Abstract
The aromatic amine carcinogen 2-aminofluorene (AF) forms covalent adducts with DNA, predominantly with guanine at the C8 position. Such lesions are bypassed by Y-family polymerases such as Dpo4 via error-free and error-prone mechanisms. We show that Dpo4 catalyzes elongation from a correct 3′-terminal C opposite [AF]G in a nonrepetitive template sequence with low efficiency. This extension leads to cognate full-length product, as well as mis-elongated products containing base mutations and deletions. Crystal structures of the Dpo4 ternary complex with the 3′-terminal primer C base opposite [AF]G in the anti conformation and with the AF-moiety positioned in the major groove, revealed both accurate and misalignment-mediated mutagenic extension pathways. The mutagenic template/primer-dNTP arrangement is promoted by interactions between the polymerase and the bulky lesion, rather than by a base pairstabilized misaligment. Further extension leads to semi-targeted mutations via this proposed polymerase-guided mechanism.
Aromatic amines are potent carcinogens, present in cigarette smoke and coal-derived synthetic fuels1, and formed during the cooking of protein-rich foods such as meat and fish2. Dye industry workers have been susceptible to bladder cancer, now known to be preventable by eliminating exposure to specific aromatic amines3. 2-acetylaminofluorene, AAF, and its deacetylated derivative aminofluorene, AF, have been intensively studied as model chemical mutagens and carcinogens4. Metabolic processing of AAF by cytochrome P450 enzymes creates reactive intermediates that predominantly bind to the C8 position of guanine, forming covalent [AF]G and [AAF]G adducts (Fig. 1a)4.
Figure 1.
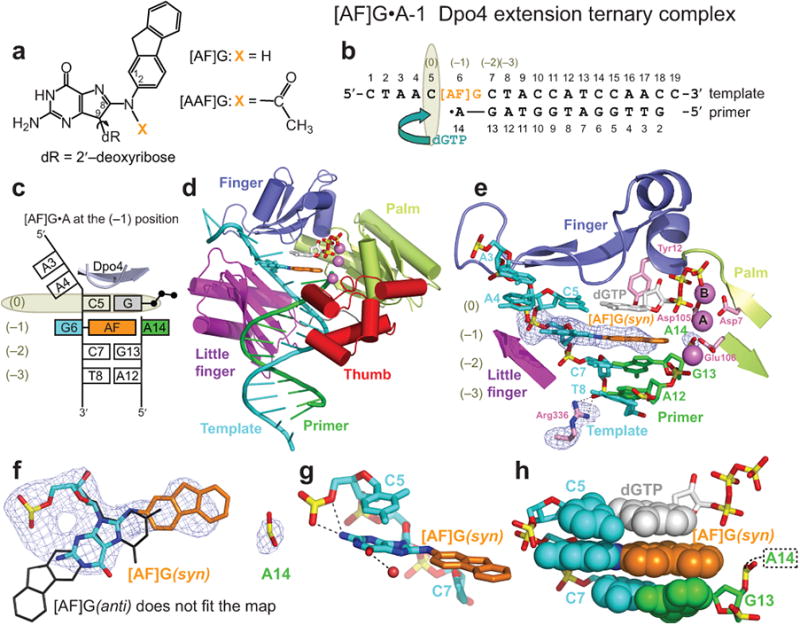
Structure of the [AF]G•A-1 Dpo4 extension ternary complex. (a) Chemical formula of 2-aminofluorene-C8-guanine, [AF]G, and 2-acetylaminofluorene-C8-guanine, [AAF]G, adducts. (b) Schematic of the expected pairing of the [AF]G-modified 19-mer template with the 13-mer primer, ending with a 2′,3′-dideoxy-A, and dGTP in the extension ternary complex with Dpo4. The insertion position at the Dpo4 active site is denoted by (0), and the post-insertion position is denoted by (–1). (c) Schematic of the observed base pairing arrangement within the Dpo4 active site. (d) Overall structure of the [AF]G•A-1 complex. (e) Structure of the active site of the [AF]G•A-1 complex. [AF]G(syn) at the (–1) position is opposite disordered 3′-terminal A14 base of the primer strand. The next template base C5 is paired with an incoming dGTP at the (0) position of the active site. The first Ca2+, cation A, is coordinated by invariant D7, D105, and E106 residues. The second Ca2+, cation B, is chelated by the phosphate groups of the incoming dGTP. Simulated annealing Fo-Fc omit map contoured at 3σ level and colored in blue (2.96 Å resolution) is shown for [AF]G, A14 and Arg336 residues. (f) [AF]G(syn) opposite A14 of the primer strand. [AF]G in the anti conformation (black lines) does not fit the map. Only the phosphate group of A14 has well-ordered density. (g) The N2 group of modified-G(syn) forms hydrogen bonds with the phosphate oxygens of C5. (h) Base stacking arrangement of the [AF]G(syn) and neighboring base pairs. The intercalated AF-moiety leaves no room for the disordered base of A14 within the template/primer helix.
The [AF]G adduct in mammalian cells and yeast is found to generate base substitutions targeted to the site of the damaged base, resulting in predominantly A and to a lesser extent T misincorporation events5-7. Furthermore, this adduct can also cause semi-targeted mutations (i.e., substitutions and frameshifts, that occur in the vicinity of the lesion site, but not at the site of the damage), with the correct C opposite the adduct. Experiments with site-specifically modified [AF]G oligonucleotides embedded in a plasmid vector replicating in simian kidney (COS-7) cells showed mutation frequencies ranging from 2-4% with C, A and T template bases positioned 5′ to the lesion site, to 70% with a G base placed 5′ to the lesion5.
High-fidelity polymerases including replicative eukaryotic Pol δ8 and E. coli Pol III9 preferentially insert correct C and misinsert some A opposite [AF]G in vitro, but are impeded by the adduct10-14. However, the bypass of DNA lesions in vivo predominantly involves specialized translesion synthesis (TLS) Y-family polymerases that replace stalled high-fidelity polymerases15-17. Y-family polymerases seem to be involved in error-free and error-prone bypass of bulky aminofluorene lesions in eukaryotic and prokaryotic systems, as illustrated below. In E. coli, the introduction of [AF]G into a plasmid reduces the efficiency of Pol III-catalyzed replication by ∼30%, suggesting that some polymerase stalling occurs18. Furthermore, induction of the SOS-response activates the Y-family polymerases and modulates [AF]G-induced mutation frequencies18-20. In simian kidney cells, the mutational spectra induced by [AF]G are consistent with the in vitro miscoding properties of eukaryotic Pol κ21. In vitro, the bypass of [AF]G lesions by human polymerase κ and E. coli Pol IV is relatively inefficient and error-prone21; in contrast, yeast Pol η predominantly inserts C opposite the [AF]G lesion and the bypass is more efficient than in the case of Pol κ22.
All known DNA polymerases have a common catalytic core formed by palm, finger and thumb domains15,23-25. High-fidelity polymerases produce tight-fitting, solvent-excluding, reaction-ready active sites, resulting from conformational changes in the finger domain upon binding of a complementary dNTP23-25. Furthermore, multiple residues are employed to proofread the minor groove of growing DNA, and terminal mismatches are displaced to an exonuclease domain. In contrast, Y-family polymerases have more spacious and solvent-accessible active sites, and have neither an exonuclease domain nor contact the minor groove edge of template/primer15. The archaeal Y-family polymerase Dpo4 and human Pol κ26 and Pol η27 rely largely on Watson-Crick base pairing for fidelity checks28. These features enable Y-family polymerases to bypass a variety of DNA lesions and concurrently cause a higher error rate and lower processivity on undamaged DNA templates15,17,29.
Crystal structures of a fragment from the high-fidelity A-family Bacillus Pol I 30 and phage T7 polymerase31 with an [AF]G adduct in the active sites showed that the adduct inhibits the finger domain closure necessary for the nucleotidyl transfer reaction to occur23-24, thus drastically reducing efficiencies of dCTP insertion opposite [AF]G 32,,10. The alignments for [AF]G opposite correct C33-34 and mismatched A35-36 in free DNA duplexes in solution have been studied by NMR, revealing three possible conformations for the AF group: with partner C, AF can be in the major groove with Watson-Crick pairing maintained, or intercalated into the helix with damaged G and its partner extruded; with partner A, AF is in the minor groove and the mismatched G•A pair positioned within the helix. Details are given in Supplementary Data.
Both replicative and Y-family polymerases can generate misalignment-mediated replication errors that lead to frameshift mutations, with or without DNA lesions21,37-43. Misalignment errors are produced via slippage of template/primer-dNTP intermediates that are stabilized by Watson-Crick base pairs surrounding looped out template bases. Recent structural work has shed light into such replication errors. Examples include the X-family polymerase λ44 and Y-family polymerase Dbh45 with misaligned unmodified template/primer complexes in repetitive sequences. Other examples are the structure of a Dpo4 complex with a looped out benzo[a]pyrene diol epoxide BPDE-N2-dG adduct46, and more recently, that of a B-family Pol II containing an abasic site47. On the other hand, the mechanism underlying semi-targeted mutagenesis remains elusive.
In this work, we aimed to explore the structural features of intermediates of Dpo4 ternary complexes about to undergo primer extension with terminal 3′-C or 3′-A primer bases opposite an [AF]G lesion. The correct dNTP was added to pair with the template base flanking the modified guanine on its 5′-side. In addition, three crystal structures of post-extension ternary complexes with [AF]G opposite C or A in primer strands, which had been elongated by one base, were also studied. These structures demonstrate an unusual mode of interaction between the bulky lesion and the little finger domain; the interactions promote template/primer-dNTP alignments leading to extension from a correct primer C base opposite the [AF]G(anti) in either an error-free or mutagenic manner, with the latter capable of producing semi-targeted replicative errors. Based on these findings, we propose a novel polymerase-stabilized mechanism of mutagenic semi-targeted [AF]G translesion bypass by Dpo4.
Results
DNA template/primer design for structure determination
In order to crystallize the Dpo4 extension ternary complexes with misinserted A and correct C opposite the [AF]G lesion (Fig. 1a) (designated [AF]G•A-1 and [AF]G•C-1 complexes), we used an [AF]G-modified 5′-CTAACG[AF]G-¨3′ 19-mer template and 2′,3′-dideoxy-A or 2′,3′-dideoxy-C terminated 13-mer primer strands (Fig. 1b). The post-extension ternary complexes with either A or C opposite the [AF]G (designated [AF]G•A-2 and [AF]G•C-2 complexes) were crystallized with 13-mer primers, which end opposite the C 5′ to the lesion with 2′,3′-dideoxy-G. The structures of the Dpo4 [AF]G-modified ternary complexes were solved by the molecular replacement method. The crystal data, together with the data collection and refinement statistics are summarized in Table 1.
Table 1.
Data collection and refinement statistics.
| [AF]G•A-1 Extension | [AF]G•C-1 Extension | [AF]G•A-2 Post-extension | [AF]G•C-2 Post-extension | |
|---|---|---|---|---|
| Data collection | ||||
| Space group | P21 | P21 | P1 | P1 |
| Cell dimensions: | ||||
| a, b, c (Å) | 52.8, 111.0, 100.8 | 54.8, 98.8, 98.6 | 52.4, 52.7, 100.1 | 52.5, 53.0, 99.9 |
| α, β, γ (°) | 90.0, 101.4, 90.0 | 90.0, 99.1, 90.0 | 82.2, 76.4, 70.1 | 81.8, 76.8, 70.1 |
| Complexes per AUa | 2 | 2 | 2 | 2 |
|
| ||||
| Resolution range (Å)b | 20-2.96 (3.07-2.96) |
20-2.70 (2.78-2.70) |
20-2.10 (2.15-2.10) |
20-2.00 (2.05-2.00) |
|
| ||||
| Rmerge (%) | 10.8 (71.0) | 8.8 (42.1) | 4.7 (40.2) | 7.5 (46.9) |
|
| ||||
| I / σI | 11.4 (1.8) | 13.6 (2.4) | 21.2 (3.4) | 15.9 (3.3) |
|
| ||||
| Completeness (%) | 96.2 (94.3) | 98.3 (97.9) | 97.6 (97.1) | 96.7 (96.3) |
|
| ||||
| Redundancy | 3.5 (3.2) | 3.4 (3.2) | 4.0 (3.9) | 4.0 (4.0) |
|
| ||||
| Refinement | ||||
|
| ||||
| Resolution range (Å) | 20-2.96 | 20-2.70 | 20-2.10 | 20-2.00 |
|
| ||||
| No. reflections | 21,625 | 26,483 | 52,487 | 59,976 |
|
| ||||
| Rfactor/Rfree | 20.5/28.2 | 19.2/25.2 | 20.5/26.3 | 20.6/24.7 |
|
| ||||
| No. atoms | ||||
|
| ||||
| Protein | 5,480 | 5,480 | 5,480 | 5,480 |
|
| ||||
| DNA | 1,026 | 1,223 | 1,305 | 1,261 |
|
| ||||
| Ligand (dNTP) | 62 | 62 | 58 | 58 |
|
| ||||
| Ion (Ca2+) | 6 | 6 | 6 | 6 |
|
| ||||
| Water | 50 | 92 | 316 | 465 |
|
| ||||
| B-factors | ||||
|
| ||||
| Protein | 34.0 | 28.0 | 30.8 | 30.2 |
|
| ||||
| DNA | 55.2 | 30.3 | 45.3 | 44.6 |
|
| ||||
| Ligand (dNTP) | 36.9 | 47.8 | 13.7 | 20.0 |
|
| ||||
| Ion (Ca2+) | 48.8 | 43.5 | 41.5 | 40.7 |
|
| ||||
| Water | 21.7 | 26.3 | 38.7 | 38.7 |
|
| ||||
| R.m.s. deviations | ||||
|
| ||||
| Bond length (Å) | 0.011 | 0.009 | 0.011 | 0.010 |
|
| ||||
| Bond angles (°) | 1.60 | 1.37 | 1.46 | 1.44 |
AU, asymmetric unit.
Values in parentheses are for highest-resolution shell.
Structure of the [AF]G•A-1 extension ternary complex
The overall structure and conformation of the [AF]G•A-1 extension ternary complex (Fig. 1c,d) is similar (Cα r.m.s.d. = 0.98 Å) to the type I unmodified ternary complex48. The Dpo4 polymerase embraces the 19-mer template/13-mer primer DNA by its four domains: palm, finger, thumb, and little finger. Dpo4 enlists three divalent cations, identified previously as Ca2+ by anomalous scattering under identical crystallization conditions49. The polymerase active site ions A and B are analogous to the metal ions present in high-fidelity polymerases50. The third cation is coordinated by the loop of the thumb domain (Fig. 1d).
At the (0) position directly below the ‘roof’ of the active site pocket, a nascent base pair is formed by the C5 template base (5′ adjacent to [AF]G) and the incoming dGTP (Figs. 1b,c, and e). The sugar ring of dGTP stacks against the aromatic ring of Tyr12 and the phosphate groups of dGTP are in the normal ‘chair’ conformation50 (Fig. 1e). The [AF]G lesion is at the expected (–1) position of the Dpo4 active site with the AF moiety inserted inside the template/primer double helix and the modified G in the syn conformation in the major groove. This base-displaced [AF]G(syn) alignment is clearly evident from the simulated annealing Fo-Fc omit map shown in Fig. 1f. The N2 group of the modified G contacts the oxygens of the phosphate group of the adjacent C5 template base and there is a water molecule near the O6 atom of the G (Fig. 1g). The face of the modified G is stacked against C5 and the hydrophobic AF-moiety is sandwiched between the base of the dGTP and the adjacent C7•G13 base pair (Fig. 1h). While only the phosphate group of the partner residue A14 is well-defined in the electron density map (Fig. 1f), there is no room for the A14 base and sugar within the AF-invaded double helix (Fig. 1h). Thus, the partner A14 base and sugar are pushed out of the double helix by the AF-moiety and apparently take multiple conformations outside the double helix; this results in the absence of ordered electron density. Hence, the [AF]G(syn) blocks primer extension from A14 and the [AF]G•A-1 complex is not catalytically competent.
‘Correct’ extension from [AF]G(anti)•C base pair
The [AF]G•C-1 extension complex (Fig. 2a) has two distinct molecules in the asymmetric unit (AU), labeled 1 and 2. These molecules reflect ‘correct’ (molecule 1) and ‘mutagenic’ (molecule 2) extension from a C base opposite the [AF]G lesion (see Discussion section). In molecule 1, the incoming dGTP forms a nascent base pair with template C5 (Fig. 2b,c), as expected. The AF-moiety is positioned outside the template/primer helix on the major groove side of the nascent duplex; the modified G is in the anti conformation and forms a Watson-Crick base pair with 3′-terminal primer base C14 (Fig. 2c,d). The dGTP is positioned in a lower alignment relative to the active site, with the sugar ring of dGTP no longer in a stacking range with Tyr12, in contrast to the alignment observed in [AF]G•A-1 (Fig. 1e). The phosphate groups of the dGTP are in the unusual ‘goat-tail’ conformation similar to that observed previously in the Dpo4 complex containing a mismatch50. Moreover, the nascent C5•dGTP base pair is shifted relative to the adjacent [AF]G(anti)•C pair, so that the base of dGTP is above the center of the modified-G•C base pair (Fig. 2e), and C5 is above the AF-moiety. Interestingly, the base of A4, 5′ to C5, resides in the same plane as the base of C5, thus forming an A4-C5 platform that stacks against the long hydrophobic face of the AF. The side chains of the Dpo4 little finger domain residues Arg331, Arg332 and Leu293 partially enclose and shield the AF-moiety from the solvent (Fig. 2e).
Figure 2.
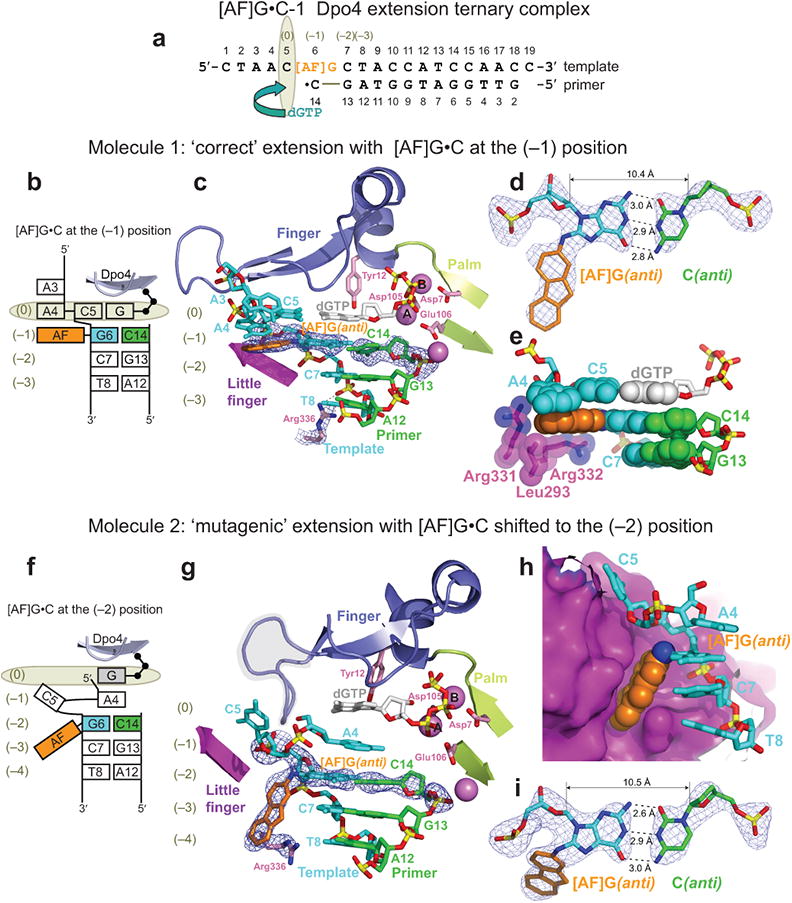
Structure of the [AF]G•C-1 Dpo4 extension ternary complex containing two distinct molecules per asymmetric unit (AU), with ‘correct’ and ‘mutagenic’ alignment for extension from the [AF]G(anti)•C base pair. (a) Schematic of the expected pairing of the [AF]G-template with the 13-mer primer, ending with a 2′,3′-dideoxy-C, and dGTP. (b) Schematic of the observed base pairing arrangement within the Dpo4 active site of molecule 1. (c) Structure of the active site of molecule 1. [AF]G(anti) at the (–1) position is opposite the primer C14 base. The simulated annealing Fo-Fc omit map contoured at 3σ level is colored in blue (2.70 Å resolution). (d) Watson-Crick base pair between the [AF]G(anti) and C14(anti) at the (–1) position. (e) Base stacking arrangement of the [AF]G(anti) opposite C14 and neighboring base pairs. The little finger domain residues Leu293, Arg331 and Arg 332 contact the AF-moiety. (f) Schematic of the observed base pairing arrangement within the Dpo4 active site of molecule 2. (g) Structure of the active site of molecule 2. The [AF]G(anti) opposite the primer C14 base are shifted to the (–2) position. Arg336 has repositioned to stack with the ‘bottom’ face of the AF-moiety. Simulated annealing Fo-Fc omit map contoured at 3σ level is colored in blue (2.70 Å resolution). The loop connecting β2 and β3 of the Dpo4 finger domain (residues Phe33 to Ala42) is highlighted by the shaded area. (h) Accommodation of the AF-moiety in a pocket on the surface of the little finger domain. (i) Watson-Crick base pair between the [AF]G(anti) and C14(anti) at the (–2) position.
‘Mutagenic’ extension from [AF]G(anti)•C base pair
In molecule 2, the template bases 5′ to the lesion site and incoming dNTP are misaligned (Fig. 2f,g), and the [AF]G(anti)•C pair is shifted to the (–2) position (Fig. 2g-i). Residue C5, which was expected to form a base pair with dGTP, is looped out of the DNA helix into the major groove, and stacks against the surface of the little finger domain (Fig. 2h). Residue A4, located 5′ to C5, is inserted back into the double helix at the (–1) position and does not have a pairing partner. The rest of the single stranded template overhang (residues C1-A3) has poor electron density.
The incoming dGTP at the (0) position also does not have a pairing partner. The sugar of dGTP is positioned too low to stack against the aromatic ring of Tyr12 (Fig. 2g), and the phosphate groups assume the ‘goat-tail’ conformation similar to that observed in molecule 1 (Fig. 2c). Furthermore, the loop connecting β2 and β3 of the Dpo4 finger domain (highlighted by the shaded area in Fig. 2g) changes its conformation, to fill the space normally taken by the sugar phosphate backbone of the template base at the (0) position and the next 5′ base of the single stranded template overhang exiting the active site (Supplementary Fig. 1).
The AF-moiety of the [AF]G(anti) is directed towards the 3′-end of the template strand and is placed in a surface pocket of the Dpo4 little finger domain (Fig. 2h). The ‘top’ wall of this pocket is formed by hydrophobic side chains of Val287 and Ile248, while the ‘bottom’ wall is created by the side chain of Arg336, which stacks against the face of the AF-moiety (Fig. 2g,h). Normally this Arg336 makes hydrogen-bonding contacts with the phosphate group of the template base at the (–3) position, as observed in [AF]G•A-1 (Fig. 1e) and molecule 1 of [AF]G•C-1 (Fig. 2c).
Translocations of the thumb and little finger domains
The contacts of the little finger and thumb domains of Dpo4 with the template/primer DNA duplex48 undergo stepwise changes that produce the alignments observed in molecule 1 and molecule 2 of [AF]G•C-1. These changes are similar to the ones observed previously with unmodified DNA after covalent nucleotide incorporation and the dNTP binding steps, respectively49. The translocations are demonstrated by changes in the contacts between the protein and the DNA and are independent of the way the structures are superimposed. The details are described in full in Supplementary Information.
The [AF]G•A-1 complex has a pattern of Dpo4 interactions with DNA backbone phosphates similar to the one in an unmodified ternary complex48-49 (Fig. 3a, top and bottom). In molecule 1 of [AF]G•C-1, the little finger domain contacts the same residues as in [AF]G•A-1 (Fig. 3b, top), while the thumb domain is shifted towards the active site (see arrow, Fig. 3b, bottom and stereo view of translocation at the full structure level in Supplementary Fig. 2a, as well as thumb domain level in Supplementary Fig. 2b). The pattern of Dpo4 interactions with DNA in molecule 1 of the [AF]G•C-1 ternary complex resembles that of an unmodified binary complex (without dNTP)49. In ‘mutagenic’ molecule 2 of [AF]G•C-1, the little finger domain has translocated from the position observed in molecule 1 (see arrow, Fig. 3c, top), while the thumb domain maintains its contacts as in molecule 1 (Fig. 3c, bottom and Supplementary Fig. 2d,e). The Dpo4-DNA interaction pattern in molecule 2 corresponds to that of a ternary complex.
Figure 3.
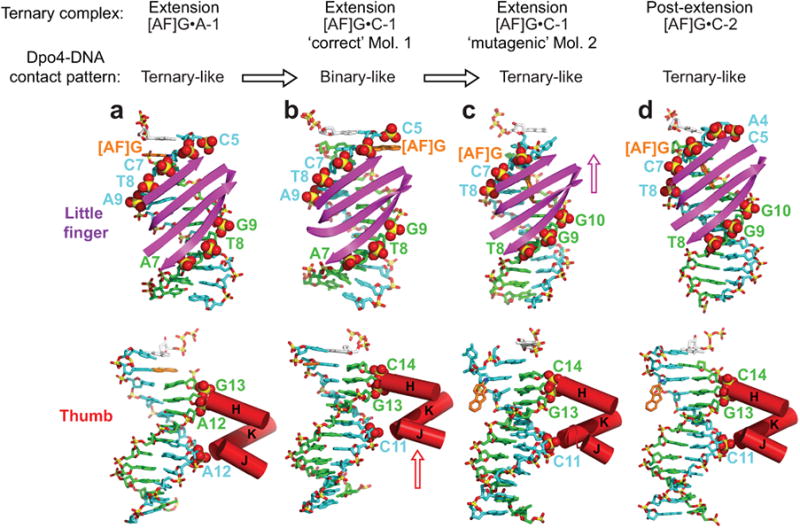
Interactions of the Dpo4 little finger and thumb domains with template/primer DNA in extension and post-extension [AF]G-modified Dpo4 ternary complexes. The backbone DNA phosphate groups in contact with Dpo4 are labeled in color and are shown by CPK spheres. The template strand is colored in cyan, the primer strand is in green. (a) The [AF]G•A-1 extension complex has a ‘ternary complex-like’ Dpo4/DNA interaction pattern. The little finger domain contacts the phosphate groups of template C5-A9 and of primer A7-G9 positions (top panel). The thumb domain contacts the phosphates of the template A12 and primer A12-G13 positions (bottom panel). The DNA segment, containing template C5-A16, primer T4-A14 bases and dGTP, is shown. (b) Molecule 1 of the [AF]G•C-1 complex with ‘correct’ template/primer-dNTP pairing alignment has a ‘binary complex-like’ Dpo4/DNA contact pattern. The little finger domain contacts the phosphate groups of template C5-A9 and primer A7-G9 positions (top panel), the thumb domain is translocated compared to the position observed in the [AF]G•A-1 complex and contacts the phosphate groups of the template C11 and primer G13-C14 positions (bottom panel). The DNA segment, containing template C5-A16, primer T4-C14 bases and dGTP is shown. (c) Molecule 2 of the [AF]G•C-1 complex with ‘mutagenic’ template/primer-dNTP pairing alignment has a ‘ternary complex-like’ Dpo4/DNA contact pattern. The little finger domain is translocated relative to the position observed in molecule 1; it contacts the phosphate groups of template [AF]G6-T8 (the C5 base is looped out and the phosphate of A4 is disordered) and primer T8-G10 positions (top panel), and the thumb domain interacts with the phosphate groups of the template C11 and primer G13-C14 positions (bottom panel). The DNA segment containing template A4-A16, primer T4-C14 bases and dGTP is shown. (d) The [AF]G•C-2 post-extension complex has a ‘ternary complex-like’ Dpo4/DNA contact pattern. The little finger domain contacts the phosphate groups of template A4-T8 and primer T8-G10 positions (top panel), and the thumb domain interacts with the phosphate groups of the template C11 and primer G13-C14 positions (bottom panel). The DNA segment containing template A4-A16, primer T4-C14 bases and dGTP is shown. The [AF]G•A-2 post-extension complex has a similar Dpo4/DNA interaction pattern to that of [AF]G•C-2.
Catalytic alignment for correct and mutagenic extension
In the ‘correct’ molecule 1, the distance between the α-phosphate of dGTP and the 3′-C of the sugar of the 3′-primer terminal C14 (the 3′-OH is absent) is ∼5.0 Å. This distance is not substantially longer than the analogous distances in unmodified complexes with a 2′, 3′-dideoxy primer terminus (∼4.2 Å, PDB ID 2AGQ50, and ∼4.3 Å, PDB ID 2ATL49). The most significant factor affecting the catalytic competence of the molecule 1 complex is the ∼4 Å repositioning (relative to the unmodified structures) of the catalytic cation A, which coordinates the 3′-OH group and activates it for reaction23. The repositioning of catalytic cation A is most likely caused by the ‘binary complex-like’ pattern of the Dpo4-template/primer DNA interactions (Fig. 3b and Supplementary Figs. 2a,b) and the improperly positioned dGTP with misaligned ‘goat-tail’ conformation of the triphosphate group (Fig. 2c). The conformation of the dNTP and the positions of the 3′-OH group, the α-phosphate of the dNTP, and the catalytic ions are often found to deviate from their ideal values in lesion-containing Dpo4 ternary complexes15. An alignment step that tunes the positions of groups essential for catalysis in the spacious active sites of the Y-family polymerases is believed to be necessary prior to covalent bond formation15.
Primer extension in the ‘mutagenic’ molecule 2 also requires reorganization: while the Dpo4/DNA contacts have a normal ‘ternary-complex-like’ pattern (Fig. 3c), the distance between the α-phosphate of dGTP and 3′-C of the sugar of C14 is ∼5.8 Å, which is greater than the ∼4.25 Å distance in the unmodified complexes. The catalytic ion A is also shifted by ∼4 Å as compared to the unmodified complexes, probably due to the C14 base being at the (–2) position (Fig. 2g), instead of the expected (–1) position and the ‘goat-tail’ conformation of the dGTP phosphate groups.
Post-extension [AF]G(anti)•C-2 complex
The [AF]G•C-2 and [AF]G•A-2 post-extension complexes with either C or A base opposite the [AF]G at the (–2) position represent a further primer extension step. The two molecules in the AU of [AF]G•C-2 (Fig. 4a) are similar to each other and have the expected ‘ternary complex-like’ Dpo4/DNA contact patterns (Fig. 3d, top and bottom). The nascent base pair is formed by the A4 template base and incoming dTTP, with its sugar stacked normally against the aromatic ring of Tyr12 (Fig. 4b,c). The AF-moiety of the [AF]G(anti), Watson-Crick paired with C14 (Fig. 4c,d), is placed in the same pocket on the surface of the little finger domain with the relocated side chain of Arg336 (Fig. 4e), as in the molecule 2 of [AF]G•C-1 (Fig. 2h). The modified-[AF]G•C base pair maintains the normal base stacking alignment with the adjacent C7•G13 and C5•G15 base pairs (Fig. 4c). The distance between the α-phosphate of dTTP and the 3′-C of the terminal G15 primer base is ∼3.9 Å and the normal alignment of the Dpo4 active site is maintained.
Figure 4.
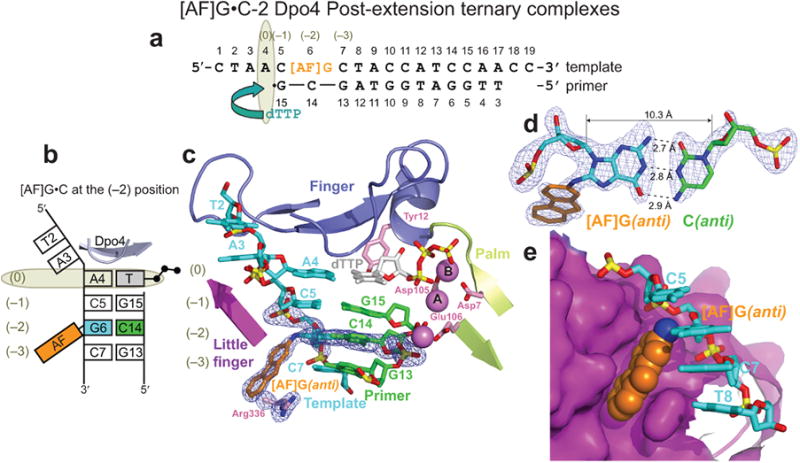
Structure of the [AF]G•C-2 Dpo4 post-extension ternary complex. (a) Schematic of the expected pairing of the [AF]G-template with the 13-mer primer, ending with a 2′,3′-dideoxy-G, and added dTTP. (b) Schematic of the observed base pairing arrangement within the Dpo4 active site. (c) Structure of the active site. Simulated annealing Fo-Fc omit map contoured at 3σ level is colored in blue (2.0 Å resolution). (d) Watson-Crick base pair between the [AF]G(anti) and C14(anti) at the (–2) position. (e) Accommodation of the AF-moiety in a pocket on the surface of the little finger domain.
Post-extension [AF]G(anti)•A-2 complex
The two molecules in the AU of the [AF]G•A-2 complex (Fig. 5a-g) are similar to the [AF]G•C-2 complex, with the AF-moiety of the [AF]G(anti) residing in the little finger domain pocket (Figs. 5c and 5f). However, the partner A14 base appears to adopt multiple conformations (Figs. 5d and 5g). Despite the high quality of the overall 2Fo-Fc electron density map at 2.1 Å for both molecules, the density for the nucleobase of A14 is poor, while the A14 phosphate and sugar moieties are well-defined (molecules 1 and 2 in Supplementary Figs. 4a and 4b, respectively). While the exact orientation of the A14 base is not clear, we are confident of A14 placement in the minor groove in molecule 1 (Fig. 5d). In molecule 2, the electron density between the phosphate group and the sugar ring of A14 is visible on a lower σ-level than it is in molecule 1; nevertheless, an inside-the-helix position of the A14 sugar is indicated (Fig. 5g). The C1′-C1′ distance between the modified-[AF]G and A14 is ∼10.6 Å, which would be appropriate for involvement of some variation of the [AF]G(anti)•A(syn) alignment51.
Figure 5.
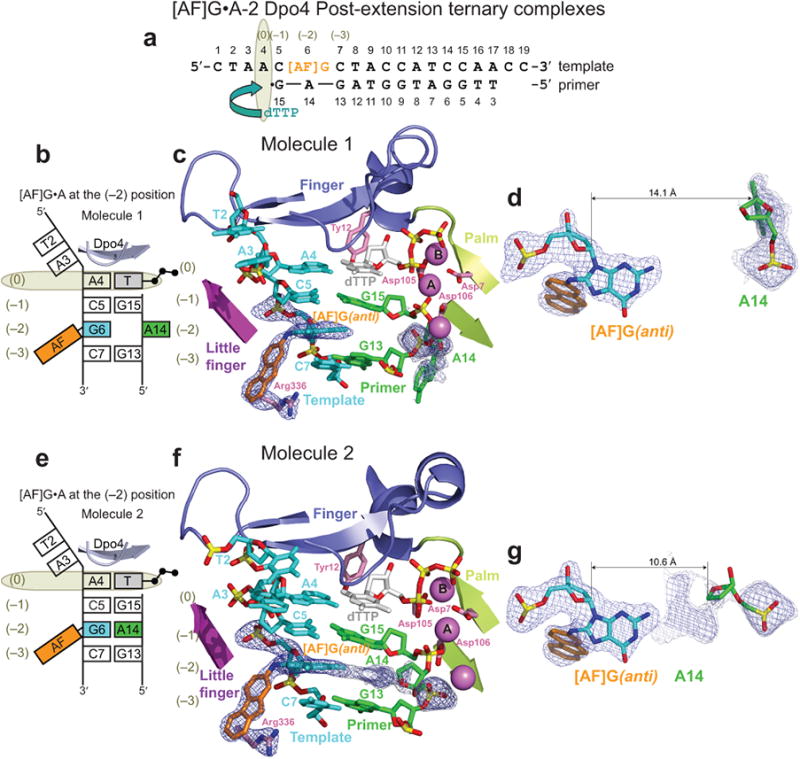
Structure of the [AF]G•A-2 Dpo4 post-extension ternary complex containing two molecules per AU with different positions of the partner A14 base. (a) Schematic of the expected template/primer-dTTP pairing. (b) Schematic of the observed base pairing arrangement within the Dpo4 active site of molecule 1, with the A14 apparently oriented outside the helix. (c) The active site of molecule 1. [AF]G(anti) at the (–2) position opposite primer A14 base. Simulated annealing Fo-Fc omit map contoured at 3σ level is colored in blue and at 2σ level is colored in gray (2.10 Å resolution). (d) The A14 base positioned opposite [AF]G(anti), appears to be oriented outside the helix on the minor groove side. (e) Schematic of the observed base pairing arrangement within the Dpo4 active site of molecule 2 with A14 apparently oriented inside the helix. (f) The active site of molecule 2. Simulated annealing Fo-Fc omit map contoured at 3σ level is colored in blue and at 2σ level it is colored in gray. (g) The A14 base positioned opposite [AF]G(anti), appears to be oriented inside the helix.
Efficiency and fidelity of Dpo4 translesion synthesis
Here we show that Dpo4 preferentially inserts a correct C base opposite the [AF]G lesion, but further extension is both error-free and error-prone. Primer extension assays clearly demonstrate that mutagenic extension occurs and includes base deletion and base substitution errors.
Dpo4 readily elongates the 13-mer primer strand using the unmodified-G DNA template, to produce predominantly full-length 19-mer extension product in the presence of all four dNTPs; only single bands corresponding to partially extended 14-, 15-, 16-, 17- and 18-mers are evident (Fig. 6a, lanes 1-5 and Supplementary Fig. 5). Dpo4 efficiently inserts a base opposite the [AF]G-adduct forming a 14-mer product, but further extension is inhibited (Fig. 6a, lanes 6-10). Moreover, additional bands for 15-, 16-, 17- and 18-mers are detected, indicating mutagenic extension, while the single band for the 19-mer suggests that the correct fully elongated product was formed (Supplementary Fig. 5). Single base incorporation experiments (Fig. 6b) and Michaelis-Menten kinetic parameters (Fig. 6c, Supplementary Fig. 6 and Supplementary Table 1) indicate that C is preferentially inserted opposite [AF]G with insertion frequency, fins, only 8-fold smaller than it is opposite unmodified G. However, the efficiency of extension from the primer 3′-terminal C•[AF]G base pair via incorporation of the next correct dGTP, is reduced by a factor of ∼150, as compared to extension from the unmodified G•C base pair (Fig. 6d lanes 3 and 7, Fig. 6e, Supplementary Fig. 7, panel a1, Supplementary Table 1).
Figure 6.
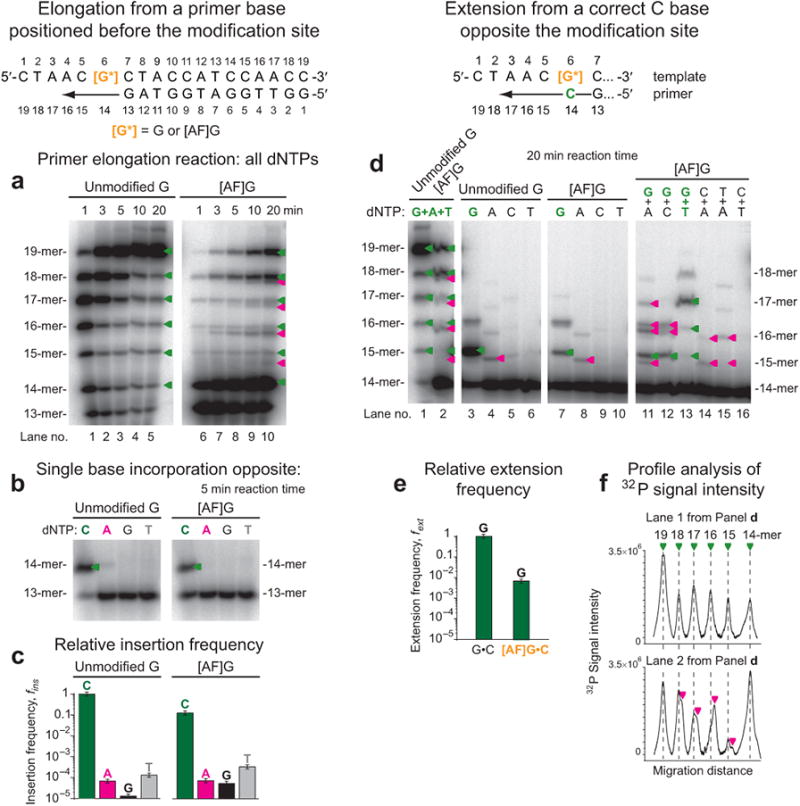
Efficiency and fidelity of base incorporation and extension of primers bound to unmodified-G and [AF]G-template by Dpo4. (a) Time course of extension of 32P 5′-endlabeled 13-mer primers bound to 19-mer templates in the presence of all four dNTPs. The 3′-end of the 13-mer primer was paired with the template base on the 3′-side of the unmodified-G, or [AF]G; the 3′-end primer base of the 14-mer is paired with G or [AF]G. In the case of the [AF]G-template, additional 15-, 16-, 17- and 18-mers bands that migrate with different mobilities than the correctly elongated bands arising from the unmodified template strand, are detected, thus indicating mutagenic extension. The green triangles represent the correctly extended products; the magenta triangles represent mutagenic extension. In the case of the [AF]G template, the fully extended 19-mer and the shorter 18-mer products comprise ∼22% and ∼20% of the overall extended and unextended primer strands, respectively, observed after a 20 min incubation time (lane 10). (b) dCTP, dATP, dGTP or dTTP single nucleotide insertion. (c) Nucleotide insertion frequencies normalized relative to the insertion of a C base opposite the unmodified-G. The Michaelis-Menten kinetic data are from the Supplementary Table 1. (d) Efficiency and fidelity of extension from a C base opposite the unmodified-G and [AF]G- by Dpo4. Lanes 1-2 demonstrate extension in the presence of a mixture of dGTP, dATP and dTTP. Lanes 3-6 and 7-10 show extension by a single nucleotide at a time (see labels), and lanes 11-16 show extension in the presence of two nucleotides at a time (see labels). The 15-mer to 18-mer primers containing mutated bases are indicated by the magenta triangles (e) Relative extension frequencies from G•C and [AF]G•C pairs in the presence of dGTP. The Michaelis-Menten kinetic data are from Supplementary Table 1. (f) Profile analysis of 32P-signal intensities shown in lanes 1 and 2 of panel d.
Primer extension experiments from correctly paired [AF]G•C termini also leads to a mixture of correctly and faulty extension products (Fig. 6d). In the presence of all dNTPs that are necessary for elongation, additional bands are detected (lane 2 in Fig. 6d, and Fig. 6f, bottom panel). However, in the presence of dGTP and dTTP, both necessary for the correct insertion opposite template bases C5, A4 and A3, only a single 17-mer extension band is detected (Fig. 6d, lane 13), indicating that error-free extension has occurred. For a dGTP mixture with dATP, the latter representing an incorrect dNTP for further extension, double bands are observed in the case of the 16-mer, indicating faulty incorporation of dATP and primer extension to 15-, 16-, and 17-mers (lane 11). Likewise, in the case of a dGTP + dCTP mixture (lane 12), the single 15-mer suggests the correct insertion of dGTP, followed by the mutagenic extension of the 15-mer to 16-mers.
We note that with the [AF]G-template Dpo4 pauses to extend the 18-mer to a 19-mer, while a significant fraction of the 18-mer remains unextended (Fig. 6a, lanes 6-10 and Fig. 6d, lane 2, and Supplementary Fig. 7a, panel a1). This suggests that this 18-mer is an extension product with a single –1 deletion. A shoulder on this band indicates that a second 18-mer extension product is present (Fig. 6a, lane 10 and Fig. 6d, lane 2) that may result from a deletion mutation as well as the mutagenic insertion of a nucleotide in the subsequent extension steps. Further evidence that the C5 base on the [AF]G-modified template can be skipped is provided in the Supplementary Information (Supplementary Fig. 8).
Primer extension via the correct dGTP from the mismatched 3′-A, G, and T opposite the [AF]G adduct is inefficient (Supplementary Table 1). Surprisingly, Dpo4 is robust in extending from an [AF]G•G terminus via addition of dTTP (Supplementary Fig. 7a, panel a4 and Supplementary Fig. 7b,c). This intermediate is stabilized by an incoming dTTP opposite template A4 and a correct terminal base pair between primer base G14 and template C5 (Supplementary Fig. 7b). The dNTP-stabilized misinsertion misalignment mechanism (Fig. 7a) has been demonstrated previously for aromatic amine adducts and other lesions21,40-42.
Figure 7.
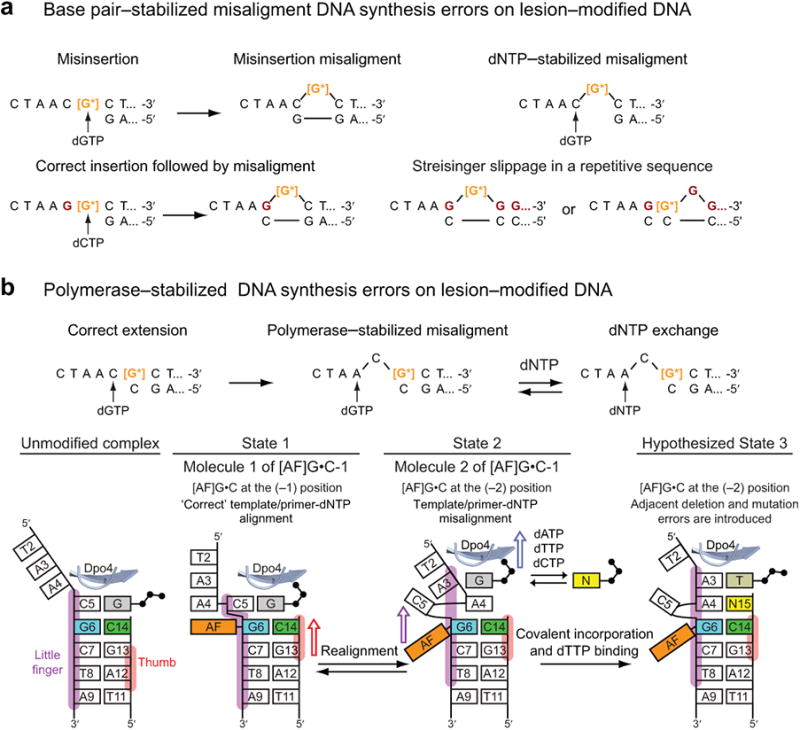
Misalignment-mediated replication errors and proposed mechanism for the [AF]G-induced semi-targeted mutagenesis. (a) Misalignment-mediated DNA synthesis errors on lesion-modified DNA. The intermediates are stabilized by Watson-Crick base-pairing. The schematics are adapted from37,39. The template bases colored in dark red are distinct from the DNA sequence in the present study. (b) Proposed mechanism for [AF]G-induced semi-targeted mutagenesis in the Dpo4 active site. The misaligned intermediates are stabilized by interactions with the DNA polymerase. Left panel: Schematics of base pairing alignment and the Dpo4 little finger (purple) and thumb domain (red) contacts with the ‘top’ portion of the template/primer DNA in an unmodified ternary complex. State 1, molecule 1 of [AF]G•C-1: C5 stacks above the AF-moiety, the base of dGTP is shifted toward the middle of the template/primer helix and the phosphate groups adopt a ‘goat-tail’ conformation50. In this state the thumb domain of Dpo4 is translocated relative to the template/primer DNA compared to the unmodified ternary complex, denoted by a red arrow. The State 1 complex has a ‘binary complex-like’ DNA/Dpo4 interaction pattern that reduces Dpo4 elongation efficiency and allows time for further realignment. State 2, molecule 2 of [AF]G•C-1: The little finger, palm and finger domains are translocated so the [AF]G(anti)•C base pair is now at the (–2) position, the AF-moiety is in a pocket on the surface of the Dpo4 little finger domain. The complex has a ‘ternary complex-like’ interaction pattern. Translocation of the little finger domain is denoted by a purple arrow, and of the finger and palm domains by a blue arrow. The unpaired dGTP can be interchanged with another dNTP. The equilibria between State 2 and State 1 would allow bringing the C5 back into the double helix for correct extension. State 3 (hypothesized): Covalent bond formation following State 2 would result in C5 base deletion, and misinsertion of G opposite template base A4. Binding of the next correct dTTP opposite A3 does not require a change in Dpo4/DNA contacts. The complex now resembles the catalytically competent [AF]G•C-2 post-extension complex.
Discussion
An intermediate poised for semi-targeted mutagenesis
Semi-targeted mutations are observed in both mammalian cells and in bacteria. The number of semi-targeted mutations is usually very low, but it reaches a significant fraction of the overall number of mutations (up to 25%) in some cases. Examples of semi-targeted mutations have been reported for various types of DNA lesions40,52-56 and the [AF]G adduct5,7. For Dpo4, in vitro observed examples include replication past (IQ)-C8-guanine42 and BPDE-N2-dG57 adducts. Details are given in Supplementary Information.
Proposed mechanisms for semi-targeted mutagenesis involve misaligned template-primer structures with the incorporation of the correct nucleotide opposite the modified base40. These mechanisms are similar to the ones proposed37-39 and observed44-45 for frameshift misalignment-mediated errors (depicted in Fig. 7a). These alternative template/primer-dNTP alignments are stabilized by Watson-Crick base pairs surrounding the looped out bases; the correctly paired terminal base pair supports further primer extension. With lesions, the looped out bases may entail the modified one21,37-43 and/or adjacent ones43,57-58. Moreover, adjacent frameshift/point mutations with two or more errors are well-documented in the case of unmodified DNA37-38 and in the presence of lesions5-6,42,52. These adjacent mutations have been attributed to sequential realignments and extension steps that proceed through base-pair stabilized slippage intermediates37-38,42.
The template/primer-dNTP alignment observed in molecule 2 of the [AF]G•C-1 Dpo4 ternary complex represents a mutagenic extension from the correct [AF]G(anti)•C base pair. In this misaligned complex, the template C5 base 5′ to the [AF]G is looped out and the next template A4 and incoming dNTP do not have pairing partners. If primer extension occurs in this template/primer-dNTP alignment, the resulting product will have deletions and/or mutated bases, causing semi-targeted mutations. Faulty primer extension products observed during Dpo4-catalyzed extension from the correct C opposite [AF[G (Fig. 6d,f) can be explained by this mutagenic intermediate. Moreover, as discussed below, this mutagenic template/primer-dNTP alignment is stabilized by interactions with the polymerase.
Catalytic competence of the [AF]G-modified complexes
In the [AF]G•C-1 extension complex, we observe two types of template/primer DNA and incoming dNTP alignments: correctly paired in molecule 1, and misaligned in molecule 2. In the Results, we have demonstrated that the active site alignments in both molecules are equivalently perturbed and require a reorganization. This is consistent with the moderate ∼150-fold overall reduction, as compared to extension from an unmodified G•C pair, in the efficiency of primer extension (Supplementary Table 1).
In the [AF]G•A-1 extension ternary complex structure, the primer 3′-OH terminus of the partner A14 base is displaced from the template/primer helix by the AF-moiety with the [AF]G adduct in the syn conformation (Fig. 1h); further primer extension from A14 in this extrahelical position is not possible. However, our kinetic data demonstrate that extension from the A14 base opposite the [AF]G adduct still occurs, although highly inefficiently, with an extension probability ∼7,000-fold smaller than from an unmodified G•C base pair (Supplementary Table 1). We hypothesize that extension from A14 might occur by rotation of the [AF]G into the anti conformation; this would place the AF-moiety into the major groove and allow the A14 to enter the helix, so that the primer 3′-OH terminus is ready for extension.
In the [AF]G•C-2 complex, the normal alignment of the Dpo4 active site is maintained, as detailed in Results, which is consistent with the nearly equal Dpo4 primer extension activity from the C5•G15 pair positioned 5′ to the unmodified-G6•C14 or the [AF]G6•C14 pairs (Supplementary Fig. 9). Thus, the presence of the [AF]G(anti) in the pocket on the surface of the little finger domain (Fig. 4e) affects the catalytic efficiency only minimally.
[AF]G conformations in Dpo4-free and Dpo4-bound states
Comparison of [AF]G adduct conformations within the Dpo4 active site in the crystalline state (this study) and in free DNA duplexes in solution33 (Supplementary Fig. 10) are described in detail under Supplementary Data. It is remarkable that the structural families observed within the polymerase in the crystal are the same as those observed by high resolution NMR studies with Watson-Crick paired duplexes, although the interactions with the Dpo4 polymerase change the conformational balance between the major groove and the base-displaced intercalated conformations of the [AF]G adduct33-36,59.
Polymerase-stabilized semi-targeted mutagenesis mechanism
Our crystal structures and primer extension data suggest a distinct and novel mechanism for generating semi-targeted mutations. We propose the following polymerase-stabilized misalignment mechanism for semi-targeted mutagenesis (Fig. 7b): (1) State 1, molecule 1 of [AF]G•C-1: The correct dNTP (dGTP) binds opposite the templating base (C5), 5′ to the lesion, for extension from a C base opposite the [AF]G adduct. The [AF]G in the anti conformation triggers rearrangements within the Dpo4 active site and causes the thumb domain of Dpo4 to be translocated relative to the DNA from the position observed in an unmodified ternary complex (left panel). Thus, the complex has a ‘binary complex-like’ DNA/Dpo4 interaction pattern. Primer extension from such a complex with a correctly aligned template/primer-dNTP will lead to the correct 17-mer upon dGTP and dTTP incorporation (Fig. 6d, lane 13); however, the extension is inefficient and allows time for additional realignments to occur. (2) State 2, molecule 2 of [AF]G•C-1: The little finger, palm and finger domains are translocated relative to the DNA, so the [AF]G(anti)•C base pair is now at the (–2) position, the C5 base is looped out, the AF-moiety is in a pocket on the surface of the Dpo4 little finger domain, and the Dpo4/DNA-dNTP complex has a ‘ternary complex-like’ interaction pattern. The unpaired dGTP can be exchanged by another dNTP present in the nucleotide pool. (3) Hypothesized State 3: Covalent bond formation in the ‘mutagenic’ template/primer-dNTP alignment of State 2 could result in a base deletion and a mutation, because C5 is looped out, and G or another base N is inserted opposite the next template base A4. Binding of the next correct dNTP (dTTP) opposite A3 does not require a change in Dpo4/DNA contacts. The alignment of the active site resembles the post-extension [AF]G•C-2 complex and is catalytically competent.
Conclusions
Our structural and biochemical data demonstrate that extension from a correct C opposite the [AF]G adduct in a non-repetitive sequence context can occur by both ‘error-free’ and ‘error-prone’ pathways; for the ‘error-prone’ case a structural rationale for semi-targeted mutations is suggested from our crystal structures. We propose that the semi-targeted mutagenesis process is driven by polymerase-template/primer-dNTP alignments in a structure where the bulky lesion is accommodated most favorably within the polymerase, but not by Watson-Crick base pairing between incorrectly aligned template/primer DNA and dNTP. Moreover, complex mutations with two errors could be achieved in one concerted step rather than by the sequential introduction of errors. Thus, a DNA lesion could trigger semi-targeted mutagenesis events that are guided by interactions with the DNA polymerase, reminiscent of the Rev1 polymerase-induced templating mechanism60.
Online Methods
Crystallization
The crystals of the Dpo4 extension ternary complexes containing [AF]G-modified 19-mer template and 13-mer primers terminated with 2′,3′-dideoxy-A (Fig. 1b) or 2′,3′-dideoxy-C (Fig. 2a) opposite [AF]G were grown in the presence of dGTP and flash-frozen in liquid nitrogen for X-ray data collection under conditions described previously49. The crystals of the post-extension complexes had a 13-mer primer terminated with 2′,3′-dideoxy-G opposite a C5 base, 5′ to the lesion site, and were grown in the presence of dTTP (Figs. 4a and 5a). Several rounds of micro seeding were employed to produce the diffraction quality crystals of the [AF]G•A-1 and [AF]G•C-1 extension ternary complexes.
Structure determination and refinement
X-ray diffraction data were collected at the NE-CAT 24-ID-C and 24-ID-E beam lines at the Advanced Photon Source (Argonne National Laboratory, Chicago). The data were collected at 1.03320 Å wavelength for [AF]G•A-1, at 0.97918 Å for [AF]G•C-1, at 0.97949 Å for [AF]G•A-2, and 0.97914 Å for [AF]G•C-2. The data were processed and scaled using the HKL2000 suite (http://www.hkl-xray.com). The structures of the extension and post-extension complexes were solved by molecular replacement method, using our published oxoGmodified insertion ternary Dpo4-DNA-dCTP structure49 as a search model. The model building, including substitution of the DNA sequence, was manually finished in TURBOFRODO (http://www.afmb.univ-mrs.fr/-TURBO-) based on the electron density maps calculated in REFMAC (http://www.ccp4.ac.uk/html/refmac5.html), and the resulting models were refined in REFMAC. The crystal data, together with the data collection and refinement statistics for all structures are summarized in Table 1. The simulated annealing omit maps were calculated in CNS (http://cns-online.org/v1.21/) with the [AF]G, partner base and Arg336 omitted from the models before they were heated to 2,000 K and then slowly cooled. Residues disordered in the electron density maps in [AF]G•A-1 molecule 2 are G2-G5 and A14 of the primer strand, and C1-A4 and C15-C19 of the template strand; in [AF]G•C-1 molecule 1 and 2 they are C1-T2 and C1-A3 of the template strand, respectively; in [AF]G•A-2 and [AF]G•C-2 they are C1 and C19 of the template strand in molecule 1, and C19 of the template in molecule 2. Figures were prepared with PyMOL (http://www.pymol.org).
Primer elongation and steady state kinetic analysis
Primer elongation assays and kinetic analysis of single base insertion and extension under steady-state single hit conditions were conducted as described by our group in the literature49. The reaction products were resolved on 20% polyacrylamide gels in the presence of 8 M urea, the gels were dried before radiography with Fuji image plates. The images were scanned on a Fuji PhosphoImager and the bands were quantified using the profile analysis mode in the ImageGauge software (Fujifilm).
Supplementary Material
Acknowledgments
We thank Dr. Yuan Cheng for help with expression and purification of Dpo4, Dr. Alexander Serganov for data collection from the [AF]G•C-1 and [AF]G•A-2 crystals, and Dr. Lihua Wang (New York University) for providing the coordinates of the [AF]G-modified DNA duplexes, shown in the Supporting Information. The research was supported by National Institutes of Health grants CA46533 to D.J.P., CA75449 to S.B., and CA99194 to N.E.G. Partial support for computational infrastructure and computer systems management was also provided to S.B. by CA28038. We would like to thank the staff at the Northeastern Collaborative Access Team beamlines of the Advanced Photon Source (APS), Argonne National Laboratory, supported by award RR-15301 from the National Center for Research Resources at the National Institute of Health, for assistance with data collection. Use of the Advanced Photon Source is supported by the U.S. Department of Energy, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357.
Footnotes
Accession codes: Protein Data Bank: Coordinates and structure factor amplitudes for the [AF]G•A-1, [AF]G•C-1, [AF]G•A-2 and [AF]G•C-2 complexes have been deposited with accession codes 3KHG, 3KHH, 3KHL, and 3KHR, respectively.
Note: Supplemental Information containing ten Figures, one Table, Supplementary Results and Discussion, and the full description of Materials and Methods is available online.
Author Contributions: O.R. conducted and interpreted all the structural and kinetic measurements under the supervision of D.J.P, S.B. and N.E.G. A.K. was responsible for the synthesis and purification of [AF]G-containing DNAs under the supervision of N.E.G. L.M. was involved in specific aspects of crystal structure refinement. The paper was written by O.R., D.J.P., S.B. and N.E.G.
References
- 1.Kadlubar FF. DNA adducts of carcinogenic aromatic amines. IARC Sci Publ. 1994;199216 [PubMed] [Google Scholar]
- 2.Turesky RJ. Formation and biochemistry of carcinogenic heterocyclic aromatic amines in cooked meats. Toxicol Lett. 2007;168:219–27. doi: 10.1016/j.toxlet.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 3.Clapp RW, Jacobs MM, Loechler EL. Environmental and occupational causes of cancer: new evidence 2005-2007. Rev Environ Health. 2008;23:1–37. doi: 10.1515/reveh.2008.23.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heflich RH, Neft RE. Genetic toxicity of 2-acetylaminofluorene, 2-aminofluorene and some of their metabolites and model metabolites. Mutat Res. 1994;318:73–114. doi: 10.1016/0165-1110(94)90025-6. [DOI] [PubMed] [Google Scholar]
- 5.Shibutani S, Suzuki N, Tan X, Johnson F, Grollman AP. Influence of flanking sequence context on the mutagenicity of acetylaminofluorene-derived DNA adducts in mammalian cells. Biochemistry. 2001;40:3717–22. doi: 10.1021/bi0027581. [DOI] [PubMed] [Google Scholar]
- 6.Watt DL, Utzat CD, Hilario P, Basu AK. Mutagenicity of the 1-nitropyrene-DNA adduct N-(deoxyguanosin-8-yl)-1-aminopyrene in mammalian cells. Chem Res Toxicol. 2007;20:1658–64. doi: 10.1021/tx700131e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tan X, Suzuki N, Grollman AP, Shibutani S. Mutagenic events in Escherichia coli and mammalian cells generated in response to acetylaminofluorene-derived DNA adducts positioned in the Nar I restriction enzyme site. Biochemistry. 2002;41:14255–62. doi: 10.1021/bi0202878. [DOI] [PubMed] [Google Scholar]
- 8.Mozzherin DJ, Shibutani S, Tan CK, Downey KM, Fisher PA. Proliferating cell nuclear antigen promotes DNA synthesis past template lesions by mammalian DNA polymerase delta. Proc Natl Acad Sci U S A. 1997;94:6126–31. doi: 10.1073/pnas.94.12.6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doisy R, Tang MS. Effect of aminofluorene and (acetylamino)fluorene adducts on the DNA replication mediated by Escherichia coli polymerases I (Klenow fragment) and III. Biochemistry. 1995;34:4358–68. doi: 10.1021/bi00013a027. [DOI] [PubMed] [Google Scholar]
- 10.Lindsley JE, Fuchs RP. Use of single-turnover kinetics to study bulky adduct bypass by T7 DNA polymerase. Biochemistry. 1994;33:764–72. doi: 10.1021/bi00169a018. [DOI] [PubMed] [Google Scholar]
- 11.Belguise-Valladier P, Fuchs RP. N-2-aminofluorene and N-2 acetylaminofluorene adducts: the local sequence context of an adduct and its chemical structure determine its replication properties. J Mol Biol. 1995;249:903–13. doi: 10.1006/jmbi.1995.0347. [DOI] [PubMed] [Google Scholar]
- 12.Miller H, Grollman AP. Kinetics of DNA polymerase I (Klenow fragment exo-) activity on damaged DNA templates: effect of proximal and distal template damage on DNA synthesis. Biochemistry. 1997;36:15336–42. doi: 10.1021/bi971927n. [DOI] [PubMed] [Google Scholar]
- 13.Shibutani S, Grollman AP. On the mechanism of frameshift (deletion) mutagenesis in vitro. J Biol Chem. 1993;268:11703–10. [PubMed] [Google Scholar]
- 14.Lone S, Romano LJ. The role of specific amino acid residues in the active site of Escherichia coli DNA polymerase I on translesion DNA synthesis across from and past an N-2-aminofluorene adduct. Biochemistry. 2007;46:2599–607. doi: 10.1021/bi061324o. [DOI] [PubMed] [Google Scholar]
- 15.Yang W, Woodgate R. What a difference a decade makes: insights into translesion DNA synthesis. Proc Natl Acad Sci U S A. 2007;104:15591–8. doi: 10.1073/pnas.0704219104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McCulloch SD, Kunkel TA. The fidelity of DNA synthesis by eukaryotic replicative and translesion synthesis polymerases. Cell Res. 2008;18:148–61. doi: 10.1038/cr.2008.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Waters LS, et al. Eukaryotic translesion polymerases and their roles and regulation in DNA damage tolerance. Microbiol Mol Biol Rev. 2009;73:134–54. doi: 10.1128/MMBR.00034-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koffel-Schwartz N, Coin F, Veaute X, Fuchs RP. Cellular strategies for accommodating replication-hindering adducts in DNA: control by the SOS response in Escherichia coli. Proc Natl Acad Sci U S A. 1996;93:7805–10. doi: 10.1073/pnas.93.15.7805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tebbs RS, Romano LJ. Mutagenesis at a site-specifically modified NarI sequence by acetylated and deacetylated aminofluorene adducts. Biochemistry. 1994;33:8998–9006. doi: 10.1021/bi00196a018. [DOI] [PubMed] [Google Scholar]
- 20.Reifferscheid G, Heil J. Validation of the SOS/umu test using test results of 486 chemicals and comparison with the Ames test and carcinogenicity data. Mutat Res. 1996;369:129–45. doi: 10.1016/s0165-1218(96)90021-x. [DOI] [PubMed] [Google Scholar]
- 21.Suzuki N, et al. Translesional synthesis past acetylaminofluorene-derived DNA adducts catalyzed by human DNA polymerase kappa and Escherichia coli DNA polymerase IV. Biochemistry. 2001;40:15176–83. doi: 10.1021/bi010702g. [DOI] [PubMed] [Google Scholar]
- 22.Vooradi V, Romano LJ. Effect of N-2-acetylaminofluorene and 2-aminofluorene adducts on DNA binding and synthesis by yeast DNA polymerase eta. Biochemistry. 2009;48:4209–16. doi: 10.1021/bi9000722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Steitz TA, Yin YW. Accuracy, lesion bypass, strand displacement and translocation by DNA polymerases. Philos Trans R Soc Lond B Biol Sci. 2004;359:17–23. doi: 10.1098/rstb.2003.1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson SJ, Beese LS. Structures of mismatch replication errors observed in a DNA polymerase. Cell. 2004;116:803–16. doi: 10.1016/s0092-8674(04)00252-1. [DOI] [PubMed] [Google Scholar]
- 25.Swan MK, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Structural basis of high-fidelity DNA synthesis by yeast DNA polymerase delta. Nat Struct Mol Biol. 2009;16:979–86. doi: 10.1038/nsmb.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lone S, et al. Human DNA polymerase kappa encircles DNA: implications for mismatch extension and lesion bypass. Mol Cell. 2007;25:601–14. doi: 10.1016/j.molcel.2007.01.018. [DOI] [PubMed] [Google Scholar]
- 27.Alt A, et al. Bypass of DNA lesions generated during anticancer treatment with cisplatin by DNA polymerase eta. Science. 2007;318:967–70. doi: 10.1126/science.1148242. [DOI] [PubMed] [Google Scholar]
- 28.Mizukami S, Kim TW, Helquist SA, Kool ET. Varying DNA base-pair size in subangstrom increments: evidence for a loose, not large, active site in low-fidelity Dpo4 polymerase. Biochemistry. 2006;45:2772–8. doi: 10.1021/bi051961z. [DOI] [PubMed] [Google Scholar]
- 29.Broyde S, Wang L, Rechkoblit O, Geacintov NE, Patel DJ. Lesion processing: high-fidelity versus lesion-bypass DNA polymerases. Trends Biochem Sci. 2008;33:209–19. doi: 10.1016/j.tibs.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hsu GW, et al. Observing translesion synthesis of an aromatic amine DNA adduct by a high-fidelity DNA polymerase. J Biol Chem. 2004;279:50280–5. doi: 10.1074/jbc.M409224200. [DOI] [PubMed] [Google Scholar]
- 31.Dutta S, et al. Crystal structures of 2-acetylaminofluorene and 2-aminofluorene in complex with T7 DNA polymerase reveal mechanisms of mutagenesis. Proc Natl Acad Sci U S A. 2004;101:16186–91. doi: 10.1073/pnas.0406516101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burnouf DY, Wagner JE. Kinetics of deoxy-CTP incorporation opposite a dGC8-N-2-aminofluorene adduct by a high-fidelity DNA polymerase. J Mol Biol. 2009;386:951–61. doi: 10.1016/j.jmb.2008.12.067. [DOI] [PubMed] [Google Scholar]
- 33.Patel DJ, et al. Nuclear magnetic resonance solution structures of covalent aromatic amine-DNA adducts and their mutagenic relevance. Chem Res Toxicol. 1998;11:391–407. doi: 10.1021/tx9702143. [DOI] [PubMed] [Google Scholar]
- 34.Meneni SR, et al. Spectroscopic and theoretical insights into sequence effects of aminofluorene-induced conformational heterogeneity and nucleotide excision repair. Biochemistry. 2007;46:11263–78. doi: 10.1021/bi700858s. [DOI] [PubMed] [Google Scholar]
- 35.Norman D, et al. NMR and computational characterization of the N-(deoxyguanosin-8-yl)aminofluorene adduct [(AF)G] opposite adenosine in DNA: (AF)G[syn].A[anti] pair formation and its pH dependence. Biochemistry. 1989;28:7462–76. doi: 10.1021/bi00444a046. [DOI] [PubMed] [Google Scholar]
- 36.Jain N, Meneni S, Jain V, Cho BP. Influence of flanking sequence context on the conformational flexibility of aminofluorene-modified dG adduct in dA mismatch DNA duplexes. Nucleic Acids Res. 2009;37:1628–37. doi: 10.1093/nar/gkn1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kunkel TA. Misalignment-mediated DNA synthesis errors. Biochemistry. 1990;29:8003–11. doi: 10.1021/bi00487a001. [DOI] [PubMed] [Google Scholar]
- 38.Ripley LS. Frameshift mutation: determinants of specificity. Annu Rev Genet. 1990;24:189–213. doi: 10.1146/annurev.ge.24.120190.001201. [DOI] [PubMed] [Google Scholar]
- 39.Tippin B, Kobayashi S, Bertram JG, Goodman MF. To slip or skip, visualizing frameshift mutation dynamics for error-prone DNA polymerases. J Biol Chem. 2004;279:45360–8. doi: 10.1074/jbc.M408600200. [DOI] [PubMed] [Google Scholar]
- 40.Lambert IB, Napolitano RL, Fuchs RP. Carcinogen-induced frameshift mutagenesis in repetitive sequences. Proc Natl Acad Sci U S A. 1992;89:1310–4. doi: 10.1073/pnas.89.4.1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shibutani S, Suzuki N, Grollman AP. Mechanism of frameshift (deletion) generated by acetylaminofluorene-derived DNA adducts in vitro. Biochemistry. 2004;43:15929–35. doi: 10.1021/bi048087e. [DOI] [PubMed] [Google Scholar]
- 42.Stover JS, Chowdhury G, Zang H, Guengerich FP, Rizzo CJ. Translesion synthesis past the C8- and N2-deoxyguanosine adducts of the dietary mutagen 2-Amino-3-methylimidazo[4,5-f]quinoline in the NarI recognition sequence by prokaryotic DNA polymerases. Chem Res Toxicol. 2006;19:1506–17. doi: 10.1021/tx0601455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bacolod MD, Krishnasamy R, Basu AK. Mutagenicity of the 1-nitropyrene-DNA adduct N-(deoxyguanosin-8-yl)-1-aminopyrene in Escherichia coli located in a nonrepetitive CGC sequence. Chem Res Toxicol. 2000;13:523–8. doi: 10.1021/tx000023r. [DOI] [PubMed] [Google Scholar]
- 44.Garcia-Diaz M, Bebenek K, Krahn JM, Pedersen LC, Kunkel TA. Structural analysis of strand misalignment during DNA synthesis by a human DNA polymerase. Cell. 2006;124:331–42. doi: 10.1016/j.cell.2005.10.039. [DOI] [PubMed] [Google Scholar]
- 45.Wilson RC, Pata JD. Structural insights into the generation of single-base deletions by the Y family DNA polymerase dbh. Mol Cell. 2008;29:767–79. doi: 10.1016/j.molcel.2008.01.014. [DOI] [PubMed] [Google Scholar]
- 46.Bauer J, et al. A structural gap in Dpo4 supports mutagenic bypass of a major benzo[a]pyrene dG adduct in DNA through template misalignment. Proc Natl Acad Sci U S A. 2007;104:14905–10. doi: 10.1073/pnas.0700717104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang F, Yang W. Structural insight into translesion synthesis by DNA Pol II. Cell. 2009;139:1279–1289. doi: 10.1016/j.cell.2009.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ling H, Boudsocq F, Woodgate R, Yang W. Crystal structure of a Y-family DNA polymerase in action: a mechanism for error-prone and lesion-bypass replication. Cell. 2001;107:91–102. doi: 10.1016/s0092-8674(01)00515-3. [DOI] [PubMed] [Google Scholar]
- 49.Rechkoblit O, et al. Stepwise translocation of Dpo4 polymerase during error-free bypass of an oxoG lesion. PLoS Biol. 2006;4:e11. doi: 10.1371/journal.pbio.0040011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vaisman A, Ling H, Woodgate R, Yang W. Fidelity of Dpo4: effect of metal ions, nucleotide selection and pyrophosphorolysis. Embo J. 2005;24:2957–67. doi: 10.1038/sj.emboj.7600786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hunter WN, Brown T, Kennard O. Structural features and hydration of d(C-G-C-G-A-A-T-T-A-G-C-G); a double helix containing two G.A mispairs. J Biomol Struct Dyn. 1986;4:173–91. doi: 10.1080/07391102.1986.10506338. [DOI] [PubMed] [Google Scholar]
- 52.Colis LC, Raychaudhury P, Basu AK. Mutational specificity of gamma-radiation-induced guanine-thymine and thymine-guanine intrastrand cross-links in mammalian cells and translesion synthesis past the guanine-thymine lesion by human DNA polymerase eta. Biochemistry. 2008;47:8070–9. doi: 10.1021/bi800529f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hendel A, Ziv O, Gueranger Q, Geacintov N, Livneh Z. Reduced efficiency and increased mutagenicity of translesion DNA synthesis across a TT cyclobutane pyrimidine dimer, but not a TT 6-4 photoproduct, in human cells lacking DNA polymerase eta. DNA Repair (Amst) 2008;7:1636–46. doi: 10.1016/j.dnarep.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bailey EA, Iyer RS, Stone MP, Harris TM, Essigmann JM. Mutational properties of the primary aflatoxin B1-DNA adduct. Proc Natl Acad Sci U S A. 1996;93:1535–9. doi: 10.1073/pnas.93.4.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bishop RE, Pauly GT, Moschel RC. O6-ethylguanine and O6-benzylguanine incorporated site-specifically in codon 12 of the rat H-ras gene induce semi-targeted as well as targeted mutations in Rat4 cells. Carcinogenesis. 1996;17:849–56. doi: 10.1093/carcin/17.4.849. [DOI] [PubMed] [Google Scholar]
- 56.Jelinsky SA, Liu T, Geacintov NE, Loechler EL. The major, N2-Gua adduct of the (+)-anti-benzo[a]pyrene diol epoxide is capable of inducing G-->A and G-->C, in addition to G-->T, mutations. Biochemistry. 1995;34:13545–53. doi: 10.1021/bi00041a034. [DOI] [PubMed] [Google Scholar]
- 57.Xu P, Oum L, Lee YC, Geacintov NE, Broyde S. Visualizing sequence-governed nucleotide selectivities and mutagenic consequences through a replicative cycle: processing of a bulky carcinogen N(2)-dG lesion in a Y-family DNA polymerase. Biochemistry. 2009;48:4677–90. doi: 10.1021/bi802363f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bresson A, Fuchs RP. Lesion bypass in yeast cells: Pol eta participates in a multi-DNA polymerase process. EMBO J. 2002;21:3881–7. doi: 10.1093/emboj/cdf363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Eckel LM, Krugh TR. 2-Aminofluorene modified DNA duplex exists in two interchangeable conformations. Nat Struct Biol. 1994;1:89–94. doi: 10.1038/nsb0294-89. [DOI] [PubMed] [Google Scholar]
- 60.Nair DT, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Rev1 employs a novel mechanism of DNA synthesis using a protein template. Science. 2005;309:2219–22. doi: 10.1126/science.1116336. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


