Abstract
Replacement of glutamate 176, the only charged amino acid in the third transmembrane helix of ExbB, with alanine (E176A) abolished ExbB activity in all determined ExbB-dependent functions of Escherichia coli. Combination of the mutations T148A in the second transmembrane helix and T181A in the third transmembrane helix, proposed to form part of a proton pathway through ExbB, also resulted in inactive ExbB. E176 and T148 are strictly conserved in ExbB and TolQ proteins, and T181 is almost strictly conserved in ExbB, TolQ, and MotA.
In Escherichia coli K-12, the functions of certain outer membrane proteins require energy derived from the cytoplasmic membrane (6, 22). These proteins serve as transporters of ferric siderophores and vitamin B12 and as receptors for infection by the phages T1 and φ80 and for killing by group B colicins (B, D, Ia, Ib, Js, M, V, 5, and 10) and microcins J25, E492, 24, H47, and M (9). Energy coupling between the outer membrane and the cytoplasmic membrane is mediated by three known proteins—TonB, ExbB, and ExbD—which seem to form a complex (22). Killing by group A colicins (A, E1 to E9, K, N, and U) also requires energy derived from the cytoplasmic membrane; this energy is mediated to outer membrane receptor proteins by TolA, TolQ, and TolR (10, 20). ExbB shares sequence similarity with TolQ, and ExbD shares sequence similarity with TolR (12). ExbB and TolQ can partially replace each other, as shown by the fact that reduced activities of exbB or tolQ mutants are abolished in exbB tolQ double mutants. Likewise, TolR and ExbD can partially replace each other, as shown by the fact that reduced activities of exbD or tolR mutants are abolished in exbD tolR double mutants (5, 8). No functional substitution between the TonB and TolA proteins has been observed.
ExbB and TolQ share the same transmembrane topology (15, 16, 27). Starting with the N terminus in the periplasm, they traverse the cytoplasmic membrane three times (transmembrane segments in ExbB between residues 16 and 39, 128 and 155, and 162 and 199; total length, 244 residues). ExbD and TolR are anchored by the N-proximal region to the cytoplasmic membrane (residues 23 to 43 in ExbD) and extend into the periplasm (total length of ExbD, 141 residues) (14, 21). Analysis of point mutants has demonstrated the importance of the predicted transmembrane segments for ExbB and ExbD and for TolQ and TolR activities. Replacement of the only charged amino acid in or close to the transmembrane region of ExbD, aspartate 25, with asparagine (D25N) abolishes ExbD activity (7), and mutations at the equivalent position in TolR, D23A and D23R, inactivate TolR (10). In TolQ, mutation A13GΔS14 in the first transmembrane region (16) and mutation A177V or G181D in the third transmembrane region (27) inactivate TolQ. Mutations V35E, V36D, and A39E in the first transmembrane region of ExbB, when it is coupled to wild-type TonB, do not alter TonB-dependent activities but suppress mutations H20Y, S16L, and ΔV17 in the transmembrane segment of TonB to different levels (18, 19).
In this study, we replaced glutamate in the highly conserved sequence of the third transmembrane segment of ExbB and TolQ. This is the only charged and polar residue in a stretch of 22 hydrophobic amino acids. E176 in ExbB and E173 in TolQ were replaced with D, Q, and A by using PCR. For exbB mutagenesis, plasmid pHE20 (8) was used, and for tolQ mutagenesis, plasmid pHE12 (8) was used. The mutagenesis primers were designed so that an XmaI restriction site was introduced close to the site to be mutagenized. Wherever possible, two nucleotides in the codons were replaced to minimize reversion. The mutated exbB or tolQ gene was carried on the low-copy-number vector pWSK29 (28), which also contained wild-type exbD or tolR, respectively. Plasmid-carried exbB and exbD or tolQ and tolR were transcribed by their own promoters. The above mutations and the lack of additional mutations were verified by nucleotide sequencing. The sensitivities of mutant and wild-type cells to the group A colicins E1, E2, and K, the group B colicins D and M, albomycin, and phage φ80 were determined (Table 1). E. coli HE2 was used as the test strain; it carries Tn10 in exbB with a polar effect on exbD expression and an amber mutation early in tolQ with a polar effect on tolR expression (8). The results with the mutants were related to the results obtained with the plasmid-borne wild-type genes. In addition, the sensitivity of strain GM1, which is the parent of HE2 and which contains chromosomally carried wild-type exbB and exbD, was determined to compare the activities of single-copy genes in their natural environment with those of plasmid-carried genes. When E. coli HE2 was transformed with the wild-type exbB and exbD genes, wild-type levels of sensitivity to colicins D and M, albomycin, and phage φ80 were obtained. The same values were obtained with the chromosomally carried wild-type exbB and exbD of E. coli GM1 (Table 1), showing that the presumed slight overexpression of exbB and exbD by the low-copy-number plasmid did not alter the ExbB- and ExbD-related activities. The E176A mutation in ExbB [ExbB(E176A)] completely abolished all activities. Mutation ExbB(E176Q) reduced by 10-fold the sensitivities to colicin M and albomycin, whereas the ExbB(E176D) mutation had no effects.
TABLE 1.
Activities of ExbB and TolQ mutants
| Protein | Sensitivity toa:
|
||||||
|---|---|---|---|---|---|---|---|
| Colicins
|
Albomycin | Phage φ80 | |||||
| E1 | E2 | K | D | M | |||
| ExbB (wild type) | 4 | 4 | 2 | 5 | 4 | 3 | 4 |
| ExbB(E176D) | 4 | 4 | R | 5 | 4 | 3 | 4 |
| ExbB(E176Q) | 3 | R | R | 5 | 3 | 2 | 4 |
| ExbB(E176A) | R | R | R | R | R | R | R |
| ExbB(T148A) | R | R | R | 5 | 3 | 2 | 4 |
| ExbB(S155A) | 4 | 4 | 2 | 5 | 3 | 3 | 4 |
| ExbB(T181A) | 3 | 1 | R | 5 | 3 | 2 | 4 |
| ExbB(T148A S155A) | R | R | R | 5 | 3 | 3 | 4 |
| ExbB(T148A T181A) | R | R | R | R | R | R | R |
| ExbB(T181A S155A) | 3 | R | R | 5 | 3 | 2 | 4 |
| TolQ (wild type) | 5 | 5 | 3 | 5 | 4 | 3 | 4 |
| TolQ(E173D) | 5 | 5 | 3 | 4 | 2 | 1 | 4 |
| TolQ(E173Q) | 4 | 4 | 2 | R | R | R | 4 |
| TolQ(E173A)b | R | R | R | R | R | R | R |
| GM1c | 5 | 5 | 2 | 5 | 4 | 3 | 4 |
E. coli HE2 exbB (exbD) tolQ (tolR) was transformed with plasmid pWSK29, on which wild-type exbD (or tolR) and wild-type exbB (or tolQ) or mutant exbB genes (or mutant tolQ genes) were cloned. The results of the sensitivity assay are presented as the last of a 10-fold dilution series that yielded a clear zone of growth inhibition. R, resistant. The assays were performed in triplicate with no deviations.
The TolQ(E173A) protein was not observed after SDS-PAGE.
E. coli GM1 is the exbB exbD wild-type parent strain of E. coli HE2 (8).
ExbB and ExbD activities partially compensate for deficient TolQ and TolR activities (8). Rather high levels of sensitivity of the E. coli HE2 exbB exbD transformants to colicins E1, E2, and K were obtained, despite the lack of tolQ and tolR activities. Sensitivity of these transformants to the group A colicins was approximately 10-fold lower than that of transformants carrying wild-type tolQ and tolR genes. The ExbB(E176D) mutation, which produced no effect on the ExbB-dependent functions, abolished sensitivity to colicin K, and the ExbB(E176Q) mutation reduced sensitivities to the group A colicins more than to the group B colicins. The residual activities of ExbB and ExbD in functions dependent on TolQ and TolR revealed the reduced activity of the ExbB(E176D) mutant and, more clearly, that of the ExbB(E176Q) mutant. As expected, the ExbB(E176A) mutant, which was inactive in the TonB-dependent functions, was also inactive in the TolA-dependent functions.
The results obtained with the TolQ mutants closely matched those obtained with the ExbB mutants (Table 1). The TolQ(E173A) mutation inactivated TolQ function. The TolQ(E173Q) and TolQ(E173D) mutations affected the activity towards the group B colicins more than the activity towards the group A colicins. Thus, the residual activities of TolQ in ExbB-dependent functions were useful in identifying the small functional effects of TolQ mutations. The sensitivities of E. coli GM1 matched those of E. coli HE2 transformed with wild-type tolQ and tolR, except for sensitivity to colicin K, which was 10-fold higher in the transformants. A higher level of expression of tolQ and tolR in the transformants probably increased colicin K sensitivity.
The transport activities of E. coli HE2 containing plasmid-carried mutant exbB genes were compared with the transport activities of E. coli HE2 transformed with the wild-type exbB gene. Cells were incubated with [55Fe3+]ferrichrome, collected on filters, and washed, and the radioactivity on the filters was measured using a liquid scintillation counter. Transport of the ExbB(E176A) mutant was totally impaired, whereas transport of ExbB(E176D) and ExbB(E176Q) mutant cells was somewhat reduced (Table 2). The transport rate of E. coli GM1 was 77% of the transport rate of the wild-type exbB transformant (Table 2); this difference may be caused by the two to three exbBD gene copies in the transformant.
TABLE 2.
Transport rates of ExbB and TolQ mutantsa
| Proteinb | Transport rate (%)d |
|---|---|
| ExbB (wild type) | 100 |
| ExbB(E176D) | 84 |
| ExbB(E176Q) | 80 |
| ExbB(E176A) | 2 |
| ExbB(T148A) | 99 |
| ExbB(S155A) | 100 |
| ExbB(T181A) | 99 |
| ExbB(T148A S155A) | 82 |
| ExbB(T148A T181A) | 6 |
| ExbB(T181A S155A) | 91 |
| TolQ (wild type) | 100 |
| TolQ(E173D) | 57 |
| TolQ(E173Q) | 48 |
| GM1c | 77 |
E. coli HE2 exbB (exbD) tolQ (tolR) was transformed with plasmid pWSK29, on which the wild-type or mutant exbB and tolQ genes were cloned. For determination of the ExbB and TolQ activities, the plasmids also carried wild-type exbD and tolR downstream of exbB and tolQ, respectively.
The TolQ(E173A) protein was not observed after SDS-PAGE.
E. coli GM1 is the wild-type exbB exbD parent strain of E. coli HE2.
Transport rates are relative to the transport rate of the wild-type transformants (100%). Results are the averages of two series of experiments with a maximum deviation of 9%.
The rate of [55Fe3+]ferrichrome transport into E. coli HE2 transformed with wild-type tolQ and tolR was approximately 60% of that into E. coli HE2 transformed with wild-type exbB and exbD. Relative to the rate of transport into E. coli HE2 expressing wild-type TolQ and TolR, the rate of transport into cells expressing the TolQ(E173D) or TolQ(E173Q) mutation was more greatly reduced than that into cells expressing the corresponding ExbB mutant proteins (Table 2).
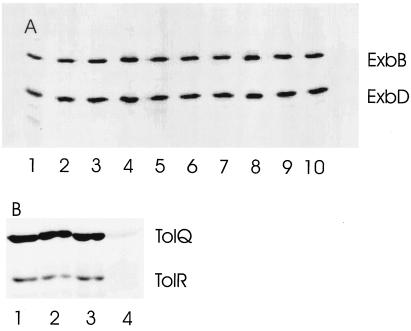
To examine whether the inactivity of the ExbB(E176A) and TolQ(E173A) mutants was caused by lack of the proteins, E. coli BL21 omp8 (23) was labeled with [35S]methionine. The exbB and exbD genes were cloned downstream of the gene 10 promoter of phage T7 on plasmid pT7-6 and transcribed by the T7 RNA polymerase encoded on the chromosome of E. coli BL21 omp8 under lacI control. In the labeling experiment, cells were treated with 0.2 μg of rifamycin/ml to inactivate the E. coli RNA polymerase. Proteins of transformants containing plasmid-carried wild-type or mutant exbB and tolQ genes were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The autoradiographs of the gels revealed similar amounts of ExbB and ExbD (Fig. 1A) and of TolQ and TolR (Fig. 1B), with the exception of TolQ(E173A), where only trace amounts of a labeled protein were observed. Examination of the nucleotide sequences of tolQ and of the region upstream of tolQ in the clone used for SDS-PAGE revealed no mutation other than the one yielding the TolQ(E173A) replacement. Different mutation strategies also yielded only transformants that contained trace amounts of the TolQ(E173A) mutant protein. Cells expressing TolQ(E173A) formed after centrifugation of sediments, which were difficult to suspend homogenously, suggesting alterations in cell surface properties. It is not clear how much the phenotype of the TolQ(E173A) mutant was influenced by the small amounts of the protein. To minimize additional mutations that down-regulate TolQ(E173A) synthesis or activity, the assays and the protein determinations were performed with freshly transformed cells. The TolQ(E173A) mutant synthesized only trace amounts of TolR also, as if the TolQ mutation destabilized not only TolQ but also the TolR protein that interacts with TolQ.
FIG. 1.
Separation of [35S]methionine-labeled proteins of E. coli BL21 omp8 by SDS-PAGE. (A) Upper bands indicate wild-type ExbB (lane 1), ExbB(E176Q) (lane 2), ExbB(E176D) (lane 3), ExbB(E176A) (lane 4), ExbB(T148A) (lane 5), ExbB(S155A) (lane 6), ExbB(T181A) (lane 7), ExbB(T148A S155A) (lane 8), ExbB(T148A T181A) (lane 9), and ExbB(S155A T181A) (lane 10); lower bands indicate wild-type ExbD. (B) Upper bands indicate wild-type TolQ (lane 1) and its derivatives TolQ(E173Q) (lane 2), TolQ(E173D) (lane 3), and TolQ(E173A) (lane 4); lower bands indicate TolR. For labeling, the transformants were grown in minimal medium, synthesis of T7 RNA polymerase was induced with 1 mM isopropyl-β-d-thiogalactopyranoside for 40 min at 37°C, and then the E. coli RNA polymerase was inhibited with 0.2 mg of rifamycin/ml for 30 min until 588 kBq of [35S]methionine per ml was added to label cells for 15 min at room temperature (15).
In previous experiments using the same methods, the ExbD band was less dense than the ExbB band (7). Different amounts of the two proteins were also determined in cells (13). We do not know the reason for this discrepancy, but the apparently greater amounts of ExbD did not affect the ExbB, ExbD, and TonB activities as revealed by comparison of chromosomally encoded and plasmid-encoded receptor activities (Table 1).
The phenotypes of the mutants indicate that E176 and E173 play important roles in the activities of ExbB and TolQ, respectively. However, E176 and E173 do not seem to be as essential in the c subunit of the ATPase of E. coli as D61, which is representative of a single aspartate residue in the hydrophobic c-subunit regions of H+-translocating ATPases. E176 is present in all 12 predicted ExbB proteins that we examined in the EMBL databank, and E173 is present in all 10 TolQ proteins examined.
Recently, two proton conduction pathways have been proposed. In one proposal, the pathway is formed by the three transmembrane helices of ExbB and the transmembrane helix of ExbD; in the other, the pathway is formed by the TonB transmembrane helix together with the three ExbB transmembrane helices and the ExbD helix (29). In the former model, the protons move along residues as follows: T181 → S155 → G151 → T148 → G144 → D25 (ExbD); in the latter model, the protons move along residues as follows: T181 → G184 → H20 (TonB) → S16 (TonB) → A188 → T148 → G144 → D25 (ExbD). Water molecules close to the G and A residues are proposed to participate in proton transfer. To test the validity of the two models, which do not include E176, we replaced amino acids carrying hydroxyl groups with alanine and expected the interruption of proton conduction along the hydroxyl groups of the serine and threonine residues and water molecules bound to the hydroxyl groups. The mutant carrying plasmid-encoded ExbB(T148A) was resistant to the group A colicins and exhibited a 10-fold-reduced sensitivity to colicin M and albomycin (Table 1). The ExbB(S155A) mutant was as sensitive to all ligands as were cells carrying wild-type ExbB. Combination of the T148A mutation with the S155A mutation resulted in resistance to the group A colicins and a 10-fold reduction in sensitivity to colicin M. The ExbB(T181A) mutant showed a reduced sensitivity to the group A colicins, colicin M, and albomycin (Table 1). Combination of the T181A mutation with the T148A mutation conferred resistance to all tested ligands (Table 1). An S155A T181A double mutant was resistant to colicins E2 and K and was only 10-fold-less sensitive to colicin E1. Sensitivity to colicin M and albomycin was reduced by 10-fold.
The rates of [55Fe3+]ferrichrome transport into cells carrying the amino acid replacements in the proposed ExbB proton conduction pathway were determined (Table 2). Only the ExbB(T148A T181A) double mutant was virtually transport inactive. The other single and double mutants showed activities between 48 and 99% of that of the wild type. As was seen with the colicin assays, the TolQ(E173D) and TolQ(E173Q) mutations reduced ferrichrome transport more severely than the corresponding ExbB mutations.
The selected residues are contained in the most conserved sequence, comprising 67 residues, of ExbB/TolQ; 57 of these are identical and 10 are conserved replacements in E. coli, Salmonella enterica serovar Typhimurium, Yersinia pestis, Pseudomonas putida, Rhizobium meliloti, and Brucella melitensis. They are part of transmembrane helices 2 and 3 and are connected by a small periplasmic loop in which amino acid variations are accumulated. When all sequences contained in the EMBL databank (46 sequences) were compared, T148 was found in all ExbB and TolQ homologs and T181 was found in all but two homologs (replaced by A or V). S155 is less conserved (nine cases) and frequently is replaced by A in TolQ. The sensitivity of the mutants coincided with the conservation of the residues. The T148A and T181A mutants were resistant to the group A colicins and showed reduced sensitivity to the group B colicins. T148A T181A double mutants were completely resistant to all agents. It is possible that the predicted proton conductance is not completely interrupted by the single mutations but is completely interrupted by the double mutation. In the single mutants, water close to alanine might bridge the gap. The S155A mutation alone and in combination with T181A showed the slightest effects. Our data do not discriminate between the two proposed proton conduction pathways since the two mutations with the strongest effects, T148A and T181A, are contained in both pathways. However, since the S155A mutation showed only slight effects and the H20R (26), H20Y, and S16L mutations in TonB inactivate TonB (18), a proton conduction pathway that includes these two residues is more consistent with the data.
The presence of similar sequences and transmembrane topologies in the flagellar motor proteins MotA and ExbB/TolQ and in MotB and ExbD/TolR has led to the proposal that there might also be functional similarities (10, 11, 17, 29). MotA forms a channel through which protons flow (2) and interacts with the motor through FliG. Together with MotB, it forms the stator of the flagellar motor. MotA traverses the cytoplasmic membrane four times and could be comparable with ExbB if the single transmembrane region of TonB is added to the three transmembrane segments of ExbB (29). MotB traverses the cytoplasmic membrane once with the same topology as ExbD and TolR. MotB contains an aspartate residue (D32) at the same position as in ExbD and TolR; D32 is strictly conserved among MotB proteins of various bacteria and replacement with asparagine abolishes motor function (3). It has been proposed that flagellar rotation is driven by cyclic conformational changes in the stator which occur as protons bind to and dissociate from D32 of MotB (1). D32 also plays a crucial role in the helix rotation model of the flagellar motor (25). It is likely that the essential roles of D25 in ExbD, D23 in TolR, and D32 in MotB reflect similar functions. Proton-driven conformational changes have also been discussed as being involved in energy-coupled outer membrane transport processes mediated by TonB and TolA (4, 10, 11, 22). If a proton conductance pathway through TonB/ExbB/ExbD and TolA/TolQ/TolR exists, E176 of ExbB and E173 of TolQ are likely to be involved. Reconstitution of these protein complexes in lipid vesicles will be required to demonstrate proton conductance and to determine the roles, which have been identified in this and other studies, that the amino acids play. MotA and MotB form large protein complexes with FliG, FliN, and FliM. It is likely that TonB, ExbB, and ExbD also form a complex. The molar ratio of the latter three proteins in cells amounts to 1:7:2 (13). If this reflects the stoichiometry of the complex, it differs from that of MotA and MotB, for which a 4:2 ratio has been proposed (1). A C-terminal fragment of TonB forms a dimer in the crystal (11) and in cells (24), and the formation of a dimer of two complete TonB molecules has been determined in vivo (24). The stoichiometry could then be 2:14:4, which results in a protein complex of 520 kDa.
This study was undertaken with two purposes in mind: (i) to determine whether the only charged and highly conserved E176 residue of ExbB and E173 of TolQ in the predicted transmembrane helix 3 plays an essential role in ExbB and TolQ activity and (ii) to determine whether experimental data can be obtained that support one or the other of the two proposed proton conductance pathways. This paper demonstrates that E176 of ExbB and probably E173 of TolQ are not essential for ExbB and TolQ activity and that the predicted proton pathways seem to function only when water bridges between nonpolar amino acid side chains are assumed. The data do not support one or the other of the two predicted proton pathways, if they exist at all.
Acknowledgments
We thank Klaus Hantke for fruitful discussions and Karen A. Brune for critical reading of the manuscript.
This work was supported by the Fonds der Chemischen Industrie.
REFERENCES
- 1.Blair, D. F. 2003. Flagellar movement driven by proton translocation. FEBS Lett. 545:86-95. [DOI] [PubMed] [Google Scholar]
- 2.Blair, D. F., and H. C. Berg. 1990. The MotA protein of E. coli is a proton-conducting component of the flagellar motor. Cell 60:439-449. [DOI] [PubMed] [Google Scholar]
- 3.Blair, D. F., and H. C. Berg. 1991. Mutations in the MotA protein of E. coli reveal domains critical for proton conduction. J. Mol. Biol. 221:1433-1442. [DOI] [PubMed] [Google Scholar]
- 4.Bradbeer, C. 1993. The proton motive force drives the outer membrane transport of cobalamin in Escherichia coli. J. Bacteriol. 175:3146-3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braun, V. 1989. The structurally related exbB and tolQ genes are interchangeable in conferring tonB-dependent colicin, bacteriophage, and albomycin sensitivity. J. Bacteriol. 171:6387-6390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braun, V. 1995. Energy-coupled transport and signal transduction through the gram-negative outer membrane via TonB-ExbB-ExbD-dependent receptor proteins. FEMS Microbiol. Rev. 16:295-307. [DOI] [PubMed] [Google Scholar]
- 7.Braun, V., S. Gaisser, C. Herrmann, K. Kampfenkel, H. Killmann, and I. Traub. 1996. Energy-coupled transport across the outer membrane of Escherichia coli: ExbB binds ExbD and TonB in vitro, and leucine 132 in the periplasmic region and aspartate 25 in the transmembrane region are important for ExbD activity. J. Bacteriol. 178:2836-2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Braun, V., and C. Herrmann. 1993. Evolutionary relationship of uptake systems for biopolymers in Escherichia coli: cross-complementation between the TonB-ExbB-ExbD and the TolA-TolQ-TolR proteins. Mol. Microbiol. 8:261-268. [DOI] [PubMed] [Google Scholar]
- 9.Braun, V., S. I. Patzer, and K. Hantke. 2002. Ton-dependent colicins and microcins: modular design and evolution. Biochimie 84:365-380. [DOI] [PubMed] [Google Scholar]
- 10.Cascales, E., R. Lloubès, and J. N. Sturgis. 2001. The TolQ-TolR proteins energize TolA and share homologies with the flagellar motor proteins MotA-MotB. Mol. Microbiol. 42:795-807. [DOI] [PubMed] [Google Scholar]
- 11.Chang, C., A. Mooser, A. Plückthun, and A. Wlodawer. 2001. Crystal structure of the dimeric C-terminal domain of TonB reveals a novel fold. J. Biol. Chem. 276:27535-27540. [DOI] [PubMed] [Google Scholar]
- 12.Eick-Helmerich, K., and V. Braun. 1989. Import of biopolymers into Escherichia coli: nucleotide sequences of the exbB and exbD genes are homologous to those of the tolQ and tolR genes, respectively. J. Bacteriol. 171:5117-5126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Higgs, P. I., R. A. Larsen, and K. Postle. 2002. Quantification of known components of the Escherichia coli TonB energy transduction system: TonB, ExbB, ExbD and FepA. Mol. Microbiol. 44:271-281. [DOI] [PubMed] [Google Scholar]
- 14.Kampfenkel, K., and V. Braun. 1992. Membrane topology of the Escherichia coli ExbD protein. J. Bacteriol. 174:5485-5487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kampfenkel, K., and V. Braun. 1993. Topology of the ExbB protein in the cytoplasmic membrane of Escherichia coli. J. Biol. Chem. 268:6050-6057. [PubMed] [Google Scholar]
- 16.Kampfenkel, K., and V. Braun. 1993. Membrane topologies of the TolQ and TolR proteins of Escherichia coli: inactivation of TolQ by a missense mutation in the proposed first transmembrane segment. J. Bacteriol. 175:4485-4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kojima, S., and D. F. Blair. 2001. Conformational change in the stator of the bacterial flagellar motor. Biochemistry 40:13041-13050. [DOI] [PubMed] [Google Scholar]
- 18.Larsen, R. A., and K. Postle. 2001. Conserved residues Ser(16) and His(20) and their relative positioning are essential for TonB activity, cross-linking of TonB with ExbB, and the ability of TonB to respond to proton motive force. J. Biol. Chem. 276:8111-8117. [DOI] [PubMed] [Google Scholar]
- 19.Larsen, R. A., M. G. Thomas, G. E. Wood, and K. Postle. 1994. Partial suppression of an Escherichia coli TonB transmembrane domain mutation (ΔV17) by a missense mutation in ExbB. Mol. Microbiol. 13:627-640. [DOI] [PubMed] [Google Scholar]
- 20.Lazzaroni, J., J.-F. Dubuisson, and A. Vianney. 2002. The Tol proteins of Escherichia coli and their involvement in the translocation of group A colicins. Biochimie 84:391-397. [DOI] [PubMed] [Google Scholar]
- 21.Müller, M., A. Vianney, J.-C. Lazzaroni, R. E. Webster, and R. Portalier. 1993. Membrane topology of the Escherichia coli TolR protein required for cell envelope integrity. J. Bacteriol. 175:6059-6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Postle, K., and R. J. Kadner. 2003. Touch and go: tying TonB to transport. Mol. Microbiol. 49:869-882. [DOI] [PubMed] [Google Scholar]
- 23.Prilipov, A., P. S. Phale, P. Van Gelder, J. P. Rosenbusch, and R. Koebnik. 1998. Coupling site-directed mutagenesis with high-level expression: large scale production of mutant porins from E. coli. FEMS Microbiol. Lett. 163:65-72. [DOI] [PubMed] [Google Scholar]
- 24.Sauter, A., P. Howard, and V. Braun. 2003. In vivo evidence for TonB dimerization. J. Bacteriol. 185:5747-5754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmitt, R. 2003. Helix rotation model of the flagellar rotary motor. Biophys. J. 85:843-852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Traub, I., S. Gaisser, and V. Braun. 1993. Activity domains of the TonB protein. Mol. Microbiol. 8:409-423. [DOI] [PubMed] [Google Scholar]
- 27.Vianney, A., T. M. Lewin, W. F. Beyer., J. Lazzaroni, R. Portalier, and R. E. Webster. 1994. Membrane topology and mutational analysis of the TolQ protein of Escherichia coli required for the uptake of macromolecules and cell envelope integrity. J. Bacteriol. 176:822-829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang, R. F., and S. R. Kushner. 1991. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100:195-199. [PubMed] [Google Scholar]
- 29.Zhai, Y. F., W. Heine, and M. H. Saier. 2003. Molecular modelling of the bacterial outer membrane receptor energizer, ExbBD/TonB, based on homology with the flagellar motor, MotAB. Biochim. Biophys. Acta 1614:201-210. [DOI] [PubMed] [Google Scholar]