Abstract
Natural isolates of Bacillus subtilis exhibit a robust multicellular behavior known as swarming. A form of motility, swarming is characterized by a rapid, coordinated progression of a bacterial population across a surface. As a collective bacterial process, swarming is often associated with biofilm formation and has been linked to virulence factor expression in pathogenic bacteria. While the swarming phenotype has been well documented for Bacillus species, an understanding of the molecular mechanisms responsible remains largely isolated to gram-negative bacteria. To better understand how swarming is controlled in members of the genus Bacillus, we investigated the effect of a series of gene deletions on swarm motility. Our analysis revealed that a strain deficient for the production of surfactin and extracellular proteolytic activity did not swarm or form biofilm. While it is known that surfactin, a lipoprotein surfactant, functions in swarming motility by reducing surface tension, this is the first report demonstrating that general extracellular protease activity also has an important function. These results not only help to define the factors involved in eliciting swarm migration but support the idea that swarming and biofilm formation may have overlapping control mechanisms.
It has long been recognized that bacteria can facilitate their growth and survival by forming cooperative, multicellular communities that are most often associated with surfaces (38). Such organized populations of microorganisms have been found in both clinical and environmental settings, where they have positive and negative impacts on human health and environmental ecology (4, 24). For these reasons, bacterial multicellularity is being actively studied, and while biofilms are most commonly ascribed to this type of behavior, swarming is another primary example of a surface-associated collective bacterial process. A form of migration, swarming facilitates the rapid colonization of surfaces by a population of bacteria. Swarming motility has been linked with biofilm formation, antibiotic resistance, and virulence factor production (1, 21, 39). Many of the virulence factors reported to be associated with the occurrence of swarming are exoenzymes and, more specifically, extracellular proteases (12, 47). While a strong link between swarming, extracellular protease production, and pathogenesis has been noted (12, 44, 47), only a few studies have focused on the requirement of proteases for swarm motility. In a recent investigation, the extracellular protease Epr was found to be essential for swarming motility in the domesticated Bacillus subtilis laboratory strain 168 (6). Since laboratory strains of B. subtilis have been shown to exhibit less-robust biofilm formation and swarm motility (3, 20), we were interested in investigating whether Epr and other extracellular proteases were involved in swarming in an undomesticated B. subtilis strain.
First described more than a century ago for the gram-negative organism Proteus, swarming motility has now been demonstrated for many different genera of bacteria (9, 10, 39). Several factors appear to be important in eliciting a swarming phenotype: cell density, nutrient content, and viscosity of the of the medium are among the most well defined (10, 14, 28). In response to these factors, cells become hyperflagellated and elongated and establish groups, or “rafts,” of cells through cell-to-cell contacts, finally migrating as microcolonies (16). The swarming phenotype has been well documented for certain Bacillus species; however, the underlying mechanism responsible is just beginning to be studied (11, 16, 20, 37). For B. subtilis, the differentiated swarmer cell is highly adapted for surface colonization, featuring a considerable increase in flagellar biosynthesis and the secretion of a slime layer, both of which aid in translocation. In this work we established that a third component, extracellular protease activity, is important for swarm motility in an undomesticated isolate of B. subtilis. Greater than 95% of the extracellular proteolytic activity in B. subtilis is contributed by the two major extracellular proteases, subtilisin (AprE) and neutral metalloprotease E (NprE) (33). Five minor extracellular or cell-wall-associated proteases, Epr, Vpr, Bpr, Mpr, and WprA, are responsible for most of the remaining activity (42). A strain compromised for the production of all of the above proteases was unable to swarm. Additionally, we observed that extracellular protease activity was critical for biofilm formation. The null phenotypes exhibited by a protease-deficient strain, under both swarming and biofilm-forming conditions, present the possibility that the mechanisms involved in regulating swarming and biofilm formation may overlap.
MATERIALS AND METHODS
Strains, growth conditions, and plasmids.
The strains and plasmids used in this study are described in Table 1. All B. subtilis strains, except 168, were derived from the wild-type strain A164 (ATCC 6051a). Escherichia coli strains were used for cloning and plasmid propagation.
TABLE 1.
B. subtilis strains and swarm motility phenotypes
| B. subtilis strain | Description and/or genotype | Swarming phenotypea | Swarming initiated (h)b | Strain source or reference |
|---|---|---|---|---|
| 168 | Lab strain, trpC2 | − | BGSC | |
| A164 | Wild-type strain | +++ | 2-4 | ATCC 6051a |
| A164Δ1 | sigF | +++ | 2-4 | 41 |
| A164Δ2 | sigF nprE | +++ | 2-4 | 41 |
| A164Δ3 | sigF nprE aprE | +++ | 2-4 | 41 |
| A164Δ4 | sigF nprE aprE amyE | +++ | 2-4 | 41 |
| A164Δ5 | sigF nprE aprE amyE srfAC | ++ | 24 | 41 |
| A164Δ6 | sigF nprE aprE amyE srfAC wprA | ++ | 24 | This work |
| A164Δ7 | sigF nprE aprE amyE srfAC wprA bpr | ++ | 24 | This work |
| A164Δ8 | sigF nprE aprE amyE srfAC wprA bpr vpr | ++ | 24 | This work |
| A164Δ9 | sigF nprE aprE amyE srfAC wprA bpr vpr mpr | ++ | 24 | This work |
| A164Δ10 | sigF nprE aprE amyE srfAC wprA bpr vpr mpr epr | − | This work | |
| A164Δ10 + srfAC | sigF nprE aprE amyE wprA bpr vpr mpr epr | +++ | 2-4 | This work |
| A164Δ5E | sigF nprE aprE amyE srfAC epr | ++ | 24 | This work |
| A164Δ8E | sigF nprE aprE amyE srfAC wprA bpr vpr epr | − | This work | |
| MBin1 | epr | +++ | 2-4 | This work |
| MBin2 | epr srfAC | ++ | 24 | This work |
| MBin3 | epr srfAC wprA | ++ | 24 | This work |
| MBin4 | epr srfAC wprA bpr | ++ | 24 | This work |
| MBin5 | epr srfAC wprA bpr vpr | + | 48 | This work |
| MBin6 | epr srfAC wprA bpr vpr mpr | + | 48 | This work |
| MBin7 | epr srfAC wprA bpr vpr mpr nprE | + | 48 | This work |
| MBin8 | epr srfAC wprA bpr vpr mpr nprE aprE | +/− | 72 | This work |
| MBin9 | epr srfAC wprA bpr vpr mpr nprE aprE amyE | +/− | 72 | This work |
| MBin10 | epr srfAC wprA bpr vpr mpr nprE aprE sigF | − | This work | |
| A164Δsfp | sfp | ++ | 24 | This work |
| A164Δhag | hag | − | This work |
Swarming phenotypes were scored as equivalent to that of A164 (+++); delayed by approximately 24 h (++), delayed by greater than or equal to 48 h (+), delayed by at least 72 h (+/−), or nonswarming (−).
The average time (from more than three independent observations) required for each strain to initiate swarming motility is noted in hours.
Both the B. subtilis and E. coli strains were propagated in Luria-Burtani (LB) medium or on LB plates containing 1.5% agar at 37°C. LB medium for E. coli was supplemented with ampicillin (100 μg/ml) when necessary. B. subtilis was made competent by the method of Anagnostopoulos and Spizizen (2). When a plasmid containing the erythromycin resistance gene (ermC) was used for transformation in B. subtilis, erythromycin resistance was induced by adding 0.2 μg of erythromycin per ml to the liquid culture and incubating for 30 min at 37°C prior to plating. Tryptose blood agar base (TBAB) (Difco, Detriot, Mich.) plates supplemented with erythromycin (1 μg/ml) and lincomycin (25 μg/ml) were used for B. subtilis transformations. Unless stated otherwise, swimming and swarming motilities were analyzed on LB plates solidified with 0.3 and 0.7% agar (Difco), respectively. When indicated, swarm motility was tested on Spizizen's minimal medium (15) solidified with 0.7% agar (Difco).
The E. coli/B. subtilis shuttle vector pNNB194 (41) was used as the backbone for all gene deletion and replacement constructs (Table 2). This plasmid has an E. coli origin of replication, a temperature-sensitive B. subtilis origin of replication, an ampicillin resistance gene (bla) for selection in E. coli, and an erythromycin resistance gene (ermC) for selection in B. subtilis. Each gene deletion construct contained approximately 800 bp of DNA homologous to the region flanking the deletion. This was achieved by first PCR amplifying 400 bp at the 5′ and 3′ ends of each gene to be deleted. The primers used for each PCR are described in Table 3. The first two primers listed for each plasmid and the last two were used in separate PCRs with A164 genomic DNA as the template. The two PCR fragments obtained were then joined using splicing by overlapping extension (17) with the first and last primers listed for each plasmid (Table 3) and subsequently cloned into pCR2.1-TOPO (Invitrogen). Finally, the fragment containing the mutant allele was subcloned into the multiple cloning site of pNNB194 to create the deletion plasmid. To generate the srfAC replacement plasmid, the srfAC gene was cloned by PCR with the 5′ and 3′ primers used for the srfAC deletion vector. This created an intact copy of the srfAC gene, which contained homology to the region flanking the deletion site. The srfAC gene was then subcloned into pNNB194 as described above.
TABLE 2.
Plasmids and E. coli strains
| Plasmid or strain | Description or genotype | Source(s) and/or reference |
|---|---|---|
| Plasmids | ||
| pCR2.1-TOPO | Cloning vector, Ampr Kanr | Invitrogen (29) |
| pNNB194 | Shuttle vector, hybrid between pE194 and pBluescript II SK+, Ermr Ampr | Bacillus Genetic Stock Center (1E18), Stratagene (40) |
| pNNB194-Δepr | epr deletion vector, Ermr Ampr | This work |
| pNNB194-ΔsrfAC | srfAC deletion vector, Ermr Ampr | 41 |
| pNNB194-ΔwprA | wprA deletion vector, Ermr Ampr | This work |
| pNNB194-Δbpr | bpr deletion vector, Ermr Ampr | This work |
| pNNB194-Δvpr | vpr deletion vector, Ermr Ampr | This work |
| pNNB194-Δmpr | mpr deletion vector, Ermr Ampr | This work |
| pNNB194-ΔnprE | nprE deletion vector, Ermr Ampr | 41 |
| pNNB194-ΔaprE | aprE deletion vector, Ermr Ampr | 41 |
| pNNB194-ΔamyE | amyE deletion vector, Ermr Ampr | 41 |
| pNNB194-ΔsigF | sigF deletion vector, Ermr Ampr | 41 |
| pNNB194-Δsfp | sfp deletion vector, Ermr Ampr | This work |
| pNNB194-Δhag | hag deletion vector, Ermr Ampr | This work |
| pNNB194-srfACwt | srfAC replacement vector, Ermr Ampr | This work |
| E. coli strains | ||
| TOP10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara-leu)7697 galU galK rpsL(Strr) endA1 nupG | Invitrogene (13) |
| JM101 | supE thi-1 D(lac-proAB) [F′ traD36 proAB lacIqZDM15] | Stratagene (48) |
TABLE 3.
Primers used for splicing by overlapping extensiona
| Plasmid | Primer ID | Sequence (5′-3′) |
|---|---|---|
| pNNB194-Δwpr | CLJe7 | GGAATTCCAAAGCTGCAGCGGCCGGCGCG |
| CLJe8 | GAAGATCTCGTATACTTGGCTTCTGCAGCTGC | |
| CLJe9 | GAAGATCTGGTCAACAAGCTGGAAAGCACTC | |
| CLJe10 | CCCAAGCTTCGTGACGTACAGCACCGTTCCGGC | |
| pNNB194-Δbpr | bpr3Eco | GAATTCAAAGTCTTTTTCCCTATTGC |
| bpr4iSfiBamNot | GCGGCCGCGGATCCGGCCCTTAAGGCCTTTTGATATTCCGTCTTAGC | |
| bpr5SfiBamNot | GGCCTTAAGGGCCGGATCCGCGGCCGCCTTCTCCTGGCACAACGGTA | |
| bpr6iKpn | GGTACCGCGGCATACGAATGGTAAAC | |
| pNNB194-Δvpr | vpr3Eco | GAATTCCAATAAATGGTCCAAATGAC |
| vpr4iSfiBamNot | GCGGCCGCGGATCCGGCCCTTAAGGCCTGTGACGTTCGGGTACACTG | |
| vpr2SfiBamNot | GGCCTTAAGGGCCGGATCCGCGGCCGCCGAAGAACCTTTCACTGTT | |
| ywcH1Kpn | GGTACCTCCGGATGTGTGGCTGCTC | |
| pNNB194-Δmpr | mpr3Eco | GAATTCATCGATACGGGTGGTTGAC |
| mpr4iSfiBamNot | GCGGCCGCGGATCCGGCCCTTAAGGCCTATCGTCGAAGCCCATCCAT | |
| mpr5SfiBamNot | GGCCTTAAGGGCCGGATCCGCGGCCGCTTGGGAACAAGGGTGACGAA | |
| mpr6iKpn | GGTACCTCCTCCGGGTTTGATTGATA | |
| pNNB194-Δsfp | Sfp1A | GAATTCCGCGTGATTCACTGATGATGC |
| Sfp1B | AGTCTGTCTGCTCGTCCTTGTCTATGACTGAGCGAACGAGCAC | |
| Sfp1C | TGTGCTCGTTCGCTCAGTCATAGACAAGGACGAGCAGACAGAC | |
| Sfp1D | GGATCCCAAGCCATTCGAACTCACAGG | |
| pNNB194-Δhag | Hag1A | GATATCGCGTTATCCAGCGATGTGATC |
| Hag1B | GCACCAAGCTTAGCACGTTGAGAGCTTGAACAACTAGCTCACG | |
| Hag1C | TCGTGAGCTAGTTGTTCAAGCTCTCAACGTGCTAAGCTTGGTG | |
| Hag1D | GGATCCCAGGTTGTAACGTAGTGAGCC | |
| pNNB194-srfACwt | srfC1 | AAGCTTTGAATGGGTGTGG |
| srfC4i | CTGACATGAGGCACTGAC |
The primers used to construct the plasmids described by A. Sloma and L. Christianson (41) are listed therein.
Strain construction.
Beginning with strain A164, ATCC 6051a, multiple gene deletions were made in a sequential manner. For the strains listed in Table 1, each deletion mutation was introduced into the chromosome by allelic exchange. The corresponding deletion plasmid was used to transform B. subtilis competent cells by selecting for erythromycin resistance on TBAB plates at the permissive temperature of 34°C. A single transformant was then streaked to TBAB supplemented with erythromycin and incubated at the nonpermissive temperature of 45°C in order to select for plasmid integration into the chromosome. To promote homologous recombination and plasmid loss from the locus targeted for deletion, LB medium was inoculated with several colonies from the TBAB plate, which were grown without selection at 34°C. The cultures were streaked to LB plates, and single colonies were patched to LB and TBAB plates containing erythromycin. Colonies that were erythromycin sensitive were screened by PCR (Extract-N-Amp Plant PCR kit; Sigma-Aldrich) to find strains in which plasmid excision had occurred with retention of the mutant allele. Gene replacement of srfAC was obtained in essentially the same manner as described above. The loss and restoration of surfactin activity were confirmed by halo formation on whole-blood agar plates (trypticase soy agar with 5% sheep blood; BD, Sparks, Md.) (41).
Motility assays.
To analyze swimming and swarming motility, cells were collected with a sterile toothpick from an overnight culture plate and used to inoculate 0.3% agar to observe swimming and 0.7% agar to observe swarming. The plates were incubated at 37°C and evaluated over time.
Protease assays.
B. subtilis cultures were grown in MRS medium (Difco) for 24 h at 37°C, and supernatants were collected following centrifugation at 10,000 × g for 10 min. Protease activity was measured by using fluorescein isothiocyanate-casein as the substrate (Sigma Chemical Co., St. Louis, Mo.). Forty microliters of fluorescein isothiocyanate-casein substrate (stock solution: 1:1 with 0.1 M Tris-10 mM CaCl2 [pH 8.0]) was mixed with 10 μl of culture sample (diluted appropriately in 0.1 M Tris-10 mM CaCl2 [pH 8]) and incubated for 1 h at 45°C. The reaction was quenched with 150 μl of 5% trichloroacetic acid and incubated at 4°C for 1 h. After centrifugation for 10 min, a 10-μl aliquot of the supernatant was mixed with 1 ml of 0.5 M borate (pH 9.0). A 200-μl aliquot of the solution was transferred to a black “U” bottom 96-well plate (ThermoLabsystems, Franklin, Mass.). Fluorescence was measured with a Fluorolite 1000 instrument (ThermoLabsystems, Franklin, Mass.), using reference channel 3 and a setting of 1176. Activity was measured in fluorescent units and expressed in units per milliliter relative to the protease standard, Subtilisin Carlsberg (Sigma).
Swarm motility assays in the presence of protease.
Proteinase K (654 U/mg), a serine protease isolated from the fungus Tritirachium album, and subtilisin Carlsberg (32.0 U/mg) from B. subtilis were obtained from Sigma-Aldrich and resuspended in 50 mM Tris (pH 7.5) at 10 mg/ml. Dispase I (6 U/mg), a neutral protease obtained from Roche Applied Science (Mannheim, Germany), was resuspended in 50 mM Tris (pH 7.5) at 5 mg/ml. Each protease was diluted in a final volume of 50 μl and then spread on the surface of a 0.7% agar plate. Plates were allowed to dry for 20 min prior to strain inoculation. The effect of various levels of protease activity on swarming was evaluated: proteinase K, 6.5, 0.65, 0.065, and 0.0065 U; subtilisin, 3.2, 0.32, 0.032, and 0.0032 U; and dispase I, 6.0, 0.6, 0.06, and 0.006 U.
Preparation of conditioned medium.
A B. subtilis A164 or A164Δ10 overnight culture was diluted 1:100 in LB and grown at 37°C to the end of log phase. The cultures were collected and centrifuged at 4,000 × g to pellet the cells. The supernatant was removed and passed through a 0.22-μm-pore-size filter. To make 0.7% agar plates, filtered conditioned medium was mixed 1:1 with fresh LB containing 1.4% agar. Heat inactivation of the conditioned medium was performed by autoclaving for 15 min. To test the effect of protease inhibitors on swarming motility, a 14-mg/ml fresh stock of phenylmethylsulfonyl fluoride (PMSF) suspended in isopropanol was used. The conditioned medium was incubated with 140 or 280 μg of PMSF/ml for 20 min prior to preparing the 0.7% agar plates.
Flagellar analysis.
In order to observe flagella produced under conditions that support swarming, cells were collected from the surfaces of 0.7% agar plates three hours after inoculation. These cells were sampled from the edge of a swarming colony or from the colony edge of a nonswarming colony as indicated. Swimming cells were obtained from cultures grown in liquid LB medium to an optical density at 600 nm of 0.5. Flagella were stained as described by Kearns and Losick (20) and examined by phase-contrast microscopy with an Axioplan Universal microscope (Carl Zeiss, Ltd., Oberkochen, Germany).
Biofilm formation.
The ability of each strain to form a pellicle on the surface of a standing culture was tested. The strains were inoculated onto minimal medium and grown according to the method described by Branda et al. (3). The minimal medium contains the following: 5 mM potassium phosphate (pH 7), 100 mM morpholinepropanesulfonic acid (pH 7), 2 mM MgCl2, 700 μM CaCl2, 50 μM MnCl2, 50 μM FeCl3, 2 μM thiamine, 0.5% glycerol, 0.5% glutamate.
RESULTS
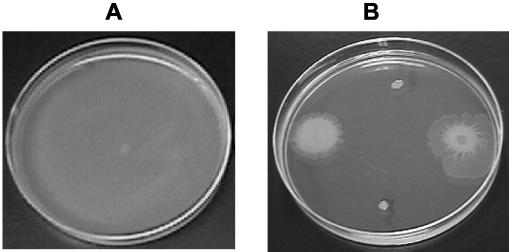
Genetic analysis of factors required for swarm motility in the wild-type strain, A164. B. subtilis strain A164 exhibits robust swarm motility on 0.7% agar (Fig. 1). When inoculated to the center of a 100-mm petri dish, A164 begins to swarm after a lag period of 2 to 4 h. Progressive expansion of the colony continues over time, resulting in complete colonization of the surface of the plate within 8 h (Fig. 1A). A set of A164 strains containing 10 sequential deletions (A164Δ1 to A164Δ10) (Table 1) was originally constructed as hosts for enzyme production; therefore, the first deletion rendered them sporulation negative, and the rest of the deletions removed unwanted extracellular activities. Although not created for the purpose of studying swarming, these strains were of interest due to published results that surfactin (a product of the srfA operon) and Epr have an established role in swarm migration (6, 25). The strains were screened for their ability to swarm, and the results were as follows: swarming was unaffected in strains A164Δ1 through A164Δ4 (Table 1). A 24-h delay in swarm migration was observed in strains A164Δ5 through A164Δ9 (Table 1 and Fig. 1B). Strain A164Δ10 exhibited a nonswarming phenotype (Table 1 and Fig. 1B).
FIG. 1.
Secreted proteases are required for swarming motility by B. subtilis A164. Swarm motility of B. subtilis A164 (A) and A164 mutants A164Δ5, MBin7, A164Δ8, and A164Δ10 (clockwise from left) (B) is shown. The absence of surfactin production in A164Δ5 and A164Δ8 results in a less-robust swarming phenotype, and a nonswarming phenotype is exhibited by A164Δ10, which is deficient in both surfactin and protease activity. Although MBin7 appears to have a nonswarming phenotype, swarming is initiated after 48 h for this strain and is not completely inhibited until deletions are made in the remaining extracellular protease, aprE, and sigF (MBin10 strain, data not shown). All strains were inoculated with a toothpick onto a 0.7% agar LB swarm plate and incubated for 20 h at 37°C.
The results of the phenotypic analysis described above were similar to those published by Dixit et al., who found Epr to be essential for swarming in the domesticated B. subtilis laboratory strain, 168 (6). However, when an epr deletion alone was introduced into A164 (MBin1), swarm motility was unaffected (Table 1). Since surfactin is important for swarm motility and B. subtilis 168 is defective in surfactin production (32), we wanted to determine the effect of mutations in both srfAC and epr on the swarming phenotype of A164. A deletion was made in the srfAC gene of strain MBin1, creating MBin2 (Table 1). Swarm motility by MBin2 was delayed but not eliminated, suggesting that one or more of the other mutations in strain A164Δ10 plays a role in swarming.
Genetic epistasis analysis was used to determine which genes are essential for swarming. This analysis showed that when an epr mutation was introduced into A164Δ5 (A164Δ5E), no additive effect on swarming was observed compared to results for A164Δ5. In contrast, deletion of epr from A164Δ8 (A164Δ8E) resulted in a nonswarming phenotype (Table 1). This analysis indicated that surfactin and some combination of minor extracellular proteases, which are the products of the genes deleted from strains A164Δ5 through A164Δ10, played a major role in swarm motility. To confirm this hypothesis, the genes encoding surfactin and each of the minor extracellular proteases were sequentially deleted from A164. The subsequent strains were tested for their ability to swarm (MBin2 through MBin6; Table 1). The results of this screen revealed that MBin6, which is deficient in surfactin and minor (but not major) extracellular protease activity, shows significantly more delay in swarm initiation than A164Δ5 (48 versus 24 h; Table 1). However, swarming motility was not completely abolished for MBin6, as was observed for A164Δ10. Since A164Δ10 contains deletions in the genes encoding both the major and minor extracellular proteases, the major extracellular protease genes were deleted from MBin6 (MBin8; Table 1). Protease activities for strains A164, A164Δ5, MBin8, and A164Δ10 were analyzed and found to be 8.36, 0.024, 0.008, and 0.011 U/ml, respectively. The protease activities for strains MBin2 through MBin6 (data not shown) were similar to that for A164. A decrease in protease activity, due to the removal of any one of the minor proteases from each of these strains, could not be assayed accurately, since the removal of one protease often results in an increase in the remaining protease activity (43). Swarming was analyzed for MBin8 to determine if a strain with levels of protease activity similar to that of A164Δ10 would have a null phenotype. Strain MBin8 exhibited a severe delay in swarm initiation (>72 h [Table 1]); however, a complete nonswarming phenotype was achieved only upon the additional deletion of sigF (MBin10; Table 1), which encodes the forespore-specific sigma factor, SigF. The deletion in sigF, as well as amyE (MBin9; Table 1), was employed in an effort to determine which deletions in A164Δ10 were responsible for the null swarming phenotype. These results demonstrate that extracellular proteases and surfactin have a major role in swarming motility in A164 and that SigF may exert a minor influence in regulating this multicellular activity.
Extracellular proteolytic activity is important for swarming.
The swarming phenotypes exhibited by the various mutant strains described above led to the hypothesis that total extracellular proteolytic activity, but not specifically activity from Epr, is important for swarm behavior. To test this idea, exogenous protease was applied to the surface of swarm medium, and surface translocation by the protease-deficient strain, A164Δ10, was determined. For this analysis, we tested three proteases originating from different sources: proteinase K, subtilisin, and dispase I (Fig. 2 and data not shown). In each case, preapplication of exogenously derived protease to the medium rescued swarming for A164Δ10. Interestingly, under these conditions, the resulting phenotype was indistinguishable from that of the wild-type strain, A164 (Fig. 2). This not only demonstrates that nonspecific proteolytic activity contributes to the multicellular process of swarming but also reveals that exogenous protease can overcome the defect in swarm initiation created by a lack of surfactin production.
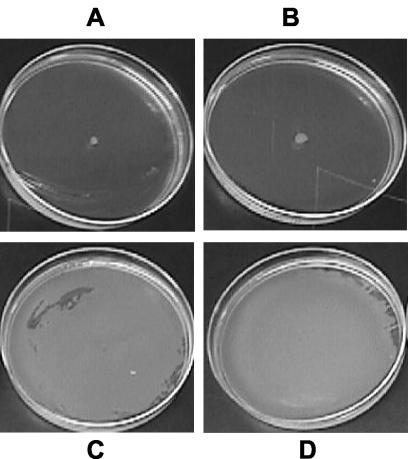
FIG. 2.
Exogenous protease rescues swarming motility in a nonswarming, protease-deficient strain. Swarming behavior of A164Δ10 untreated control (A) or with 0.065 (B), 0.65 (C), or 6.5 (D) U of proteinase K is shown. Swarm plates were incubated at 37°C for 24 h. Swarming motility was also rescued by treatment with subtilisin and dispase I treatment (data not shown) at all concentrations except the lowest (see Materials and Methods for concentrations tested and inoculation method).
As an alternative test of the hypothesis that secreted proteases play an important role in swarming motility, we tested whether conditioned medium obtained from cultures of selected bacterial strains could suppress the swarming-defective phenotype of A164Δ10. Conditioned medium was obtained from either A164 or A164Δ10 and used to prepare swarm plates. A164Δ10 was able to swarm on plates containing conditioned medium from A164 but not from A164Δ10. To determine if this observation was directly related to the protease activity, medium conditioned by A164 cells was either heat inactivated or incubated with the nonspecific protease inhibitor PMSF prior to use (Fig. 3 and data not shown). Previous work has shown that AprE and most of the minor extracellular proteases are sensitive to PMSF (33). Consistent with the hypothesis, A164Δ10 did not swarm well in the presence of PMSF-treated, A164-conditioned medium (Fig. 3). In addition, heat-inactivated conditioned medium was not able to restore the ability of A164Δ10 to swarm (data not shown).
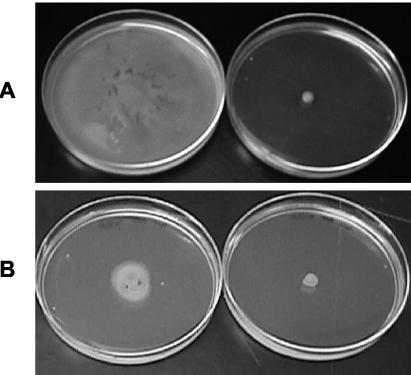
FIG. 3.
Conditioned medium from A164, but not A164Δ10, rescues swarm motility of A164Δ10. A164Δ10 was inoculated onto swarm plates prepared with conditioned medium and incubated for 24 h at 37°C. Conditioned medium incubated with isopropanol alone had no effect on swarming (data not shown). (A) Conditioned medium obtained from A164 (left) or A164Δ10 (right). (B) Conditioned medium obtained from A164 and preincubated with PMSF at a final concentration of 140 (left) or 280 (right) μg/ml, respectively.
Minimal medium supports swarming motility.
An obvious role for extracellular degradative enzymes, such as proteases, is the acquisition of nutrients from the surrounding environment. Potentially, proteolysis could be required to modify or degrade small peptides in LB medium that supply the energy necessary to initiate and sustain the process of swarm migration. To assess swarm motility in the absence of protein substrate, A164 was inoculated onto 0.7% agar Spizizen's minimal medium plates. Except for a decrease in the thickness of the swarming colony, motility was otherwise identical to that exhibited on 0.7% LB agar plates (data not shown). Furthermore, exogenous protease applied to the minimal medium plates rescued swarming for A164Δ10 (data not shown). These results imply that the role of extracellular proteases is not to fulfill a nutritional requirement necessary to drive swarm migration. Additionally, this observation suggests that the proteases are acting on either proteins secreted by the cell or proteins present on the bacterial cell itself.
Protease-deficient strains exhibit swim motility and display hyperflagellation on swarm agar plates.
Flagellar biosynthesis is necessary for swarm motility (39). In addition, cells isolated from the edge of a swarming colony often exhibit a dramatic increase in the number of flagella produced compared to cells isolated from liquid culture (8, 11, 49). To determine if flagellar formation was affected for A164Δ10, both swim motility assays and microscopic analysis were performed. Swim motility of A164Δ10 and other mutants, as assessed by cell movement through the water-filled channels of a 0.3% agar plate, was equivalent to that of A164 (Fig. 4). To evaluate whether A164Δ10 exhibited any differences in the number of flagellar organelles, microscopic examination of cells obtained from mid-log-phase cultures grown either in LB broth or on 0.7% LB agar plates was performed. Flagellar formation under both conditions appeared to be unaffected for A164Δ10 and resembled that of A164 (Fig. 4).
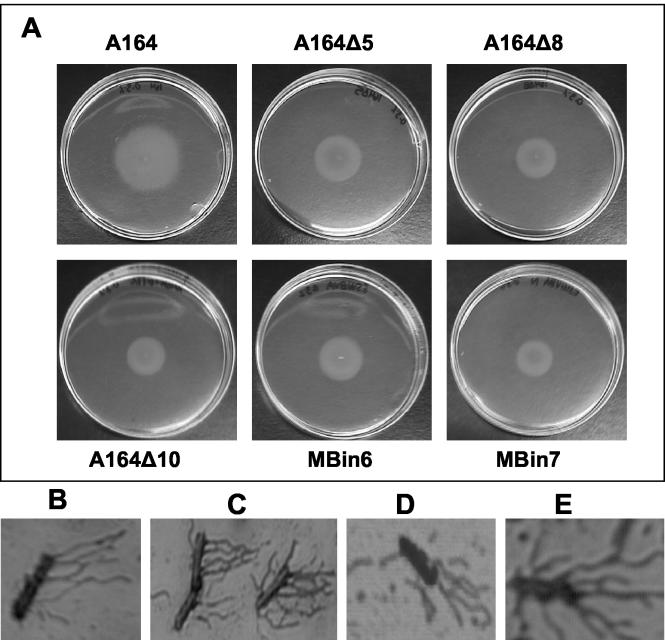
FIG. 4.
Protease activity is not required for swimming motility or hyperflagellation on swarm plates. (A) Swim motility assay at 6 h following inoculation. Swimming motility for all A164 mutants was equivalent to that of A164. The appearance of slightly increased motility for A164 is due to the small amount of swarming, which still occurs at this agar concentration and at a faster rate than swimming. Cells were stained for flagella from mid-log A164 cultures (B), A164 swarm plates (C), A164Δ10 mid-log cultures (D), and A164Δ10 swarm plates (E). The absence of surfactin and protease activity had no effect on flagellar formation in liquid medium or on swarm plates. All cells were collected from swarm plates 3 h after inoculation. Microscopic analysis was performed at a magnification of ×100.
Surfactin can rescue swarming motility in a protease-deficient strain.
Biosurfactants, such as surfactin produced by B. subtilis, aid in surface translocation by reducing surface tension. In several bacterial species, defects in swarm motility have been observed in mutants lacking surfactant production (5, 9, 31). Surfactin is a cyclic lipopeptide whose synthesis is catalyzed by the Srf complex (34). Deletion of srfAC eliminates surfactin production and delays the initiation of swarming for B. subtilis A164. Since swarm initiation is completely blocked for A164Δ10, a strain deficient for the production of both surfactin and most secreted proteases, we wanted to assess how the introduction of wild-type levels of surfactin would affect the nonswarming phenotype. To do this, the ca. 800-bp deletion at the srfAC locus was replaced with an intact copy of the gene. Restoration of surfactin production was evaluated and confirmed by testing B. subtilis A164Δ10+srfAC for hemolytic activity when grown on whole-blood agar (data not shown). When inoculated to 0.7% agar, A164Δ10+srfAC initiated and exhibited swarm motility identical to that of A164 (data not shown), demonstrating that wild-type levels of surfactin, from the restored srfAC locus, can reestablish swarming motility in A164Δ10. In addition, this result revealed that surfactin, as well as secreted proteases, affects swarming by B. subtilis A164.
Secreted proteases function in biofilm formation.
Our present evidence suggests that extracellular protease, in combination with surfactin, plays a role in swarming. This led us to consider whether secreted proteases might have a role in other types of multicellular behaviors. A few studies have found that certain factors which affect swarm motility also inhibit other multicellular behaviors, such as biofilm formation (18, 36, 39). To determine if proteolytic activity is involved in the formation of a biofilm, protease-deficient strains were tested for the ability to form a biofilm or pellicle on the surface of a standing culture. While the wild-type strain A164 formed a thick pellicle with prominent surface structures, strains unable to produce surfactin due to a deletion in either the sfp or srfAC gene (A164sfp and MBin6, respectively) formed a thin, flat pellicle (Fig. 5). Sfp is a phosphopantetheinyl transferase which primes the nonribosomal surfactin peptide synthetase (encoded by the srfA operon). Sfp and the srfAC operon are both required for the production of surfactin (34). Besides surfactin, MBin6 is also deficient in minor extracellular proteolytic activity. While MBin6 maintained the ability to form a biofilm, A164Δ10 exhibited a null phenotype. In addition to surfactin and minor extracellular proteolytic activity, A164Δ10 is also deficient in major extracellular protease activity. Significantly, biofilm formation in A164Δ10 was rescued when exogenous protease (dispase I, subtilisin, or proteinase K) was added to the media at all concentrations tested (2 to 500 U) (data not shown). Additionally, strain A164Δ10+srfAC was able to form a biofilm (data not shown). These results suggest that extracellular factors central for swarm motility, such as proteases and surfactin, are also important for biofilm formation.
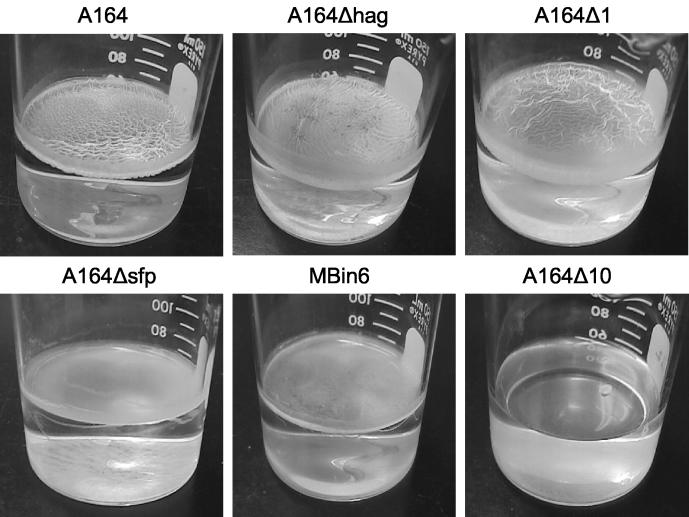
FIG. 5.
Extracellular protease activity is required for biofilm formation. A164 deletion strains were analyzed for their ability to form biofilms. While mutants defective in flagellar formation (A164Δhag), sporulation (A164Δ1), or surfactin production (A164Δsfp and MBin6) retained the ability to produce biofilm, a mutant (A164Δ10) deficient in both surfactin and extracellular protease activity, as well as sporulation, did not form a biofilm.
DISCUSSION
B. subtilis produces eight characterized extracellular or cell wall-associated proteases. Seven of these have been detected in the growth medium, while the remaining protease, a product of the nprB gene, is not secreted at detectable levels (26, 33, 45, 46). The alkaline serine protease subtilisin (AprE), along with the neutral metalloprotease E (NprE), account for more than 95% of the extracellular proteolytic activity of B. subtilis and are commonly referred to as the major extracellular proteases. The remainder of the protease activity has been attributed to minor extracellular proteases, with the serine protease Epr being a principal contributor (42). None of these proteases has been found to be essential either for growth or for sporulation of B. subtilis. However, transcription of each is tightly controlled, with the highest level of expression almost always demonstrated to occur at the end of log phase (19, 33). In a study focused on determining how epr is regulated, Dixit et al. found that Epr function was necessary for swarming for the laboratory strain B. subtilis 168 (6). These results were consistent with the nonswarming phenotype we observed for the wild-type strain, A164, deficient in extracellular protease activity (A164Δ10). However, they conflicted with our observation that a deletion of epr alone in A164 had no effect on swarming. While it appeared that Epr did function in swarming, it was apparent that other extracellular factors are also important.
The published investigations of swarming in strain 168 reveal a phenotype that appears less vigorous than that exhibited by A164 (6, 30, 31). While the wild-type strain, A164, is able to produce surfactin, the domesticated laboratory strain 168 has lost this ability due to a frameshift mutation in the sfp gene, which is essential for surfactin synthesis (32). Based on the established importance of biosurfactants in swarm motility and the delayed initiation of swarming displayed by A164 strains that are deficient for surfactin production (Table 1; Fig. 1), it seemed plausible that abolishing the production of both surfactin and Epr in A164 would result in a nonswarming phenotype. A strain containing deletions in both epr and srfAC (MBin2) was tested and found to have maintained a somewhat reduced ability to swarm. This suggested that while Epr and surfactin production appeared to be important for swarming motility, additional proteins were involved. Genetic epistasis analysis was performed, and the results suggested that while sigF, nprE, aprE, and amyE had a small role, if any, some of the minor extracellular proteases, in addition to Epr, were important for swarming (Table 1). However, only a strain containing deletions in all of the extracellular proteases, as well as srfAC and sigF, exhibited a completely nonswarming phenotype (Table 1; Fig. 1).
Taken together, the above results led us to hypothesize that total extracellular proteolytic activity contributes to the initiation of this multicellular behavior. To investigate this hypothesis, swarming motility of strain A164Δ10 (deficient in extracellular protease activity, surfactin, SigF, and AmyE) was analyzed in the presence of exogenous protease (Fig. 2). Regardless of the source of the protease or whether a rich or minimal medium was used, A164Δ10 displayed a capacity for swarming that was identical to that observed for A164. Additionally, swarming of A164Δ10 was supported on plates made with medium preconditioned with strain A164 and was inhibited when the conditioned medium was treated with the protease inhibitor, PMSF (Fig. 3). Finally, a strain containing deletions in all seven extracellular proteases, as well as srfAC and sigF, exhibited a nonswarming phenotype.
These observations support the hypothesis that the multicellular behavior of swarming motility is stimulated by the presence of extracellular protease. However, elucidating the nature of the role that proteolytic activity plays in swarming will require further study. Significantly, the ability of exogenous protease to rescue swarming on minimal medium excludes the obvious idea that proteolytic digestion of media components alone encourages swarm motility. A second theory is that proteins secreted by the cells require cleavage before they can function to promote swarming. This type of regulation is not foreign to Bacillus and other gram-positive bacteria. Several processes are controlled through cell-cell signaling mediated by peptides that are proteolytically processed into signaling molecules (7, 35). However, the ability of wild-type levels of surfactin to complement a deficiency in proteolytic activity indicates that extracellular protease does not function by providing an essential signal for swarmer cell development or initiation of cellular migration. It is conceivable, however, that the proteases cleave cell surface proteins, leading to cell surface modifications that aid in bacterium-bacterium or bacterium-surface interactions which promote surface translocation. Microscopic inspection of the nonswarming protease-deficient strain, A164Δ10, revealed a random and rapid movement of isolated cells at the periphery of the colony when inoculated to the surface of a swarm plate, but no multicellular “rafts” were observed (data not shown). Further investigation will be needed to determine the significance of this observation.
It should be noted that the nonswarming phenotype was observed with a strain deficient for both protease and surfactin activity. Since surfactin can promote spreading motility of bacteria by enhancing surface fluidity, even in nonflagellated mutants (22, 27), it is probable that the ability of surfactin to compensate for the loss of proteolytic activity during swarming is due to its surface tension-reducing properties. However, the mechanism by which exogenous protease restores swarming motility to wild-type levels in a surfactin-minus strain is unclear.
It has been suggested that bacterial colonization of surfaces through swarming motility plays a role in biofilm formation (23). Recently, Branda et al. showed that wild-type isolates of B. subtilis form biofilms with complex architectures on the surfaces of standing liquid cultures. Additionally, the investigators observed structural defects in this architecture for mutants deficient in surfactin production (3). Since it appeared that surfactin was required for both efficient swarming and structured pellicle-type biofilms, we wanted to determine if a similar relationship existed for extracellular proteases. To ascertain whether or not extracellular proteases function in biofilm formation, we evaluated A164Δ10 under conditions that stimulate pellicle formation. Intriguingly, we found that in addition to the loss of swarm motility, A164Δ10 was incapable of forming any pellicle structure under the tested conditions (Fig. 5). Furthermore, when exogenous protease was added to the medium, A164Δ10 produced a pellicle (data not shown), which supports the idea that a correlation exists between swarming and biofilm formation.
Acknowledgments
We thank Mike Thomas for providing strains, plasmids, and primers and Brian Clancy-Gorre for protease assay data. We are also grateful to Mike Thomas, Régine Behr, and Dan Kearns for technical support and Bill Widner and Randy Berka for critical reading of the manuscript.
REFERENCES
- 1.Allison, C., H. C. Lai, and C. Hughes. 1992. Co-ordinate expression of virulence genes during swarm-cell differentiation and population migration of Proteus mirabilis. Mol. Microbiol. 6:1583-1591. [DOI] [PubMed] [Google Scholar]
- 2.Anagnostopoulos, C., and J. Spizizen. 1961. Requirements for transformation of Bacillus subtilis. J. Bacteriol. 81:741-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Branda, S. S., J. E. Gonzalez-Pastor, S. Ben-Yehuda, R. Losick, and R. Kolter. 2001. Fruiting body formation by Bacillus subtilis. Proc. Natl. Acad. Sci. USA 98:11621-11626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Costerton, J. W., K. J. Cheng, G. G. Geesey, T. I. Ladd, J. C. Nickel, M. Dasgupta, and T. J. Marrie. 1987. Bacterial biofilms in nature and disease. Annu. Rev. Microbiol. 41:435-464. [DOI] [PubMed] [Google Scholar]
- 5.Deziel, E., F. Lepine, S. Milot, and R. Villemur. 2003. rhlA is required for the production of a novel biosurfactant promoting swarming motility in Pseudomonas aeruginosa: 3-(3-hydroxyalkanoyloxy)alkanoic acids (HAAs), the precursors of rhamnolipids. Microbiology 149:2005-2013. [DOI] [PubMed] [Google Scholar]
- 6.Dixit, M., C. S. Murudkar, and K. K. Rao. 2002. epr is transcribed from a final σD promoter and is involved in swarming of Bacillus subtilis. J. Bacteriol. 184:596-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dunny, G. M., and B. A. Leonard. 1997. Cell-cell communication in gram-positive bacteria. Annu. Rev. Microbiol. 51:527-564. [DOI] [PubMed] [Google Scholar]
- 8.Eberl, L., G. Christiansen, S. Molin, and M. Givskov. 1996. Differentiation of Serratia liquefaciens into swarm cells is controlled by the expression of the flhD master operon. J. Bacteriol. 178:554-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eberl, L., S. Molin, and M. Givskov. 1999. Surface motility of Serratia liquefaciens MG1. J. Bacteriol. 181:1703-1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fraser, G. M., and C. Hughes. 1999. Swarming motility. Curr. Opin. Microbiol. 2:630-635. [DOI] [PubMed] [Google Scholar]
- 11.Ghelardi, E., F. Celandroni, S. Salvetti, D. J. Beecher, M. Gominet, D. Lereclus, A. C. Wong, and S. Senesi. 2002. Requirement of flhA for swarming differentiation, flagellin export, and secretion of virulence-associated proteins in Bacillus thuringiensis. J. Bacteriol. 184:6424-6433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Givskov, M., L. Eberl, and S. Molin. 1997. Control of exoenzyme production, motility and cell differentiation in Serratia liquefaciens. FEMS Microbiol. Lett. 148:115-122. [Google Scholar]
- 13.Grant, S. G., J. Jessee, F. R. Bloom, and D. Hanahan. 1990. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc. Natl. Acad. Sci. USA 87:4645-4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harshey, R. M. 1994. Bees aren't the only ones: swarming in gram-negative bacteria. Mol. Microbiol. 13:389-394. [DOI] [PubMed] [Google Scholar]
- 15.Harwood, C. R., and A. R. Archibald. 1990. Growth maintenance and general techniques, p. 1-19. In C. R. Harwood and S. M. Cutting (ed.), Molecular biological methods for Bacillus. John Wiley & Sons, Chichester, England.
- 16.Henrichsen, J. 1972. Bacterial surface translocation: a survey and a classification. Bacteriol. Rev. 36:478-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Horton, R. M., H. D. Hunt, S. N. Ho, J. K. Pullen, and L. R. Pease. 1989. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77:61-68. [DOI] [PubMed] [Google Scholar]
- 18.Izquierdo, L., N. Abitiu, N. Coderch, B. Hita, S. Merino, R. Gavin, J. M. Tomas, and M. Regue. 2002. The inner-core lipopolysaccharide biosynthetic waaE gene: function and genetic distribution among some Enterobacteriaceae. Microbiology 148:3485-3496. [DOI] [PubMed] [Google Scholar]
- 19.Kallio, P. T., J. E. Fagelson, J. A. Hoch, and M. A. Strauch. 1991. The transition state regulator Hpr of Bacillus subtilis is a DNA-binding protein. J. Biol. Chem. 266:13411-13417. [PubMed] [Google Scholar]
- 20.Kearns, D. B., and R. Losick. 2003. Swarming motility in undomesticated Bacillus subtilis. Mol Microbiol. 49:581-590. [DOI] [PubMed] [Google Scholar]
- 21.Kim, W., T. Killam, V. Sood, and M. G. Surette. 2003. Swarm-cell differentiation in Salmonella enterica serovar Typhimurium results in elevated resistance to multiple antibiotics. J. Bacteriol. 185:3111-3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kinsinger, R. F., M. C. Shirk, and R. Fall. 2003. Rapid surface motility in Bacillus subtilis is dependent on extracellular surfactin and potassium ion. J. Bacteriol. 185:5627-5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kirov, S. M. 2003. Bacteria that express lateral flagella enable dissection of the multifunctional roles of flagella in pathogenesis. FEMS Microbiol. Lett. 224:151-159. [DOI] [PubMed] [Google Scholar]
- 24.Kolter, R., and R. Losick. 1998. One for all and all for one. Science 280:226-227. [DOI] [PubMed] [Google Scholar]
- 25.Lindum, P. W., U. Anthoni, C. Christophersen, L. Eberl, S. Molin, and M. Givskov. 1998. N-acyl-l-homoserine lactone autoinducers control production of an extracellular lipopeptide biosurfactant required for swarming motility of Serratia liquefaciens MG1. J. Bacteriol. 180:6384-6388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Margot, P., and D. Karamata. 1996. The wprA gene of Bacillus subtilis 168, expressed during exponential growth, encodes a cell-wall-associated protease. Microbiology 142:3437-3444. [DOI] [PubMed] [Google Scholar]
- 27.Matsuyama, T., A. Bhasin, and R. M. Harshey. 1995. Mutational analysis of flagellum-independent surface spreading of Serratia marcescens 274 on a low-agar medium. J. Bacteriol. 177:987-991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCarter, L., and M. Silverman. 1990. Surface-induced swarmer cell differentiation of Vibrio parahaemolyticus. Mol. Microbiol. 4:1057-1062. [DOI] [PubMed] [Google Scholar]
- 29.Mead, D. A., N. K. Pey, C. Herrnstadt, R. A. Marcil, and L. M. Smith. 1991. A universal method for the direct cloning of PCR amplified nucleic acid. Biotechnology (N. Y.) 9:657-663. [DOI] [PubMed] [Google Scholar]
- 30.Mendelson, N. H., A. Bourque, K. Wilkening, K. R. Anderson, and J. C. Watkins. 1999. Organized cell swimming motions in Bacillus subtilis colonies: patterns of short-lived whirls and jets. J. Bacteriol. 181:600-609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mendelson, N. H., and B. Salhi. 1996. Patterns of reporter gene expression in the phase diagram of Bacillus subtilis colony forms. J. Bacteriol. 178:1980-1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakano, M. M., N. Corbell, J. Besson, and P. Zuber. 1992. Isolation and characterization of sfp: a gene that functions in the production of the lipopeptide biosurfactant, surfactin, in Bacillus subtilis. Mol. Gen. Genet. 232:313-321. [DOI] [PubMed] [Google Scholar]
- 33.Pero, J., and A. Sloma. 1993. Proteases, p. 939-952. In J. A. Hoch, R. Losick, and A. L. Sonenshein (ed.), Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. American Society for Microbiology, Washington, D.C.
- 34.Peypoux, F., J. M. Bonmatin, and J. Wallach. 1999. Recent trends in the biochemistry of surfactin. Appl. Microbiol. Biotechnol. 51:553-563. [DOI] [PubMed] [Google Scholar]
- 35.Pottathil, M., and B. A. Lazazzera. 2003. The extracellular Phr peptide-Rap phosphatase signaling circuit of Bacillus subtilis. Front. Biosci. 8:d32-45. [DOI] [PubMed] [Google Scholar]
- 36.Ren, D., J. J. Sims, and T. K. Wood. 2002. Inhibition of biofilm formation and swarming of Bacillus subtilis by (5Z)-4-bromo-5-(bromomethylene)-3-butyl-2(5H)-furanone. Lett. Appl. Microbiol. 34:293-299. [DOI] [PubMed] [Google Scholar]
- 37.Senesi, S., F. Celandroni, S. Salvetti, D. J. Beecher, A. C. Wong, and E. Ghelardi. 2002. Swarming motility in Bacillus cereus and characterization of a fliY mutant impaired in swarm cell differentiation. Microbiology 148:1785-1794. [DOI] [PubMed] [Google Scholar]
- 38.Shapiro, J. A. 1998. Thinking about bacterial populations as multicellular organisms. Annu. Rev. Microbiol. 52:81-104. [DOI] [PubMed] [Google Scholar]
- 39.Sharma, M., and S. K. Anand. 2002. Swarming: a coordinated bacterial activity. Curr. Sci. 83:707-715. [Google Scholar]
- 40.Short, J. M., J. M. Fernandez, J. A. Sorge, and W. D. Huse. 1988. Lambda ZAP: a bacteriophage lambda expression vector with in vivo excision properties. Nucleic Acids Res. 16:7583-7600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sloma, A., and L. Christianson. April 1999. Nucleic acids encoding a polypeptide having protease activity. U.S. patent 5,891,701.
- 42.Sloma, A., C. F. Rudolph, G. A. Rufo, Jr., B. J. Sullivan, K. A. Theriault, D. Ally, and J. Pero. 1990. Gene encoding a novel extracellular metalloprotease in Bacillus subtilis. J. Bacteriol. 172:1024-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sloma, A., G. A. Rufo, Jr., K. A. Theriault, M. Dwyer, S. W. Wilson, and J. Pero. 1991. Cloning and characterization of the gene for an additional extracellular serine protease of Bacillus subtilis. J. Bacteriol. 173:6889-6895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sonnleitner, E., S. Hagens, F. Rosenau, S. Wilhelm, A. Habel, K. E. Jager, and U. Blasi. 2003. Reduced virulence of a hfq mutant of Pseudomonas aeruginosa O1. Microb. Pathog. 35:217-228. [DOI] [PubMed] [Google Scholar]
- 45.Tran, L., X. C. Wu, and S. L. Wong. 1991. Cloning and expression of a novel protease gene encoding an extracellular neutral protease from Bacillus subtilis. J. Bacteriol. 173:6364-6372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Valbuzzi, A., E. Ferrari, and A. M. Albertini. 1999. A novel member of the subtilisin-like protease family from Bacillus subtilis. Microbiology 145:3121-3127. [DOI] [PubMed] [Google Scholar]
- 47.Walker, K. E., S. Moghaddame-Jafari, C. V. Lockatell, D. Johnson, and R. Belas. 1999. ZapA, the IgA-degrading metalloprotease of Proteus mirabilis, is a virulence factor expressed specifically in swarmer cells. Mol. Microbiol. 32:825-836. [DOI] [PubMed] [Google Scholar]
- 48.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 49.Young, G. M., M. J. Smith, S. A. Minnich, and V. L. Miller. 1999. The Yersinia enterocolitica motility master regulatory operon, flhDC, is required for flagellin production, swimming motility, and swarming motility. J. Bacteriol. 181:2823-2833. [DOI] [PMC free article] [PubMed] [Google Scholar]