Abstract
The alternative sigma factor σB of the food pathogen Bacillus cereus is activated upon stress exposure and plays a role in the adaptive response of vegetative cells. This study describes the identification of σB-dependent genes in B. cereus. Two-dimensional gel electrophoresis was performed with protein extracts from a σB-overproducing B. cereus strain. Nine protein spots, which were absent from the negative control, were identified by matrix-assisted laser desorption ionization-time of flight mass spectrometry or N-terminal sequencing. The σB-dependent expression of the corresponding genes was confirmed by Northern blot analysis with RNA isolated from B. cereus ATCC 14579 and its sigB null mutant. Northern blot analysis also revealed that six other genes were part of σB-dependent operons. The proteins that are predicted to be encoded by the σB-dependent genes include an intracellular protease, a Mg2+ transporter, and a thiamine biosynthesis protein (ThiG). Highly conserved promoter sites were found to precede all σB-dependent genes, with the exception of thiG. By searching the B. cereus genome for this conserved promoter sequence, five more candidate σB-dependent genes were identified. Northern blot analysis and in vitro transcription experiments with a reconstituted B. cereus σB-RNA polymerase holoenzyme confirmed the σB dependency of two of these genes and strongly suggested that two other genes, encoding an oligopeptide-binding OppA-like protein and subunit II of the cytochrome d ubiquinol oxidase, are also σB dependent. In conclusion, σB of B. cereus not only regulates genes directly involved in the stress response but may also control specific metabolic rearrangements.
Bacillus cereus is a common food-borne pathogen which can cause outbreaks of food-borne disease. Symptoms are generally mild and self-limiting, ranging from diarrhea to vomiting (17, 31). Occasionally, however, the symptoms of B. cereus food-borne disease can be life-threatening. For instance, in a B. cereus food-poisoning outbreak in France in 1998, one of the symptoms was bloody diarrhea, which resulted in the deaths of three persons (35). B. cereus is also an emerging pathogen in clinical settings, where it can cause severe infections, especially in immunocompromised patients (8, 14, 20).
B. cereus is closely related to Bacillus anthracis, the causative agent of anthrax, and Bacillus thuringiensis, which is widely used as a biological pest control agent. These organisms, together with Bacillus weihenstephanensis and Bacillus mycoides, form the B. cereus group (for a recent review, see reference 24). Whole-genome sequencing of B. cereus and B. anthracis showed a remarkably conserved core set of genes. The presence of virulence-associated plasmids and subtle chromosomal differences may explain the phenotypic differences between the different members of the B. cereus group (23, 47, 48).
For both B. cereus and B. anthracis, the alternative sigma factor σB has been studied in considerable detail. For B. anthracis, σB was shown to be upregulated upon heat shock and during the stationary growth phase. Furthermore, a sigB null mutant had attenuated virulence in a mouse model (13, 34). In B. cereus, heat shock also had a strong σB-activating effect, but other stresses (such as ethanol shock, osmotic shock, and acid stress) were also found to lead to the activation of σB. There was no detectable effect of the sigB deletion on the production of B. cereus virulence factors such as hemolysins, lecithinases, and the nonhemolytic enterotoxin Nhe, but σB was shown to play a role in the adaptive heat stress response of B. cereus (52).
The role of σB in the stress response of vegetative cells has been studied in the human pathogens Staphylococcus aureus and Listeria monocytogenes, but most extensively in Bacillus subtilis. A. B. subtilis sigB null mutant has an increased sensitivity to a wide variety of stresses, including acid, ethanol, heat, salt, and oxidative stress (9, 53). The set of σB-dependent genes (the σB regulon) has been identified by a number of techniques, including two-dimensional gel electrophoresis, σB-promoter consensus searching, and transcriptome profiling by DNA microarray analysis. This resulted in a set of approximately 200 σB-dependent genes. Relatively few of these genes seem to have a role in actively protecting the cell against environmental stress. The majority of σB-dependent genes code for proteins that seem to be involved in a metabolic rearrangement that can confer passive stress resistance (for a recent review, see reference 45). In the human pathogen S. aureus, σB plays a role in both stress resistance and the expression of virulence determinants (21). In a proteomics study, 23 σB-dependent genes were identified in S. aureus. Several of the encoded proteins were predicted to have a function in the generation of NADH or in membrane transport mechanisms (15). For the food-borne pathogen L. monocytogenes, σB has been shown to be involved in protection against osmotic stress and oxidative stress and in the acid tolerance response (2, 11, 12). The σB regulon of L. monocytogenes was recently identified by DNA microarray analysis. Several stress response genes and genes involved in virulence were thus identified as being σB dependent (29).
A comparison between the σB regulons of B. subtilis, S. aureus, and L. monocytogenes revealed a considerable overlap in the functions of the σB-dependent genes in these organisms. However, the divergence of the σB regulons suggests that the σB regulon has evolved to serve different roles among gram-positive bacteria (29). The natural niche of B. cereus has been proposed to be the nutrient-rich environment of the insect intestine (23, 24, 36), and as a consequence the σB regulon of B. cereus may have evolved to serve specific functions in this environment. Furthermore, the identification of the σB regulon of B. cereus may provide an explanation of the weakened heat stress response of the sigB null mutant of B. cereus (52) and give clues about further roles for σB in B. cereus.
In this paper, we describe the identification of a total of 15 σB-dependent proteins by a two-dimensional gel electrophoresis (2D-E) approach upon σB overproduction in B. cereus, followed by a Northern blot analysis. By performing a σB promoter consensus search of the B. cereus genome, we identified five more candidate σB-dependent genes. Northern blot analysis and in vitro transcription experiments with a reconstituted B. cereus σB-RNA polymerase (RNAP) holoenzyme confirmed the σB dependency of two of these genes and strongly suggested that two other genes are also σB dependent. Several of the identified σB-dependent proteins do not have a clearly defined function, but others may have roles in the turnover of misfolded proteins or in influencing metabolic fluxes through the cell.
MATERIALS AND METHODS
Bacterial strains, culture media, growth conditions, and genetic methods.
B. cereus ATCC 14579 and its sigB null mutant FM1400 (52) were cultured in brain heart infusion (BHI) medium at 30°C, with aeration at 200 rpm. The growth of the culture was monitored by measurement of the optical density at 600 nm (OD600). Escherichia coli MC1061 (7) was used as a host for the vectors of the nisin inducible controlled expression (NICE) system and was cultured in Luria broth at 37°C with aeration at 200 rpm (49). The antibiotics used were chloramphenicol at 10 μg/ml and erythromycin at 150 μg/ml (for E. coli) or 5 μg/ml (for B. cereus).
Plasmid DNAs were purified from E. coli with a Qiaprep Spin Miniprep kit (Westburg, Leusden, The Netherlands). B. cereus was transformed with plasmid DNA by electroporation, as described previously for B. thuringiensis (3). For the purification of plasmids from B. cereus, 5 ml of a culture in the mid-exponential growth phase was spun down, resuspended in 250 μl of THMS (30 mM Tris-HCl [pH 8.0], 3 mM MgCl2, 25% sucrose) plus 2 mg of lysozyme/ml, and incubated for 1 h at 37°C before proceeding with the standard plasmid purification protocol. Pwo polymerase (Roche Diagnostics, Almere, The Netherlands) was used for all PCRs in this study. Radiochemicals were obtained from Hartmann Analytic GmbH, Braunschweig, Germany. Other genetic methods have been described previously (52).
Inducible overproduction of σB in B. cereus.
sigB was amplified by a PCR employing the primers OBcSigBF (GCAGCCATGGTGGAAATCCAATCTCAACCT) and OBcSigBR (GCAGCTGCAGTGTATCTAAAAATGCGGCTTG), which introduced an NcoI and a PstI site (underlined), respectively. The PCR product was cloned into pNZ8048 (32), and the resulting vector, pFM100T, was transformed into E. coli MC1061. From this strain, the plasmid DNA was purified, and after sequencing to check for the absence of mutations in the insert, the vector was transformed into B. cereus ATCC 14579, which already carried pNZ9520 (30). The overproduction of σB was induced by the addition of nisin to a mid-exponential-phase culture (OD600 = 0.4 to 0.5) to a final concentration of 10 ng/ml. The cells were then cultured for a further 90 min before being harvested.
Total RNA isolation and Northern blotting techniques.
RNA was isolated from B. cereus by the use of RNAwiz (Ambion, Huntingdon, United Kingdom). After precipitation of the nucleic acid, residual DNA was removed with 20 U of RNase-free DNase I (Ambion). After phenol-chloroform extraction and precipitation, the RNA was quantified by measuring the OD260. All RNA samples had an OD260/OD280 ratio of ≥1.9.
Five micrograms of total RNA was separated in a 1.2% agarose-0.66 M formaldehyde-morpholinepropanesulfonic acid (MOPS) gel which was run at a 40-V constant voltage and blotted onto a Zeta-Probe membrane (Bio-Rad, Richmond, Calif.). Internal PCR fragments of open reading frames were used as probes. The PCR fragments were radiolabeled with [α-32P]dATP by nick translation. After hybridization with ULTRAhyb hybridization buffer (Ambion) and stringent washing according to the manufacturer's instructions, the blots were exposed to a phosphorimager screen. After an exposure time of 16 to 24 h, the screen was scanned on a Storm 840 system (Amersham Biosciences, Roosendaal, The Netherlands). ImageQuant TL software (Amersham Biosciences) was used for image analysis. A 0.24- to 9.5-kb RNA ladder (Invitrogen, Breda, The Netherlands) was used to determine the transcript sizes.
Protein extraction, Western blotting techniques, and 2D-E.
Protein extraction and Western blotting with an anti-σB antiserum were performed as described previously (52). 2D-E was performed as described previously (43). In brief, equal amounts of protein (40 μg for analytical gels and 800 μg for preparative gels) were first separated in 11-cm-wide Immobiline DryStrip gels (Amersham Biosciences) at pHs 4 to 7 and subsequently separated in ExcelGel precast sodium dodecyl sulfate (SDS)-12 to 14% polyacrylamide gradient gels (Amersham Biosciences). The Precision Plus protein standard (Bio-Rad) was used as a molecular weight standard. The gels were silver stained and analyzed with PD-Quest software (version 7.1; Bio-Rad). Experiments were performed at least in triplicate, and representative gels are shown in the figures.
Identification of protein spots by MALDI-TOF and N-terminal sequencing.
Matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) analysis of manually excised spots was performed at the Maastricht Proteomics Center (Department of Human Biology, University of Maastricht, Maastricht, The Netherlands) by trypsin digestion in a MassPrep station (Micromass, Almere, The Netherlands) and subsequent analysis with a MALDI-TOF LR mass spectrometer (Micromass). Peptide mass fingerprints were analyzed with Mascot software (Matrix Science Ltd., London, United Kingdom).
For determination of the N-terminal sequences of specific spots, gels were blotted onto Hybond-P polyvinylidene difluoride membranes (Amersham Biosciences) in a Trans-Blot unit (Bio-Rad) with 10 mM CAPS buffer (pH 11.0) plus 15% methanol at 50 V for 30 min and were then stained with Coomassie blue. Protein spots were cut from the blot and analyzed by consecutive Edman degradation with the model 476A protein sequencing system (Applied Biosystems) at the Sequence Center, University Utrecht (Utrecht, The Netherlands).
Purification of B. cereus RNAP and in vitro transcription techniques.
B. cereus cells (20 g of wet weight) from a culture grown in BHI medium at 30°C to an OD600 of 1 were homogenized in 25 ml of lysis buffer (0.05 M Tris-HCl [pH 8.0], 5% glycerol, 2 mM EDTA, 0.1 mM dithiothreitol, 1 mM β-mercaptoethanol, 0.23 M NaCl, and 23 μg of phenylmethylsulfonyl fluoride/ml) and lysed by two passages through a French press at 10,000 lb/in2. Subsequently, RNAP was purified by following established protocols for the purification of E. coli RNAP (5, 16), using Polymin-P fractionation, heparin-Sepharose affinity chromatography, A5M gel filtration, and phosphocellulose chromatography. The RNA core and holoenzyme were eluted from the phosphocellulose column with P50 buffer (40 mM potassium phosphate buffer [pH 8.0], 1 mM dithiothreitol, 0.1 mM EDTA, 50% glycerol) supplemented with 0.2 and 0.5 M KCl, respectively. Aliquots from eluted fractions were analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) in a 15% polyacrylamide gel followed by silver staining. The peqGOLD protein marker (Peqlab Biotechnologie GmbH, Erlangen, Germany) was used as a molecular weight standard.
For in vitro transcription experiments, a reaction mixture containing 600 nM σB (52), 30 nM B. cereus core RNAP, and a 30 nM PCR-generated template DNA in transcription buffer (20 mM Tris-HCl [pH 8.0], 50 mM K-glutamate, 10 mM MgCl2, 0.5 mM dithiothreitol, 20 μM EDTA, 5% glycerol) in a final volume of 10 μl was incubated on ice for 30 min. In control experiments, σB was replaced with an equal volume of σB dialysis buffer (50 mM sodium phosphate buffer [pH 7.8], 0.3 M NaCl, 50% glycerol). The mixture was subsequently incubated at 30°C for 5 min, followed by the addition of 10 μl of 1 mM (each) ATP, CTP, GTP, and [α-32P]UTP (3,000 Ci/mmol) in transcription buffer supplemented with 0.1 M NaCl. After a 30-min incubation at 30°C, 5 μl of chase mix (10 mM [each] ATP, CTP, GTP, and UTP plus 400 μg of heparin/ml) was added, and the mix was incubated for a further 10 min to finish the transcription reaction and to prevent reinitiation. The transcription reaction was stopped by the addition of 25 μl of formamide loading buffer (95% formamide, 18 mM EDTA, 0.025% SDS, xylene cyanol, and bromophenol blue). After denaturation of the samples by heating at 95°C for 5 min, 5-μl aliquots were analyzed on a 7 M urea-8% polyacrylamide gel which was run at 90 W. The transcripts were visualized by autoradiography using phosphorimager screens and a Storm 840 system. Size estimates of the runoff transcription products were made by using a 32P-labeled low-range RNA ladder (MBI Fermentas GmbH, St. Leon-Rot, Germany).
RESULTS
Inducible overproduction of σB in B. cereus.
The σB-regulated genes of B. cereus were initially identified by a proteomics-based approach. First, protein profiles of the sigB null mutant and its parent during exponential growth at 30°C and upon heat shock to 42°C for 30 min were determined (data not shown). Only two σB-dependent protein spots, corresponding to the previously identified stress-induced proteins YflT and RsbV (43), could be identified. To get a more complete overview of the σB regulon, the protein profile of a B. cereus strain in which overproduction of σB could be induced was determined by 2D-E. To confirm the σB-dependent expression of the genes corresponding to the proteins that were identified by 2D-E, we performed a Northern blot analysis, using RNA samples from B. cereus ATCC 14579 and the sigB null mutant FM1400.
The NICE system was used to obtain inducible overproduction of σB in B. cereus. The NICE system consists of two vectors, one of which (pNZ9520) contains the genes for NisR and NisK. NisK senses the presence of nisin in the medium and phosphorylates NisR, which in its turn activates the nisA promoter on the second vector (pNZ8048) (32). The sigB gene was cloned downstream of the nisA promoter on pNZ8048, resulting in pFM100T. The addition of subinhibitory amounts of nisin to a culture of B. cereus carrying pNZ9520 and pFM100T led to the overproduction of σB. This was assayed by Western blotting with an anti-σB antiserum. In cultures harboring pNZ9520 and the empty vector pNZ8048, σB was present at low levels. In noninduced cultures of B. cereus carrying pNZ9520 and pFM100T, elevated σB levels were present, which increased further when nisin was added to the culture to activate the overproduction system (Fig. 1A).
FIG. 1.
Inducible overproduction of σB in B. cereus. (A) Intracellular σB levels in B. cereus during inducible overproduction of σB. Immunoblotting with an anti-σB antiserum was performed on protein extracts from B. cereus harboring pNZ9520 and pNZ8048 or pNZ9520 and pFM100T. Proteins were extracted 90 min after the addition of nisin (final concentration, 10 ng/ml) to a mid-exponential-phase culture. (B) Northern blot analysis of inducible σB overproduction. The total RNA was isolated from B. cereus harboring pNZ9520 and pFM100T at mid-exponential phase (t = 0) and 5, 15, 30, 60, and 90 min after the addition of 10 ng of nisin/ml. The blot was probed with a 32P-labeled internal PCR product of sigB (left panel). After visualization of the hybridized probe, the blot was stripped and probed with a 32P-labeled internal PCR product of orf4 (right panel). The sizes of the transcripts (in kilobases) are indicated.
To analyze the kinetics of σB overproduction and the biological activity of the overproduced σB, we isolated total RNA at different time points during the induction of the σB overproduction system and performed Northern blotting with these RNA samples. The blots were probed with a sigB- and orf4-specific probe (Fig. 1B) and showed the transcription of the sigB overexpression vector and the sigB operon during σB overproduction and the activation of the σB-dependent promoter upstream of orf4 (52). For the RNA isolated from a B. cereus strain carrying pNZ9520 and the empty vector pNZ8048, barely detectable levels of sigB and orf4 expression were observed under the conditions tested (data not shown), corresponding with the Western blot data.
In the blot probed with the sigB-specific probe, a weak signal at 1.0 kb, corresponding to the mRNA of the sigB overexpression plasmid pFM100T, could be visualized before the addition of nisin to the culture. This indicates that the nisA promoter is somewhat leaky, which explains why the 2.1-kb transcript that was observed in both blots and which corresponds to the chromosomal rsbV-rsbW-sigB-orf4 mRNA is already present before the induction of the NICE system. Even though the NICE system is not particularly leaky, even a low-level production of σB is presumably enough to switch on transcription of the sigB operon, because it is autoregulated by σB (52). However, after the induction of σB overproduction by the addition of nisin to the culture, the σB levels were highly increased (Fig. 1A), and a Northern blot analysis of the σB-dependent 0.5-kb orf4 transcript under these conditions showed that the overproduced σB protein was biologically active. The transcript levels of the orf4 mRNA increased steadily over time, presumably because some time is needed for the production of functional σB protein after the induction of σB overproduction.
These data show that chromosomal σB-dependent promoters of B. cereus are activated to high levels upon σB overproduction. Subsequently, 2D-E was used to map the protein profiles of B. cereus during σB overproduction.
2D-E of proteins extracted from B. cereus upon σB overproduction.
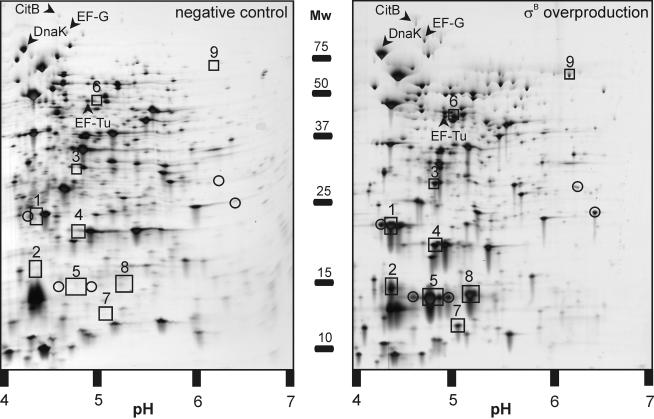
Protein profiling was performed on protein extracts of B. cereus cells carrying pNZ9520 and pNZ8048 (the negative control) or pNZ9520 and pFM100T (the σB-overproducing strain) that were isolated 90 min after the addition of 10 ng of nisin/ml to a mid-exponential-phase culture. Clear differences between the protein profiles could be observed (Fig. 2). Fourteen spots for the protein sample isolated upon σB overproduction could not be matched with spots in the control gel. These proteins were prime candidates for being σB dependent. Nine of these 14 spots could be identified by MALDI-TOF analysis, or if no satisfactory peptide mass fingerprint was obtained, by N-terminal sequencing (Table 1). A subsequent Northern blot analysis showed that all nine of these proteins are indeed σB dependent (see below).
FIG. 2.
2D-E of extracts of B. cereus cells carrying pNZ9520 and pNZ8048 (left panel) or pNZ9520 and pFM100T (right panel). Proteins were extracted 90 min after the addition of nisin (final concentration, 10 ng/ml) to a mid-exponential-phase culture. The molecular masses (in kilodaltons) of the markers and the pI range are indicated. Proteins that were upregulated, but were not σB dependent, as shown by a subsequent Northern blot analysis, are indicated by arrows and their respective identifications. Unidentified proteins are circled. Identified proteins in the σB overproduction sample which could not be matched with a protein in the control sample and which were confirmed to be σB dependent by a subsequent Northern blot analysis are boxed and numbered, corresponding to the data in Table 1.
TABLE 1.
Identification of σB-dependent proteins of B. cereus after 2D-E analysis
| Spot no. | N-terminal sequence or MALDI-TOF resulta | Identified proteinb | Predicted function | Molecular mass (kDa)/pI | Predicted localizationc |
|---|---|---|---|---|---|
| 1 | 44 (6) | YfkM (RZC01423) | Intracellular protease I | 19/4.46 | C |
| 2 | MSHDVKEL | Orf4 (RZC05127) | Putative bacterioferritin | 17/4.53 | C |
| 3 | 38 (7) | YflT (RZC05134) | General stress protein | 15/4.69 | C |
| 4 | 68 (9) | RZC04880 | Unknown | 18/4.85 | M |
| 5 | 65 (12) | RZC04727 | Unknown | 39/4.91 | M |
| 6 | MNLAINIL | RsbV (RZC05131) | Anti-σB antagonist | 13/5.17 | C |
| 7 | 67 (10) | RZC04730 | General stress protein | 15/5.04 | C |
| 8 | 113 (18) | KatE (RZC01424) | Catalase | 75/6.62 | C |
| 9 | MLNIGPF | ThiG (RZC02927) | Thiazole biosynthesis protein | 27/4.76 | C |
For proteins identified by N-terminal sequencing, the derived sequence is shown. For proteins identified by MALDI-TOF analysis, the probability based MOWSE score and the number of peptides matched (in parentheses) are indicated. A score of >63 is considered significant. Identifications of proteins with scores of <63 were confirmed by matching the predicted molecular mass and pI with the position of the spot on the 2D-E gel.
Protein designations were based on homologous proteins from other bacteria. The B. cereus genome sequence codes are also specified (in parentheses).
The subcellular localization of the proteins was predicted with the PSORT server (http://psort.nibb.ac.jp). C, predicted cytoplasmic protein; M, predicted membrane protein.
Four proteins that were moderately (two- to threefold) upregulated were also identified (Fig. 2, arrows). These proteins, the chaperone DnaK, the protein elongation factors EF-Tu and EF-G, and the aconitase CitB, were shown not to be σB dependent by subsequent Northern blotting (data not shown), and their upregulation may be explained by the stress caused by the artificial overproduction of σB. Their upregulation reflects findings for E. coli, in which the expression of many genes, including genes for central metabolism and heat shock response genes, is upregulated upon protein overproduction (39).
Identification of σB-dependent proteins in B. cereus and their possible functions in the B. cereus stress response.
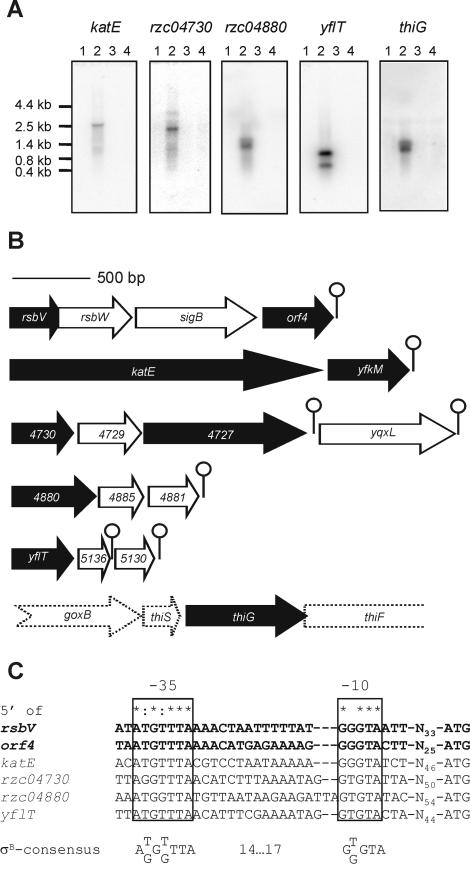
The upregulation upon σB overproduction of the proteins listed in Table 1 strongly suggested that they are members of the σB regulon of B. cereus. This was confirmed by Northern blotting experiments in which blots containing total RNA from cultures of the B. cereus sigB deletion mutant and its parent strain, isolated during the mid-exponential growth phase and after a 10-min exposure to 42°C, were hybridized with probes specific for the structural genes of all nine identified proteins (Fig. 3A). The genes orf4 and rsbV were previously shown to be σB dependent (52) and were not included in this experiment. Northern blot analysis with probes corresponding to yfkM and rzc04727 gave results that were identical to those of Northern blot analysis with probes corresponding to katE and rzc04730, respectively (data not shown). This was expected, because these open reading frames are in the same operons. In all cases, transcripts were only visualized in RNA samples isolated from B. cereus ATCC 14579 upon a heat shock from 30 to 42°C, not in RNA samples from the sigB null mutant, indicating that all of the examined genes are σB dependent. Northern blotting also revealed the operon structure of the different σB-dependent genes and resulted in the identification of six additional σB-dependent genes in B. cereus (Fig. 3B and Table 2). The operons were further examined for the presence of predicted stem-loop structures, which can function as terminators, and for the presence of predicted σB-dependent promoters upstream of the operon (Fig. 3C). These promoters were subsequently experimentally proven to be σB dependent by in vitro transcription experiments (see below).
FIG. 3.
Transcriptional analysis of σB-dependent genes in B. cereus. (A) Northern blot analysis of transcription of σB-dependent genes in B. cereus. Total RNAs were extracted from B. cereus ATCC 14579 and B. cereus FM1400 cells during mid-exponential growth in BHI medium (lanes 1 and 3, respectively) and after a 10-min exposure to 42°C (lanes 2 and 4, respectively). 32P-labeled internal PCR products of the different genes were used as probes. Northern blot analyses with probes corresponding to yfkM and rzc04727 gave identical results as the Northern blot analyses with probes corresponding to katE and rzc04730, respectively (data not shown). Marker sizes (in kilobases) are indicated. (B) Operon structure of σB-dependent genes in B. cereus. The arrows represent open reading frames and indicate their orientations and sizes. Black arrows correspond to genes that were identified on the basis of 2D-E experiments. White arrows denote genes that are cotranscribed. Predicted stem-loop structures are indicated as lollipop structures. The three-letter codes of the genes, or when no such code exists, the B. cereus genome sequence code is indicated. The common part (RZC0) of the B. cereus genome sequence designations was omitted because of lack of space. The dashed arrows of the genes surrounding thiG indicate that it is not clear if these genes are cotranscribed with thiG. (C) Alignment of predicted σB-dependent promoter sequences. The −35 and −10 regions are indicated. The spacing to the start codon is also specified. The promoters 5′ of rsbV and orf4 are shown in bold to indicate that these have been experimentally defined (52). Completely conserved residues are indicated with asterisks. Residues that are conserved in five of the six promoter sites are indicated with colons. The extracted σB promoter consensus sequence is also indicated.
TABLE 2.
New σB-dependent B. cereus genes that are part of σB-dependent operons
| Cotranscribed genea | Predicted protein function | Molecular mass (kDa)/pI | Predicted localizationb |
|---|---|---|---|
| rzc04729 | Unknown | 15/10.14 | M |
| yqxL (rzc01861) | CorA-like Mg2+ and Co2+ transporter protein | 37/8.69 | M |
| rzc04885 | Unknown | 11/9.89 | C |
| rzc04881 | Unknown | 11/4.05 | C |
| ywmG (rzc05136) | Unknown | 7/10.17 | C |
| rzc05130 | Conserved membrane protein | 9/9.67 | M |
Gene designations were based on homologous genes in other bacteria. The B. cereus genome sequence codes are also specified (in parentheses).
The subcellular localization of the proteins was predicted with the PSORT server (http://psort.nibb.ac.jp). C, predicted cytoplasmic protein; M, predicted membrane protein.
The functions of the 15 σB-dependent proteins which were identified by 2D-E analysis upon σB overproduction are summarized in Tables 1 and 2. Eight of the σB-dependent proteins do not have a clearly defined function in B. cereus, and their role in the physiology, and more specifically, the stress response of B. cereus remains unclear. One of these is YflT, which was previously found to be a heat-shock-inducible protein of B. cereus (43). The B. subtilis homologue of YflT is also heat inducible and σB dependent and responds strongly to σB activation (19, 46), suggesting that this protein has a significant role in the σB-regulated component of the stress response in bacilli. RZC04881, whose structural gene is cotranscribed with rzc04880, is highly homologous (90% amino acid identity) with the protein encoded by the open reading frame pX02-45 from the capsule plasmid of B. anthracis, reflecting the previously reported spread of B. anthracis virulence plasmid genes throughout the B. cereus group (40, 41). Finally, RZC04727 is a protein that is unique to the B. cereus group. Its C-terminal part, however, has a low-level homology (23% amino acid identity) with a predicted ATPase of the HSP70 class in Clostridium acetobutylicum, suggesting that RZC04727 may function as a protein with chaperone activity in B. cereus.
The σB-dependent proteins that have predicted functions in B. cereus may serve a variety of roles in the stress response. YfkM is annotated as an intracellular protease and belongs to the Pfam (1) DJ-1/PfpI family. Proteins from this family are widespread throughout all kingdoms of life and have a wide range of functions, but most importantly they possess chaperone and proteolytic activities (33). YfkM may therefore also function as a chaperone during heat stress in B. cereus and may contribute to the correct folding of proteins or the breakdown of misfolded proteins under this condition. The catalase KatE is one of three predicted catalases in the B. cereus genome sequence. Its homologue in B. subtilis is also σB dependent, but its role in stress resistance is thought to be insignificant, as the deletion of katE had no detectable effect on the resistance of B. subtilis against oxidative stress (9). yqxL is transcribed from the promoter 5′ of rzc04730. Although there is a stem-loop structure present downstream of rzc04727, this structure is not strong enough to completely stop transcription, as shown by the weak readthrough transcript at approximately 3.2 kb visualized on the Northern blot that was probed with a rzc04730-specific DNA (Fig. 3A). A subsequent Northern blot analysis of the transcription of yqxL showed that its expression is completely σB dependent (data not shown). YqxL is one of three predicted CorA-type transporters in B. cereus. CorA-type transporters have been suggested to be the major constitutive Mg2+ uptake system of both the Bacteria and the Archaea (37, 50). These findings suggest that the σB-dependent activation of YqxL influences the flux of Mg2+ ions over the cytoplasmic membrane during stress.
The mechanism of the σB-dependent transcriptional activation of thiG is not immediately apparent. We were unable to locate a candidate σB-dependent promoter or a clear downstream stem-loop structure which could match the approximately 1.3-kb transcript visualized on the Northern blot, and consequently we could not identify the genes that are cotranscribed with thiG with any certainty. Possibly a promoter is involved which is different from the other proposed σB-dependent promoters (Fig. 3C), or the expression of thiG may be indirectly regulated by σB. On the basis of its homology (80% amino acid identity) with ThiG of B. subtilis, we can assume that ThiG catalyzes the formation of the phosphate ring in the thiazole moiety of thiamine, which is one of the last steps in thiamine biosynthesis (42). Thiamine pyrophosphate (or vitamin B1) is an essential cofactor for several enzymes in carbohydrate metabolism, and its mechanistic role is to stabilize the acyl carbanion (25). In B. cereus, the biosynthesis of thiamine is not always needed, because it can take up thiamine from the medium (51). This explains why thiG is not expressed during exponential growth in rich BHI broth (Fig. 3A). The stress-induced upregulation of thiG may be explained by a disturbance of the thiamine uptake system under stress conditions, after which the biosynthesis of thiamine is required for further cellular growth.
Identification of candidate σB-dependent promoters and promoter consensus search.
In Fig. 3C, an alignment of the candidate σB-dependent promoters that could be identified upstream of the set of σB-dependent genes in B. cereus (with the exception of thiG) is shown. These sites were identified because they are practically identical to the experimentally defined σB-dependent promoters 5′ of rsbV and orf4 (52). The alignment of these promoters revealed a preliminary σB promoter consensus sequence with a −35 region sequence of AKGKTTA (K = T or G) and a −10 region sequence of GKGTA, with a spacing of 14 to 17 nucleotides. σB promoter consensus sequences have also been determined for B. subtilis and L. monocytogenes. For B. subtilis, the σB promoter consensus sequences for the −35 and −10 regions were defined as rGGwTTrA and GGgtAt, respectively (capital letters indicate highly conserved residues and lowercase letters indicate less conserved residues [R = A or G and W = A or T]), with a spacing of 12 to 15 nucleotides (18). In L. monocytogenes, the σB-dependent promoter consensus is GTTT for the −35 region and GGGWAT for the −10 region, with a spacing of 13 to 17 nucleotides (29). This indicates that there may be some differences, both in the sequences of the −35 and −10 regions and in the spacing between these regions, in the promoter sequences that are recognized by σB in B. cereus, B. subtilis, and L. monocytogenes. However, more σB-dependent promoters in B. cereus should be identified before definitive conclusions can be drawn.
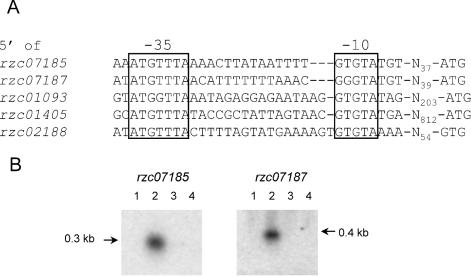
The B. cereus σB promoter consensus, extracted from the alignment of the predicted promoter sites upstream of the four σB-dependent operons identified in this study and of the sigB operon, was used to further search the B. cereus genome sequence to identify other candidate σB-dependent genes. Five hits were found in an intergenic region with an open reading frame within 1 kb of the predicted promoter site (Fig. 4A). Northern blot analysis was subsequently performed to study if these open reading frames were transcribed in a σB-dependent fashion.
FIG. 4.
Identification of σB-dependent genes in B. cereus by σB-dependent promoter consensus search. (A) Alignment of predicted σB-dependent promoter sequences, identified by a σB-dependent promoter consensus search. The −35 and −10 regions are indicated. The spacing to the start codon is also specified. (B) Northern blot analysis of transcription of rzc07185 and rzc07187. Total RNA was extracted from B. cereus ATCC 14579 and B. cereus FM1400 cells during mid-exponential growth in BHI medium (lanes 1 and 3, respectively) and after a 10-min exposure to 42°C (lanes 2 and 4, respectively). 32P-labeled internal PCR products of rzc07185 and rzc07187 were used as probes. The transcript sizes (in kilobases) are indicated.
Of these five hits, rzc07185 and rzc07187 were found to be dependent on σB for their expression (Fig. 4B). Both of these genes code for small (7.6 and 9.5 kDa, respectively, with pIs of 7.68 and 10.47) proteins that are monocistronically transcribed. RZC07185 and RZC07187 are predicted to be a membrane-associated and a cytoplasmic protein, respectively. Northern blot analysis of the other predicted σB-dependent genes (rzc01093, encoding one of 15 oligopeptide-binding OppA proteins of B. cereus; rzc01405, encoding a sporulation kinase; and rzc02188, encoding subunit II of the cytochrome d ubiquinol oxidase) showed weak transcripts under all conditions tested (data not shown). Therefore, these genes are expressed at low levels in a σB-independent fashion. However, a subsequent in vitro transcription analysis showed that the promoter sites upstream of rzc01093 and rzc02188 can be recognized by σB-RNAP (see below), indicating that the σB-dependent transcription of these genes may be relevant under conditions other than the ones used for the isolation of RNAs for Northern blot analysis in this study.
rzc07185 and rzc07187 lie in an approximately 10-kb region of the B. cereus genome where a number of σB-regulated genes are clustered. This region starts with yflT (at position 982113), ends with rzc04881 (at position 992843), and encompasses a total of 15 open reading frames, including the sigB operon, of which 12 have been experimentally proven to be σB dependent. A clustering of σB-dependent transcriptional units has also been reported for B. subtilis (19), but the significance of this observation in both B. cereus and B. subtilis is still unclear.
Determination of σB dependency of promoters by in vitro transcription with reconstituted B. cereus σB-RNAP.
There may be several reasons for the apparent inactivity of the promoters upstream of rzc01093, rzc01405, and rzc02188. Because their −35 and −10 sequences fit the σB promoter consensus that was defined previously, a σB-dependent effect on transcription was expected. However, there may be additional control mechanisms which cause the apparently σB-independent transcription under the conditions that we tested. Additional proof for the σB dependency of these upstream promoter sites was obtained by in vitro transcription experiments with a reconstituted σB-RNAP holoenzyme. Furthermore, we obtained biochemical evidence of the σB dependence of the candidate σB-dependent promoters that were defined previously (Fig. 3C). A similar methodology has also been used for B. subtilis, e.g., for the determination of σW-dependent promoter sites (6, 22). From 20 g of wet B. cereus cells, we were able to purify approximately 5.2 mg of RNAP. In the final phosphocellulose step, RNAP was eluted with buffers containing two concentrations (0.2 and 0.5 M) of KCl. This allowed us to separate fractions corresponding to approximately 3 mg of core RNAP (which eluted at 0.2 M KCl) and 2.2 mg of holoenzyme (which eluted at 0.5 M KCl). This distinction between the core and holoenzyme forms was first based on data resulting from an SDS-PAGE analysis of the purified fractions (Fig. 5). In the fractions eluted with 0.2 M KCl, we could not visualize a protein corresponding to the size of σA, whereas such a protein was seen in the fractions that were eluted with 0.5 M KCl. This was confirmed when an aliquot of the first fraction was used for in vitro experiments, in which an extraneously added sigma factor was needed to start transcription (Fig. 6), while an aliquot from the latter fraction could start transcription by itself (data not shown). In all fractions, proteins with sizes corresponding to the α, β, and β′ subunits of RNAP were present. The 21-kDa δ subunit of RNAP was not seen on SDS-PAGE gels and was thus presumably lost during our purification protocol. For B. subtilis, it was observed that the presence of this subunit in multiple-round in vitro transcription reactions can increase the amount of RNA synthesized but that it is not essential for the transcription process (27). A weak band corresponding to the size of the ω subunit was visible in both the core and holoenzyme preparations (data not shown).
FIG. 5.
Purification of B. cereus RNAP. An SDS-PAGE analysis of 10-μl aliquots of fractions after phosphocellulose chromatography with P50 buffer containing 0.2 M KCl (core RNAP) and 0.5 M KCl (holo-RNAP) is shown. The proteins were visualized by silver staining. Purified E. coli RNAP holoenzyme (Ec RNAP) was also loaded for comparison. The predicted sizes of the β, β′, α, and σA subunits of the B. cereus RNAP are indicated. The fraction used as the core RNAP in subsequent in vitro transcription experiments is indicated with an arrow. The sizes of the molecular weight markers (in kilodaltons) are specified.
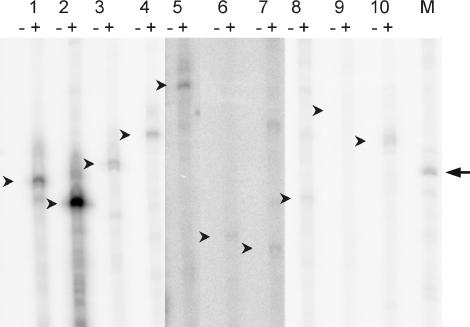
FIG. 6.
In vitro transcription of predicted σB-dependent promoters. PCR products containing the promoter regions of rsbV (lane 1), orf4 (lane 2), katE (lane 3), rzc04730 (lane 4), yflT (lane 5), rzc07185 (lane 6), rzc07187 (lane 7), rzc01093 (lane 8), rzc01405 (lane 9), and rzc02188 (lane 10) were incubated with B. cereus RNAP in the absence (−) or presence (+) of σB. After electrophoresis, runoff transcription products were visualized by exposure to a phosphorimager screen and scanning with a Storm scanner. The expected sizes of the σB-dependent transcription products are indicated with small arrows. The band in the marker lane (M), indicated with an arrow, corresponds to an RNA size of 200 nucleotides. All data are from a single, representative experiment. The signals for samples 5, 6, and 7 were weaker than the other samples and were enhanced with ImageQuant TL software for optimal clarity.
For in vitro transcription experiments, the different proposed σB-dependent promoter regions were amplified by PCR. The primers were chosen in such a way that the σB-dependent transcripts were sized between 150 and 240 nucleotides. For in vitro transcription reactions with these templates, no transcripts were observed when σB was not added (Fig. 6), showing that the purified RNAP fraction used behaves as a core enzyme. When σB was included in the reaction mixture, resulting in a reconstituted σB-RNAP holoenzyme, transcripts with the expected sizes could be visualized for the promoter sites upstream of rsbV and orf4 (Fig. 6, samples 1 and 2), which were previously experimentally determined to be σB dependent by primer extension analysis (52). When templates with the promoter regions upstream of the σB-dependent genes katE, rzc04730, yflT, rzc07185, and rzc07187 were included in the in vitro transcription reaction mix, transcripts with sizes that matched those expected for the predicted σB-dependent promoter sites were also formed (Fig. 6, samples 3 to 7). A template with the candidate σB-dependent promoter site upstream of rzc04880 was also used in this experiment, but for this sample the result was that many strong nonspecific transcripts were formed in the reaction (data not shown).
Interestingly, we could identify σB-dependent transcription of the promoter sites upstream of rzc01093 and rzc02188, but not of that upstream of rzc01405. These sites were predicted to be σB dependent in our promoter consensus search, but this could not be confirmed by Northern blot analysis. These data suggest that σB may play a role in the transcription of both rzc01093 and rzc02188 under different growth conditions than the ones used for this study.
DISCUSSION
For this study, we used two different approaches, 2D-E upon σB overproduction and in vitro transcription with a reconstituted σB-RNAP holoenzyme, to identify σB-dependent genes in B. cereus. We have introduced an inducible σB overproduction system in B. cereus, and have shown that upon induction of this system, a strong σB response was triggered, resulting in the de novo production of 14 proteins, as visualized by 2D-E. Nine of these 14 proteins were identified by MALDI-TOF or N-terminal sequencing. The transcription of the structural genes coding for these nine proteins was confirmed to be σB dependent by subsequent Northern blot analyses.
Some of the σB-dependent proteins are predicted to have a role in degrading incorrectly folded proteins (YfkM, and possibly, RZC04727). These proteins may thus have a role in protecting vegetative cells of B. cereus against high temperatures. Their σB dependency may explain the attenuated heat shock response of the sigB null mutant of B. cereus. Other proteins do not have such obvious roles in the stress response, but they may subtly tweak the cellular metabolism, which could lead to an increased passive stress resistance. An example of this metabolic rerouting is most apparent in the case of the σB-dependent upregulation of ThiG, which may serve to increase the thiamine pool in B. cereus. The fact that only ThiG and not the whole thiamine biosynthesis pathway is upregulated may be explained by the finding that for Paenibacillus alvei (originally named Bacillus paraalvei), which is closely related to the B. cereus group (26), a lack of thiamine can be overcome by the addition of specific amino acids (phenylalanine, alanine, valine, isoleucine, and cysteine) to the medium (28). Indeed, in B. subtilis, ThiG alone can catalyze the formation of the thiazole moiety of thiamine, thereby bypassing the rest of the thiamine biosynthesis pathway (42). The σB-dependent upregulation of the expression of thiG may be explained by a metabolic rearrangement which shuttles amino acids into the thiamine biosynthesis pathway. By following this metabolic route, only ThiG, and not the entire thiamine biosynthesis pathway, would be needed for the generation of the thiazole moiety.
All of the σB-dependent genes that were identified on the basis of 2D-E experiments, with the exception of thiG, were preceded by a highly conserved promoter motif, which fits the experimentally determined σB-dependent promoter sequence in the sigB operon (52). An alignment of these promoter sites resulted in a σB promoter consensus sequence, and subsequently the B. cereus genome was searched with this sequence. Of the five genes that were directly preceded by a promoter site that matched the σB promoter consensus sequence, two (rzc07185 and rzc07187) were found to be σB dependent by Northern blot analysis. The fact that we did not detect their corresponding proteins in the 2D-E experiments can be explained by their small sizes and their pIs, which lie outside the range tested in our experiments. The three other open reading frames, even though they were preceded by a conserved σB-dependent promoter, were not dependent on σB for their expression under the tested conditions.
By performing in vitro transcription experiments with a reconstituted B. cereus σB-RNAP holoenzyme, we were able to identify the σB dependency of genes in a system that was independent from the growth phase or other regulatory factors. Furthermore, these experiments provided further proof for the σB dependency of the predicted promoter sites. Purification of both core and holoenzyme forms of the RNAP of B. cereus was performed and yielded an active enzyme. Two genes, encoding an oligopeptide permease subunit (rzc01093) and subunit II of the cytochrome d ubiquinol oxidase (rzc02188), were preceded by promoters that could be recognized in vitro by σB-RNAP. Northern blot analysis showed that these genes were not transcribed in a σB-dependent fashion under our experimental stress conditions. This may, however, be explained by additional control mechanisms that operate under the conditions in which RNA was extracted from B. cereus. For B. cereus, cytochrome d is not expressed in a rich medium with a fermentable sugar substrate, but is only expressed when fermentable sugars are not present or under conditions of oxygen limitation or anaerobiosis (10). Because RNA was isolated from aerobically growing cells in the mid-exponential growth phase, there may not have been a trigger for the cell to produce cytochrome d, and therefore no σB-dependent transcription of rzc02188 was observed. A similar mechanism may operate for the expression of rzc01093, which encodes one of the oligopeptide-binding OppA proteins of B. cereus. In other bacteria, OppA expression is tightly regulated, and this subunit of the oligopeptide permease system is generally expressed only under specific circumstances, which include changes in the intracellular amino acid pools (38). Further studies incorporating other growth conditions may reveal situations in which the σB-dependent expression of rzc01093 and rzc02188 is physiologically relevant.
A comparison of the σB-dependent genes in B. cereus that have been described in this study with the σB regulons of B. subtilis (19, 44, 46) and L. monocytogenes (29) reveals a considerable overlap in functionality among the three organisms, with the overlap being the largest between B. cereus and B. subtilis, as can be expected because of the relatively close phylogenetic relationship between the two. Of the 19 known σB-dependent genes of B. cereus, 8 have a homologue in B. subtilis and all of these are also σB dependent in B. subtilis. There is considerably less homology with L. monocytogenes. Five σB-dependent genes from B. cereus have a homologue in L. monocytogenes. Of these five, only rsbV, rsbW, and sigB are σB dependent in both organisms. However, this may be an underestimation, because a DNA microarray with a partial covering of the L. monocytogenes genome was used to define the σB regulon of this organism, so more σB-dependent genes in L. monocytogenes may be identified in the future. A comparison with the known σB-dependent genes of S. aureus (15) revealed that only YfkM is σB dependent in both B. cereus and S. aureus. This indicates that the σB regulon of B. cereus has probably evolved to serve specific functions in the B. cereus group. This may reflect differences in the ecological niches of these organisms. The ecological niche of B. cereus is quite different from those of the other organisms discussed here, as it may be an important symbiont in the nutrient-rich environment of the insect gut (23, 24, 36). In other bacteria, σB is not directly involved in vitamin metabolism, so the σB-dependent upregulation of ThiG may be specifically coupled to the particular environments in which B. cereus lives. Furthermore, the possible σB-dependent proteins Rzc01093 (an oligopeptide-binding OppA protein) and Rzc02188 (subunit II of the cytochrome d ubiquinol oxidase) may also be important during growth in the nutrient-rich environment of the insect gut, where oxygen concentrations are less than atmospheric (4). More definitive conclusions about the role of σB in the lifestyle of B. cereus can, however, only be drawn when more σB-dependent genes are identified. For the identification of more σB-dependent genes, transcriptome profiling of the B. cereus sigB null mutant and its parent strain may be necessary. In addition, further in vitro transcription analysis using the reconstituted σB-RNAP holoenzyme may also shed further light on the σB regulon of B. cereus. We have demonstrated, however, that in B. cereus the σB regulon can play a role in protecting the cell against stress by upregulating chaperone activity in the cell and by adjusting its metabolism.
Acknowledgments
We thank Rolf Wagner and Reinhild Wurm (Institut fur Physikalische Biologie, Heinrich-Heine-Universität Düsseldorf, Düsseldorf, Germany) for invaluable assistance in the purification of the RNAP of B. cereus and for critical reading of the manuscript. We thank Johan W. Renes and Freek Bouwman (Maastricht Proteomics Center, University Maastricht, Maastricht, The Netherlands) for the MALDI-TOF analysis of protein spots.
REFERENCES
- 1.Bateman, A., E. Birney, L. Cerruti, R. Durbin, L. Etwiller, S. R. Eddy, S. Griffiths-Jones, K. L. Howe, M. Marshall, and E. L. Sonnhammer. 2002. The Pfam protein families database. Nucleic Acids Res. 30:276-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Becker, L. A., M. S. Cetin, R. W. Hutkins, and A. K. Benson. 1998. Identification of the gene encoding the alternative sigma factor σB from Listeria monocytogenes and its role in osmotolerance. J. Bacteriol. 180:4547-4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bone, E. J., and D. J. Ellar. 1989. Transformation of Bacillus thuringiensis by electroporation. FEMS Microbiol. Lett. 49:171-177. [DOI] [PubMed] [Google Scholar]
- 4.Brune, A., D. Emerson, and J. A. Breznak. 1995. The termite gut microflora as an oxygen sink: microelectrode determination of oxygen and pH gradients in guts of lower and higher termites. Appl. Environ. Microbiol. 61:2681-2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burgess, R. R., and J. J. Jendrisak. 1975. A procedure for the rapid, large-scale purification of Escherichia coli DNA-dependent RNA polymerase involving Polymin P precipitation and DNA-cellulose chromatography. Biochemistry 14:4634-4638. [DOI] [PubMed] [Google Scholar]
- 6.Cao, M., P. A. Kobel, M. M. Morshedi, M. F. Wu, C. Paddon, and J. D. Helmann. 2002. Defining the Bacillus subtilis σW-regulon: a comparative analysis of promoter consensus search, run-off transcription/macroarray analysis (ROMA), and transcriptional profiling approaches. J. Mol. Biol. 316:443-457. [DOI] [PubMed] [Google Scholar]
- 7.Casadaban, M. J., and S. N. Cohen. 1980. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J. Mol. Biol. 138:179-207. [DOI] [PubMed] [Google Scholar]
- 8.Drobniewski, F. A. 1993. Bacillus cereus and related species. Clin. Microbiol. Rev. 6:324-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Engelmann, S., and M. Hecker. 1996. Impaired oxidative stress resistance of Bacillus subtilis sigB mutants and the role of katA and katE. FEMS Microbiol. Lett. 145:63-69. [DOI] [PubMed] [Google Scholar]
- 10.Escamilla, J. E., R. Ramirez, I. P. Delarenal, G. Zarzoza, and V. Linares. 1987. Expression of cytochrome oxidases in Bacillus cereus—effects of oxygen-tension and carbon source. J. Gen. Microbiol. 133:3549-3555. [Google Scholar]
- 11.Ferreira, A., C. P. O'Byrne, and K. J. Boor. 2001. Role of σB in heat, ethanol, acid, and oxidative stress resistance and during carbon starvation in Listeria monocytogenes. Appl. Environ. Microbiol. 67:4454-4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferreira, A., D. Sue, C. P. O'Byrne, and K. J. Boor 2003. Role of Listeria monocytogenes σB in survival of lethal acidic conditions and in the acquired acid tolerance response. Appl. Environ. Microbiol. 69:2692-2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fouet, A., O. Namy, and G. Lambert. 2000. Characterization of the operon encoding the alternative σB factor from Bacillus anthracis and its role in virulence. J. Bacteriol. 182:5036-5045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaur, A. H., C. C. Patrick, J. A. McCullers, P. M. Flynn, T. A. Pearson, B. I. Razzouk, S. J. Thompson, and J. L. Shenep. 2001. Bacillus cereus bacteremia and meningitis in immunocompromised children. Clin. Infect. Dis. 32:1456-1462. [DOI] [PubMed] [Google Scholar]
- 15.Gertz, S., S. Engelmann, R. Schmid, A. K. Ziebandt, K. Tischer, C. Scharf, J. Hacker, and M. Hecker. 2000. Characterization of the σB regulon in Staphylococcus aureus. J. Bacteriol. 182:6983-6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gonzalez, N., J. Wiggs, and M. J. Chamberlin. 1977. A simple procedure for resolution of Escherichia coli RNA polymerase holoenzyme from core polymerase. Arch. Biochem. Biophys. 182:404-408. [DOI] [PubMed] [Google Scholar]
- 17.Granum, P. E. 2001. Bacillus cereus, p. 327-336. In M. P. Doyle, L. R. Beuchat, and T. J. Montville (ed.), Food microbiology: fundamentals and frontiers, 2nd ed. American Society for Microbiology, Washington, D.C.
- 18.Helmann, J. D., and C. P. Moran, Jr. 2002. RNA polymerase and sigma factors, p. 289-312. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, D.C.
- 19.Helmann, J. D., M. F. Wu, P. A. Kobel, F. J. Gamo, M. Wilson, M. M. Morshedi, M. Navre, and C. Paddon. 2001. Global transcriptional response of Bacillus subtilis to heat shock. J. Bacteriol. 183:7318-7328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hilliard, N. J., R. L. Schelonka, and K. B. Waites. 2003. Bacillus cereus bacteremia in a preterm neonate. J. Clin. Microbiol. 41:3441-3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Horsburgh, M. J., J. L. Aish, I. J. White, L. Shaw, J. K. Lithgow, and S. J. Foster. 2002. σB modulates virulence determinant expression and stress resistance: characterization of a functional rsbU strain derived from Staphylococcus aureus 8325-4. J. Bacteriol. 184:5457-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang, X., A. Gaballa, M. Cao, and J. D. Helmann. 1999. Identification of target promoters for the Bacillus subtilis extracytoplasmic function sigma factor, σW. Mol. Microbiol. 31:361-371. [DOI] [PubMed] [Google Scholar]
- 23.Ivanova, N., A. Sorokin, I. Anderson, N. Galleron, B. Candelon, V. Kapatral, A. Bhattacharyya, G. Reznik, N. Mikhailova, A. Lapidus, L. Chu, M. Mazur, E. Goltsman, N. Larsen, M. D'Souza, T. Walunas, Y. Grechkin, G. Pusch, R. Haselkorn, M. Fonstein, E. S. Dusko, R. Overbeek, and N. Kyrpides. 2003. Genome sequence of Bacillus cereus and comparative analysis with Bacillus anthracis. Nature 423:87-91. [DOI] [PubMed] [Google Scholar]
- 24.Jensen, G. B., B. M. Hansen, J. Eilenberg, and J. Mahillon. 2003. The hidden lifestyles of Bacillus cereus and relatives. Environ. Microbiol. 5:631-640. [DOI] [PubMed] [Google Scholar]
- 25.Jordan, F. 2003. Current mechanistic understanding of thiamin diphosphate-dependent enzymatic reactions. Nat. Prod. Rep. 20:184-201. [DOI] [PubMed] [Google Scholar]
- 26.Joung, K. B., and J. C. Cote. 2002. A single phylogenetic analysis of Bacillus thuringiensis strains and bacilli species inferred from 16S rRNA gene restriction fragment length polymorphism is congruent with two independent phylogenetic analyses. J. Appl. Microbiol. 93:1075-1082. [DOI] [PubMed] [Google Scholar]
- 27.Juang, Y. L., and J. D. Helmann. 1994. The delta subunit of Bacillus subtilis RNA polymerase. An allosteric effector of the initiation and core-recycling phases of transcription. J. Mol. Biol. 239:1-14. [DOI] [PubMed] [Google Scholar]
- 28.Katznelson, H. 1947. Substitution of thiamine by certain amino acids in the nutrition of Bacillus paraalvei. J. Biol. Chem. 167:615-616. [PubMed] [Google Scholar]
- 29.Kazmierczak, M. J., S. C. Mithoe, K. J. Boor, and M. Wiedmann. 2003. Listeria monocytogenes σB regulates stress response and virulence functions. J. Bacteriol. 185:5722-5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kleerebezem, M., M. M. Beerthuyzen, E. E. Vaughan, W. M. de Vos, and O. P. Kuipers. 1997. Controlled gene expression systems for lactic acid bacteria: transferable nisin-inducible expression cassettes for Lactococcus, Leuconostoc, and Lactobacillus spp. Appl. Environ. Microbiol. 63:4581-4584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kotiranta, A., K. Lounatmaa, and M. Haapasalo. 2000. Epidemiology and pathogenesis of Bacillus cereus infections. Microbes Infect. 2:189-198. [DOI] [PubMed] [Google Scholar]
- 32.Kuipers, O. P., P. De Ruyter, M. Kleerebezem, and W. M. de Vos. 1998. Quorum sensing-controlled gene expression in lactic acid bacteria. J. Biotechnol. 64:15-21. [Google Scholar]
- 33.Lee, S. J., S. J. Kim, I. K. Kim, J. Ko, C. S. Jeong, G. H. Kim, C. Park, S. O. Kang, P. G. Suh, H. S. Lee, and S. S. Cha. 2003. Crystal structures of human DJ-1 and Escherichia coli Hsp31, which share an evolutionarily conserved domain. J. Biol. Chem. 278:44552-44559. [DOI] [PubMed] [Google Scholar]
- 34.Liu, H., N. H. Bergman, B. Thomason, S. Shallom, A. Hazen, J. Crossno, D. A. Rasko, J. Ravel, T. D. Read, S. N. Peterson, J. Yates III, and P. C. Hanna. 2004. Formation and composition of the Bacillus anthracis endospore. J. Bacteriol. 186:164-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lund, T., M. L. De Buyser, and P. E. Granum. 2000. A new cytotoxin from Bacillus cereus that may cause necrotic enteritis. Mol. Microbiol. 38:254-261. [DOI] [PubMed] [Google Scholar]
- 36.Margulis, L., J. Z. Jorgensen, S. Dolan, R. Kolchinsky, F. A. Rainey, and S. C. Lo. 1998. The Arthromitus stage of Bacillus cereus: intestinal symbionts of animals. Proc. Natl. Acad. Sci. USA 95:1236-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moncrief, M. B., and M. E. Maguire. 1999. Magnesium transport in prokaryotes. J. Biol. Inorg. Chem. 4:523-527. [DOI] [PubMed] [Google Scholar]
- 38.Monnet, V. 2003. Bacterial oligopeptide-binding proteins. Cell. Mol. Life Sci. 60:2100-2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oh, M. K., and J. C. Liao. 2000. DNA microarray detection of metabolic responses to protein overproduction in Escherichia coli. Metab. Eng. 2:201-209. [DOI] [PubMed] [Google Scholar]
- 40.Pannucci, J., R. T. Okinaka, R. Sabin, and C. R. Kuske. 2002. Bacillus anthracis pXO1 plasmid sequence conservation among closely related bacterial species. J. Bacteriol. 184:134-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pannucci, J., R. T. Okinaka, E. Williams, R. Sabin, L. O. Ticknor, and C. R. Kuske. 2002. DNA sequence conservation between the Bacillus anthracis pXO2 plasmid and genomic sequence from closely related bacteria. BMC Genomics 3:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Park, J. H., P. C. Dorrestein, H. Zhai, C. Kinsland, F. W. McLafferty, and T. P. Begley. 2003. Biosynthesis of the thiazole moiety of thiamin pyrophosphate (vitamin B1). Biochemistry 42:12430-12438. [DOI] [PubMed] [Google Scholar]
- 43.Periago, P. M., W. van Schaik, T. Abee, and J. A. Wouters. 2002. Identification of proteins involved in the heat stress response of Bacillus cereus ATCC 14579. Appl. Environ. Microbiol. 68:3486-3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Petersohn, A., M. Brigulla, S. Haas, J. D. Hoheisel, U. Volker, and M. Hecker. 2001. Global analysis of the general stress response of Bacillus subtilis. J. Bacteriol. 183:5617-5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Price, C. W. General stress response, p. 369-384. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, D.C.
- 46.Price, C. W., P. Fawcett, H. Ceremonie, N. Su, C. K. Murphy, and P. Youngman. 2001. Genome-wide analysis of the general stress response in Bacillus subtilis. Mol. Microbiol. 41:757-774. [DOI] [PubMed] [Google Scholar]
- 47.Rasko, D. A., J. Ravel, O. A. Okstad, E. Helgason, R. Z. Cer, L. Jiang, K. A. Shores, D. E. Fouts, N. J. Tourasse, S. V. Angiuoli, J. Kolonay, W. C. Nelson, A. B. Kolsto, C. M. Fraser, and T. D. Read. 2004. The genome sequence of Bacillus cereus ATCC 10987 reveals metabolic adaptations and a large plasmid related to Bacillus anthracis pXO1. Nucleic Acids Res. 32:977-988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Read, T. D., S. N. Peterson, N. Tourasse, L. W. Baillie, I. T. Paulsen, K. E. Nelson, H. Tettelin, D. E. Fouts, J. A. Eisen, S. R. Gill, E. K. Holtzapple, O. A. Okstad, E. Helgason, J. Rilstone, M. Wu, J. F. Kolonay, M. J. Beanan, R. J. Dodson, L. M. Brinkac, M. Gwinn, R. T. DeBoy, R. Madpu, S. C. Daugherty, A. S. Durkin, D. H. Haft, W. C. Nelson, J. D. Peterson, M. Pop, H. M. Khouri, D. Radune, J. L. Benton, Y. Mahamoud, L. Jiang, I. R. Hance, J. F. Weidman, K. J. Berry, R. D. Plaut, A. M. Wolf, K. L. Watkins, W. C. Nierman, A. Hazen, R. Cline, C. Redmond, J. E. Thwaite, O. White, S. L. Salzberg, B. Thomason, A. M. Friedlander, T. M. Koehler, P. C. Hanna, A. B. Kolsto, and C. M. Fraser. 2003. The genome sequence of Bacillus anthracis Ames and comparison to closely related bacteria. Nature 423:81-86. [DOI] [PubMed] [Google Scholar]
- 49.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 50.Smith, R. L., E. Gottlieb, L. M. Kucharski, and M. E. Maguire. 1998. Functional similarity between archaeal and bacterial CorA magnesium transporters. J. Bacteriol. 180:2788-2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Toburen-Bots, I., and H. Hagedorn. 1977. Studies on the thiamine transport system in Bacillus cereus. Arch. Microbiol. 113:23-31. [DOI] [PubMed] [Google Scholar]
- 52.Van Schaik, W., M. H. Tempelaars, J. A. Wouters, W. M. De Vos, and T. Abee. 2004. The alternative sigma factor σB of Bacillus cereus: response to stress and role in heat adaptation. J. Bacteriol. 186:316-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Volker, U., B. Maul, and M. Hecker. 1999. Expression of the σB-dependent general stress regulon confers multiple stress resistance in Bacillus subtilis. J. Bacteriol. 181:3942-3948. [DOI] [PMC free article] [PubMed] [Google Scholar]