Abstract
The transcription factors σF and σB are related RNA polymerase sigma factors that govern dissimilar networks of adaptation to stress conditions in Bacillus subtilis. The two factors are controlled by closely related regulatory pathways, involving protein kinases and phosphatases. We report that insulation of the σF pathway from the σB pathway involves the integrated action of both the cognate kinase and the cognate phosphatase.
In the gram-positive bacterium Bacillus subtilis, the transcriptional control proteins σB and σF are closely related RNA polymerase sigma factors that help to govern alternative responses to conditions of environmental and physiological stress. The σB factor is a general stress response sigma factor that activates a regulon of more than 100 genes in response to a variety of adverse conditions, including impaired energy production and exposure to certain toxic agents, such as ethanol (reviewed in references 12 and 27). This σB-mediated stress response is usually a transient adaptation to unfavorable conditions in which expression of the σB regulon is elevated for only a limited period of time. The σF factor, in contrast, is part of a highly elaborate and irreversible sporulation response to adverse environmental conditions (in this case, nutrient limitation). Sporulation is a multistage process of differentiation that culminates in the formation of a dormant cell type, the endospore, that is capable of resisting environmental extremes, such as heat, desiccation, and radiation (reviewed in reference 26). A conspicuous feature of this sporulation process is the formation of an asymmetrically positioned septum that divides the differentiating bacterium (the sporangium) into a small forespore cell and a larger mother cell. Sporulation involves the activation of a cascade of developmental sigma factors in which σF plays a central role, becoming activated selectively in the forespore compartment of the sporangium and helping to render the process irreversible (J. Dworkin and R. Losick, unpublished results).
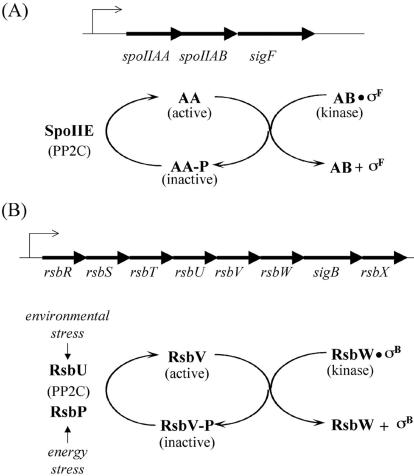
Interestingly, σB and σF are controlled in a similar manner by three interacting regulatory proteins that are paralogous between the two pathways. As shown in Fig. 1, RsbW and SpoIIAB (hereafter referred to as AB) are dual-function proteins that act both as antisigma factors and as protein kinases; RsbV and SpoIIAA (hereafter referred to as AA) are antiantisigma factors; and last, RsbP, RsbU, and SpoIIE are PP2C-like protein phosphatases. In both the σB and σF regulatory pathways, the activity of the sigma factor is negatively regulated by the cognate antisigma factor, which holds the transcription factor in an inactive complex. Release from the complex is mediated by the antiantisigma factors, RsbV and AA. The action of the antiantisigma factors is counteracted by the kinase activity of the dual-function RsbW and AB proteins, which phosphorylate and thereby inactivate their respective antiantisigma factors. Finally, the phosphorylated RsbV and AA proteins are reactivated by the action of a phosphatase: RsbU and RsbP dephosphorylate RsbV-P, and SpoIIE dephosphorylates SpoIIAA-P (AA-P) (reviewed in references 12, 26, and 27). A distinctive feature of the σB pathway is an upstream extension of the regulatory circuit involving a paralog of RsbW/AB called RsbT, a paralog of RsbV/AA called RsbS, and a paralog of RsbU/SpoIIE called RsbX (17, 31).
FIG. 1.
σF and σB activity are regulated by paralogous pathways. (A) The structure of the spoIIA operon and the σF regulatory circuit. (B) The structure of the sigB operon and the σB regulatory circuit.
It is generally assumed that the σB and σF pathways are insulated from each other despite the close similarity of the corresponding paralogs in the two regulatory systems. For example, it is known that each antisigma factor binds preferentially with its cognate sigma factor and antiantisigma factor (1). We were therefore surprised to discover a low but significant level of phosphorylation of AA in the absence of its cognate kinase, AB. Furthermore, high levels of phosphorylation of AA were observed in cells lacking both AB and the phosphatase SpoIIE. These findings prompted us to investigate the extent to which the σF regulatory pathway is subject to “cross talk” by components of the σB pathway and, conversely, whether the σF pathway exerts any influence on the expression of genes in the σB regulon.
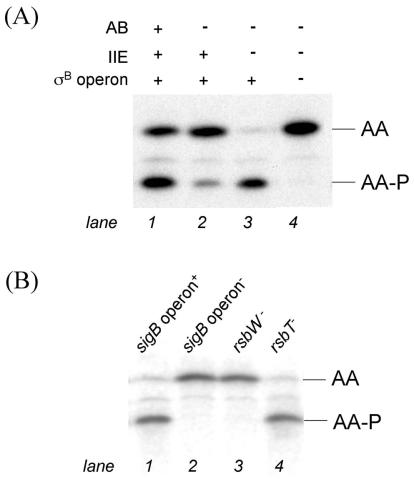
To monitor the phosphorylation state of AA, we took advantage of a recently improved procedure for resolving AA and AA-P by isoelectric focusing and determining the ratio of the two forms of the phosphoprotein quantitatively by immunoblotting with anti-AA antibodies (6). Because σF is toxic to cells lacking AB, all of our experiments were carried out with cells lacking the sporulation transcription factor. Figure 2A (lane 1) shows that in cells that were 90 min into the sporulation process, the ratio of AA to AA-P was approximately 0.8, and other results showed that this value was not measurably affected by the presence or absence of σF (6; also data not shown). Next, we investigated the dependence of the accumulation of AA-P on the AB kinase. We were surprised to discover that a small amount of AA was present in its phosphorylated form in cells lacking the cognate kinase (Fig. 2A, lane 2). Strikingly, AA was almost entirely in its phosphorylated form when the cells lacked both the AB kinase and the AA-P phosphatase SpoIIE (Fig. 2A, lane 3). The simplest interpretation of these results is that AB is not the only kinase that is capable of phosphorylating AA during sporulation and that the activity of this other kinase is normally masked by the SpoIIE-mediated dephosphorylation of AA-P.
FIG. 2.
RsbW phosphorylates AA in the absence of AB. (A) The in vivo ratio of AA to AA-P in a strain lacking σF (KC461 [lane 1]), a strain lacking σF and AB (KC454 [lane 2]), a strain lacking σF, AB, and SpoIIE (KC472 [lane 3]), and a strain lacking σF, AB, SpoIIE, and the entire sigB operon (KC476 [lane 4]). (B) The in vivo ratio of AA to AA-P in strains that lack σF, AB, and SpoIIE and harbor either an intact sigB operon (KC472 [lane 1]), an insertion-deletion of the sigB operon (KC476 [lane 2]), a Campbell disruption of rsbW (KC486 [lane 3]), or an in-frame deletion of rsbT (KC487 [lane 4]). Lysates of cells harvested 90 min after the start of sporulation were subjected to isoelectric focusing followed by immunoblotting with anti-SpoIIAA antibodies.
The most attractive candidates for the other kinase were the paralogs of AB, RsbW and RsbT, which are responsible for phosphorylating the AA-related phosphoproteins RsbV and RsbS, respectively, in the pathway governing the activity of σB (Fig. 1B). In support of the idea that one or both of these kinases was responsible for phosphorylating AA during sporulation, introduction of a deletion of the eight-gene sigB operon (see Fig. 1B) into cells with double mutations for AB and SpoIIE largely, if not entirely, eliminated the accumulation of AA-P (Fig. 2B, lane 2). This sigB operon deletion was created by double-crossover integration of pHC10 (Table 1) to insert a kanamycin-resistance cassette in place of the region extending from codon 145 of the first gene in the operon (rsbR) to codon 188 of the last gene (rsbX). A similar result was obtained by introducing a disruption of rsbW into the double mutant (Fig. 2B, lane 3). The disruption was created by single-crossover recombination between the chromosomal rsbW gene and a kanamycin resistance plasmid (pKC61) with an insert that extended from bp 149 to 366 of rsbW. In contrast, little or no effect was seen when an in-frame deletion of the gene for RsbT (17) was introduced via cotransduction of pKC60 (see Table 1) with SPP1 (19) into cells lacking AB and SpoIIE (Fig. 2B, lane 4). These results are consistent with the idea that RsbW (and not RsbT) was responsible for phosphorylating AA during sporulation. However, it remained possible that the phenotype of the Campbell integration into rsbW was due to an indirect, polar effect on the expression of the downstream gene sigB (encoding σB) or rsbX (encoding a PP2C-like phosphatase for RsbS-P) (31).
TABLE 1.
Strains and plasmids
| Strain or plasmid | Genotype | Reference |
|---|---|---|
| Strains | ||
| PY79 | Wild-type parent | 32 |
| RL1274 | spoIIAA::spc | 23 |
| RL2182 | spoIIAA::cat | 23 |
| MO3418 | spoIIE::phleo | 2 |
| PB198 | amyE::ctc-lacZ cat trp2C | 5 |
| PB423 | As PB198, rsbTΔ1 | 17 |
| PB642 | As PB198, pHC10(Δ(rsbR rsbS rsbT rsbU rsbV rsbW sigB rsbX)::kan) | This study |
| KC454 | spoIIA::spc amyE::pKC33(spoIIAA cat) | This study |
| KC461 | spoIIA::spc amyE::pAB54(spoIIAA-spoIIAB kan) | This study |
| KC472 | spoIIE::phleo spoIIA::spc amyE::pKC33(spoIIAA cat) | This study |
| KC476 | As KC472, pHC10(Δ(rsbR rsbS rsbT rsbU rsbV rsbW sigB rsbX)::kan) | This study |
| KC479 | amyE::ctc-lacZ cat | This study |
| KC482 | As KC479, pHC10(Δ(rsbR rsbS rsbT rsbU rsbV rsbW sigB rsbX)::kan) | This study |
| KC486 | As KC472, rsbWΩpKC61(rsbW 146-366 kan) | This study |
| KC483 | As PB423, ydcDEΩpKC60(kan) | This study |
| KC487 | As KC472, ydcDEΩpKC60(kan) rsbTΔ1 | This study |
| KC495 | amyE::pKC62(Pctc-gfp spc) | This study |
| KC496 | As KC495, pHC10(Δ(rsbR rsbS rsbT rsbU rsbV rsbW sigB rsbX)::kan) | This study |
| KC502 | As KC479, spoIIA::spc | This study |
| KC503 | As KC482, spoIIA::spc | This study |
| KC504 | As KC495, spoIIA::cat | This study |
| KC505 | As KC496, spoIIA::cat | This study |
| Plasmids | ||
| pAB54 | bla kan amyE::spoIIAA+-spoIIAB+ | 7 |
| pKC33 | bla cat amyE::spoIIAA | 6 |
| pHC10 | bla rsbR(nt− 143-392)-kan-rsbX(nt 545-1158) | This study |
| pKC60 | bla kan ydcDE | This study |
| pKC61 | bla kan rsbW(nt149-366) | This study |
| pKC62 | bla spc amyE::Pctc-gfp | This study |
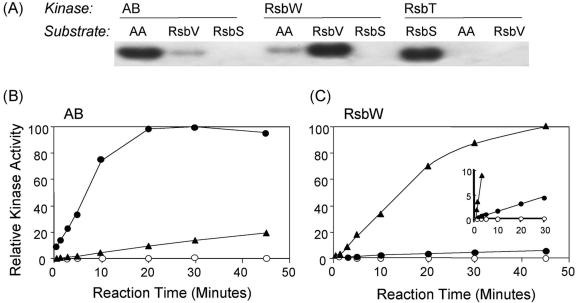
To investigate further the possibility that RsbW was directly responsible for phosphorylating AA, we carried out experiments to measure the capacity of RsbW to phosphorylate AA in vitro. We performed these assays using purified, His-tagged versions of all three of the AB-like kinases—AB itself, RsbW, and RsbT—and all three potential substrates—AA, RsbV, and RsbS (9, 18, 31). Phosphorylation was carried out in 100-μl reaction mixtures containing 1 mM unlabeled ATP, 50 μCi of [γ-32P]ATP, 20 μM protein substrate, 2 μM kinase, 50 mM Tris (pH 7.6), 50 mM KCl, 10 mM MgCl2, 1 mM dithiothreitol, and 0.1 mM EDTA. Reactions were terminated by the addition of 2.5 μl of 5× sodium dodecyl sulfate (SDS) sample buffer (60 mM Tris-HCl [pH 6.8], 14.4 mM 2-mercaptoethanol, 2% SDS, 25% glycerol, 0.1% bromphenol blue) per 10 μl of reaction mix, after which samples were subjected to electrophoresis in a 15% SDS polyacrylamide gel. As shown in Fig. 3A, all three kinases were, as expected, highly and preferentially active with their cognate substrates: AB with AA, RsbW with RsbV, and RsbT with RsbS. However, whereas the RsbT kinase showed no detectable activity with AA (or RsbV) as a substrate, the RsbW kinase exhibited a low but significant level of activity with the noncognate substrate AA. Conversely, the AB kinase exhibited a low but significant level of activity with the noncognate substrate RsbV.
FIG. 3.
SpoIIAA is phosphorylated in vitro by RsbW. (A) The purified substrate AA, RsbV, or RsbS (20 μM) was incubated with [γ-32P]ATP and the purified kinase indicated, either AB, RsbW, or RsbT (2 μM). After 30 min at 37°C, the reaction mixtures were analyzed by SDS-polyacrylamide gel electrophoresis and autoradiography. (B) The AA (•), RsbV (▴), or RsbS (○) substrate was incubated with [γ-32P]ATP and the AB kinase. Reactions were sampled after the indicated times at 37°C and analyzed by SDS-polyacrylamide gel electrophoresis. Amounts of labeled product were determined by phosphorimaging and expressed as percentages of the cognate AA substrate labeled at 30 min. (C) The AA (•), RsbV (▴), or RsbS (○) substrate was incubated with [γ-32P]ATP and the RsbW kinase. Reactions were treated as described for panel B, with the amount of each labeled product expressed as a percentage of the cognate RsbV substrate labeled at 45 min.
To measure this cross-phosphorylation quantitatively, we performed the time course experiment shown in Fig. 3B and C. By using the initial slopes of the reactions as a measure of relative activity, we found that RsbW manifested 4% as much activity with AA as a substrate than with its cognate substrate and that AB exhibited 7% as much activity with RsbV as a substrate than with its cognate substrate. In contrast, neither RsbW nor AB had any detectable activity with RsbS, the cognate substrate for the RsbT kinase. We conclude that RsbW exhibits a low but significant capacity to phosphorylate the noncognate substrate AA in vitro.
We next wanted to know if elimination of RsbW had a measurable effect on σF activity or sporulation. We monitored σF activity by use of a lacZ fusion to a σF-dependent promoter in an otherwise wild-type strain and also in a strain lacking the entire sigB operon. There was no significant difference in σF activity between the two strains (data not shown). We also detected no significant difference in the relative levels of AA and AA-P between the two strains, nor any difference in sporulation efficiency as determined by the production of heat-resistant spores (data not shown). We conclude that RsbW is present and active in sporulating cells and is capable of phosphorylating AA, but that under the conditions used in this investigation any such cross talk has no significant physiological consequence. It is possible that an RsbW-mediated impact on σF activity would become evident if σB was highly induced by some additional stress applied early in sporulation. σB induction would be expected to increase the cellular concentration of RsbW relative to that of AB due to the autocatalytic activation of the last four downstream genes in the σB operon (16). In support of this possibility, Mendez et al. recently reported results that suggest induction of σB by low temperature does reduce σF activity (20).
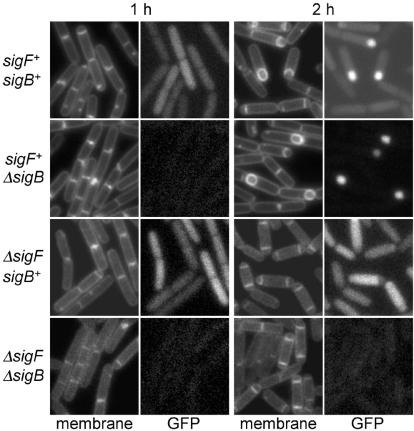
Whereas the σB pathway does not seem to materially contribute to σF-directed gene expression under standard sporulation conditions, σF may contribute to extending the expression of certain genes under the control of σB, as we now explain. To determine if σB was active during sporulation, we created a fusion of the gene for the green fluorescent protein to the promoter for ctc, a standard reporter for σB activity (14, 15, 21). A PCR fragment containing the ctc promoter region (from 199 nucleotides upstream of the ctc start codon to 19 nucleotides upstream of the start codon, thereby excluding the endogenous ribosome-binding site) was ligated into pMF22 (8) digested with EcoRI and HindIII to create pKC62 harboring the reporter construct (see Table 1). The results of fluorescence microscopy (carried out as described in reference 10) showed that the ctc-gfp fusion was active at the start of sporulation and that a further-enhanced level of fluorescence could be observed in the forespore after asymmetric division (Fig. 4). The use of mutants lacking σB or σF demonstrated that expression of the fusion in predivisional sporangia was dependent upon the general stress-response factor σB but that expression in the forespore was dependent upon the sporulation regulatory factor σF (Fig. 4). Consistent with this interpretation, expression of a ctc-lacZ fusion (5) as monitored by the accumulation of β-galactosidase (assay performed as described in reference 11) could be detected at hour 1 of sporulation in cells that were wild type for σB but not until hour 2 in cells that were mutant for σB. Little or no expression of ctc-lacZ could be detected at either time in cells that lacked both σB and σF (Table 2). The simplest explanation for these results is that σF is capable of recognizing the promoter for ctc and sustaining its transcription during sporulation. A second example of a gene whose expression is influenced by both σF and σB is katX, which encodes a catalase active during spore germination (4). Like ctc, katX is transcribed in a σB-dependent manner under stress conditions and in a σF-dependent, σB-independent manner during sporulation (4, 25).
FIG. 4.
Transcription from the ctc promoter is directed by both σB and σF. Fluorescence of green fluorescent protein (GFP) expressed under the control of the ctc promoter was monitored by fluorescence microscopy 1 and 2 h after the start of sporulation in cells wild type for sigB and sigF (KC495), with a deletion of sigB (KC496), with a deletion of sigF (KC504), or with a deletion of both sigF and sigB (KC505). Cell membranes were stained with trimethylammonium diphenyl-hexatriene p-toluenesulfonate. One-second exposures were captured with Metamorph version 4.6 (Universal Imaging) with autoscaling on.
TABLE 2.
Transcription from the ctc promoter is directed by both σB and σF
| Strain (genotype) | Activity (Miller units) aftera:
|
|
|---|---|---|
| 1 h | 2 h | |
| KC479 (sigF+sigB+) | 35 | 37 |
| KC482 (sigF+ ΔsigB) | 1 | 9 |
| KC502 (ΔsigF sigB+) | 28 | 27 |
| KC503 (ΔsigF ΔsigB) | 1 | 1 |
Activity of β-galactosidase expressed under the control of the ctc promoter was assayed after 1 and 2 h of sporulation.
The ctc and katX promoter sequences are similar to each other and to the consensus sequences for both σF- and σB-dependent promoters (25, 29). It is not readily apparent what promoter characteristics might prevent transcription of the σB-regulated genes by σF-associated RNA polymerase. To date there are no other confirmed examples of overlap between the two regulons. However, the known σF regulon remains quite small (13), and it is possible that ctc and katX are representatives of a larger class of genes that is regulated by both sigma factors.
The chief conclusion from the present investigation is that under normal conditions the σF pathway is well insulated from the regulatory components of the σB pathway. We propose that this insulation is achieved both by the action of the SpoIIE phosphatase and by the substrate specificity of the RsbW kinase. RsbW can phosphorylate AA and, as we have shown, even allow AA-P to accumulate to high levels, but only under conditions in which the SpoIIE phosphatase is absent. Thus, tight segregation of one regulatory system from the other is a composite consequence of both inefficient phosphorylation by the noncognate kinase and efficient dephosphorylation by the cognate phosphatase. In this view, the absence of cross talk is achieved by the integrated activity of the entire regulatory system, rather than by selective substrate specificity alone.
This principle, namely, that the composite action of both kinases and phosphatases is responsible for preventing cross talk, may apply to other regulatory systems as well. These include eukaryotic signaling pathways that function by phosphorylation and dephosphorylation and also bacterial two-component regulatory systems, for which multiple paralogs are commonly present within a single cell. In two-component systems, the sensor kinase is often both a kinase and a phosphatase that acts on the phosphorylated form of the cognate response regulator (22, 24, 28). In some cases a sensor kinase for one two-component system can be seen to phosphorylate the response regulator for a second two-component system when the cognate sensor kinase, and hence its phosphatase, is absent. For example, activation of the response regulator that governs the response to conditions of nitrogen limitation, NtrC, has been observed in cells that lack the cognate histidine kinase, NtrB (30). Absent the NtrB kinase/phosphatase, NtrC is evidently able to undergo cross-activation by other, noncognate histidine kinases (3, 30).
Our results and those of others demonstrate that interactions that can occur between an enzyme and a substrate in isolation may not occur under physiological conditions due to the presence of competing reactions that serve to insulate signal transduction pathways. Such interactions pose a challenge to current efforts to determine the topologies of cellular networks with high-throughput assays. These endeavors must ultimately include some way to determine which of the many interactions observed in a given assay are spurious or neutral and which are physiologically meaningful and adaptive.
Acknowledgments
We thank Hélène Ceremonie for constructing pHC10 and strain PB642.
K.C. was supported by a National Science Foundation predoctoral fellowship. This work was supported by NIH grants GM18458 to R.L. and GM42077 to C.W.P.
REFERENCES
- 1.Alper, S., A. Dufour, D. A. Garsin, L. Duncan, and R. Losick. 1996. Role of adenosine nucleotides in the regulation of a stress-response transcription factor in Bacillus subtilis. J. Mol. Biol. 260:165-177. [DOI] [PubMed] [Google Scholar]
- 2.Arigoni, F., A. M. Guerout-Fleury, I. Barak, and P. Stragier. 1999. The SpoIIE phosphatase, the sporulation septum and the establishment of forespore-specific transcription in Bacillus subtilis: a reassessment. Mol. Microbiol. 31:1407-1415. [DOI] [PubMed] [Google Scholar]
- 3.Armitage, J. P., C. J. Dorman, K. Hellingwerf, R. Schmitt, D. Summers, and B. Holland. 2003. Thinking and decision making, bacterial style: bacterial neural networks, Obernai, France, 7th-12th June 2002. Mol. Microbiol. 47:583-593. [DOI] [PubMed] [Google Scholar]
- 4.Bagyan, I., L. Casillas-Martinez, and P. Setlow. 1998. The katX gene, which codes for the catalase in spores of Bacillus subtilis, is a forespore-specific gene controlled by σF, and KatX is essential for hydrogen peroxide resistance of the germinating spore. J. Bacteriol. 180:2057-2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boylan, S. A., A. Rutherford, S. M. Thomas, and C. W. Price. 1992. Activation of Bacillus subtilis transcription factor σB by a regulatory pathway responsive to stationary-phase signals. J. Bacteriol. 174:3695-3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carniol, K., P. Eichenberger, and R. Losick. 2004. A threshold mechanism governing activation of the developmental regulatory protein σF in Bacillus subtilis. J. Biol. Chem. 279:14860-14870. [DOI] [PubMed] [Google Scholar]
- 7.Decatur, A. L., and R. Losick. 1996. Three sites of contact between the Bacillus subtilis transcription factor σF and its antisigma factor SpoIIAB. Genes Dev. 10:2348-2358. [DOI] [PubMed] [Google Scholar]
- 8.Fujita, M., and R. Losick. 2003. The master regulator for entry into sporulation in Bacillus subtilis becomes a cell-specific transcription factor after asymmetric division. Genes Dev. 17:1166-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gaidenko, T. A., X. Yang, Y. M. Lee, and C. W. Price. 1999. Threonine phosphorylation of modulator protein RsbR governs its ability to regulate a serine kinase in the environmental stress signaling pathway of Bacillus subtilis. J. Mol. Biol. 288:29-39. [DOI] [PubMed] [Google Scholar]
- 10.Gueiros-Filho, F. J., and R. Losick. 2002. A widely conserved bacterial cell division protein that promotes assembly of the tubulin-like protein FtsZ. Genes Dev. 16:2544-2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harwood, C. R., and S. M. Cutting (ed.). 1990. Molecular biological methods for Bacillus. John Wiley & Sons, New York, N.Y.
- 12.Hecker, M., and U. Volker. 2001. General stress response of Bacillus subtilis and other bacteria. Adv. Microb. Physiol. 44:35-91. [DOI] [PubMed] [Google Scholar]
- 13.Helman, J., and C. P. Moran. 2002. Polymerase and sigma factors, p. 289-312. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, D.C.
- 14.Igo, M., M. Lampe, C. Ray, W. Schafer, C. P. Moran, Jr., and R. Losick. 1987. Genetic studies of a secondary RNA polymerase sigma factor in Bacillus subtilis. J. Bacteriol. 169:3464-3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Igo, M. M., and R. Losick. 1986. Regulation of a promoter that is utilized by minor forms of RNA polymerase holoenzyme in Bacillus subtilis. J. Mol. Biol. 191:615-624. [DOI] [PubMed] [Google Scholar]
- 16.Kalman, S., M. L. Duncan, S. M. Thomas, and C. W. Price. 1990. Similar organization of the sigB and spoIIA operons encoding alternate sigma factors of Bacillus subtilis RNA polymerase. J. Bacteriol. 172:5575-5585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kang, C. M., M. S. Brody, S. Akbar, X. Yang, and C. W. Price. 1996. Homologous pairs of regulatory proteins control activity of Bacillus subtilis transcription factor σB in response to environmental stress. J. Bacteriol. 178:3846-3853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kang, C. M., K. Vijay, and C. W. Price. 1998. Serine kinase activity of a Bacillus subtilis switch protein is required to transduce environmental stress signals but not to activate its target PP2C phosphatase. Mol. Microbiol. 30:189-196. [DOI] [PubMed] [Google Scholar]
- 19.Kearns, D. B., and R. Losick. 2003. Swarming motility in undomesticated Bacillus subtilis. Mol. Microbiol. 49:581-590. [DOI] [PubMed] [Google Scholar]
- 20.Mendez, M. B., L. M. Orsaria, V. Philippe, M. E. Pedrido, and R. R. Grau. 2004. Novel roles of the master transcription factors Spo0A and σB for survival and sporulation of Bacillus subtilis at low growth temperature. J. Bacteriol. 186:989-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moran, C. P., Jr., W. C. Johnson, and R. Losick. 1982. Close contacts between sigma 37-RNA polymerase and a Bacillus subtilis chromosomal promoter. J. Mol. Biol. 162:709-713. [DOI] [PubMed] [Google Scholar]
- 22.Ninfa, E. G., M. R. Atkinson, E. S. Kamberov, and A. J. Ninfa. 1993. Mechanism of autophosphorylation of Escherichia coli nitrogen regulator II (NRII or NtrB): trans-phosphorylation between subunits. J. Bacteriol. 175:7024-7032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pan, Q., D. A. Garsin, and R. Losick. 2001. Self-reinforcing activation of a cell-specific transcription factor by proteolysis of an anti-sigma factor in B. subtilis. Mol. Cell 8:873-883. [DOI] [PubMed] [Google Scholar]
- 24.Parkinson, J. S., and E. C. Kofoid. 1992. Communication modules in bacterial signaling proteins. Annu. Rev. Genet. 26:71-112. [DOI] [PubMed] [Google Scholar]
- 25.Petersohn, A., S. Engelmann, P. Setlow, and M. Hecker. 1999. The katX gene of Bacillus subtilis is under dual control of σB and σF. Mol. Gen. Genet. 262:173-179. [DOI] [PubMed] [Google Scholar]
- 26.Piggot, P. J., and R. Losick. 2002. Sporulation genes and intercompartmental regulation, p. 483-518. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, D.C.
- 27.Price, C. 2002. General stress response, p. 369-384. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relative: from genes to cells. ASM Press, Washington, D.C.
- 28.Stock, J. B., A. J. Ninfa, and A. M. Stock. 1989. Protein phosphorylation and regulation of adaptive responses in bacteria. Microbiol. Rev. 53:450-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tatti, K. M., and C. P. Moran, Jr. 1984. Promoter recognition by sigma-37 RNA polymerase from Bacillus subtilis. J. Mol. Biol. 175:285-297. [DOI] [PubMed] [Google Scholar]
- 30.Verhamme, D. T., J. C. Arents, P. W. Postma, W. Crielaard, and K. J. Hellingwerf. 2002. Investigation of in vivo cross-talk between key two-component systems of Escherichia coli. Microbiology 148:69-78. [DOI] [PubMed] [Google Scholar]
- 31.Yang, X., C. M. Kang, M. S. Brody, and C. W. Price. 1996. Opposing pairs of serine protein kinases and phosphatases transmit signals of environmental stress to activate a bacterial transcription factor. Genes Dev. 10:2265-2275. [DOI] [PubMed] [Google Scholar]
- 32.Youngman, P., J. B. Perkins, and R. Losick. 1984. Construction of a cloning site near one end of Tn917 into which foreign DNA may be inserted without affecting transposition in Bacillus subtilis or expression of the transposon-borne erm gene. Plasmid 12:1-9. [DOI] [PubMed] [Google Scholar]