Abstract
Here, we tested seven 2-acylated-1,4-hydronaphthoquinones for their cytotoxic effects on a panel of cancer lymphoma/leukemia cells and compared to a non-cancer origin cell line. Several naphthohydroquinones exhibited selective cytotoxic effects on lymphoma/leukemia cells with lowest activity on non-cancer cells. The mode of cell death induced by an acylated naphthohydroquinone, which has a long alkyl chain, was found to be via apoptosis. Furthermore, the naphthohydroquinone provoked mitochondria depolarization and activation of its downstream effector, caspase-3, thus implicating the intrinsic apoptotic pathway as its mechanism to exert cell death.
Keywords: Apoptosis, Cytotoxicity, Acylated hydroquinones, Anti-lymphoma, Photochemistry
1. Introduction
Quinones are widely distributed in bacteria, plants, and even the human body.1,2 Naphthoquinones are important members of the quinone family. They can generate reactive oxygen species such as superoxides and hydroxyl radicals3–6 and strongly induce cell death.7 Two of the most well studied naphthoquinones, plumbagin and menadione (VitK3), exhibit desirable cytotoxic activities against a variety of cancer cell lines and appear to be relatively safe in vivo.8–10 Although it is believed that the cytotoxic effects of naphthoquinones are due to the induction of apoptosis,8,10 some of these compounds can also promote necrosis through free-radical formation.11,12 In previous work, we tested a variety of 1,4-naphthoquinones with a similar chemical structure to plumbagin and menadione. They were found to induce cell death primarily via apoptosis while others were primarily necrotic in nature.13,14 The reduced form 1,4-naphthohydroquinones are p-hydroquinones, and they are usually thermodynamically less stable than naphthoquinones. However, p-hydroquinones can be formed by electron transfers15,16 not only enzymatically, but also disproponation reaction of semiquinones, which is actually the species to generate superoxide by reaction of O2.15 Hydroquinones are a family of hydroaromatic compounds, and ketone moiety with hydroxyaromatic structure have been found to have mutagenic properties.17 In addition, it has been reported that p-hydroquinone with carbonyl compounds at ortho position reduce the viability of tumor cells.18–20 It was proposed that the mechanism for the toxic effect was an inhibition of oxygen consumption in intact tumor cells.18,19 Most recently, a tricyclic compounds, 9,10-dihydroxy-4,4-dimethyl-1,4,5,8-tetrahydroanthracen-1-one, was found to block the electron flow through the NADH dehydrogenase leading to inhibition of mitochondrial bioenergetics and induction of G2/M-phase arrest.21 The results of cytotoxicity study indicated that the activity of cell growth inhibition increases with the increase in free radical stability.18 They also mentioned that cell penetration resulting from the lipophilicity of the compounds contributed to the inhibition in cell growth.19 We have synthesized simple 2-acylated-1,4-naphthohydroquinones, which possess different acyl groups, and herein we report the cytotoxic effects of the compounds on selective lymphoma/leukemia cell lines.
2. Results and discussion
2.1. Chemistry
The photochemistry of quinones has been investigated extensively and has a long history. A number of reviews have been dedicated to the photochemistry and the photochemical oxidation–reduction processes of quinones.1,22–24 Under the irradiation of light, quinones undergo a wide variety of chemical reactions. Mostly the primary step in these photochemical reactions of quinones is an electron transfer, and electron donor–acceptor interactions and hydrogen bond formation play an important role in these processes.
The solar-driven photo-Friedel–Crafts acylations has been studied in the last decade.25 Synthesis of 2-acylated naphthohydroquinone was carried out by photochemical reaction of 1,4-naphthoquinone with appropriate aldehydes in benzene under sunlight (Eq. 1). We used sunlight as an energy source for the synthesis of the compounds. After several days under sunlight and under constant stirring, the novel compounds were obtained in good yields. As solvents, benzene, t-BuOH and acetone/t-BuOH were tested, and benzene provided the best results; these results are depicted in Table 1. All compounds were purified by column chromatography and identified by NMR spectroscopy. These NMR spectra indicated that one of the hydroxyl groups has the intramolecular hydrogen bonding to carbonyl oxygen in the acyl group.
 |
(1) |
Table 1.
2-Acyl-1,4-naphthohydroquinone synthesis from 1,4-naphthoquinone and aldehydes
| Entry | R | Time (days) | Yield (%) |
|---|---|---|---|
| 1 | p-Me–C6H4 | 4 | 68 |
| 2 | p-MeO–C6H4 | 5 | 55 |
| 3 | C11H23 | 6 | 76 |
| 4 | Et | 3 | 76 |
| 5 | Me | 4 | 70 |
| 6 | Ph | 6 | 84 |
| 7 | CH=CH—CH3 | 6 | 72 |
2.2. Biological activity
2.2.1. MTS assay
To better assess the biochemical cytotoxicity of acylated naphthohydroquinones, a commercial enzymatic assay was used to quantify viability (see Section 2 for details). The inhibitory concentration 50% (IC50) in µM is defined as the concentration of compound required to inhibit 50% of the conversion of 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium) (MTS) to formazan of cell population, as compared with the absorbance produced by untreated cells after 72 h of incubation (see Table 2). Cellular metabolic activity was measured via NAD(P)H-dependent cellular oxidoreductase enzyme. Four different leukemia/lymphoma human cell lines were utilized to assess the activity of the compounds (CEM, Jurkat, Nalm-6, and Ramos) and compared to non-cancerous fibroblast human cell line (Hs27).
Table 2.
IC50 values (µM) of naphthohydroquinones against human cells
| Cell line | Chemical compounds | ||||||
|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | |
| CEM | 0.74 | 0.01 | 5.99 | 3.95 | 5.24 | 5.44 | 3.19 |
| Jurkat | 3.35 | 2.78 | 6.56 | 4.54 | 5.60 | 6.35 | 6.24 |
| Nalm-6 | 2.41 | 0.88 | 5.80 | 2.02 | 5.09 | 2.58 | 4.63 |
| Ramos | 0.52 | 0.66 | 0.98 | 2.49 | 0.75 | 4.13 | 4.25 |
| Hs27 | 4.37 | 4.97 | 43.75 | 18.03 | 9.87 | 8.30 | 37.14 |
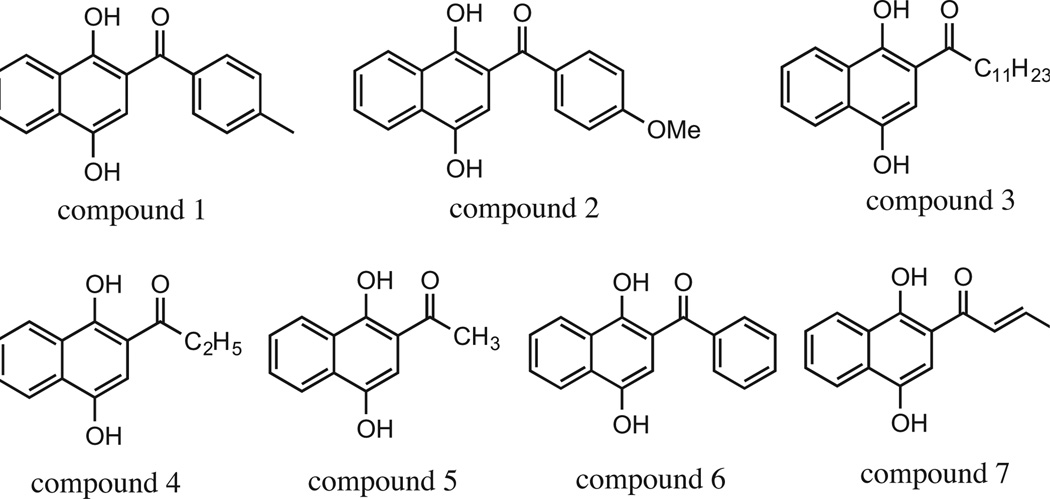
Treatment of cells with seven acylated naphthohydroquinones (structures are shown in Fig. 1) was administrated in a series of doubling increments from 1.56, 3.125, 6.25, 12.5, 25, 50, to 100 µM. As shown in Table 2, the IC50 was obtained and recorded for all cell lines. At this point, three compounds, 3, 4 and 7 were selected to perform further analysis since they exhibited little cytotoxicity to the non-cancerous cell line (IC50 >10 µM; Hs27). Araya-Maturana et al. reported that an increase of free radical stability lead to an increase in the activity of cell growth inhibition in a cytotoxicity study of phenols having carbonyl group at ortho position.18 They also mentioned that lipophilicity is also a factor that affects the inhibitory activity of the compounds on cell growth.19 Recently, they reported the anti-tumor mechanism of an ortho-carbonyl substituted hydroquinone, 9,10-dihydroxyl-4,4-dimethyl-1,4,5,8-tetrahydroanthracen-1-one, which inhibit mitochondrial bioenergetics.21 We tested compounds 3, 4 and 7, however, compound 4 generated insoluble crystals when added to the cell culture medium (aqueous environment) and was not analyzed further. The human CEM cell line was found to be more sensitive to the experimental compounds among the other cell lines and was selected to perform more rigorous cell death and apoptosis experiments.
Figure 1.
Seven naphthohydroquinones assayed for activity.
2.2.2. 2-Acyl-1,4-naphthohydroquinone induces phosphatidylserine translocation on T-lymphocyte cell line
Phosphatidylserine (PS) is predominantly located on the inner side of the plasma membrane leaflet facing the cytosol but is externalized to the outer membrane leaflet in cells undergoing apoptosis. Annexin V has high affinity for PS and fluorescent-labeled (annexin V-FITC) is routinely used for detection of PS translocation in live cells via flow cytometry. To identify if cytotoxicity is induced via apoptosis or necrosis, cells were treated with chemical compounds for 72 h followed by annexin-V FITC staining (see Section 4).
Compounds 3 (1,4-dihydroxy-2-dodecanoyl-naphthelene) and 7 (1,4-dihydroxy-2-crotonyl-napthalene) were tested at final concentration of 10 and 50 µM, respectively (Fig. 2A–C). Apoptotic cell distribution is annotated as the sum of percentages of both early and late phases of apoptosis (white bars), emitting green fluorescence signal, annexin V-FITC positive; whereas cells that were stained just with PI due to the damage of their plasma membrane, but without FITC signal, were considered necrotic cells (black bars; Fig. 2A). Analysis using the two-tailed Student’s paired t-test of experimental compounds against untreated and DMSO controls was consistently P <0.001, respectively. Each bar represents the average of three independent experimental values, and error bars the standard deviation of the mean. Untreated cells, as well as cells treated with 1% v/v DMSO and 1 mM H2O2, were also examined. Panels B–E are representative flow cytometric dot plots utilized to estimate the distribution of apoptosis/necrosis cells, where the FL1 or FL2 detectors settings were accommodated on the x- and y-axis, respectively (Fig. 2). Independent of their fluorescence, the color of dots in the plots depicts a density gradient transitioning from low blue to high red. Data analysis from quadrant regions in the dot plots are interpreted as follows: the bottom left quadrant are unstained viable cells, with a healthy membrane; the top left quadrant shows necrotic cells, PI positive; the top right quadrant are cells in late apoptosis, annexin V-FITC and propidium iodide double-positive; and the right bottom quadrant indicate cells in early apoptosis, positive to annexin V-FITC, but negative to propidium iodide. Using this assay, compound 3 was found to preferentially induce cell death via apoptosis (annexin V-positive), rather than necrosis (Fig. 2B). As can be seen in Figure 2C, compound 7 (1,4-dihydroxy-2-crotonyl-napthalene) elicited a substantial amount of necrosis instead of apoptosis when compared with solvent and untreated controls. Compound 3 induced significantly more apoptosis (95.35%) than compound 7 (1.7%; Fig. 2A–C). Solvent and untreated control cells did not display any substantial enhancement of apoptosis or necrosis (Fig. 2D and E). This data confirms that compound 3 caused PS externalization on the CEM cell line, suggesting that this compound induces cell death via apoptosis.
Figure 2.
Compound 3, but no compound 7, induce PS translocation on human T-leukemic CEM cells.
2.2.3. 2-Acyl-1,4-naphthohydroquinone-induces cytotoxicity via a mitochondrial-dependent pathway
In order to determine if the intrinsic/mitochondrial pathway is involved in the initiation of cell death process, CEM cells were treated for 6 and 8 h with compound 3 and changes in the mitochondrial Δψm were determined by staining with the aggregate-forming lipophilic cationic fluorophore JC-1 and monitored via flow cytometry. After dissipation of mitochondrial Δψm, the JC-1 reagent emitted a green fluorescence signal, which is a very early facet in apoptosis,26 whereas cells with polarized mitochondrial membrane emitted a red signal. In panel A, cells emitting green fluorescence signal (y-axis) versus treatment type (x-axis) are depicted (Fig. 3). Panels B–E, representative flow cytometric dot plots used to determine the results depicted in A. Analysis using the two-tailed Student’s paired t-test of experimental compounds against untreated and DMSO controls was consistently P <0.001, respectively. Each bar represents the average of triplicates and error bars the standard deviation of mean. Cells exposed to the mitochondrial stressor CCCP (50 µM) were used as positive controls. DMSO solvent and untreated controls were also analyzed in parallel. Approximately 10,000 events were captured and analyzed per sample using CXP software (Beckman Coulter). In Figure 3A compound 3 showed a pattern resembling cells treated with the proton ionophore CCCP, which strongly induced mitochondrial depolarization. As can be seen in Figure 3, compound 3 induced a significant increase in mitochondrial depolarization from 6 to 8 h (P = 0.014). These findings suggested that compound 3 induced cytotoxicity via mitochondrial depolarization on T-lymphocyte CEM cells in a time-dependent manner.
Figure 3.
Compound 3-mediated cytotoxicity appeared to be initiated via mitochondrial Δψm disruption on T-lymphocyte CEM cells. CEM cells were treated for 6 and 8 h with compound 3 and changes in the mitochondrial Δψm were determined by staining with the aggregate-forming lipophilic cationic fluorophore JC-1 and monitored via flow cytometry. In panel A, cells emitting green fluorescence signal (y-axis) versus treatment type (x-axis) are depicted. Panels B–E, representative flow cytometric dot plots used to determine the results depicted in A.
2.2.4. Compound 3 inflicts cytotoxicity via caspase-3 activation
To examine whether caspase-3 activation is involved in the cytotoxicity provoked by compound 3, a cell permeable fluorogenic substrate was used to detect cells with active caspase-3. This caspase-3 substrate allows its detection in live-cell mode by flow cytometry. Cells labeled with Nucview 488 caspase-3 substrate were examined via flow cytometry as compared with untreated and solvent controls (Fig. 4). In panel A, the total numbers of cells with active caspase-3 are graphed along the y-axis, whereas different treatments are plotted along the x-axis. Each bar represents average of triplicate values, and error bars the standard deviation. Analysis using the two-tailed Student’s paired t-test of experimental compounds against untreated and DMSO controls was consistently P <0.01, respectively. Panels B–E are representative flow cytometric dot plots used to determine the distribution of cells with active caspase-3, where the FL1 and FL2 detectors were set on the x- and y-axis, respectively. Cells exposed to 50 µM Carbonyl cyanide m-chlorophenyl hydrazone (CCCP) mitochondrial stressor were used as positive controls. Also, untreated and solvent DMSO treated cells were analyzed concurrently. These results suggest that compound 3 has a tendency to induce cytotoxicity via caspase-3 activation on the CEM cell line. In agreement with the previous experiments, these findings confirm that 2-acyl-1,4-naphthohydroquinone induced activation of caspase-3 indicating that apoptosis was preferentially involved in the pathway to induce its cell death. These results suggest that compound 3 was provoking mitochondrial depolarization, as an initial signal to inflict its anti-cancer cell death activity, and furthermore, followed by the downstream activation of caspase-3, a crucial effector in the execution of the intrinsic apoptosis pathway.
Figure 4.
Compound 3 exerts its toxic activity via caspase-3 activation. To examine whether caspase-3 activation is involved in the cytotoxicity provoked by compound 3, a cell permeable fluorogenic substrate was used for detection. This caspase-3 substrate allows the detection of caspase-3 activity in living cells via flow cytometry. In panel A, the total numbers of cells with active caspase-3 are graphed along the y-axis, whereas different treatments are plotted along the x-axis. Panels B–E are representative flow cytometric dot plots used to determine the distribution of cells with active caspase-3, where the FL1 and FL2 detectors were set on the x- and y-axis, respectively.
3. Conclusions
In summary, the MTS assay was used to screen the cytotoxicity of seven of the 2-acyl-1,4-naphthohydroquinone compounds synthesized from1,4-naphthoquinone and aldehydes. Additionally, MTS method was used to determine the IC50 values. Compounds 3 and 7 were identified to have cytotoxic selectivity against human lymphoma/leukemia at low micromolar concentrations, as compared with the non-cancer Hs-27 cells. To increase the reliability of our assumptions, we used several methods to decipher the potential pathway utilized by the chemical compounds to perturb the cellular homeostasis. In CEM cells, compound 3 induced cell death by apoptotic mechanism, provoking PS externalization, as indicated by the annexin V-FITC assay. In contrast, compound 7 utilized the necrotic pathway. These results were corroborated with the use of the JC-1 reagent, demonstrating that compound 3 caused mitochondrial depolarization. Moreover, the compound 3-mediated cytotoxicity was subsequently confirmed to be via the activation of caspase-3 pathway, confirming that it induces apoptosis. Hence, photochemically synthesized acylated naphthohydroquinones may represent a promising new alternative in the treatment of hematological malignancies and warrants additional evaluation in an in vivo animal model.
4. Experimental
4.1. General procedure for synthesis of 2-acyl naphthohydroquinones
In a 35 ml Pyrex sealable reaction tube, a solution of 1 mmol of 1,4-naphthoquinone and 10 mmol of an aldehyde with 20 ml benzene was prepared and degassed with nitrogen. The reaction tube was then sealed and placed on the roof for exposure to direct sunlight. A magnetic stir plate was used to allow constant stirring of the solution. The reaction mixture was then checked by TLC. Column chromatography using ethyl acetate/hexanes or methanol/dichloromethane mixture as the eluent afforded the desired products. The structure of the obtained products, 2-acyl-1,4-naphthohydroquinones, was confirmed by 1H and 13C NMR, and IR spectroscopy. Melting points are uncorrected.
4.1.1. 2-(4-Methylbenzoyl)-1,4-naphthohydroquinone (1)
Mp = 167–169 °C. 1H NMR (600 MHz, CDCl3): δ 13.56 (s, 1H), 8.50 (d, J = 8.3 Hz, 1H), 8.11 (d, J = 8.2 Hz, 1H), 7.70 (t, J = 7.4, 1H), 7.61–7.59 (m, 3H), 7.30 (d, J = 7.4, 2H), 6.90 (s, 1H), 4.9 (s, 1H), 2.45 (s, 3H). 13C NMR (150 MHz, CDCl3): δ 200.58, 158.54, 142.45, 135.43, 129.89, 129.03, 126.50, 126.01, 124.61, 121.64, 111.57, 108.23, 21.53. IR (neat, cm−1) νmax 3295.48 (OH), 3072, 2921 (CH), 1632, 1607, 1583, 1553, 1507, 1470, 1425, 1394, 1360, 1288, 1200, 1185, 1161, 1116, 1077, 1030, 999, 977, 954, 856, 832, 781, 763, 721.
4.1.2. 2-(4-Methoxybenzoyl)-1,4-naphthohydroquinone (2)
Mp = 130–132 °C. 1H NMR (600 MHz, CDCl3): δ 13.51 (s, 1H), 8.5 (d, J = 8.3 Hz, 1H), 8.12 (d, J = 8.2 Hz, 1H), 7.73–7.68 (m, 3H), 7.59 (t, J = 7.4 Hz, 1H), 6.99 (d, J = 9.1 Hz, 2H), 6.94 (s, 1H), 4.89 (s, 1H), 3.88 (s, 3H). 13C NMR (150 MHz, CDCl3): δ 199.45, 162.58, 158.24, 142.50, 132.20, 131.45, 130.71, 129.78, 129.20, 126.47, 126.15, 124.57, 121.64, 113.67, 111.63, 108.33, 55.47. IR (neat, cm−1) νmax 3465.88 (OH), 3054, 2925 (CH), 1631, 1590, 1566, 1511, 1462, 1451, 1418, 1359, 1326, 970, 949, 867, 779, 763, 720.
4.1.3. 2-Dodecanoyl-1,4-naphthohydroquinone (3)
Mp = 95–97 °C. 1H NMR (600 MHz, CDCl3): δ 13.78 (s, 1H), 8.45 (d, J = 8.3 Hz, 1H), 8.09 (d, J = 8.3 Hz, 1H), 7.67 (t, J = 8.2 Hz, 1H), 7.58 (t, J = 8.2 Hz, 1H), 7.01 (s, 1H), 4.95 (s, 1H), 2.96 (t, J = 7.4 Hz, 2H), 1,77 (m, 2H), 1.40–1.25 (m, 16H), 0.87 (t, J = 6.7 Hz, 3H). 13C NMR (150 MHz, CDCl3): δ 206.19, 157.42, 142.90, 129.66, 129.39, 126.19, 124.62, 121.52, 111.70, 105.51, 38.76, 31.89, 29.61, 24.64, 22.66, 14.07. IR (neat, cm−1) νmax 3336 (OH), 3069, 2919, 2851, 1634, 1591, 1515, 1468, 1431, 1401, 1378, 1239, 1213, 1191, 1154, 1137, 1076, 1054, 1029, 960, 879, 819, 787.
4.1.4. 2-Propanoyl-1,4-naphthohydroquinone (4)
Mp = 178–179 °C. 1H NMR (600 MHz, CDCl3): δ 13.70 (s, 1H), 8.46 (d, J = 8.3, 1H), 8.15 (d, J = 8.3 Hz, 1H), 7.67 (td, J = 8.2, 1.6 Hz, 1H), 7.57 (td, J = 8.2, 1.6 Hz, 1H), 7.02 (s, 1H), 4.90 (s, 1H), 3.03 (q, J = 7.4 Hz, 2H), 1.27 (t, J = 7.4 Hz, 3H). 13C NMR (150 MHz, CDCl3): δ 206.5, 155.2, 144.8, 129.9, 128.9, 126.0, 125.7, 123.9, 122.3, 112.0, 104.3, 31.8, 8.3. IR (neat, cm−1) νmax 3349 (OH), 2983, 2915 (CH), 1634, 1594, 1513, 1475, 1459, 1414, 1398, 1383, 1306, 1210, 1138, 1073, 1040, 1024, 990, 969, 852, 805, 773, 723, 681.
4.1.5. 2-Acetyl-1,4-naphthohydroquinone (5)
Mp = 211–213 °C. 1H NMR (600 MHz; DMSO-d6) δ: 13.63 (s,1H), 8.45 (d,J = 8.3,1H), 8.1 (d, J = 8.2, 1H), 7.68 (t, J = 8.3, H), 7.58 (t, J = 8.3, 1H), 6.98 (s,1H), 4.94 (s, 1H), 2.64 (s, 3H). 13C NMR (150 MHz; DMSO-d6) δ: 205.2, 154.7, 145.1, 130.0, 129.8, 126.9, 125.5, 124.1, 122.7, 113.0, 105.5, 27.7. IR (neat, cm−1) νmax 3245 (OH), 2921, 1636, 1597, 1525, 1471, 1439, 1403, 1365, 1318, 1232, 1204, 1152, 1076, 1019, 982, 878, 829, 725, 657.
4.1.6. 2-Benzoyl-1,4-naphthohydroquinone (6)
Mp = 114–116 °C. 1H NMR (600 MHz; CDCl3) δ: 13.56 (s, 1H), 8.51 (d, J = 8.9, 1H), 8.12 (d, J = 8.2, 1H), 7.72–7.65 (m, 3H), 7.60 (t, J = 8.0, 1H), 7.55 (t, J = 6.8, 1H), 7.48 (m, 2H), 6.85 (s, 1H). 13C NMR (150 MHz; CDCl3) δ: 200.5, 154.9, 144.6, 137.9, 131.7, 129.5, 129.3, 128.7, 128.4, 126.5, 125.3, 123.7, 122.2, 112.5, 106.6. IR (neat, cm−1) νmax 3293 (OH), 3075, 2924, 1630, 1585, 1556, 1508, 1495, 1470, 1445, 1394, 1298, 1253, 1073, 1026, 996, 959, 813, 768, 701.
4.1.7. 2-Crotonyl-1,4-naphthohydroquinone (7)
Mp = 164–166 °C. 1H NMR (600 MHz; CDCl3) δ: 14.29 (s, 1H), 8.46 (d, J = 8.2 Hz, 1H), 8.09 (d, J = 8.2 Hz, 1H), 7.68 (td, J = 8.3, 6.9, 1.3 Hz, 1H), 7.57 (td, J = 8.2, 6.9, 1.2 Hz, 1H), 7.23 (m, 1H), 7.08 (s, 1H), 7.03 (dd, J = 15.0, 1.7 Hz, 1H), 4.92 (s, 1H), 2.05 (dd, J = 6.9, 1.6 Hz, 3H). 13C NMR (150 MHz; CDCl3) δ: 193.07, 159.12, 145.36, 142.80, 129.76, 129.45, 126.44, 126.22, 125.87, 124.62, 121.51, 111.76, 105.33, 29.70, 18.73. IR (neat, cm−1) νmax 3303 (OH), 3073, 2918, 2850, 1630, 1577, 1515, 1474, 1435, 1309, 1298, 1221, 1141, 1076, 1063, 959, 893, 809.
4.2. Biological activity
4.2.1. Dilutions of experimental chemical compounds
Synthetic chemical compounds stock solutions and their dilutions were prepared in dimethyl sulfoxide (DMSO; Sigma–Aldrich, St. Louis, MO) and as necessary, aliquots were added directly to 96 and 24-well plates containing cells in complete media.
4.2.2. Cell line and culture conditions
A panel of four human leukemia/lymphoma cell lines was utilized: T lymphoblast acute lymphoblastic leukemia CCRF-CEM (CEM)27 mature-T acute lymphoblastic leukemia Jurkat,28 pre-B acute lymphoblastic leukemia Nalm-6,29 and B lymphoblast Burkitt’s lymphoma Ramos.30 For comparison purposes, a non-cancer origin cell line, human dermal neonatal foreskin Hs27 fibroblasts (Hs27; ATCC, Manassas, VA), was used as a control. The culture media for leukemia/lymphoma (CEM, Jurkat, Nalm-6 and Ramos) and fibroblast (Hs27) cells was RPMI and DMEM (HyClone, Logan UT), respectively. Both of these growth media were contained 10% heat-inactivated fetal bovine serum (HyClone, Logan, UT), 100 U/ml penicillin and 100 µg/ml streptomycin (HyClone). Exponentially growing cells with 60–75% confluency were counted and seeded into 96-well plate at a density of 10,000 cells in 100 µl culture media. For the apoptosis assays, 24-well plates were used at the density of 50,000 cells in 500 µl culture media per well. The incubation conditions were consistently maintained at 37 °C in a humidified 5% CO2 atmosphere at all times. To guarantee high viability, cells were prepared as previously described.31 Each experimental point and controls were assessed in eight replicates for 96-well plates, and four replicates for 24-well plates.
4.2.3. MTS assay
Human cells were cultured and prepared in 96-well plates as described above and then exposed to the experimental compounds at a gradient of concentration from 1.56 to 100 µM. After 72 h of incubation, 20 µl of the MTS reagent (CellTiter 96 AQueousOne Solution Cell Proliferation Assay; Promega, Madison, WI) were added to each well and subsequently incubated for an additional 4 h,32 following the manufacturer’s instructions. The colored formazan product was measured by absorbance at 490 nm with a reference wavelength of 650 nm using a microplate reader (SPECTRA MAX 190, Molecular Devices, Sunnyvale, CA).33 Control wells containing the same volumes of culture medium and-MTS reagent without cells were utilized to subtract background absorbance. Additionally, cells treated with 1 mM of hydrogen peroxide (H2O2; Sigma–Aldrich, St. Louis, MO) were used as positive controls for cytotoxicity. Solvent controls (1% DMSO) and untreated cells were also included concurrently in each experimental plate. Data were expressed as the percentage of cell viability relative to solvent treated cells. In these assays, each sample was performed in replicates of eight and the mean and standard deviation values are depicted.
4.2.4. Annexin V/PI apoptosis/necrosis assay
After seeding, CEM cells were incubated for an hour, and then treated with the experimental chemical compounds at final concentration of 10 µM, followed by additional incubation of 72 h. Afterwards, cells from each individual well were collected in a pre-chilled ice-water tubes, washed and processed essentially as detailed previously.34 Briefly, cells were stained with a solution containing a mixture of annexin V-FITC and PI in 100 µl of binding buffer (Beckman Coulter, Miami, FL). After 15 min of incubation on ice in the dark, 400 µl of ice-cold binding buffer was added to the cell suspensions and immediately analyzed via flow cytometry (Cytomics FC 500; Beckman Coulter, Miami, FL). The total percentage of apoptotic cells was defined as the sum of both early and late stages of apoptosis (annexin V-FITC positive), bottom and top right quadrants in flow cytometric dot plots, respectively. For each sample, approximately 10,000 individual events were acquired per sample and analyzed with CXP software (Beckman Coulter, Miami, FL).
4.2.5. Mitochondrial membrane potential (Δψm) polychromatic analysis
CEM cells were treated at a final concentration of 1 µM of the experimental compound, incubated for 8 h and stained with tetraethylbenzimidazolylcarbocyanine iodide (JC-1) fluorophore, an aggregate-forming lipophilic cationic reagent, following the manufacturer’s protocol (MitoProbe; Life Technologies, Grand Island, NY). Cells with polarized mitochondria allow JC-1 aggregation, which emits a red signal; whereas cells with depolarized mitochondria only permit JC-1 monomer formation, emitting a green signal. As a positive control of mitochondrial Δψm disruptor, a proton ionophore, carbonyl cyanide 3-chlorophenylhydrazone (CCCP; 50 µM) was utilized. In addition, solvent controls (1% v/v DMSO) and untreated controls were analyzed in parallel. Approximately, 10,000 events were acquired and analyzed using CXP software (Beckman Coulter).
4.2.6. Live-cell and real-time detection of intracellular caspase-3 activation
CEM cells were seeded on a 24-well plates as described above and treated with 1 µM of experimental compounds for 8 h.35 Afterwards, cysteine-aspartic proteases (caspase)-3 activation was identified by the usage of a fluorogenic NucView 488 Caspase-3/7 substrate for live cells (Biotium, Hayward, CA), following the vendor’s instructions. Permeability of the substrate to cells with intact plasma membrane allows for the detection of caspase-3 activation in live cells. Cells displaying a green fluorescence signal, indicative of caspase-3 activation, were monitored via flow cytometry (Cytomics FC500). The same positive and negative controls were used as described in 4.2.5. Data acquisition was also described in the previous section.
4.2.7. Statistical analysis
Each experimental point, as well as controls, was performed using at least four replicates. Experimental variability was specified using data presented as average with its corresponding standard deviation. The statistical significance of the differences between experimental samples and their corresponding controls were performed via two-tailed paired Student’s t-tests. To designate whether comparisons of two-group independent samples have statistical significance, P <0.05 was considered significant.
Supplementary Material
Acknowledgments
We thank the Cytometry, Screening and Imaging Core Facilitiy supported by a Research Centers for Minority Institutions Grant 8G12MD007592 from the National Institutes on Minority Health and Health Disparities (NIMHD) a component of NIH. We also acknowledge support from the NIGMS SCORE Grant 1SC3GM10 3713 to RJA. D.P. and F.D. were supported by the RISE program (NIGMS/NIH 2R25GM069621-10), and F.D. was also supported by the University of Texas at El Paso (University Research Institute Award 2012).
Footnotes
Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.bmc.2013.12.007.
References and notes
- 1.Maruyama K, Osuka A. In: The Chemistry of the Quinonoid Compounds. Patai S, Rappoport Z, editors. Vol. 2. New York: John Wiley & Sons; 1988. p. 759. [Google Scholar]
- 2.Thomson RH. Naturally Occurring Quinones IV. Springer; 1997. [Google Scholar]
- 3.Iyanagi T, Yamazaki I. Biochim. Biophy. Acta (BBA)—Bioenerg. 1970;216:282. doi: 10.1016/0005-2728(70)90220-3. [DOI] [PubMed] [Google Scholar]
- 4.Kitamura S, Tatsumi K. Drug Metab. Dispos. 1999;27:98. [PubMed] [Google Scholar]
- 5.Lind C, Hochstein P, Ernster L. Arch. Biochem. Biophys. 1982;216:178. doi: 10.1016/0003-9861(82)90202-8. [DOI] [PubMed] [Google Scholar]
- 6.Ohnishi T, Yamazaki H, Iyanagi T, Nakamura T, Yamazaki I. Biochim. Biophys. Acta (BBA)—Bioenerg. 1969;172:357. doi: 10.1016/0005-2728(69)90132-7. [DOI] [PubMed] [Google Scholar]
- 7.Siegel D, Reigan P, Ross D. In: Advances in Bioactivation Research. Elfarra A, editor. IX. New York: Springer; 2008. p. 1. [Google Scholar]
- 8.Krishnaswamy M, Purushothaman KK. Indian J. Exp. Biol. 1980;18:876. [PubMed] [Google Scholar]
- 9.Munday R, Smith BL, Munday CM. Chem. Biol. Interact. 1998;108:155. doi: 10.1016/s0009-2797(97)00105-1. [DOI] [PubMed] [Google Scholar]
- 10.Lamson DW, Plaza SM. Altern. Med. Rev. 2003;8:303. [PubMed] [Google Scholar]
- 11.Kaminski M, Karbowski M, Miyazaki Y, Kedzior J, Spodnik JH, Gil A, Wozniak M, Wakabayashi T. Folia Morphol. (Warsz) 2002;61:217. [PubMed] [Google Scholar]
- 12.Inbaraj JJ, Chignell CF. Chem. Res. Toxicol. 2004;17:55. doi: 10.1021/tx034132s. [DOI] [PubMed] [Google Scholar]
- 13.Montoya J, Varela-Ramirez A, Estrada A, Martinez LE, Garza K, Aguilera RJ. Biochem. Biophys. Res. Commun. 2004;325:1517. doi: 10.1016/j.bbrc.2004.10.196. [DOI] [PubMed] [Google Scholar]
- 14.Montoya J, Varela-Ramirez A, Shanmugasundram M, Martinez LE, Primm TP, Aguilera RJ. Biochem. Biophys. Res. Commun. 2005;335:367. doi: 10.1016/j.bbrc.2005.07.086. [DOI] [PubMed] [Google Scholar]
- 15.Brunmark A, Cadenas E. Free Radical Biol. Med. 1989;7:435. doi: 10.1016/0891-5849(89)90126-3. [DOI] [PubMed] [Google Scholar]
- 16.O’Brien PJ. Chem.-Biol. Interact. 1991;80:1. doi: 10.1016/0009-2797(91)90029-7. [DOI] [PubMed] [Google Scholar]
- 17.Elliger CA, Henika PR, MacGregor JT. Mutat. Res./Genet. Toxicol. 1984;135:77. doi: 10.1016/0165-1218(84)90159-9. [DOI] [PubMed] [Google Scholar]
- 18.Araya-Maturana R, Delgado-Castro T, Gárate M, Ferreira J, Pavani M, Pessoa-Mahana H, Cassels BK. Bioorg. Med. Chem. 2002;10:3057. doi: 10.1016/s0968-0896(02)00154-2. [DOI] [PubMed] [Google Scholar]
- 19.Araya-Maturana R, Cardona W, Cassels BK, Delgado-Castro T, Ferreira J, Miranda D, Pavani M, Pessoa-Mahana H, Soto-Delgado J, Weiss-López B. Bioorg. Med. Chem. 2006;14:4664. doi: 10.1016/j.bmc.2006.02.011. [DOI] [PubMed] [Google Scholar]
- 20.Rodríguez J, Olea-Azar C, Cavieres C, Norambuena E, Delgado-Castro T, Soto-Delgado J, Araya-Maturana R. Bioorg. Med. Chem. 2007;15:7058. doi: 10.1016/j.bmc.2007.07.013. [DOI] [PubMed] [Google Scholar]
- 21.Urra FA, Martínez-Cifuentes M, Pavani M, Lapier M, Jaña-Prado F, Parra E, Maya JD, Pessoa-Mahana H, Ferreira J, Araya-Maturana R. Toxicol. Appl. Pharmacol. 2013;267:218. doi: 10.1016/j.taap.2012.12.023. [DOI] [PubMed] [Google Scholar]
- 22.Levin PP, Kuz’min VA. Usp. Khim. 1987;56:527. [Google Scholar]
- 23.Mukherjee T. Proc. Indian Natl. Sci. Acad.—Part A. 2000;66:239. [Google Scholar]
- 24.Abraham I, Joshi R, Pardasani P, Pardasani RT. J. Braz. Chem. Soc. 2011;22:385. [Google Scholar]
- 25.Oelgemöller M, Mattay J. In: Handbook of Organic Photochemistry and Photobiology. 2nd ed. Horspool W, Lenci F, editors. Boca Raton: CRC Press; 2003. p. 88. [Google Scholar]
- 26.Li H, Zhu H, Xu CJ, Yuan J. Cell. 1998;94:491. doi: 10.1016/s0092-8674(00)81590-1. [DOI] [PubMed] [Google Scholar]
- 27.Foley GE, Lazarus H, Farber S, Uzman BG, Boone BA, McCarthy RE. Cancer. 1965;18:522. doi: 10.1002/1097-0142(196504)18:4<522::aid-cncr2820180418>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 28.Wendler I, Jentsch KD, Schneider J, Hunsmann G. Med. Microbiol. Immunol. 1987;176:273. doi: 10.1007/BF00190533. [DOI] [PubMed] [Google Scholar]
- 29.Hurwitz R, Hozier J, Lebien T, Minowada J, Gajl-Peczalska K, Kubonishi I, Kersey J. Int. J. Cancer. 1979;23:174. doi: 10.1002/ijc.2910230206. [DOI] [PubMed] [Google Scholar]
- 30.Klein G, Giovanella B, Westman A, Stehlin JS, Mumford D. Intervirology. 1975;5:319. doi: 10.1159/000149930. [DOI] [PubMed] [Google Scholar]
- 31.Lema C, Varela-Ramirez A, Aguilera RJ. Curr. Cell. Biochem. 2011;1:1. [PMC free article] [PubMed] [Google Scholar]
- 32.Xu W, Osei-Prempeh G, Lema C, Davis Oldham E, Aguilera RJ, Parkin S, Rankin SE, Knutson BL, Lehmler HJ. Carbohydr. Res. 2012;349:12. doi: 10.1016/j.carres.2011.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oldham ED, Seelam S, Lema C, Aguilera RJ, Fiegel J, Rankin SE, Knutson BL, Lehmler HJ. Carbohydr. Res. 2013;379C:68. doi: 10.1016/j.carres.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Varela-Ramirez A, Costanzo M, Carrasco Y, Pannell K, Aguilera RJ. Cell Biol. Toxicol. 2011;27:159. doi: 10.1007/s10565-010-9178-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robles-Escajeda E, Lerma D, Nyakeriga AM, Ross JA, Kirken RA, Aguilera RJ, Varela-Ramirez A. PLos ONE. 2013;8:e73508. doi: 10.1371/journal.pone.0073508. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.