessler KA, Sanford JR, Sawyer RH, Schmidt CJ, Triplett EW, Tuberville TD, Venegas-Anaya M, Howard JT, Jarvis ED, Guillette LJ, Glenn TC, Green RE, Ray DA. Sequencing three crocodilian genomes to illuminate the evolution of archosaurs and amniotes. Genome Biol. 2012;13:415. doi: 10.1186/gb-2012-13-1-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford DL. PAUP* Phylogenetic Analysis Using Parsimony (* and Other Methods) v.4.0b10 for Macintosh. Sinauer Associates; Sunderland: 2002. [Google Scholar]
- Tarver JE, Donoghue PCJ, Peterson KJ. Do miRNAs have a deep evolutionary history? Bioessays. 2012;34:857–866. doi: 10.1002/bies.201200055. [DOI] [PubMed] [Google Scholar]
- Tarver JE, Sperling EA, Nailord A, Heimberg AM, Robinson JM, King BL, Pisani D, Donoghue PCJ, Peterson KJ. miRNAs: small genes with big potential in metazoan phylogenetics. Mol Biol Evol. 2013;30:2369–2382. doi: 10.1093/molbev/mst133. [DOI] [PubMed] [Google Scholar]
- Thorne JL, Kishino H, Painter IS. Estimating the rate of evolution of the rate of molecular evolution. Mol Biol Evol. 1998;15:1647–1657. doi: 10.1093/oxfordjournals.molbev.a025892. [DOI] [PubMed] [Google Scholar]
- Tzika AC, Helaers R, Schramm G, Milinkovitch M. Reptilian-transcriptome v1.0, a glimpse in the brain transcriptome of five divergent Sauropsida lineages and the phylogenetic position of turtles. EvoDevo. 2011;2:19. doi: 10.1186/2041-9139-2-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Pascual-Anaya J, Zadissa A, Li W, Niimura Y, Huang Z, Li C, White S, Xiong Z, Fang D, Wang B, Ming Y, Chen Y, Zheng Y, Kuraku S, Pignatelli M, Herrero J, Beal K, Nozawa M, Li Q, Wang J, Zhang H, Yu L, Shigenobu S, Wang J, Liu J, Flicek P, Searle S, Wang J, Kuratani S, Yin Y, Aken B, Zhang G, Irie N. The draft genomes of soft-shell turtle and green sea turtle yield insights into the development and evolution of the turtle-specific body plan. Nat Genetics. 2013;45:701–706. doi: 10.1038/ng.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werneburg I, Sánchez-Villagra MR. Timing of organogenesis support basal position of turtles in the amniote tree of life. BMC Evol Biol. 2009;9:82. doi: 10.1186/1471-2148-9-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler BM, Heimberg AM, Moy VN, Sperling EA, Holstein TW, Heber S, Peterson KJ. The deep evolution of metazoan microRNAs. Evol Dev. 2009;11:50–68. doi: 10.1111/j.1525-142X.2008.00302.x. [DOI] [PubMed] [Google Scholar]
- Zardoya R, Meyer A. Complete mitochondrial genome suggests diapsid affinities of turtles. Proc Nat Acad Sci USA. 1998;95:14226–14231. doi: 10.1073/pnas.95.24.14226. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.