Abstract
The centrosome cycle is most often coordinated with mitotic cell division through the activity of various essential cell cycle regulators, consequently ensuring that the centriole is duplicated once, and only once, per cell cycle. However, this coupling can be altered in specific developmental contexts; for example, multi-ciliated cells generate hundreds of centrioles without any S-phase requirement for their biogenesis, while Drosophila follicle cells eliminate their centrosomes as they begin to endoreduplicate. In order to better understand how the centrosome cycle and the cell cycle are coordinated in a developmental context we use the endoreduplicating intestinal cell lineage of C. elegans to address how novel variations of the cell cycle impact this important process. In C. elegans, the larval intestinal cells undergo one nuclear division without subsequent cytokinesis, followed by four endocycles that are characterized by successive rounds of S-phase. We monitored the levels of centriolar/centrosomal markers and found that centrosomes lose their pericentriolar material following the nuclear division that occurs during the L1 stage and is thereafter never re-gained. The centrioles then become refractory to S phase regulators that would normally promote duplication during the first endocycle, after which they are eliminated during the L2 stage. Furthermore, we show that SPD-2 plays a central role in the numeral regulation of centrioles as a potential target of CDK activity. On the other hand, the phosphorylation on SPD-2 by Polo-like kinase, the transcriptional regulation of genes that affect centriole biogenesis, and the ubiquitin/proteasome degradation pathway, contribute collectively to the final elimination of the centrioles during the L2 stage.
Introduction
In many animal cells, the centrosome acts as the major microtubule organization center (MTOC) playing a key role in defining cell shape, cell division and overall microtubule geometry [1]. This MTOC function is of special importance in proliferating cells where the two centrosomes are responsible for accurately establishing the bipolar spindle. Therefore, centrosome function and its numeral integrity are essential for many organisms. Altering their numbers can lead to genomic instability and/or tumorigenesis [2]–[3].
The centrosome consists of a pair of barrel-shaped centrioles surrounded by pericentriolar material (PCM) [4]. During the centrosome cycle, the centrioles must disengage, duplicate, separate and undergo centrosome maturation, while each event takes place at an appropriate stage of the cell cycle under the control of stage-specific cell cycle kinases. A Polo-like kinase-1 (PLK-1) contributes to disengagement between the parental centrioles during M phase and consequently licenses centriole duplication in response to Cyclin-Dependent Kinase (CDK)-2 in the subsequent G1/S [5]–[6]. During G2/M, the parental centrioles further separate under CDK-1 and PLK-1 control [7]–[9]. The two centrosomes undergo maturation by recruiting PCM, consequently increasing the microtubule organizing capacity of the centrosome. This process is regulated by M phase kinases, such as PLK-1 and Aurora A kinases [10]–[11].
Centriole assembly is rate-limiting during duplication. In C. elegans it involves the sequential recruitment of many proteins that are conserved from invertebrates to humans [12]–[15]. Among these, a coiled-coil protein called SPD-2 plays a critical role in the process being the first of several proteins that localize to the mother centriole during centriole biogenesis. It is thereafter joined by ZYG-1, a protein kinase that is likely analogous to PLK-4 [16]–[19]. SAS-6, a probable ZYG-1 target [20]–[21] joins the complex thereafter with SAS-5 to assemble the “central tube” structure. Finally SAS-4 will be recruited to regulate microtubule attachment onto the central tube [12]–[13], [22]–[24].
Because various enzyme activities that drive cell division are also required for centriole duplication, the two processes are considered to be “coupled”. However, such coupling can be altered in various contexts. For example, in some respiratory epithelia hundreds of centriole-derived organelles that are critical for ciliogenesis called basal bodies are generated spontaneously without any requirement for DNA replication [25]–[26]. The converse is also true in the endocycling follicle cells of the Drosophila melanogaster egg chamber, wherein the centriole does not duplicate with each round of S phase and is eventually eliminated [27]–[28]. In each of these developmental contexts centriole duplication must be uncoupled from the cell cycle, yet how this uncoupling occurs remains poorly understood.
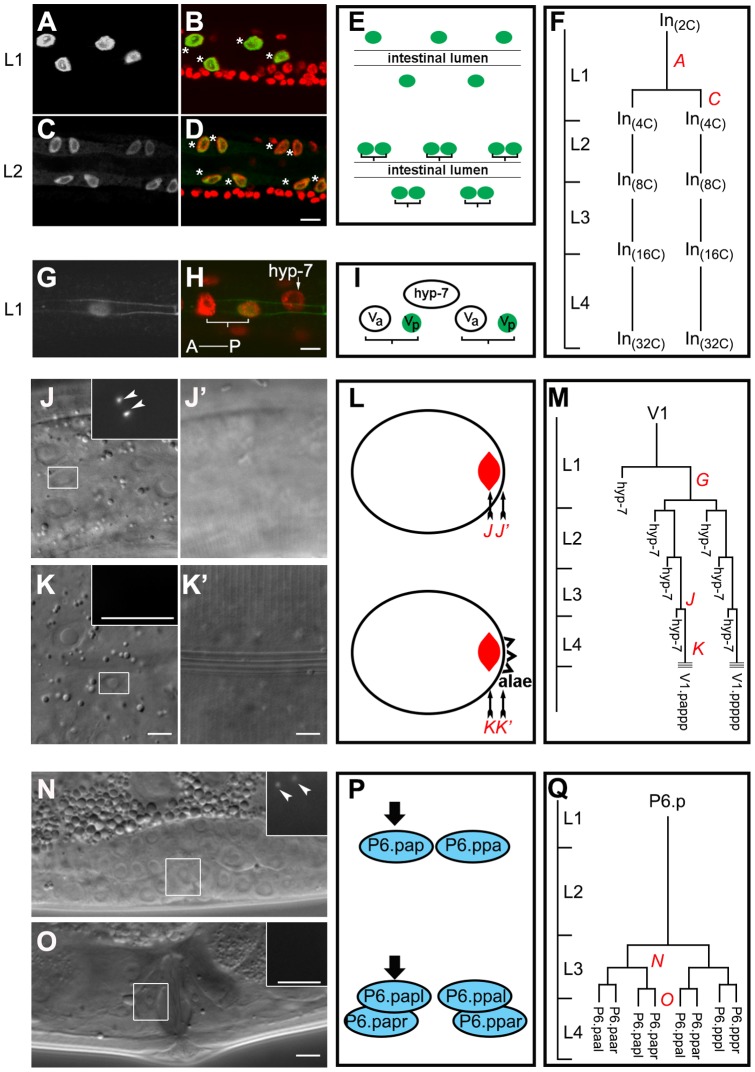
In C. elegans both the intestine and the lateral hypodermal cells execute endocycles during larval development, giving rise to polyploid cells in the adult [29]. The intestinal nuclei undergo a single round of nuclear division in the absence of cytokinesis at the end of the first larval stage (L1) to become binucleate (Figure 1A–1E), followed by a single endocycle at the end of each larval stage [29] (Figure 1F). In the hypodermal V cell lineage, an anterior daughter cell is generated that undergoes endoreduplication and will eventually fuse with the hyp7 syncytium, while the posterior seam cell daughter will divide once during the L1 (Figure 1G–1I, 1M). After an equational division at the L1/L2 transition the V cell lineage repeats its L1 pattern of cell division in each subsequent larval stage, yielding one anterior endocycling cell that fuses with the hypodermis and its sister that will continue to execute a mitotic stem cell division [29] (Figure 1M).
Figure 1. Centrioles are eliminated in many somatic cells of C. elegans following the completion of mitosis.
(A–D) Larvae expressing intestine-specific elt-2::GFP were stained with DAPI (red) and anti-GFP (green) [44]. (A and C) show anti-GFP signal alone. Asterisks mark the intestinal nuclei. (E) A schematic diagram shows the relative position of intestinal nuclei before and after the nuclear division at the L1/L2 transition. Lineage brackets indicate two daughter cells from a common intestinal cell mother. Green ovals, intestinal nuclei. (F) A representative map of the postembryonic intestinal cell lineage: C refers to haploid DNA content in the nuclei. L1–L4 on the y axis indicate developmental timing showing the different larval stages; In, intestinal cells [29]–[30]. (G and H) Animals co-expressing the adherens junction marker ajm-1::GFP and seam cell marker scm-1::GFP [76] were stained with DAPI (red) and anti-GFP (green). (G) shows anti-GFP signal alone. Lineage brackets indicate two daughter cells from a common V cell mother. hyp7 and the arrow marks a hyp7 nucleus. A, anterior; P, posterior. (I) A schematic diagram summarizes the relative positions of V cells shown in (G) and (H). (J–K′) SPD-2::GFP was observed in the seam cells before, but not after the final cell division. White rectangles indicate the seam cells from the V1 lineage and the insets represent the magnified views of GFP signal in the corresponding white rectangles in J and K. J′ and K′ show the focal plane of the cuticle to indicate the presence or absence of adult alae (which indicates terminally differentiated seam cells in J and K), respectively. (L) A schematic diagram indicates the cross section view of a C. elegans body. Red spindles, V cell nuclei. The red italic letters and the black arrows together indicate the focal planes in the corresponding micrographs. (M) A map of the V1 lineage. The parallel lines indicate the alae/terminal differentiation. (N and O) SPD-2::GFP can be seen in the vulva cell lineage (P6.p) before (N) but not after (O) the completion of cell division. White rectangles highlight P6.p descendants and the insets represent the magnified views of GFP signal in the corresponding white rectangles. (P) A schematic diagram highlights later P6.p cell divisions a-anterior; p-posterior; l-left; r-right. Blue ovals depict nuclei of P6.p descendants. Black arrows point out the boxed nuclei in (N) or (O). (Q) A map of the P6.p cell lineage. The arrowheads indicate the SPD-2 foci. Scale bar, 5 µm. Red italicized letters in the lineage maps F, M, and Q show the precise time when the cells represented in the corresponding panels (non-italicized bold letters) were imaged.
Because the endocycling cells undergo reiterative rounds of DNA replication, it is unclear how the centrioles would respond to these successive rounds of S-phase-associated enzyme activity. We therefore used the postembryonic intestinal cell lineage as a model to determine the fate of centrioles in these endocycling cells and found that the centrioles lose their PCM following the nuclear division that occurs during the L1 stage and never regain it thereafter. Centriole duplication then becomes uncoupled from the first S-phase of the endocycles (endo-S), which precedes their elimination later during the L2 stage. We show that SPD-2, an important centriolar and pericentriolar component, may play a central role in the numeral regulation of centriole duplication, while transcriptional regulation of genes that affect centriole biogenesis, concomitant with the timely function of the ubiquitin/proteasome degradation pathway, contribute to the final elimination of the centrioles during the L2 stage.
Results
The centriole is eliminated in endocycling cells
During post-embryonic development in C. elegans, cells in both the hypodermal V cell and the intestinal E lineage execute endocycles to generate polyploid cells [29]–[30]. The successive cycles of DNA replication that are characteristic of the endocycle are driven by canonical S-phase regulators, many of which have previously been shown to trigger centriole duplication during the mitotic cell cycle [5], [31]–[32]. If centrosome duplication remains sensitive to the S-phase CDK activity the centrioles could potentially duplicate at each S-phase, resulting in an accumulation of centrioles in the polyploid adult cells. Alternatively, the centrioles might behave as they do in the Drosophila follicle cells, and become “uncoupled” from the endo-S-phase activities to be subsequently eliminated [28]. We therefore determined the centriole numbers/fate in the polyploid cells of C. elegans to distinguish between these possibilities.
We monitored the levels of two centriolar proteins in the intestinal cells throughout postembryonic development: SPD-2, which is associated both with the centriole and the PCM, and a highly conserved centriolar component called SAS-4 that is associated exclusively with centrioles [13], [33]. We first fused SPD-2 to GFP and found that it is most prominently expressed in the distal, mitotic region of the adult germ line (Figure 2A, a and a′), yet was notably absent from the adult intestinal cells (Figure 2A, b and b′), suggesting that SPD-2 was either not expressed in the adult intestinal lineage, and/or it was eliminated during development.
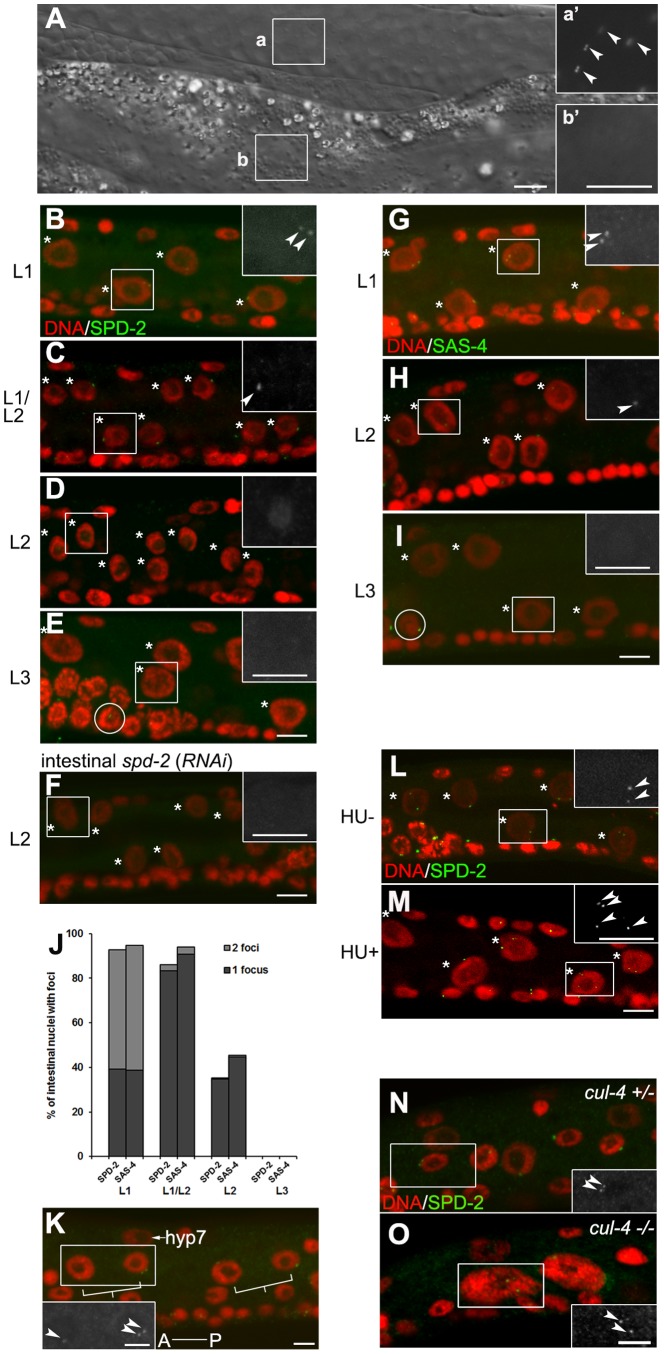
Figure 2. Centrioles no longer duplicate in endocycling cells prior to their elimination.
(A) SPD-2::GFP signal can be seen throughout the germ line until oogenesis in (a), while in (b) it is undetectable in the intestinal cells in adult hermaphrodites. a′ and b′ are high magnification images of the GFP signal in the field identified by the two white rectangles a and b. (B–F) and (G–I) SPD-2 and SAS-4 foci are detectable up to the L1/L2 transition. The signal is no longer detectable by the L3 stage. The nuclear localized SPD-2 is absent following spd-2(RNAi) in (F). Animals were stained with DAPI (red) and anti-SPD-2 (green) in (B–F) or anti-SAS-4 (green) in (G–I). Arrowheads indicate centrioles, while asterisks indicate intestinal nuclei. The insets represent magnified SPD-2 or SAS-4 signal of the highlighted regions (white rectangles). The circles in (E) and (I) highlight the germ cells with either SPD-2 foci or SAS-4 foci. (J) Quantification of centriole numbers and described in (B–E) and (G–I) based on SPD-2 or SAS-4 detection. n≥56 for each stage. (K) Centriole duplication is uncoupled from the endocycles in the lateral hypodermal V cells in the late L1 stage. The centriole appears to be uncoupled from S-phase in the anterior endoreduplicating daughter cell but remains coupled to DNA replication in the mitotic posterior daughter. The square brackets indicate the daughter cells from a common mother V cell, while arrowheads indicate the centrioles. The inset is a magnified view of the region delineated by the white rectangle. A, anterior; P, posterior. hyp7 marks the hyp7 nucleus. (L and M) Supernumerary centrioles are detected in HU-treated L1 animals (M) but not in the control (L). The inset is a magnified SPD-2 signal of the region delineated by the white rectangle. Arrowheads indicate centrioles, while asterisks indicate intestinal nuclei. (N and O) Centriole duplication occurs once in response to S phase, but centrioles do not overduplicate and are not eliminated during un-quantized DNA re-replication. Heterozygous in (N) and homozygous cul-4 (gk434) mutants in (O) were stained with DAPI (red) and SPD-2 (green) respectively. The insets show the SPD-2 signal in the regions outlined by the white rectangles. Arrowheads indicate centrioles. Scale bar, 5 µm.
We therefore examined SPD-2 expression at each stage in the postembryonic intestine using the L1 nuclear division as a developmental landmark to discern between the L1 and L2 stage, since it represents the end of the mitotic cell cycle program. In wild type larvae, throughout the L1 stage and until ∼3 hours after the nuclear division, SPD-2 was present at the centrioles in the majority of the intestinal cells (Figure 2B, 2C, 2J). However, SPD-2 became undetectable during progression through the L2 stage: only half of the intestinal nuclei possessed SPD-2 foci 6–8 hours after the nuclear division (Figure 2J). This progressive loss of SPD-2 precedes its complete elimination by the L3 stage (Figure 2E, 2J).
In parallel, like SPD-2, SAS-4 was associated with intestinal nuclei throughout the L1 and during the L1/L2 transition before becoming undetectable by the end of the L2 (Figure 2G–2I, 2J), suggesting that the loss of SPD-2 signal during the L2 faithfully represents the fate of the centriole. Taken together, our observations reveal that the centrioles are still present until the completion of the nuclear division in the L1, but are progressively eliminated during the L2 stage in the intestinal cells.
Centriole duplication becomes uncoupled from S phase in endocycling cells
The intestinal nuclear division is therefore a temporal landmark that not only marks the end of the mitotic cell division cycle in this lineage but also the onset of centriole elimination. Prior to the intestinal nuclear division, two SPD-2-positive foci are detectable in more than half of the intestinal nuclei 10–12 h into the L1 stage (Figure 2B, 2J), a period that corresponds to the final S-phase before the onset of the endocycle program [29]. The same intestinal nuclei also harbor two SAS-4-positive foci likely representing two pairs of centrioles that arose from a single round of duplication (Figure 2G, 2J). In order to confirm if the two SPD-2 foci result from centriole duplication during the final mitotic S-phase, we labeled cells with propidium iodide (PI) to quantify the DNA content in the intestinal nuclei, providing us with a pre-/post-S-phase reference point [34]. From our analysis we noted that single SPD-2 foci, most likely representing a pair of centrioles, were predominant in 2C intestinal cells (Figure S1A; average intestinal cell C value = 1.98); whereas the majority of 4C intestinal cells contain SPD-2 doublets corresponding to two pairs of centrioles (Figure S1B; average intestinal cell C value = 4.02), suggesting that centriole duplication is appropriately coupled during the final mitotic S-phase in the L1 intestinal cells.
After the nuclear division, only a single SPD-2 focus (1 centriole pair) is detectable adjacent to the majority of the 4C nuclei that are generated after the first endo-S phase (Figure 2C, 2J, S1C; average intestinal cell C value = 3.98). Moreover, the numbers of SAS-4 foci are similar to the numbers of SPD-2 foci observed following the intestinal nuclear division (Figure 2H, 2J), suggesting that the centrioles do not duplicate after the division despite the initiation of initial S-phase associated with the first endocycle. Although we never see two SPD-2/SAS-4 foci after the nuclear division, we cannot formally rule out that the centrioles indeed duplicate but never separate, making it impossible to resolve them by standard confocal microscopy. The intestinal nuclear division is therefore a critical landmark that is associated with a potential change in the ability of the centriole to duplicate in response to the surrounding S-phase CDK activity. This apparent refractoriness may be a general feature of endoreduplicating cells, since a similar SPD-2 singlet was present in the anterior endoreduplicating daughter cells of the V cell lineage, whereas a doublet was observed in the posterior seam cell nuclei that do not undergo endocycles, but rather execute mitotic cell divisions (Figure 2K).
We also noticed that the majority of the SPD-2 signal transiently diffuses throughout the nuclei beginning at the L2, before the signal becomes undetectable later during the L2 stage (Figure 2D). This nuclear signal disappears following spd-2(RNAi), demonstrating that this signal is indeed SPD-2-specific (Figure 2F). Similarly, in the germ line, where centrioles are eliminated at the onset of oogenesis [35]–[36], SPD-2 also becomes diffuse prior to the loss of centriolar markers (Figure S2). This transient change in SPD-2 localization appears to precede the elimination of the centrioles and may reflect specific modifications of SPD-2 that determine centriole stability in both the intestine and the germ line.
The intestinal nuclear division is followed by a failure to recruit PCM to the centriole
One of the major functions of the centrosome is to organize the mitotic spindle during mitosis through recruitment of the γ-tubulin complex and other components of the PCM. The changes in centriole duplication and stability during the mitosis-endocycle transition led us to examine if any functional modification of the centrioles might also occur as a consequence of this process. Previous studies indicated that γ-tubulin recruitment is subject to cell cycle-dependent regulation in mitotic cells [37]. During centrosome maturation γ-tubulin accumulates around the centrioles, resulting in substantially enlarged γ-tubulin foci at metaphase. The intensity of γ-tubulin gradually returns to baseline levels at the onset of the next interphase [37]. We observed similar baseline levels of γ-tubulin expression during the L1 interphase (Figure 3A, 3B). The intensity of the γ-tubulin foci increases substantially when the metaphase chromosomes become discernable (Figure 3C, 3D; n = 15). During anaphase, most of the centrosomal γ-tubulin rapidly disperses prior to the onset of the following S-phase (Figure 3C, 3D). The anaphase dispersal of γ-tubulin is not due to the disappearance of the centriole (Figure S3A, a-a” and b-b”), since centriole elimination only begins later during the L2 stage (Figure 2J). Moreover, the γ-tubulin levels around the centrioles never recover thereafter (Figure 3E–3H). This change likely compromises the ability of the centrioles to function as a MTOC in intestinal cells following this stage (Figure S3B). This anaphase dispersal of γ-tubulin does not occur in the mitotically proliferating cells in the ventral hypodermis (Figure 3I, 3J), or in the germ cells that are simultaneously undergoing mitotic divisions (Figure 3K, 3L). Not surprisingly, centrioles still act as an MTOC in these mitotic cells (Figure S3C). Overall, our observations indicate that just subsequent to the intestinal cell nuclear division that occurs at late L1 stage, the γ-tubulin that is associated with the centriole disperses, thereafter compromising the ability of the centriole to act as a MTOC.
Figure 3. Centrioles lose their capacity to recruit γ-tubulin following the intestinal nuclear division that precedes the onset of endoreduplication.
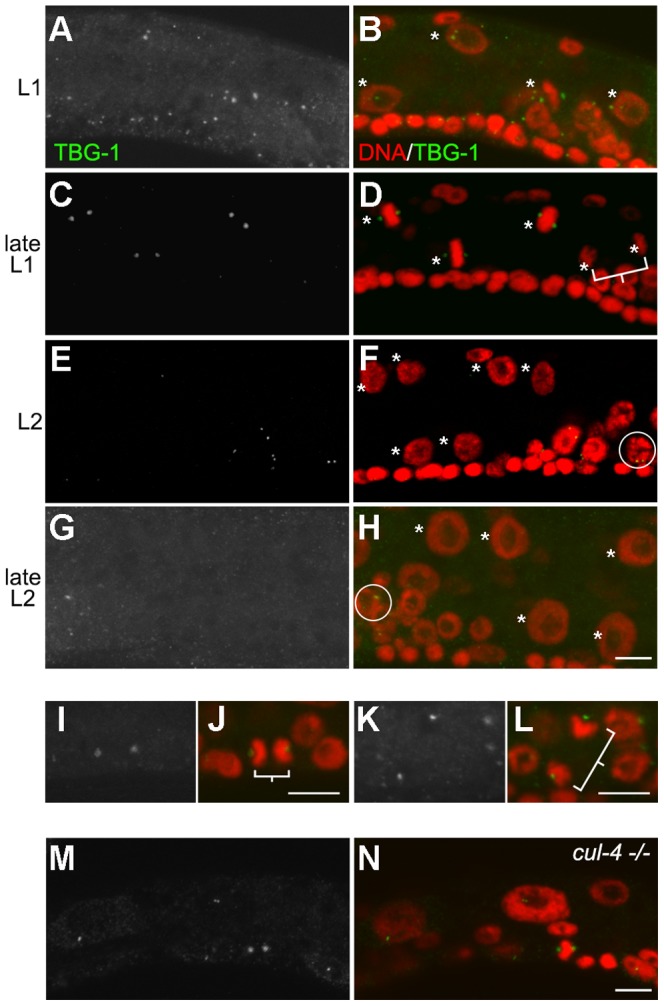
(A–H) Wild type larvae were stained for γ-tubulin (TBG-1) before, during, and after the intestinal nuclear division. (A), (C), (E) and (G) represent the TBG-1 only; (B), (D), (F), and (H) TBG-1 with DAPI. The asterisks indicate the intestinal nuclei and the square bracket highlights a pair of sister intestinal nuclei. The circles in (F) and (H) highlight germ cells with γ-tubulin foci. (I–L) High resolution micrograph of cells following division during the L1 stage. (I and K) show TBG-1 signal only; (J and L) TBG-1 with DAPI. (I and J), dividing P cell in the ventral hypodermis; (K and L), germ line precursor cell. Square brackets highlight sister cells. (M and N) Similar staining was performed in cul-4 (gk434) homozygous animals. All cells were stained with DAPI (red) and anti-γ-tubulin (green). Scale bar, 5 µm.
Centriole response during unscheduled DNA synthesis
We noted that centrioles do not duplicate with each round of DNA replication in endocycling cells but are essentially refractory to the endo-S and are subsequently eliminated during the L2 stage. To determine whether the uncoupling of centriole duplication from S-phase activity and subsequent elimination is unique to endocycling cells, or alternatively a general response to reiterative or prolonged phases of DNA replication, we examined centriole behaviour during contexts of extended or misregulated S phase. First we used hydroxyurea (HU) to arrest DNA replication in cells during the L1 stage prior to the final mitotic S phase and then we tested the effects of HU just prior to the onset of the endocycle program [38]–[39]. Similar to what is observed in HU-treated vertebrate cells or in the blastomeres of the C. elegans embryo, a small population (∼15%) of intestinal cells that were HU-arrested before the nuclear division did indeed possess supernumerary centrioles (Figure 2L, 2M; Table 1), [38]–[40], while the centriole numbers indicate that only a single extra duplication event per centriole took place during this period (Figure 2L, 2M). Furthermore, the number of HU-blocked cells that possess supernumerary centrioles increased with time (data not shown) suggesting that a critical constraint to centriole duplication may be progressively relieved during prolonged S phase.
Table 1. % of supernumerary centrioles in HU treated animals.
The frequency of supernumerary centrioles is quantified by the number of intestinal nuclei with more than two SPD-2 foci during the L1 arrest or more than one SPD-2 focus during the L2 arrest.
*6-hour HU treatment on synchronized L1 animals.
**6-hour HU treatment on post L1 nuclear division larvae.
In contrast, when the same treatment was performed for a comparable duration after the nuclear division and prior to the onset of the endocycles, no detectable centriole amplification was observed compared to the control intestinal cells (Table 1). In all cases however, whether DNA replication was blocked before or after the nuclear division, following the release from HU treatment the centrioles were eliminated during the L2 stage.
Since the majority of the centrioles in the intestinal cells did not over-duplicate during a prolonged S phase we were curious whether the centrioles in other cell types might react in a similar way when the cycles of DNA replication and mitosis are disrupted. A mutation in cul-4 stabilizes a positive regulator of a DNA replication licensing factor that causes the hypodermal cells of cul-4 mutants to undergo un-quantized DNA replication yielding nuclei that contain >100 C DNA content (Figure 2O) [41]. We were therefore curious whether the centrioles would duplicate under conditions of re-replication in these hypodermal cells or whether the cells would respond to the re-replication by eliminating the centrioles. We monitored centriole numbers in the hypodermal seam cells of wild type and cul-4 mutant larvae and found that like wild type animals the centrioles duplicate normally once at the first S-phase (Figure 2N), but are neither over-duplicated nor eliminated thereafter during the numerous phases of un-quantized re-replication (Figure 2O). Furthermore, the γ-tubulin levels in the cul-4 hypodermal cells remain restricted to centriolar foci: they no longer fluctuate and are maintained at basal levels typical of interphase cells (Figure 3M, 3N). These observations suggest that the centrioles in cul-4 cells undergo duplication normally in response to the first S phase, but do not duplicate in response to the signals associated with the inappropriate re-replication events that occur thereafter, nor do the centrioles mature to accumulate γ-tubulin. Because these hypodermal cells do not transit through mitosis after their initial S phase, they probably do not become properly licensed for subsequent centriole duplication [6], [42]. However, despite their inability to duplicate and or recruit PCM, these centrioles are never eliminated in the cul-4 mutant hypodermal cells.
The elimination process we observe in the endocycling cells (both the anterior hypdodermal daughters and the intestinal cells) must therefore be dependent on the developmental fate of these cells and independent of the S phase program per se since the cul-4 seam cell centrioles persist into later stages, while the centrioles in the intestinal cells are gradually eliminated following the onset of the endocycle program to become undetectable by the L3 stage, even in situations where the cells are blocked in S phase.
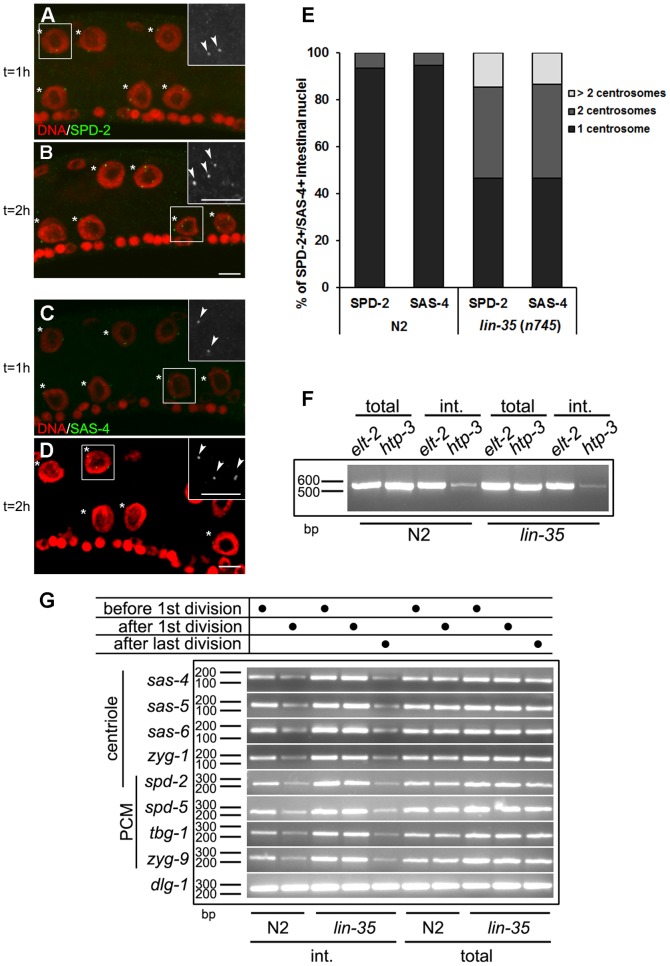
Since the elimination of the centrioles occurs specifically during the L2 stage, we next wondered whether elimination might be under temporal control, or whether it is part of the endocycle program and therefore contingent on the transition from the mitotic divisions to the endocycles. To distinguish between these possibilities, we used a lin-35 (n745) mutant that is mutated for the C. elegans orthologue of Rb [43]. These animals repeat the nuclear divisions, thus giving rise to supernumerary intestinal nuclei prior to their eventual switch to the endocycle program later in L2 stage [44], but developmental timing is otherwise normal. Interestingly, although centriole numbers were unaffected in lin-35 (n745) animals during the L1 stage (data not shown), two SPD-2 foci per 4C nucleus were frequently visualized in lin-35/Rb mutants 1-2 hours after the first nuclear division (Figure 4A, 4E, S1D). Similarly, this was reflected in the number of SAS-4 foci during the L2 stage (Figure 4C, 4E), presumably as a consequence of failing to uncouple centriole duplication from the first endo-S phase. Some animals occasionally even possessed more than two foci (Figure 4B, 4D, 4E). Because we see numeral defects in lin-35/Rb mutants that do not initiate the endocycle program in a timely manner, our data suggest that centriole duplication will be re-licensed as long as cells undergo a mitotic nuclear division and do not execute the endocycle program. However, following the onset of the endocycle program after the nuclear division(s) have terminated, the cells become refractory to the endo-S and will eliminate the centrioles shortly thereafter. Taken together, our data suggest that the apparent uncoupling of centriole duplication from S phase and subsequent centriole elimination relies on the initiation of the endocycle program, and does not result as a consequence of prolonged or aberrant S-phase, or chronological developmental time per se.
Figure 4. lin-35/Rb mutants undergo additional rounds of centriole duplication.
(A and B) lin-35/Rb mutant larvae were stained with DAPI (red) and anti-SPD-2 (green) to monitor centriole dynamics at the nuclear division. The panel (A) was obtained by staining animals approximately one hour after the nuclear division (t = 1 h), while (B) was acquired two hours after the division (t = 2 h). Asterisks indicate the intestinal nuclei and the arrowheads indicate SPD-2 foci. (C and D) lin-35/Rb mutant larvae were stained with DAPI (red) and anti-SAS-4 (green) to monitor centriole numbers after the nuclear division. The insets in (A–D) represent magnified views of regions highlighted by the white rectangles. Scale bar, 5 µm. (E) Quantification of SPD-2 or SAS-4 foci in intestinal nuclei in both wild type and lin-35/Rb mutants two hours after the first intestinal nuclear division. n = 75. (F) RT-PCR analysis of cell-specific transcripts from N2 and lin-35 (n745). elt-2 is intestinal specific, while htp-3 is expressed exclusively in the germ line. (G) The expression of spd-2, -5, zyg-1, -9, sas-4, -5, -6, tbg-1 and dlg-1 (control) [77] was quantified using RT-PCR from total or intestine-enriched mRNA from wild type (N2) or from lin-35 (n745) larvae before or after the first nuclear division, and 6–8 hours after the last nuclear division in lin-35 (n745) mutants. int., intestinal. bp, base pair.
Centriole elimination is preceded by transcriptional attenuation of genes that drive duplication
lin-35/Rb mutants exhibit a plethora of defects that arise due to the misexpression of genes that would normally be silenced [45]. Because we observed aberrant centriole duplication at the L1 nuclear division in the lin-35/Rb mutants, we reasoned that some of the misregulated gene targets in these animals might include genes involved in centriole duplication. In order to precisely analyze the expression levels of these gene products exclusively in the intestine we performed mRNA tagging to enrich for intestinal-specific transcripts following immunoprecipitation (Figure 4F) [46]. Taking advantage of this enriched fraction of intestine-specific mRNA, we monitored the levels of PCM gene products or those required for centriole duplication, at both the pre- and post L1 nuclear division.
During the L1 stage, all the known genes that affect centriole duplication were expressed at higher levels in lin-35 (n745) mutants than in wild type larvae, consistent with previous analyses (Figure 4G) [45]. Elevated expression of these genes is also observed in intestinal cells, which may drive the centriole duplication during the L2 stage.
In wild type larvae, this suite of centriole duplication genes were rapidly reduced in the intestinal mRNA fraction following the nuclear division (L1/L2 transition), while their expression levels remained unchanged in mRNA obtained from whole animals (Figure 4G). This was also reflected in the expression of PCM components [37], [47]–[48]. We then compared the expression levels of these genes between wild type and lin-35/Rb mutants and found that they were not appropriately attenuated in the intestinal cells following the first intestinal nuclear division in lin-35/Rb animals (Figure 4G). However, despite their inability to downregulate these genes immediately following the nuclear division, their expression eventually drops to near wild type levels 6–8 hours after the final observed nuclear divisions take place in the lin-35/Rb mutants (Figure 4G). This results in a significant delay in their attenuation, similar to what is observed with the cyclin genes prior to the onset of the endocycle program [44]. These transcriptional data are further corroborated by our immunostaining results (Figure 4A–4D) and suggest that many of the key genes that regulate centriole duplication are transcriptionally attenuated at, or around, the time that the intestinal cells begin to endoreduplicate. Although this correlation is intriguing our data do not unequivocally confirm that transcriptional attenuation plays a direct role in the eventual elimination of the centrioles by the L3 stage.
SPD-2 phosphorylation in cell cycle uncoupling and stability: CDK and PLK
Centriole duplication is controlled by many cell cycle kinases during mitotic cell division. On the other hand, kinase activities that drive G2 and M phase events are most likely reduced after the final nuclear division in the intestine which precedes centriole elimination. We therefore wondered whether any of the well-characterized cell cycle kinases might modify SPD-2 to affect this process, particularly since recent biochemical and genetic analyses have revealed that direct phosphorylation of centriolar components can impact both duplication and maturation [49]–[51].
Using a bioinformatic Group-based Prediction System (GPS) [52]–[53], we identified a number of predicted phosphorylation sites on SPD-2 (Figure 5A). In order to experimentally test whether these amino acids affect centriole fate in the endocycling intestinal cells we mutated each of these sites to either Alanine (A) or Glutamic Acid (E) to convert the wild type SPD-2 sites into non-phosphorylable or phosphomimetic variants, respectively. The variants, or the wild type SPD-2, were integrated as single copies into the genome [54] and the resulting transgenic animals were crossed into the spd-2 (oj29) mutant background [55].
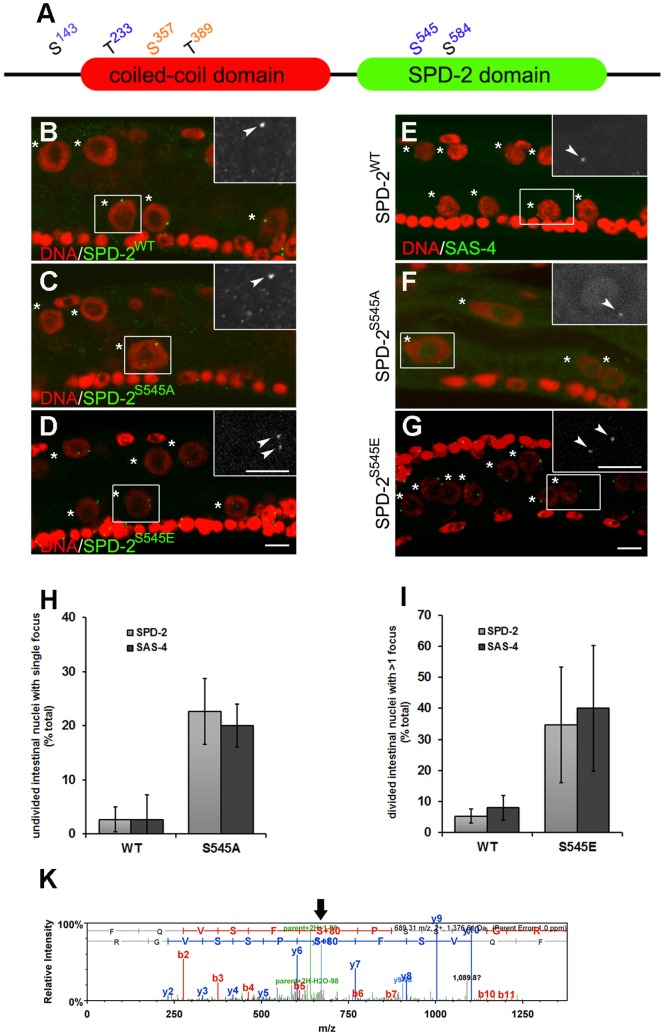
Figure 5. Phosphorylation of SPD-2 affects numeral regulation of centrioles in the intestinal cells.
(A) Diagram of SPD-2 and its potential phosphorylated sites. Numbers represent amino acid position. S, Serine. T, Threonine. Orange numbers: predicted consensus PLK phosphorylation site. Blue numbers: predicted consensus CDK phosphorylation site. Blue or Orange S indicates experimentally-confirmed phosphorylated Serine [58], [78]. (B–D) and (E–G) Early L2 spd-2 (oj29) animals carrying transgenic WT or the S545-variant SPD-2 following the intestinal nuclear division. DAPI (red) and SPD-2 (green) in (B–D) or SAS-4 (green) in (E–G). Asterisks indicate the intestinal nuclei and arrowheads show SPD-2 or SAS-4 foci. The insets show high magnification of the regions within the white rectangles. Scale bar, 5 µm. (H) The frequency of centriole duplication failure is represented by quantifying undivided intestinal nuclei associated with single SPD-2 or SAS-4 foci. (I) The frequency of supernumerary centriole duplication is indicated by the number of divided intestinal nuclei with more than one SPD-2 or SAS-4 focus after the nuclear division. Error bar, standard deviation; n≥75; P<0.05 (t-test). (K) Mass spectrometric analysis of SPD-2. +80 indicates the phosphorylated amino acid and the arrow highlights S545 in red.
We found that the wild type transgene (SPD-2WT) or the SPD-2 variants could rescue spd-2 (oj29)-associated embryonic lethality at 25°C from 100% to less than 4.5% (n≥110), suggesting that these transgenes possess essential SPD-2 function. However, among all the variants, only SPD-2S545A transgenic animals, where a predicted CDK-targeted Serine residue at 545 was mutated to Alanine, demonstrated apparent defects whereby the intestinal nuclei frequently failed to divide at the L1/L2 transition (Figure 5C), compared to SPD-2WT animals (Figure 5B). We monitored centriole numbers in the SPD-2S545A variants at the time of the nuclear division and found that only one SPD-2 focus was detectable adjacent to the undivided nuclei at the L2 stage (Figure 5C). The absence of centriole duplication is not due to the lack of S phase in these intestinal cells since they possess twice the DNA content of their divided neighboring intestinal nuclei (Figure S1E, C value 4.27 vs. average 2.11), suggesting a problem in centriole duplication. This occurred in approximately 25% of the intestinal nuclei in SPD-2S545A animals (Figure 5H). Again, we observed similar defects in centriole duplication when we monitored SAS-4 levels in the SPD-2S545A background (Figure 5E, 5F). A single SAS-4 focus (1 centriole pair) was frequently observed adjacent to a failed nuclear division in the L2 stage (Figure 5H), indicating that the disruption of SPD-2 phosphorylation on S545 compromises centriole duplication and renders the intestinal nuclei incapable of dividing as they transition into the L2 and the associated endocycle program.
CDK-2 activity, paired with either cyclin E or cyclin A, plays a central role in centriole duplication in many species [5]. The defect in centriole duplication in the SPD-2S545A variants led us to further examine if CDK-2 is required for centriole duplication in the intestine during the L1. We performed cdk-2(RNAi) using an intestine-specific RNAi-sensitive strain circumventing any essential function of the gene during development [56]-[57]. Consistent with the centriole duplication defects in the SPD-2S545A variants, less than 20% of the intestinal cdk-2(RNAi) animals had duplicated their centrioles after 13 h into the L1 stage (Figure S4B, S4C), while nearly 70% of the intestinal nuclei were associated with two SPD-2 foci in control animals (Figure S4A, S4C), suggesting that SPD-2S545A may be an important CDK-2 target in regulating timely centriole duplication.
Conversely, 35% of the SPD-2S545E post-division nuclei were associated with the appearance of supernumerary SPD-2 foci in the intestinal cells during the L2 stage (Figure 5D, 5I), whereas centriole duplication was not noticeably affected during the L1 stage (data not shown). Moreover, following the intestinal nuclear division, the C value of intestinal nuclei range from 4.08 to 4.14 in mid-L2 stage (Figure S1F; n = 20; p>0.1), suggesting that intestinal nuclei divide normally in the phosphomimetic SPD-2S545E variants and the supernumerary SPD-2 foci appear during the L2. Moreover SPD-2S545E was not sufficient to rescue the centriole duplication phenotype in cdk-2(RNAi) animals indicating that S545 is most probably not the only critical CDK2 target on SPD-2 (data not shown). Supernumerary SAS-4 foci were also detectable in the SPD-2S545E variants suggesting that this single amino acid change does not uniquely affect SPD-2 levels and or function, but rather its effects impinge on the entire centriole (Figure 5G, 5I). This phenotype is distinct from the lin-35/Rb mutant since the intestinal nuclei do not undergo additional divisions, indicating that SPD-2S545E specifically affects the numeral control by affecting centriole duplication without impinging directly on cell cycle progress.
Mass spectrometric analysis confirmed that these residues were indeed phosphorylated in vivo in the intestinal cells at the time of the L1/L2 transition (Figure 5K). We also found using in vitro kinase assays that S545 could be phosphorylated by human CDK-2/cyclin A and CDK-1/cyclin B (Figure S4D, S4E), but not by CDK-2/cyclin E (data not shown). Taken together, S545 is likely to be a physiologically relevant CDK target site on SPD-2 that affects centriole duplication.
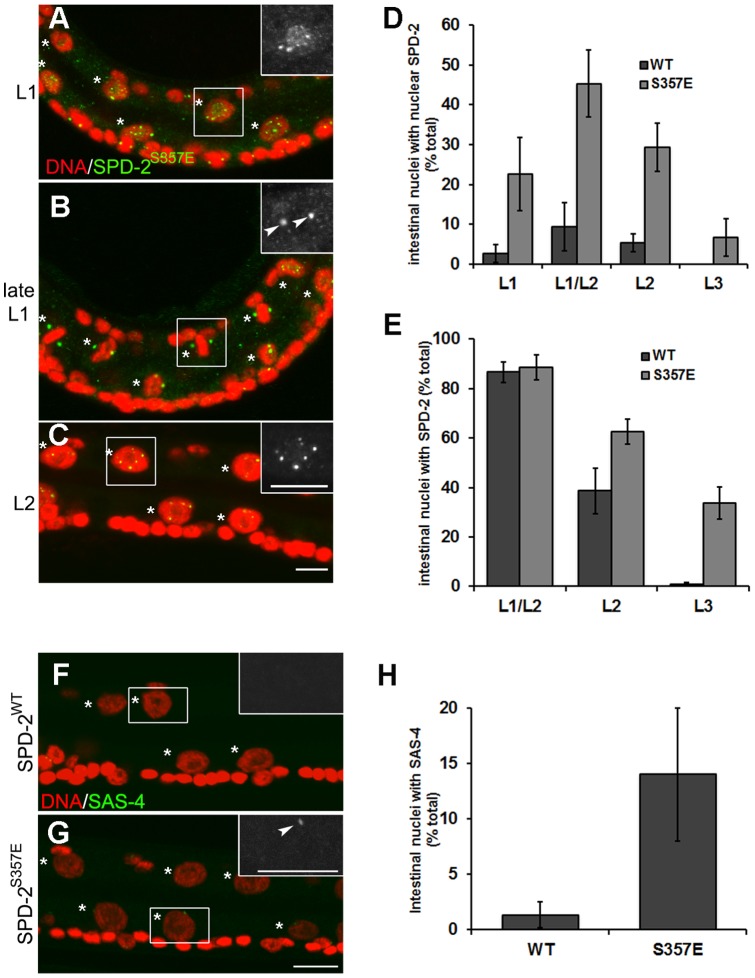
The SPD-2S357 phosphomimetic variant stabilizes SPD-2 and affects centriole stability in the endocycling intestinal cells
SPD-2 was previously reported to be phosphorylated on a consensus PLK phosphorylation site at Serine S357 [58]. We also identified S357 in our MS/MS analysis following an in vitro kinase assay performed with human PLK-1 and GST::SPD-2, although we were unable to detect the phosphorylation on S357 in vivo (Figure S5A and data not shown). Consistent with a functional role for this site, the phosphomimetic modification of Serine S357 (SPD-2S357E) caused SPD-2 to accumulate in aggregate-like puncta, paralleled by an increased frequency of cells with nuclear-localized SPD-2 prior to the nuclear division (Figure 6A, 6D), whereas the nuclear localization of wild type SPD-2 usually precedes its elimination during the L2 stage (Figure 2D). This SPD-2 accumulation has no apparent effect on centriole function in MTOC, since SPD-2 is normally present on the opposing poles of the condensing nuclei prior to nuclear division (Figure 6B). SPD-2S357E is still detectable in a number of intestinal cells even by the end of L2, long after SPD-2WT normally disappears (Figure 6C, 6E). This persistence of SPD-2S357E was not always reflected by the SAS-4 levels, but nonetheless 14% of the SPD-2S357E variant intestinal cells still possessed SAS-4 foci in the late L2 stage (Figure 6F-6H), suggesting that a portion of the centrioles are stabilized in this variant background. Therefore the phosphomimetic SPD-2S357E modification is sufficient to stabilize some SPD-2, causing it to accumulate in aggregate-like nuclear puncta. The same modification of SPD-2 also delays the elimination of some, but not all centrioles based on the persistence of the centriolar marker SAS-4.
Figure 6. Phosphorylation of S357 on SPD-2 affects appropriate localization and stability.
(A–C) spd-2 (oj29) animals carrying the SPD-2S357E variant were stained with DAPI (red) and anti-SPD-2 (green) in the L1, L1/L2 and L2. The asterisks indicate the intestinal nuclei. Scale bar, 5 µm. (D) SPD-2 staining was monitored in intestinal cells and the percentage of intestinal nuclei that demonstrate nuclear-localized SPD-2 in strains carrying either wild type SPD-2 or SPD-2S357E variant were determined. (E) The frequency of SPD-2 persistence is quantified by counting the number of intestinal cells that continue to show any SPD-2 signal at later larval stages. Error bar, standard deviation; n≥50; P<0.05 (t-test). (F) Late L2 spd-2 (oj29) animals expressing the SPD-2WT or (G) the SPD-2S357E variant were stained with DAPI (red) and anti-SAS-4 (green). The number of SAS-4 foci were quantified and indicated in (H). The asterisks indicate the intestinal nuclei while arrowheads show SAS-4 foci. Error bar, standard deviation; n = 50; P<0.05 (t-test). Scale bar, 5 µm.
In order to confirm that PLK-1 might stabilize SPD-2 in these cells we monitored the levels of SPD-2 at the L1/L2 transition using intestinal-specific plk-1(RNAi) animals. In these animals, despite that two SPD-2 foci are present, about 15% of the intestinal nuclei fail to divide (Figure S5B–S5D), suggesting that PLK-1 is required for the intestinal nuclear division, but not centriole duplication. Moreover, SPD-2 was not abnormally destabilized in plk-1(RNAi) animals, or in the non-phosphorylable SPD-2S357A variants, suggesting other factors may regulate the stability of SPD-2 in addition to PLK-1.
Ubiquitin mediated degradation of SPD-2
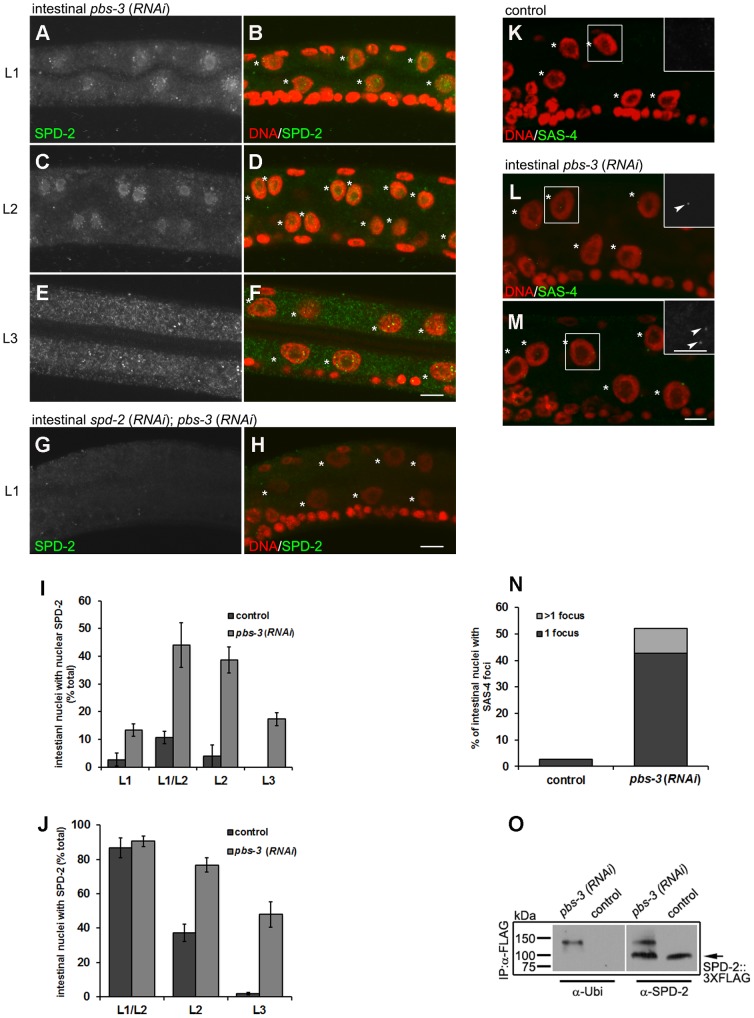
Recent data have implicated ubiquitin-mediated proteolytic degradation in the appropriate regulation of centrosomal components [59]–[61]. If SPD-2 abundance is controlled by ubiquitin-mediated degradation then the absence of essential ubiquitylation/proteasome components may also affect the elimination of the intestinal centrioles. proteasome b-subunit 3 (pbs-3) is essential for proteasome function [62] and in pbs-3(RNAi) animals, SPD-2 shows substantial nuclear accumulation (Figure 7A–7D, 7I), while SPD-2 signal is still detectable in the L3 stage, considerably later than in control animals (Figure 7E, 7F, 7J). Both the accumulation and persistence of SPD-2 are greatly reduced in pbs-3(RNAi); spd-2(RNAi) animals, confirming that the signals are indeed SPD-2-specific (Figure 7G, 7H). Similar effects on SPD-2 were also observed in pbs-5(RNAi) animals alone (data not shown), consistent with a role of proteasome-mediated degradation in the timely elimination of the centrioles after the intestinal cells commence endoreduplication. Intriguingly, in addition to the delayed centriole elimination phenotype, anti-SAS-4 staining revealed that centriole duplication was occasionally observed in pbs-3(RNAi) animals (Figure 7K–7N), which could reflect the stabilization of additional proteins involved in centriole duplication, or other key effectors of numeral regulation. Furthermore, by compromising proteasome function we were able to detect a higher molecular weight band in our FLAG-tagged SPD-2 immunoprecipitates. This band was recognized by both anti-SPD-2 and anti-Ubiquitin, consistent with the ubiquitylation of SPD-2 in vivo, which likely precedes its degradation (Figure 7O).
Figure 7. SPD-2 is ubiquitylated and its localization and stability are dependent on proteasome function.
(A–F) Larvae were subjected to pbs-3(RNAi) and subsequently stained with DAPI (red) and anti-SPD-2 (green) in the L1, L2 and L3 stages, respectively. (A), (C) and (E) show anti-SPD-2 alone. (G–H) Larvae were subjected to pbs-3(RNAi); spd-2(RNAi) and subsequently stained with DAPI (red) and anti-SPD-2 (green) in the L1. (G) shows anti-SPD-2 alone. Asterisks indicate the intestinal nuclei. Scale bar, 5 µm. (I) SPD-2 nuclear localization was monitored and the number of intestinal cells that demonstrate diffuse nuclear SPD-2 staining was compared in control and pbs-3(RNAi). (J) The effects of pbs-3(RNAi) on SPD-2 stability were quantified by determining the number of intestinal cells that express SPD-2 at later larval stages. Error bar, standard deviation; n≥50; P<0.05 (t-test). (K–M) SAS-4 levels were stained in the intestinal cells of late L2 pbs-3(RNAi) animals and were quantified as above (N). The asterisks indicate the intestinal nuclei, while arrowheads point to SAS-4 foci. Scale bar, 5 µm. n≥50; P<0.05 (t-test). (O) Homogenates obtained from animals expressing 3XFLAG tagged SPD-2 subjected to pbs-3(RNAi) or gfp(RNAi) (control), were incubated with anti-FLAG antisera and associated proteins were immunoprecipitated with Protein A-agarose. The pellets were blotted with anti-ubiquitin or anti-SPD-2 respectively. kDa, kilo Dalton.
Overall, our data suggest that during the switch to the endocycle program SPD-2 is ubiquitylated and subsequently degraded by the proteasome. Mimicking the potential PLK-1-dependent phosphorylation on S357 enhances nuclear localization and aggregation of SPD-2, which may interfere with destabilizing modifications that signal the removal of SPD-2. However, other cues that over-ride this modification must occur after the nuclear division to destabilize the centriole-associated SPD-2 and other centriolar components during the elimination process.
Discussion
The intimate links between the cell division cycle and the centrosome cycle ensure that the duplication and maturation of the centrosome occur in synchrony with the formation of the central spindle during mitosis. Consistent with this, many of the key enzymatic activities that drive the events specific to each stage of the cell cycle simultaneously affect aspects of the centrosome cycle [63].
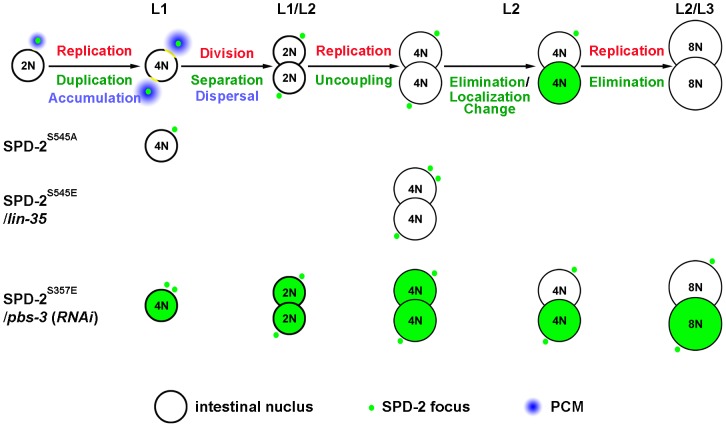
During the development of many organisms however, the mitotic cell cycle is replaced by the endocycle: an alternative means to provide tissue mass or to increase nuclear output/volume [32]. We were interested in how centrioles would respond to this modified cell cycle. We found that the centrioles appear to undergo a cell cycle uncoupling event shortly after the intestinal nuclear division. This uncoupling is contingent on the transition from mitotic cell division to an endocycle program at the end of the L1 stage. Following this division the centrioles are no longer duplicated and are eliminated during the L2 stage; an event that is an intrinsic developmental fate in these endocycling cells (Figure 8).
Figure 8. A model to depict centriole uncoupling and elimination in the postembryonic C. elegans intestine.
The complete postembryonic lineage of single intestinal cell in the wide type animals is shown. Each SPD-2 focus (green) represents a centriole pair. Centrioles undergo duplication and separation during the L1 nuclear division. The centrioles then lose their PCM during anaphase and undergo cell cycle “uncoupling” whereby centrioles are unaffected by the oscillations of successive endocycle S-phases. These events precede the diffusion of SPD-2 into the intestinal nuclei followed by its eventual elimination. The effects of SPD-2 variants or genetic backgrounds on centriole duplication/stability are highlighted at the relevant developmental stages. Substituting Serine 545 with Alanine on SPD-2 results in centriole duplication failure, whereas replacing Serine 545 with Glutamic Acid or, alternatively in lin-35 mutants, centrioles overduplicate. SPD-2 becomes stabilized when pbs-3 is abrogated or if SPD-2 S357 is converted to a phosphomimetic residue.
In cycling cells, the numeral integrity of centriole numbers is maintained through the regulation of centriole duplication driven by various cell cycle kinases including CDK2, in addition to a licensing mechanism that restrains the duplicated centrioles from responding to S-phase activities until after anaphase [42], [64]. Recent observations suggest that alternative mechanisms may also contribute to the numeral integrity and may be context-specific [65]–[66]. Although we cannot formally exclude that the centrioles do indeed duplicate during the endo-S but simply cannot be resolved due to their proximity or tight engagement, that we were able to observe supernumerary centrioles in the SPD-2S545E variant would suggest that this residue may be a target of the licensing mechanism per se in the intestinal cells. It is further tempting to implicate CDK activity in this step since both CDK1 and CDK2 can phosphorylate this residue in vitro. When the intestinal cells terminate the nuclear division, this residue may be unphosphorylated and persist in that state in the resulting daughter nuclei that will never execute a typical G2 or M-phase after that point. This modification could potentially contribute to licensing centriole duplication in the daughter cells.
γ-tubulin, a key PCM component that also plays important roles in centriole duplication [22], undergoes rapid anaphase dispersal from the centrosome at the end of the L1 stage in intestinal cells. Interestingly, centriole duplication still occurs in the SPD-2S545E variant after this point, perhaps because in C. elegans γ-tubulin is not essential for the initial recruitment of SAS-4 to the centriole [33], a crucial step for centriole duplication. Consistent with this, ultra-structural studies in C. elegans revealed that reducing γ-tubulin may result in abnormal centriole structure but does not necessarily block duplication [67]. At this point we cannot rule out that the ultra-structure of the supernumerary centrioles in our SPD-2S545E variants is not altered, nor can we exclude that the SPD-2S545E may recruit SAS-4 more efficiently so that the elevated levels of SAS-4 can compensate for the subthreshold levels of cytoplasmic γ-tubulin.
Centrosome/cell cycle uncoupling is not unique to endocycling cells as this occurs in other developmental contexts as well. During spermatogenesis in several different organisms the haploid sperm fertilizes the oocyte with a pair of centrioles, indicating that centriole duplication had taken place during meiosis II in the absence of DNA replication [68]. Similarly, in the multiciliated cells of the trachea, many centrioles arise spontaneously, independent of cell division [26]. In C. elegans, centrosome/cell cycle uncoupling also occurs in the endocycling anterior daughter of the V cells, but the effects we observed with the SPD-2S545E variants are restricted to the centrioles of the intestinal cells. How the centriole is generally sensitized to the various activities associated with cell cycle progression, or how it duplicates independently of these influences may be entirely cell-type specific.
The final nuclear division is a critical decision point, after which the centrioles become insensitive to the first endocycle S-phase and are eliminated shortly thereafter. Their eventual elimination is not dependent on their state of licensing nor a response to aberrant S phase activity, but rather a defined developmentally-controlled process typical of the intestinal endocycle program.
The PLK-1 site may be important to block centriole turnover in that the phosphomimetic SPD-2S357E replacement stabilizes SPD-2 and enhances its aggregation (Figure 8). Our data suggest however that the centrioles are not destabilized in the non-phosphorylable SPD-2S357A variant or in plk-1(RNAi) animals, indicating that additional mechanisms must function redundantly with PLK-1 to regulate centriole stability. How this phosphorylation affects stability is still questionable, but it may affect recognition by the proteasomal degradation pathway, since this pathway plays a key role in the timely elimination of the centrioles, not only in the intestinal endocycles, but also in mammalian cells and Drosophila [59], [61] (Figure 8),
It is tempting to generalize a role for of SPD-2 in all developmental contexts that might include uncoupling and/or elimination. In germ cells that initiate oogenesis, SPD-2 also changes its cellular distribution prior to the elimination of the centrioles in leptotene (Figure. S2). However, in the germ line, modification of S357 on SPD-2 appears to have little or no consequence. Moreover, gene products that have been identified to affect the timing of centriole elimination in the germ line [36] have no effect on centrosome/cell cycle uncoupling and/or elimination in the gut (data not shown). Despite the differences between the two tissues, the pathways may nevertheless converge upon a common component, SPD-2, where both modes of regulation independently impinge on the cellular SPD-2 distribution to ultimately affect its stability.
Although centriole elimination during oogenesis is common in many organisms, it also occurs in various other contexts, namely after the cell commits to a terminally differentiated state. In most mammalian cells the centriole pair will undergo specific modifications to generate a basal body to contribute to the primary cilium following the last mitotic division [69], while in Drosophila and C. elegans this is not the case, and only a small collection of cells are ciliated [70]–[71]. Interestingly, the epithelial cells of C. elegans, such as the seam cells (Figure 1J–1K′, 1L, 1M) and the vulva cells (Figure 1N–1Q), do not possess primary cilia. In these cells SPD-2::GFP remains visible until they finish their final division, suggesting that SPD-2 may also be targeted for elimination upon terminal differentiation. It is not clear why it would be advantageous to remove the centrioles from a differentiated cell. Perhaps its presence could sensitize the cell to regenerate the mitotic spindle through some aberrant acquisition of MTOC capacity, potentially a critical step toward unscheduled/unequivalent divisions that could drive hyper- or neoplasia. In this light, in addition to its co-opted role in signaling, the role of the primary cilium may be to sequester the centriole for future entry into the mitotic cell cycle, such as during wound healing for example. Our interrogation of how such organelles are removed in a developmentally-regulated manner will be informative to identify genes that are generally required to control this process in a coordinated manner in diverse cell types.
Materials and Methods
Strains and alleles
C. elegans Bristol strain N2 was used as the wild-type strain and was cultured as described previously [72]. Strains carrying a single copy of tagged wild type SPD-2, or variants thereof were generated as described [54]. L1/L2 transition is defined with the intestinal nuclear division, whereas L2/L3 with the larval molt. All strains with the temperature sensitive spd-2 (oj29) allele were maintained at the permissive temperature and all experiments were performed on larvae grown previously for 6 h at 25°C. The following alleles were used in this study: spd-2 (oj29), lin-35 (n745) and cul-4 (gk434). MR0156 (rrIS01[elt-2::GFP]X), MR1567 (rrIS1567[spd-2::SPD-2::3XFLAG; unc-119(+)] II; unc-119(ed3)III), MR1657 (unc-13 [e1091] spd-2[oj29] I; (rrIS1495 [spd-2::SPD-2::GFP; unc-119(+)]II; unc-119(ed3)III), MR1672 (unc-13[e1091] spd-2[oj29] I; (rrIS1488 [spd-2::SPD-2(S545A)::GFP; unc-119(+)]II; unc-119(ed3)III), MR1667 (unc-13[e1091] spd-2[oj29] I; (rrIS1514 [spd-2::SPD-2(S545E)::GFP; unc-119(+)]II; unc-119(ed3)III), MR1652 (unc-13[e1091] spd-2[oj29] I; (rrIS1587 [spd-2::SPD-2(S357E)::GFP; unc-119(+)]II; unc-119(ed3)III), MR1778 (rrIS1778 [elt-2::FLAG::PAB-1; unc-119(+)]II; unc-119(ed3)III), MR1779 (lin-35(n745)I; rrIS1778 [elt-2::FLAG::PAB-1; unc-119(+)]II; unc-119(ed3)III), MR1785 (rrEx1785 [elt-2::RDE-1; inx-6::GFP]; rde-1(ne219)V).
DNA constructs, site-directed mutagenesis, RNAi and HU treatment
For the transgenes encoding either the wild type or the GFP-tagged SPD-2 variant, a 2433 bp sequence upstream of the full length spd-2 but lacking its natural stop codon was first amplified from N2 and cloned in frame upstream of GFP in pPD95.79 following digestion with SalI and XmaI pMR812. Subsequently a fragment containing spd-2::SPD-2::GFP::unc-54 3′UTR was removed from pMR812 and then cloned into pCFJ151 to yield pMR831. PCR for site-directed mutagenesis was performed using Gene-Tailor site-directed mutagenesis (Invitrogen) on pMR831 in order to generate SPD-2 variants including pMR832 (S545A), pRM833 (S545E) and pMR857 (S357E).
The 3XFLAG including a stop codon was first cloned into pPD49.26 to generate pMR837. Subsequently a 2433 bp sequence upstream to the start codon of spd-2 and the genomic DNA encoding full length spd-2 lacking its natural stop codon was amplified and cloned in frame into pMR837 so that 3XFLAG would be C-terminal thus generating pMR841. Eventually, a spd-2::SPD-2::3XFLAG::unc-54 3′UTR was removed from pMR837 and then cloned into pCFJ151 to yield pMR850.
For the intestinal mRNA enrichment experiment, the elt-2 promoter and FLAG::PAB-1 [46], were flanked with sequences compatible for Gateway cloning (Invitrogen) and cloned into pCFJ150 to generate pMR869. For tissue-specific RNAi, the elt-2::RDE-1::unc-54 3′UTR was cloned into pMR542. For recombinant GST::SPD-2, spd-2 cDNA was cloned into pGEX-6p-2 to generate pMR873 (GE Healthcare).
The cdk-2(RNAi), plk-1(RNAi) and pbs-3(RNAi) presented in the microscopy were performed with our tissue-specific RNAi system, whereas the pbs-3(RNAi) presented in the western blot (Figure 7O) was performed by conventional RNAi using the Ahringer feeding RNAi library [62].
HU treatment was performed as described elsewhere [73]. For the HU block of the final mitotic intestinal cell cycle, synchronized L1 animals were cultured under standard conditions for five hours [72], followed by a 6-hour HU treatment. DNA content was unchanged over the course of our experiment indicating the HU block was efficient. For the HU block in the endoreduplicating intestinal cells, animals were allowed to develop under standard conditions until the completion of intestinal nuclei division [72] and then treated with HU for six hours. Centrioles and DNA content were monitored accordingly.
Microscopy
With the exception of Figures 1J–1K′, 1N–1O, 2A and S2, other indirect microscopy was performed using a 63× oil-immersion objective lens on a LSM510 META Confocal microscope (Zeiss) 1–4 µm optical sections were acquired with a 109.6 µm pinhole and 1024×1024 pixel resolution. For each experiment, intestinal nuclei samples were taken from no less than 15 different animals to avoid any bias. Focal planes containing centrioles were projected using an LSM510 Version 3.2 SP2. Images were processed using Adobe Photoshop, preserving relative image intensities within a series. The microscopic work of Figure S2 was performed using 100× oil-immersion objective lens in a DeltaVision Image Restoration System (Applied Precision). Data were collected as a series of 13–27 optical sections in increments of 0.2 µm under standard parameters with the softWoRx 3.0 software (Applied Precision). The microscopic work of Figure 1J–1K′, 1N-1O, and 2A were performed using 63× oil-immersion objective lens in a Zeiss AX10 microscopy under stander parameters with the AxioVision 4.8 software (Zeiss). All microscopies were performed at 20°C.
Purification of GST::SPD-2 and in vitro Kinase Assay
SPD-2 was overexpressed as a N-terminal glutathione-S-transferase (GST) fusion protein in BL21 bacteria following a 6-hour IPTG-induction at 37°C. Protein was purified with Glutathione Sepharose 4B (GE Healthcare) and 50 µl of eluted GST::SPD-2 fusion protein was incubated with human CDK2/cyclin E, CDK2/cyclin A, CDK1/cyclin B, or PLK-1 (Millipore) respectively for 30 minutes. Reactions were subjected to 8% SDS-PAGE and the corresponding bands were excised and analyzed by MS.
PI staining, Antibodies and Immunological methods
PI staining
PI (Sigma P-4170) staining and the measurement of DNA content were performed as described elsewhere [34].
Immunofluorescence
The following primary antibodies and dilutions were used: 1∶100 anti-SPD-2 rabbit polyclonal [74] (a gift from Dr. K. O'Connell, National Institutes of Health, Bethesda, MD), 1∶100 anti-SAS-4 rabbit or goat polyclonal (Santa Cruz Biotechnology, SC98949 and SC20418), 1∶100 rabbit anti-γ-tubulin (Sigma-Aldrich, T1450), 1∶100 mouse monoclonal anti-α-tubulin (Sigma-Aldrich T9026) and 1∶100 mouse monoclonal anti-GFP (Abcam, ab1218). 1∶250 secondary antibodies were anti-mouse or anti-rabbit AlexaFluor 488 and anti-rabbit AlexaFluor 555 (Molecular Probe). DAPI (Sigma-Aldrich) was used to reveal DNA. L1, L2 or L3 larvae were fixed and stained as described previously [75].
Immunoprecipitation, purification and Immunoblotting
To confirm in vivo phosphorylation, 30 ml of packed young larvae were collected and re-suspended in one volume of lysis buffer [20]. Animals were lysed using repetitive 15-second sonication for 15 cycles on ice with 30-second intervals between each cycle. The lysate was then centrifuged at 4°C for 10 minutes. Supernatants were incubated with 250 µl anti-FLAG affinity gel (Sigma Aldrich) for 2 hours at 4°C. The gel was washed with lysis buffer and the beads were resuspended in 0.1 M Glycine buffer (pH2.5), centrifuged, and 50 µl of the eluate was subjected to 8% SDS-PAGE, followed by coomassie blue staining described elsewhere (http://www.mass-spec.siu.edu/CoomassieStainingProtocol). The pieces of polyacrylamide gel corresponding to SPD-2::3XFLAG predicted size were cut for MS analysis.
To detect SPD-2 ubiquitylation, proteins were purified from 10 ml of packed worms of each genotype as described above. 40 µl of the eluate was then subjected to 8% SDS-PAGE then transferred to a nitrocellulose membrane (Bio-Rad) and blotted as described elsewhere: (http://www.cellsignal.com/support/protocols/Western_BSA.html).
The following primary and secondary antibodies were used: 1∶1000 anti-SPD-2 rabbit polyclonal, 1∶500 anti-Ubiquitin mouse monoclonal (Santa Cruz Biotechnology, SC-8017), and 1∶2500 goat anti-rabbit or mouse HRP conjugated IgG (Bio-Rad). Protein bands were detected using chemifluoresence (Clarity Western ECL Substrate, Bio-Rad).
Intestinal mRNA enrichment and RT-PCR
The intestinal mRNA tagging protocol was described elsewhere [46]. The amount of RNA from each sample was calibrated to generate comparable amplicon levels for the control genes dlg-1 and glo-1. PCR was performed for 15 cycles with ProtoScript M-MuLV Taq RT-PCR Kit (NEB, E6400S) and gene-specific primers for each query gene. The primers were designed and analyzed with on-line tool (www.basic.northwestern.edu/biotools/oligocalc.html). Two pairs of primers were designed for each tested gene except for elt-2 and htp-3 (one pair). The primer sequences are listed in the Form S1.
Supporting Information
The coupling of centriole duplication with S-phase is regulated developmentally and can be genetically altered. (A–C) spd-2 (oj29) animals carrying the wild type SPD-2::GFP were stained with PI (Red) in the L1 and L2. (D) lin-35 (n745) animals were stained with PI (red) and anti-SPD-2 (green) in the L2. (E) spd-2 (oj29) animals carrying the SPD-2S545A variant were stained with PI (Red) at the L1/L2 transition. (F) spd-2 (oj29) animals carrying the SPD-2S545E variant were stained with PI (Red) in mid-L2 stage. The asterisks indicate the intestinal nuclei and the arrow heads point out the SPD-2 foci. The arrows indicate the muscle cells that are used as the 2C reference and the numbers under the intestinal nuclei indicates their DNA content. Scale bar, 5 µm.
(TIF)
SPD-2 diffuses into nuclei before its elimination in the germ line. The gonad of a N2 young adult was dissected and stained with DAPI (red) and SPD-2 (green). Scale bar, 5 µm.
(TIF)
γ-tubulin dispersal from the centriole affects its MTOC function. (A) N2 at late L2 were stained with DAPI (blue), anti-SAS-4 (red) and anti-γ-tubulin (green). (a) highlights a metaphase nucleus while the lineage bracket in (b) shows anaphase nuclei. (a′) and (a′′) indicate the magnified SAS-4 or γ-tubulin signal of a, respectively; whereas (b′) and (b′′) correspond to the magnified SAS-4 or γ-tubulin signal of b, respectively. Asterisks, intestinal nuclei; arrowheads, SAS-4 or γ-tubulin foci. Scale bar 5 µm. (B) and (C) L2 wild type animal was stained with DAPI (blue), anti-SAS-4 (green) and anti-α-tubulin (red). (B) shows the intestinal nuclei; whereas lateral hypodermal cells of the same animal in (C). The insets show the α-tubulin signal alone in the framed region in the corresponding panels. Asterisks, intestinal nuclei. Scale bar, 5 µm.
(TIF)
CDK-2 affects centriole duplication during L1. (A) The intestinal-specific RNAi sensitized animals were subjected to a control RNAi or (B) cdk-2(RNAi) and stained with DAPI (red) or anti-SPD-2 (green). Asterisks indicate the intestinal nuclei and arrowheads point to SPD-2 foci. The insets show high magnification of the regions within the white rectangles. Scale bar, 5 µm. (C) The frequency of centriole duplication failure is represented by quantifying the number of intestinal cells that possess two SPD-2 foci 10 hours into the L1 stage. Error bar, standard deviation; n≥50; P<0.05 (t-test). (D and E) Mass spectrometric analysis of GST::SPD-2 incubated with human CDK2/cyclin A or CDK1/cyclin B, respectively. +80 indicates the phosphorylated amino acid and the arrow highlights S545 in red.
(TIF)
PLK-1 affects the intestinal nuclear division, but not centriole duplication during the L1 stage. (A) Mass spectrometric analysis of GST::SPD-2 incubated with human PLK-1. +80 indicates the phosphorylated amino acid and the arrow indicates the position of S357 in red. (B) The intestinal-specific RNAi sensitized animals were subjected to control RNAi or (C) plk-1(RNAi) and stained with DAPI (red) or anti-SPD-2 (green). Asterisks indicate the intestinal nuclei and arrowheads point to SPD-2 foci. The insets show high magnification of the regions within the white rectangles. Scale bar, 5 µm. (D) The frequency of nuclear division failure is represented by quantifying undivided nuclei with two SPD-2 foci. Error bar, standard deviation; n≥50; P<0.05 (t-test).
(TIF)
The primers used for quantifying the relative mRNA abundance of the genes essential for centriole duplication or centrosomal function. Two pairs of primers were designed for each tested gene except for elt-2 and htp-3 (one pair). The primer sequences were designed and analyzed with the on-line tool available at www.basic.northwestern.edu/biotools/oligocalc.html.
(XLS)
Acknowledgments
We are very grateful to Dr. Kevin O'Connell for reagents; to Shaolin Li and Amanda Stout for technical assistance; and to the Caenorhabditis Genetics Center for strains. We also thank Dr. Eric Bonneil at IRIC for Mass Spectrometric analysis, and Dr. Judith Lacoste as well as Dr. Elke Küster-Schöck (CIAN, Dept. of Biology, McGill university) for help with microscopy.
Funding Statement
This work was funded by the Canadian Institutes of Health Research (CIHR) and the Natural Sciences and Engineering Research Council (NSERC). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Nigg EA, Stearns T (2011) The centrosome cycle: Centriole biogenesis, duplication and inherent asymmetries. Nat Cell Biol 13: 1154–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Basto R, Brunk K, Vinadogrova T, Peel N, Franz A, et al. (2008) Centrosome amplification can initiate tumorigenesis in flies. Cell 133: 1032–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ganem NJ, Godinho SA, Pellman D (2009) A mechanism linking extra centrosomes to chromosomal instability. Nature 460: 278–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nigg EA, Raff JW (2009) Centrioles, centrosomes, and cilia in health and disease. Cell 139: 663–678. [DOI] [PubMed] [Google Scholar]
- 5. Hinchcliffe EF, Sluder G (2002) Two for two: Cdk2 and its role in centrosome doubling. Oncogene 21: 6154–6160. [DOI] [PubMed] [Google Scholar]
- 6. Tsou MF, Wang WJ, George KA, Uryu K, Stearns T, et al. (2009) Polo kinase and separase regulate the mitotic licensing of centriole duplication. Dev Cell 17: 344–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bahe S, Stierhof YD, Wilkinson CJ, Leiss F, Nigg EA (2005) Rootletin forms centriole-associated filaments and functions in centrosome cohesion. J Cell Biol 171: 27–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Crasta K, Huang P, Morgan G, Winey M, Surana U (2006) Cdk-1 regulates centrosome separation by restraining proteolysis of microtubule-associated proteins. EMBO 25: 2551–2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Smith E, Hegarat N, Vesely C, Roseboom I, Larch C, et al. (2011) Differential control of Eg5-dependent centrosome separation by Plk1 and Cdk-1. EMBO J 30: 2233–2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hannak E, Kirkham M, Hyman AA, Oegema K (2001) Aurora-1 kinase is required for centrosome maturation in Caenorhabditis elegans . J Cell Biol 155: 1109–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lee K, Rhee K (2011) PLK1 phosphorylation of pericentrin initiates centrosome maturation at the onset of mitosis. J Cell Biol 195: 1093–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Delattre M, Canard C, Gonczy P (2006) Sequential protein recruitment in C. elegans centriole formation. Curr Biol 16: 1844–1849. [DOI] [PubMed] [Google Scholar]
- 13. Pelletier L, O'Toole E, Schwager A, Hyman AA, Muller-Reichert T (2006) Centriole assembly in Caenorhabditis elegans . Nature 444: 619–623. [DOI] [PubMed] [Google Scholar]
- 14. Rodrigues-Martins A, Bettencourt-Dias M, Riparbelli M, Ferreira C, Ferreira I, et al. (2007) DSAS-6 organizes a tube-like centriole precursor, and its absence suggests modularity in centriole assembly. Curr Biol 17: 1465–1472. [DOI] [PubMed] [Google Scholar]
- 15. Zhu F, Lawo S, Bird A, Pinchev D, Ralph A, et al. (2008) The mammalian SPD-2 ortholog cep192 regulates centrosome biogenesis. Curr Biol 18: 136–141. [DOI] [PubMed] [Google Scholar]
- 16. Bettencourt-Dias M, Rodrigues-Martins A, Carpenter L, Riparbelli M, Lehmann L, et al. (2005) SAK/PLK4 is required for centriole duplication and flagella development. Curr Biol 15: 2199–2207. [DOI] [PubMed] [Google Scholar]
- 17. Habedank R, Stierhof YD, Wilkinson CJ, Nigg EA (2005) The polo kinase Plk4 functions in centriole duplication. Nat Cell Biol 7: 1140–1146. [DOI] [PubMed] [Google Scholar]
- 18. O'Connell KF, Caron C, Kopish KR, Hurd DD, Kemphues KJ, et al. (2001) The C. elegans zyg-1 gene encodes a regulator of centrosome duplication with distinct maternal and paternal roles in the embryos. Cell 105: 547–558. [DOI] [PubMed] [Google Scholar]
- 19. Pelletier L, Ozlu N, Hannak E, Cowan C, Habermann B, et al. (2004) The Caenorhabditis elegans centrosomal protein SPD-2 is required for both pericentriolar material recruitment and centriole duplication. Curr Biol 14: 863–873. [DOI] [PubMed] [Google Scholar]
- 20. Kitagawa D, Busso C, Fluckiger I, Gonczy P (2009) Phosphorylation of SAS-6 by ZYG-1 is critical for centriole formation in C. elegans embryo. Dev Cell 17: 900–907. [DOI] [PubMed] [Google Scholar]
- 21. Leidel S, Delattre M, Baumer K, Gonczy P (2005) SAS-6 defines a protein family required for centrosome duplication in C. elegans and human cells. Nat Cell Biol 7: 115–125. [DOI] [PubMed] [Google Scholar]
- 22. Dammermann A, Muller-Reichert T, Pelletier L, Habermann B, Desai A, et al. (2004) Centriole assembly requires both centriolar and pericentriolar material proteins. Dev Cell 7: 818–829. [DOI] [PubMed] [Google Scholar]
- 23. Delattre M, Leidel S, Wani K, Baumer K, Bamat J, et al. (2004) Centriolar SAS-5 is required for centrosome duplication in C. elegans . Nat Cell Biol 6: 656–664. [DOI] [PubMed] [Google Scholar]
- 24. Kirkham M, Muller-Reichert T, Oegema K, Grill S, Hyman AA (2003) SAS-4 is a C. elegans centriolar protein that controls centrosome size. Cell 112: 575–587. [DOI] [PubMed] [Google Scholar]
- 25. Carvalho-Santos Z, Azimzadeh J, Pereira-Leal JB, Bettencourt-Dias M (2011) Tracing the origins of centrioles, cilia and flagella. J Cell Biol 194: 165–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Vladar EK, Stearns T (2007) Molecular characterization of centriole assembly in ciliated epithelial cells. J Cell Biol 178: 31–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bolivar J, Huynh JR, Lopez-Schier H, Gonzalez C, St Johnston D, et al. (2001) Centrosome migration into the Drosophila oocyte is independent of BicD and egl, and of the organization of the microtubule cytoskeleton. Development 128: 1889–1897. [DOI] [PubMed] [Google Scholar]
- 28. Mahowald AP, Caulton JH, Edwards MK, Floyd AD (1979) Loss of centrioles and polyploidization in follicle cells of Drosophila melanogaster . Exp Cell Res 118: 404–410. [DOI] [PubMed] [Google Scholar]
- 29. Hedgecock EM, White JG (1985) Polyploid tissues in the nematode Caenorhabditis elegans . Dev Biol 107: 128–133. [DOI] [PubMed] [Google Scholar]
- 30. Kipreos ET (2005) C. elegans cell cycles: invariance and stem cell division. Nat Rev 6: 766–776. [DOI] [PubMed] [Google Scholar]
- 31. Hemerly AS, Prasanth SG, Siddiqui K, Stillman B (2009) Orc1 controls centriole and centrosome copy number in human cells. Science 323: 789–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lee HO, Davidson JM, Duronio RJ (2009) Endoreplication: polyploidy with purpose. Genes Dev 23: 2461–2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dammermann A, Maddox PS, Desai A, Oegema K (2008) SAS-4 is recruited to a dynamic structure in newly forming centrioles that is stabilized by the γ-tubulin-mediated addition of centriolar microtubules. J Cell Biol 180: 771–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Boxem M, Srinivasan DG, van den Heuvel S (1999) The Caenorhabditis elegans gene ncc-1 encodes a cdc2-related kinase required for M phase in meiotic and mitotic cell divisions, but not for S phase. Development 126: 2227–2239. [DOI] [PubMed] [Google Scholar]
- 35. Kim DY, Roy R (2006) Cell cycle regulators control centrosome elimination during oogenesis in Caenorhabditis elegans . J Cell Biol 174: 751–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mikeladze-Dvali T, von Tobel L, Strnad P, Knott G, Leonhardt H, et al. (2012) Analysis of centriole elimination during C. elegans oogenesis. Development 139: 1670–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bobinnec Y, Fukuda M, Nishida E (2000) Identification and characterization of Caenorhabditis elegans γ-tubulin in dividing cells and differentiated tissues. J Cell Sci 21: 3747–3759. [DOI] [PubMed] [Google Scholar]
- 38. Khodjakov A, Rieder CL, Sluder G, Cassels G, Sibon O, et al. (2002) De novo formation of centrosomes in vertebrate cells arrested during S phase. J Cell Biol 158: 1171–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Brauchle M, Baumer K, Gonczy P (2003) Differential activation of the DNA replication checkpoint contributes to asynchrony of cell divisions in C. elegans embryos. Curr Biol 13: 819–827. [DOI] [PubMed] [Google Scholar]
- 40.Balczon R, Bao L, Zimmer WE, Brown K, Zinkowski RP, et al. (1995) Dissociation of centrosome replication events from cycles of DNA synthesis and mitotic division in hydroxyurea-arrested Chinese hamster ovary cells. J Cell Biol 130: : 105–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhong W, Feng H, Santiago FE, Kipreos ET (2003) CUL-4 ubiquitin ligase maintains genome stability by restraining DNA-replication licensing. Nature 423: 885–889. [DOI] [PubMed] [Google Scholar]
- 42. Tsou MF, Stearns T (2006) Mechanism limiting centrosome duplication to once per cell cycle. Nature 442: 947–951. [DOI] [PubMed] [Google Scholar]
- 43. Lu X, Horvitz R (1998) lin-35 and lin-53, two genes that antagonize a C. elegans Ras pathway, encode proteins similar to Rb and its binding protein RbAp48. Cell 95: 981–991. [DOI] [PubMed] [Google Scholar]
- 44. Ouellet J, Roy R (2007) The lin-35/Rb and RNAi pathways cooperate to regulate a key cell cycle transition in C. elegans . BMC Dev Biol 7: 38 Available: http://www.biomedcentral.com/1471-213X/7/38. Accessed 2012 Sept 09.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kirienko NV, Fay DS (2007) Transcriptome profiling of the C. elegans Rb ortholog reveals diverse developmental roles. Dev Biol 305: 674–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Roy PJ, Stuart JM, Lund J, Kim SK (2002) Chromosomal clustering of muscle-expressed genes in Caenorhabditis elegans . Nature 418: 975–979. [DOI] [PubMed] [Google Scholar]
- 47. Hamill DR, Severson AF, Carter JC, Bowerman B (2002) Centrosome maturation and mitotic spindle assembly in C. elegans requires SPD-5, a protein with multiple coiled-coil domain. Dev Cell 3: 673–684. [DOI] [PubMed] [Google Scholar]
- 48. Matthews LR, Carter P, Thierry-Mieg D, Kemphues K (1998) ZYG-9, a Caenorhabditis elegans protein required for microtubule organization and function, is a component for meiotic and mitotic spindle poles. J Cell Biol 141: 1159–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Decker M, Jaensch S, Pozniakovsky A, Zinke A, O'Connell KF, et al. (2011) Limiting amounts of centrosome material set centrosome size in C. elegans embryos. Curr Biol 21: 1259–1267. [DOI] [PubMed] [Google Scholar]
- 50. Kitagawa D, Fluckiger I, Polanowska J, Keller D, Reboul J, et al. (2011) PP2A phosphatase acts upon SAS-5 to ensure centriole formation in C. elegans embryos. Dev Cell 20: 550–562. [DOI] [PubMed] [Google Scholar]
- 51. Song MH, Liu Y, Anderson DE, Jahng WJ, O'Connell KF (2011) Protein phosphatase 2A-SUR-6/B55 regulates centriole duplication in C. elegans by controlling the levels of centriole assembly factors. Dev Cell 20: 563–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Xue Y, Ren J, Gao X, Jin C, Wen L, et al. (2008) GPS 2.0, a Tool to Predict Kinase-specific Phosphorylation Sites in Hierarchy. Mol Cell Proteomics 7: 1598–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhou F, Xue Y, Chen G, Yao X (2004) GPS: a novel group-based phosphorylation predicting and scoring method. Biochem Biophys Res Commun 325: 1443–1448. [DOI] [PubMed] [Google Scholar]
- 54. Frøkjær-Jensen C, Davis MW, Hopkins CE, Newman BJ, Thummel JM, et al. (2008) Single-copy insertion of transgenes in Caenorhabditis elegans . Nat Genet 40: 1375–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. O'Connell KF, Leys CM, White JG (1998) A genetic screen for temperature-sensitive cell-division mutants of Caenorhabditis elegans . Genetics 149: 1303–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Chotard L, Mishra AK, Sylvain MA, Tuck S, Lambright DG, et al. (2010) TBC-2 regulates RAB-5/RAB-7 mediated endosomal trafficking in Caenorhabditis elegans . Mol Biol Cell 21: 2258–2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. McGhee JD, Fukushige T, Krause MW, Minnema SE, Goszczynski B, et al. (2009) ELT-2 is the predominant transcription factor controlling differentiation and function of the C. elegans intestine, from embryo to adult. Dev Biol 327: 551–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Bodenmiller B, Campbell D, Gerrits B, Lam H, Jovanovic M, et al. (2008) PhosphoPep–a database of protein phosphorylation sites in model organisms. Nat Biotechnol 26: 1339–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Cunha-Ferreira I, Rodrigues-Martins A, Bento I, Riparbelli MM, Zhang W, et al. (2009) The SCF/Slimb ubiquitin ligase limits centrosome amplification through degradation of SAK/PLK-4. Curr Biol 19: 43–49. [DOI] [PubMed] [Google Scholar]
- 60. Kais Z, Parvin JD (2008) Regulation of centrosomes by the BRCA1-dependent ubiquitin ligase. Cancer Biol Ther 7: 1540–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Rogers GC, Rusan NM, Roberts DM, Peifer M, Rogers SL (2009) The SCF Slimb ubiquitin ligase regulates Plk4/Sak levels to block centriole re-duplication. J Cell Biol 184: 225–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, et al. (2003) Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421: 231–237. [DOI] [PubMed] [Google Scholar]
- 63. Doxsey S, Zimmerman W, Mikule K (2005) Centrosome control of the cell cycle. Trends Cell Biol 15: 303–311. [DOI] [PubMed] [Google Scholar]
- 64. Wong C, Stearns T (2003) Centrosome number is controlled by a centrosome-intrinsic block to reduplication. Nat Cell Biol 5: 539–544. [DOI] [PubMed] [Google Scholar]
- 65. Cabral G, Sans SS, Cowan CR, Dammermann A (2013) Multiple mechanisms contribute to centriole separation in C. elegans . Curr Biol 23: 1380–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Schvarzstein M, Pattabiraman D, Bembenek JN, Villeneuve AM (2013) Meiotic HORMA domain proteins prevent untimely centriole disengagement during Caenorhabditis elegans spermatocyte meiosis. Proc Natl Acad Sci U S A 110: 898–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. O'Toole E, Greenan G, Lange KI, Srayko M, Muller-Reichert T (2012) The role of γ-tubulin in centrosomal microtubule organization. PLoS ONE 7: e29795 Available: http://www.plosone.org/article/info%3Adoi%2F10.1371%2Fjournal.pone.0029795. Accessed 2013 May 01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Schatten G (1994) The centrosome and its mode of inheritance: the reduction of the centrosome during gametogenesis and its restoration during fertilization. Dev Biol 165: 299–335. [DOI] [PubMed] [Google Scholar]
- 69. Dawe HR, Farr H, Gull K (2007) Centriole/basal body morphogenesis and migration during ciliogenesis in animal cells. J. Cell Sci 120: 7–15. [DOI] [PubMed] [Google Scholar]
- 70. Enjolras C, Thomas J, Chhin B, Cortier E, Duteyrat JL, et al. (2012) Drosophila chibby is required for basal body formation and ciliogenesis but not for Wg signaling. J Cell Biol 197: 313–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Inglis PN, Ou G, Leroux MR, Scholey JM (2006) The sensory cilia of Caenorhabditis elegans WormBook, Available: http://www.wormbook.org/chapters/www_ciliumbiogenesis.2/ciliumbiogenesis.html. Accessed 2013 Dec 01. [DOI] [PMC free article] [PubMed]
- 72. Brenner S (1974) The genetics of Caenorhabditis elegans . Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Ambros V (1999) Cell cycle-dependent sequencing of cell fate decisions in Caenorhabditis elegans vulva precursor cells. Development 126: 1947–1956. [DOI] [PubMed] [Google Scholar]
- 74. Kemp CA, Kopish KR, Zipperlen P, Ahringer J, O'Connell KF (2004) Centrosome maturation and duplication in C. elegans requires the coiled-coil protein SPD-2. Dev Cell 6: 511–523. [DOI] [PubMed] [Google Scholar]
- 75. Finney M, Ruvkun G (1990) The unc-86 gene product couples cell lineage and cell identity in C. elegans . Cell 63: 895–905. [DOI] [PubMed] [Google Scholar]
- 76. Pepper ASR, McCane JE, Kemper K, Yeung DA, Lee RC, et al. (2004) The C. elegans heterochronic gene lin-46 affects developmental timing at two larval stages and encodes a relative of the scaffolding protein gephyrin. Development 131: 2049–2059. [DOI] [PubMed] [Google Scholar]
- 77. McMahon L, Legouis R, Vonesch J, Labouesse M (2001) Assembly of C. elegans apical junctions involves positioning and compaction by LET-413 and protein aggregation by the MAGUK protein DLG-1. J Cell Sci 114: 2265–2277. [DOI] [PubMed] [Google Scholar]
- 78. Zielinska DF, Gnad F, Jedrusik-Bode E, Wisniewski JR, Mann M (2009) Caenorhabditis elegans has a phosphoproteome atypical for metazoans that is enriched in developmental and sex determination proteins. J. Proteome Res 8: 4029–4049. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The coupling of centriole duplication with S-phase is regulated developmentally and can be genetically altered. (A–C) spd-2 (oj29) animals carrying the wild type SPD-2::GFP were stained with PI (Red) in the L1 and L2. (D) lin-35 (n745) animals were stained with PI (red) and anti-SPD-2 (green) in the L2. (E) spd-2 (oj29) animals carrying the SPD-2S545A variant were stained with PI (Red) at the L1/L2 transition. (F) spd-2 (oj29) animals carrying the SPD-2S545E variant were stained with PI (Red) in mid-L2 stage. The asterisks indicate the intestinal nuclei and the arrow heads point out the SPD-2 foci. The arrows indicate the muscle cells that are used as the 2C reference and the numbers under the intestinal nuclei indicates their DNA content. Scale bar, 5 µm.
(TIF)
SPD-2 diffuses into nuclei before its elimination in the germ line. The gonad of a N2 young adult was dissected and stained with DAPI (red) and SPD-2 (green). Scale bar, 5 µm.
(TIF)
γ-tubulin dispersal from the centriole affects its MTOC function. (A) N2 at late L2 were stained with DAPI (blue), anti-SAS-4 (red) and anti-γ-tubulin (green). (a) highlights a metaphase nucleus while the lineage bracket in (b) shows anaphase nuclei. (a′) and (a′′) indicate the magnified SAS-4 or γ-tubulin signal of a, respectively; whereas (b′) and (b′′) correspond to the magnified SAS-4 or γ-tubulin signal of b, respectively. Asterisks, intestinal nuclei; arrowheads, SAS-4 or γ-tubulin foci. Scale bar 5 µm. (B) and (C) L2 wild type animal was stained with DAPI (blue), anti-SAS-4 (green) and anti-α-tubulin (red). (B) shows the intestinal nuclei; whereas lateral hypodermal cells of the same animal in (C). The insets show the α-tubulin signal alone in the framed region in the corresponding panels. Asterisks, intestinal nuclei. Scale bar, 5 µm.
(TIF)
CDK-2 affects centriole duplication during L1. (A) The intestinal-specific RNAi sensitized animals were subjected to a control RNAi or (B) cdk-2(RNAi) and stained with DAPI (red) or anti-SPD-2 (green). Asterisks indicate the intestinal nuclei and arrowheads point to SPD-2 foci. The insets show high magnification of the regions within the white rectangles. Scale bar, 5 µm. (C) The frequency of centriole duplication failure is represented by quantifying the number of intestinal cells that possess two SPD-2 foci 10 hours into the L1 stage. Error bar, standard deviation; n≥50; P<0.05 (t-test). (D and E) Mass spectrometric analysis of GST::SPD-2 incubated with human CDK2/cyclin A or CDK1/cyclin B, respectively. +80 indicates the phosphorylated amino acid and the arrow highlights S545 in red.
(TIF)
PLK-1 affects the intestinal nuclear division, but not centriole duplication during the L1 stage. (A) Mass spectrometric analysis of GST::SPD-2 incubated with human PLK-1. +80 indicates the phosphorylated amino acid and the arrow indicates the position of S357 in red. (B) The intestinal-specific RNAi sensitized animals were subjected to control RNAi or (C) plk-1(RNAi) and stained with DAPI (red) or anti-SPD-2 (green). Asterisks indicate the intestinal nuclei and arrowheads point to SPD-2 foci. The insets show high magnification of the regions within the white rectangles. Scale bar, 5 µm. (D) The frequency of nuclear division failure is represented by quantifying undivided nuclei with two SPD-2 foci. Error bar, standard deviation; n≥50; P<0.05 (t-test).
(TIF)
The primers used for quantifying the relative mRNA abundance of the genes essential for centriole duplication or centrosomal function. Two pairs of primers were designed for each tested gene except for elt-2 and htp-3 (one pair). The primer sequences were designed and analyzed with the on-line tool available at www.basic.northwestern.edu/biotools/oligocalc.html.
(XLS)