Abstract
In this study, we report on the performance of an automated approach to discovery of potential prostate cancer drugs from the biomedical literature. We used the semantic relationships in SemMedDB, a database of structured knowledge extracted from all MEDLINE citations using SemRep, to extract potential relationships using knowledge of cancer drugs pathways. Two cancer drugs pathway schemas were constructed using these relationships extracted from SemMedDB. Through both pathway schemas, we found drugs already used for prostate cancer therapy and drugs not currently listed as the prostate cancer medications. Our study demonstrates that the appropriate linking of relevant structured semantic relationships stored in SemMedDB can support the discovery of potential prostate cancer drugs.
Keywords: prostate cancer, drug discovery, natural language processing, SemRep, SemMedDB, semantic predication, MEDLINE
Introduction
The American Cancer Society estimates that 233,000 out of 855,220 new cases of cancer in the United States will be prostate cancer and that prostate cancer will cause approximately 29,480 deaths, making it the second deadliest cancer for men.1 Treatment options for prostate cancer include surveillance, removal of the prostate and surrounding tissue, radiation therapy, hormonal therapy including removal of the testicles or suppression of testosterone production, stabilization of bone to limit metastases, and chemotherapeutic or immunotherapeutic agents.2 Removal of the prostate often results in significant morbidity, including urinary and sexual dysfunction3 or potentially fecal incontinence.4 Hormonal treatment of prostate cancer, although standard, has been shown to significantly decrease quality of life in the domains of mental and general health and activity and energy.5 Chemotherapy and immunotherapy are generally used for recurrent prostate cancer. A list of drugs used for treatment and palliation of prostate cancer are included in Table 1.
Table 1.
Standard drugs for prostate cancer.
| HORMONAL THERAPY | IMMUNOTHERAPEUTICS | ||
|---|---|---|---|
| Estrogens and Progestins | Antiandrogens | Antiadrenal agents | Prednisone |
| Diethylstilbestrol | Enzalutamide | Ketoconazole | Sipuleucel-T |
| Chlorotrianisene | Buserelin | Aminoglutethimide | CHEMOPREVENTION |
| Ethinyl estradiol | Flutamide | Finasteride | |
| Conjugated estrogens | Bicalutamide | RADIATION THERAPY | Dutasteride |
| Megestrol acetate | Cyproterone acetate | Radium-223 | ANTI-METASTATIC THERAPY |
| LH-RH agonists | Nilutamide | CHEMOTHERAPEUTICS | Bisphosphonates |
| Goserelin | Abiraterone | Docetaxel | Sodium clodronate |
| Leuprolide | LH-RH antagonists | Cabazitaxel | Antiosteoclast agents |
| GR agonists | Degarelix | Paclitaxel | Denosumab |
| Dexamethasone | |||
With the high impact of prostate cancer in the United States and around the world, the continued development of effective therapeutic options is of utmost importance. However, the average cost for bringing a new drug to the market has been estimated to be nearly $1 billion in the US.6 The whole discovery process requires years of development and experimentation, including costly and time-consuming clinical trials. Thus, the development of an efficient and accurate informatics system for drug repurposing, which can leverage the literature without significant manual effort, is needed. We propose to use semantic predications extracted from the literature to expedite drug discovery and potentially to reduce development time and cost.
In this paper, we report on a system built on natural language processing (NLP) that can find potential prostate cancer drugs based on the knowledge contained within the biomedical literature. Specifically, the system extracts all relevant semantic predications from SemMedDB7 (a database of semantic relationships generated by SemRep8) and identifies candidate prostate cancer drugs based on proposed pathway schemas and manual filtering by a physician. Using this approach, our methodology discovers potential prostate cancer drugs that are supported by evidence in the biomedical literature.
Background
This study leverages several publicly available NLP tools that have been developed at the National Library of Medicine (NLM) including Unified Medical Language System (UMLS), SemRep, and SemMedDB.
UMLS
The UMLS provides biomedical domain knowledge for researchers and includes the Metathesaurus, Semantic Network, and SPECIALIST Lexicon.9 The Metathesaurus integrates concepts from over 100 vocabularies, classifications, and coding systems into one structure. The Semantic Network provides a hierarchy of semantic types assigned to Metathesaurus concepts as well as relationships between those semantic types. The SPECIALIST Lexicon10 includes lexical information (such as part-of-speech, morphology, and object structure of verbs) to support NLP systems.
SemRep
SemRep is an NLP application that extracts semantic predications from the biomedical research literature. The system relies on all components of the UMLS. For underspecified syntactic analysis, the SPECIALIST Lexicon provides input to the MedPost part-of-speech tagger11 and subsequent syntactic rules. MetaMap12 is used to map noun phrases in the syntactic structure to Metathesaurus concepts, and indicator rules map syntactic components to relationships in an extended version of the Semantic Network.
Each semantic predication, a subject–PREDICATE–object triple, consists of a semantic relationship from the extended version of the Semantic Network as a predicate and arguments from the Metathesaurus concepts. SemRep predicates cover genetic etiology of disease (eg, ASSOCIATED_WITH, CAUSES), substance interactions (eg, INTERACTS_ WITH, STIMULATES), clinical medicine (eg, TREATS, DIAGNOSES), and pharmacogenomics (eg, AFFECTS, AUGMENTS).13 For example, SemRep interprets the biomedical text in (1) as the semantic predication in (2), identifying the word “linked” as an indicator of the semantic relationship ASSOCIATED_WITH:
-
(1)
Extracellular matrix associated protein CYR61 is linked to prostate cancer development (PMID: 20172544).
-
(2)
CYR61 ASSOCIATED_WITH Malignant neoplasm of prostate (MNP).
SemMedDB
All MEDLINE citations have been processed with SemRep, and extracted predications stored in a database, SemMedDB.7 The version of SemMedDB used for this study is based on citations published as of September 30, 2013. The database maintains links from each predication to its source sentence along with the citation identifier (PMID). It also includes positional information regarding arguments and predicates in a given sentence as well as the distance between an argument and its indicator. We have recently exploited SemMedDB as a structured knowledge resource for discovering drug–drug interactions in clinical data.14
Discovery patterns
In the earlier work,14 we used discovery patterns to identify pairs of drugs that have a shared association with specific genes and biological functions, suggesting that the drugs interact. The patterns we used take the form Drug1→Gene→Drug2 or Drug1→Gene1→ Biological Function←Gene2←Drug2. In this paper, we modified these patterns for the new goal of identifying candidate drugs for prostate cancer. The idea of discovery patterns was first introduced by Hristovski et al.15,16 The authors suggested that specific combinations of semantic predication patterns could provide plausible hypotheses for biomedical phenomena. This idea was applied to drug repurposing for cancer by defining a discovery pattern that links antipsychotic agents to cancer through a common gene.17 Cohen et al. developed a vector space model-based method to automatically detect discovery patterns to predict candidate targets for repurposed drugs using SemRep predications that contain a drug–gene and gene-disorder predication combination.18 An example provided by Cohen and authors includes several intermediate genes linking thalidomide to multiple myeloma.
Related work
Other authors have used a number of techniques to extract cancer-related information from biomedical resources, leveraging both the literature and structured data sources. For example, Chun et al. developed a maximum entropy-based named entity recognizer and a topic-classified relation recognizer to extract information from MEDLINE abstracts on prostate cancer.19 They had biologists annotate a corpus consisting of gene and prostate cancer relations to train the machine learning tools. Epstein used statistical association rules primarily applied to co-occurring words in MEDLINE citations to explore how text mining can be exploited to reduce cost and enhance effectiveness in cancer research. They provide examples in several areas, which include designing therapeutic strategies, clinical trial design, and targeted drug efficacy for different cancer subtypes.20 Deng et al. developed a statistical method to select prostate cancer biomarkers from mass spectrometry and microarray datasets; the authors then used text mining from Online Mendelian Inheritance in Man (OMIM) to validate results.21 Finally, Lu et al. used an order-prediction model to predict cancer drug indications based on chemical–chemical interactions.22
Methods
Our approach (Fig. 1) included four basic components: (1) identifying possible UMLS concepts (with MetaMap) related to prostate cancer, (2) extracting all semantic predications relevant to prostate cancer concepts as well as the genes and drugs that are in a relationship with those concepts from SemMedDB, (3) discovering all possible cancer drugs based on combinations of semantic predications according to pathway schemas, and (4) providing potential unknown prostate cancer drugs after human review and exclusion of known drugs. These components are achieved through a series of steps detailed below.
Figure 1.
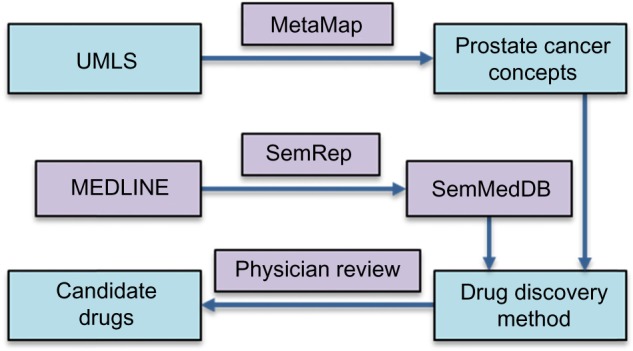
Prostate cancer concepts are found from the UMLS using MetaMap. SemRep extracts semantic predications from the MEDLINE database and stores them in SemMedDB. Predications from SemMedDB are found containing the prostate cancer concepts as objects and genes as subjects and more predications are found that contain drugs as subjects and genes as objects. Additional predications are selected that contain genes as both subject and object. These predications are lined up in either the Drug→Gene→Cancer pathway schema or the Drug→ Gene1→ Gene2→Cancer pathway schema to produce a list of potential drugs and their mechanism of action in treating prostate cancer. A physician selects the best candidates based on the source citations and other relevant knowledge.
Step 1: Prostate cancer concept extraction. We retrieved relevant prostate cancer concepts from UMLS Metathesaurus. Two concepts were found and used for this study: C0376358: prostate cancer (MNP) [neoplastic process] and C0600139: prostate cancer (prostate carcinoma) [neoplastic process]. Note that numbers starting with a “C” are concept unique identifiers in UMLS Metathesaurus, and their corresponding semantic types (eg, neoplastic process) are given in square brackets.
Step 2: Semantic predication extraction from SemMedDB. We extracted three types of predications from SemMedDB: gene–cancer (ie, predications with a gene as the subject and a cancer concept as the object), gene–gene, and drug–gene. We first find all predications describing an influence between a gene and one of the prostate cancer UMLS concepts (Step 1). Specifically, predications having a gene as the subject, one of the prostate cancer concepts as the object, and one of the six restricted predicate types – AFFECTS, ASSOCIATED_WITH, AUGMENTS, CAUSES, DISRUPTS, and PREDISPOSES – were extracted as gene–cancer predications. Additionally, drug–gene predications were extracted by finding those that contained a drug as the subject and a gene as the object with any of the following predicates: INHIBITS, STIMULATES, or INTERACTS_WITH. We also extracted gene–gene predications. These were required to have a gene as both the subject and object and STIMULATES, INHIBITS, or INTERACTS_WITH as the predicate.
Step 3: Prostate cancer discovery pathways (Fig. 2)
Figure 2.
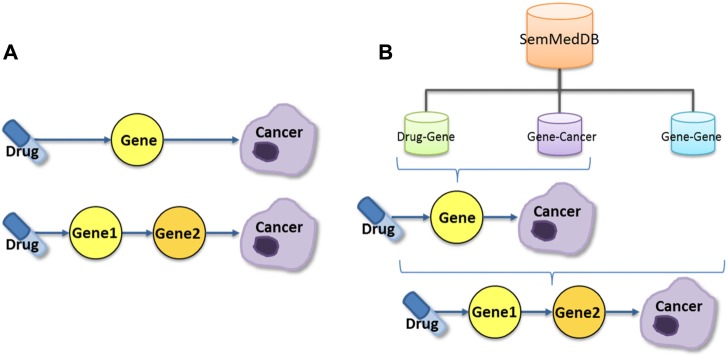
(A) Two pathway schemas are utilized. The first connects a drug–gene predication with a gene–cancer predication and the second connects a drug–gene predication to a gene–gene predication and then the object gene of the gene–gene predication to a gene–cancer predication. (B) Drug–gene, gene–cancer, and gene–gene predications are all retrieved from SemMedDB. While all three types are used for the Drug→Gene1→Gene2→Cancer pathway, only the drug–gene and gene–cancer predications are used for the Drug→Gene→Cancer pathway.
Drug→Gene→Cancer (DGC) pathway. We identified the potential drugs using the drug–gene and gene–cancer predications previously extracted in Step 2. Potential prostate cancer drug candidates were generated by matching the object gene in a drug–gene predication with the subject gene in a gene–cancer predication. For example, combining dexamethasone INHIBITS EGR1 with EGR1 PREDISPOSES MNP produces the pathway dexamethasone→EGR1→MNP. Note that an inhibitory effect on a gene that promotes cancer suggests the possibility of treating cancer as does a stimulatory effect on a gene that suppresses cancer.
Drug→Gene1→Gene2→Cancer (DGGC) pathway. We also identified the potential drugs by adding the gene–gene predications as an extension to the DGC pathway. Potential drug candidates were generated when the following two matches were satisfied: (1) The object gene in drug–gene predication is the same as the subject gene in gene–gene predication; (2) the object gene in gene–gene predication and the subject gene in gene–cancer predication are the same. As an example, three predications (quercetin INHIBITS FAS, FAS STIMULATES NFKB1, and NFKB1 ASSOCIATED_WITH MNP) can be combined to form the pathway quercetin→FAS→NFKB1→MNP.
Step 4: Physician selection of semantic predications. We first retrieved the MEDLINE sentences that produced drug candidates based on DGC and DGGC pathways from SemMedDB. One author (MJC, a physician) then selected the most promising candidates from the semantic predications matching each of the pathways. The selection considered the logical implications of the combination of predications. For instance, if the gene in a DGC pathway contributed to prostate cancer, the drug would need to reduce the abundance or activity of the gene. For the non-specific predicates INTERACTS_WITH and ASSOCIATED_WITH, the actual nature of the interaction or association needed to be ascertained from the abstract or full text article. Consideration was also given to the validity of the component predications relative to their source sentence.
Results
Drugs discovered through DGC pathway schema
Step 2 of our method resulted in 6511 predications containing 853 drug terms, 1107 gene terms, and 2 cancer terms. The break down for each type of predication is given in Table 2.
Table 2.
Counts of predications and unique subjects, predicates, and objects for each type of predication.
| PREDICATIONS | UNIQUE SUBJECTS | UNIQUE PREDICATES | UNIQUE OBJECTS | |
|---|---|---|---|---|
| Drug–gene | 2255 | 853 | 3 | 88 |
| Gene–gene | 2621 | 775 | 3 | 117 |
| Gene–cancer | 1635 | 513 | 7 | 2 |
Using the DGC pathway schema (Step 3i), we found 18 potential prostate cancer drugs and 3 drugs with some established usage (Table 3). For a gene that promotes growth or impact of cancer, the example drug is inhibitory; whereas for a gene that decreases cancer progression, the drug is stimulatory. Note that ASSOCIATED_WITH can either indicate a promoting or decreasing effect and requires exploration of the source text. For example, FAS is pro-apoptotic, and so in this case the association with prostate cancer is a decreasing effect that suggests therapeutic potential. Many drugs share the same pathway, for example, No. 1–4, No. 5–6, No. 7–12, No. 13–16, and No. 17–19 (Table 3). In the first example, simvastatin inhibits the gene CYR61, which has been associated with prostate cancer (MNP). With further inspection, the specific association is that CYR61 expression is increased in prostate cancer. This chain indicates simvastatin may have potential to inhibit MNP to some degree.
Table 3.
Resulting drug candidates through DGC pathway.
| NO. | DRUG | → | GENE | → | CANCER | ESTABLISHED USE |
|---|---|---|---|---|---|---|
| 1 | Simvastatin | INH | CYR61 | ASC | MNP | No |
| 2 | Tamoxifen | INH | CYR61 | ASC | MNP | No |
| 3 | Lovastatin | INH | CYR61 | ASC | MNP | No |
| 4 | Pimagedine | INH | CYR61 | ASC | MNP | No |
| 5 | Dexamethasone | INH | EGR1 | PRE | MNP | Yes |
| 6 | Sch-23390 | INH | EGR1 | PRE | MNP | No |
| 7 | Adriamycin | STI | FAS | ASC | MNP | No |
| 8 | Carboplatin | STI | FAS | ASC | MNP | No |
| 9 | Carvedilol | STI | FAS | ASC | MNP | No |
| 10 | Curcumin | STI | FAS | ASC | MNP | No |
| 11 | Hydralazine | STI | FAS | ASC | MNP | No |
| 12 | Paclitaxel | STI | FAS | ASC | MNP | Yes |
| 13 | Hydroxamate | INH | HDAC9 | ASC | MNP | No |
| 14 | Bortezomib | INH | HDAC9 | ASC | MNP | No |
| 15 | Bryostatin 1 | INH | HDAC9 | ASC | MNP | No |
| 16 | Valproic acid | INH | HDAC9 | ASC | MNP | No |
| 17 | Catechin | INH | HSPA5 | ASC | MNP | No |
| 18 | Metformin | INH | HSPA5 | ASC | MNP | No |
| 19 | Nicotinic Acid | INH | HSPA5 | ASC | MNP | No |
| 20 | 5,6-dimethylxanthenoneacetic acid | STI | NFKB1 | ASC | MNP | No |
| 21 | Ketoconazole | INH | CYP3A5 | ASC | MNP | Yes |
Abbreviations: ASC, ASSOCIATED_WITH; INH, INHIBITS; PRE, PREDISPOSES; STI, STIMULATES; MNP, Malignant neoplasm of prostate.
Drugs discovered through Drug→Gene1→Gene2→Cancer (DGGC) pathway schema
Applying the DGGC pathway schema (Step 3ii) to our predication set and the subsequent physician selection of semantic predications (Step 4) yielded two unknown drug candidates (Sch-23390 and quercetin) and the known prostate cancer drug dexamethasone (Table 4). In the pathway to cancer for the compound quercetin (Table 4, No. 3), FAS stimulates NFkappaB, which is further described in the source (PMID: 15289496) as an inflammatory response instead of a proapoptotic signal, and activation of NFkappaB is then associated with prostate cancer progression. Therefore, inhibition of FAS by quercetin might reduce prostate cancer progression.
Table 4.
Resulting drug candidates discovered through DGGC pathway.
| NO. | DRUG | → | GENE1 | → | GENE2 | → | CANCER |
|---|---|---|---|---|---|---|---|
| 1 | Dexamethasone | INH | EGR1 | STI | SIRT1 | ASC | MNP |
| 2 | Sch-23390 | INH | EGR1 | STI | SIRT1 | ASC | MNP |
| 3 | Quercetin | INH | FAS | STI | NFKB1 | ASC | MNP |
Abbreviations: STI, STIMULATES; INH, INHIBITS; ASC, ASSOCIATED_WITH; MNP, Malignant neoplasm of prostate.
Literature evidence for cancer drugs generated from DGC and DGGC pathway schemas
Some example predications and their source sentences from those that resulted in selected pathways are listed in Table 5. The source of the sentences, including PMID and title/abstract are also extracted. The underlined words in sentences are related to subjects and objects in the predications. Bold and italic words in the sentences indicate the relationships (predicates) between two biomedical concepts. Predicates (eg, STIMULATES) in the semantic predications can be generated from verbs (eg, induce, promote) or nouns (eg, induction, upregulation, stimulation). All biomedical concepts were mapped to UMLS concepts. For example, NFkappaB was mapped to the gene NFKB1 (Table 4, No. 1), zif268 mapped to EGR1 (Table 5, No. 4).
Table 5.
Sentence citations for selected drug–gene, gene–gene, and gene–cancer semantic predications.
| NO. | SEMANTIC PREDICATIONS | SENTENCE (PMID, TITLE/ABSTRACT) | |
|---|---|---|---|
| Drug → Gene predications | |||
| 1 | 5,6-dimethylxanthenoneacetic acid STIMULATES NFKB1 |
Induction of STAT and NFKappaB activation by the antitumor agents 5,6-dimethylxanthenone-4-acetic acid and flavone acetic acid in a murine macrophage cell line. (10484075, title) | |
| 2 | Adriamycin STIMULATES FAS |
DR5, FAS, Bax, Bad, Puma and Bnip3L were induced by 5-FU and adriamycin (ADR) in HCT116 cells in a p53-dependent manner. (21709442, abstract) | |
| 3 | Simvastatin INHIBITS CYR61 |
Simvastatin inhibits cytokine-stimulated Cyr61 expression in osteoblastic cells: a therapeutic benefit for arthritis. (20191585, title) | |
| 4 | Catechin INHIBITS HSPA5 |
Our results show that catechin reduces the expression level of GRP78/BiP, leads to cell proliferation rates of GD cells similar levels to normal cells, and improves wound healing activity. (21884680, abstract) | |
| 5 | Carboplatin STIMULATES FAS |
Carboplatin induces Fas (APO-1/CD95)-dependent apoptosis of human tongue carcinoma cells: sensitization for apoptosis by upregulation of FADD expression. (12740905, title) | |
| 6 | Curcumin STIMULATES FAS |
Curcumin also promoted the levels of Fas and FADD, Bax, cytochrome c release, but decreased the levels of Bcl-2 causing changes of DeltaPsim. (19513510, abstract) | |
| 7 | Dexamethasone INHIBITS EGR1 |
Inhibition of EGR-1 and NF-kappa B gene expression by dexamethasone during phorbol ester-induced human monocytic differentiation. (1417981, title) | |
| 8 | Carvedilol STIMULATES EGR1 |
Immunocytochemical analysis of rabbit hearts demonstrated an upregulation of Fas protein in ischemic cardiomyocytes, and treatment with carvedilol reduced both the intensity of staining as well as the area stained. (9468187, abstract) | |
| 9 | Hydralazine STIMULATES FAS |
VPA did not increase the expression of Fas on the surface of osteosarcoma cells, while hydralazine did, and the combination of VPA with hydralazine increased the expression of cell-surface Fas. (22576685, abstract) | |
| 10 | Lovastatin INHIBITS CYR61 |
Lovastatin also completely inhibited arecoline-induced Cyr61 synthesis and the inhibition is dose-dependent. (21317023, abstract) | |
| 11 | Metformin INHIBITS HSPA5 |
Metformin reduced the GRP78 mRNA expression in HM rats. (22445233, abstract) | |
| 12 | Nicotinic Acid INHIBITS HSPA5 |
NA and NAM also decreased constitutive levels of both activated NF-kappaB and GRP78, two proteins that respond to oxidative stress. (10745276, abstract) (Note: NA is the abbreviation of nicotinic acid) | |
| 13 | Paclitaxel STIMULATES FAS |
Therefore, paclitaxel enhances the thermochemotherapy of the osteosarcoma cell line and this is primarily accomplished by the upregulation of FAS expression and the induction of apoptosis. (22948360, abstract) | |
| 14 | Pimagedine INHIBITS CYR61 |
Treatment with aminoguanidine inhibited Cyr61 and Ctgf expression in diabetic rats, with reductions of 31 and 36%, respectively, compared with untreated animals. (17333105, abstract) | |
| 15 | Quercetin INHIBITS FAS |
FAS gene expression was significantly inhibited by quercetin but not enalapril, losartan, or curcumin compared with the control. (10925121, abstract) | |
| 16 | Sch-23390 INHIBITS EGR1 |
The dopamine D1 receptor antagonist SCH-23390 decreases the mRNA levels of the transcription factor zif268 (krox-24) in adult rat intact striatum–an in situ hybridization study. (1491805, title) | |
| 17 | Tamoxifen INHIBITS CYR61 |
Induction of Cyr61 mRNA was blocked by tamoxifen and ICI182,780, inhibitors of the estrogen receptor. (11297518, abstract) | |
| 18 | Ketoconazole INHIBITS CYP3A5 |
we demonstrated a modulatory role of cytochrome b(5) mostly for the metabolism of domperidone and confrmed selective inhibition of CYP3A4 over CYP3A5 by Ketoconazole. (21281268, abstract) | |
| Gene1 → Gene2 predications | |||
| 19 | EGR1 STIMULATES SIRT1 |
An autoregulatory loop reverts the mechanosensitive Sirt1 induction by EGR1 in skeletal muscle cells. (22820707, title) | |
| 20 | FAS STIMULATES NFKB1 |
NFkappaB activation by Fas is mediated through FADD, caspase-8, and RIP and is inhibited by FLIP. (15289496, title) | |
| Gene → Prostate Cancer predications (MNP: Malignant neoplasm of prostate) | |||
| 21 | EGR1 PREDISPOSES MNP |
These results suggest that Egr-1 may promote prostate cancer development by modulating the activity of factors NF-kappaB and AP-1, which are involved in cell proliferation and apoptosis. (21743958, abstract) | |
| 22 | HSPA5 ASSOCIATED_WITH MNP |
GRP78 regulates clusterin stability, retrotranslocation and mitochondrial localization under ER stress in prostate cancer. (22689054, title) | |
| 23 | FAS ASSOCIATED_WITH MNP |
The decreased expression of Fas in a large fraction of prostate cancers compared with their normal cells suggested that loss of Fas expression might play a role in tumorigenesis in some prostate cancers possibly by inhibiting apoptosis mediated by Fas. (19161534, abstract) | |
| 24 | SIRT1 ASSOCIATED_WITH MNP |
Overexpressed SIRT1 in advanced prostate cancer may play an important role in prostate cancer progression. (23038275, abstract) | |
| 25 | CYR61 ASSOCIATED_WITH MNP |
Extracellular matrix associated protein CYR61 is linked to prostate cancer development. (20172544, title) | |
| 26 | NFKB1 ASSOCIATED_WITH MNP |
BACKGROUND: Cell line models suggest that activation of NFkappaB is associated with progression of prostate cancer. (23093296, abstract) | |
Discussion
Our method of identifying cancer drugs from the biomedical literature is novel since it makes use of knowledge from the entire MEDLINE database (via semantic predications extracted by SemRep). Moreover, we design the two different pathway schemas to allow for linking knowledge from different citations and potentially even different fields of biomedical science. This preliminary work is not intended to provide an exhaustive list of candidate prostate cancer drugs, but it provides a significant starting point for future exploration.
Clinical implications
Both of our pathway schemas provided both drugs already used for prostate cancer therapy and drugs not currently associated with its treatment. One of the known drugs, dexamethasone, is part of standard combined therapy for certain prostate cancer patients, whereas ketoconazole and paclitaxel are less common in standard protocols but exist in studies of experimental treatment. In general, the drugs not currently used are obvious candidates because they are standard or experimental treatments for other cancers, for instance simvastatin has been investigated for pancreatic cancer,23 leukemia,24 and lung cancer.25 Tamoxifen is a somewhat unexpected candidate since it is an estrogen receptor antagonist, but it has been suggested in the literature that it may inhibit prostate cell proliferation.26 Adriamycin is included in the resulting therapeutic candidates and has already been investigated for use in prostate cancer, although clinical trials results have been controversial suggesting its activity is limited.27
Advantages of SemMedDB predications in finding unknown cancer drugs
Our methodology uses semantic predications extracted from all of MEDLINE. In addition to providing broad access to biomedical knowledge in the literature, SemRep predications identify the nature of the relationships between entities, going beyond techniques that use concept co-occurrence. The semantic predications are not only machine readable and computable, but they are also human readable and intuitive. In our method, we are able to take advantage of this by specifying predicates and semantic types of subjects and objects. This is an essential component to the construction of our pathway schemas that significantly facilitates the automatic generation of meaningful candidate pathways.
Drug discovery guidance
Our method facilitates the search for new prostate cancer drugs by focusing on likely candidates that already have supporting evidence in the literature and provide not only a candidate list but a specific mechanism of action. This facilitates preclinical investigation necessary before clinical trials may be considered. This method has the potential to find candidates that may not have been considered since the semantic predications are derived from any of the journals included in MEDLINE, which are not limited to cancer research but come from a wide range of biomedical research fields.
Evaluation of semantic predications
SemRep output has been evaluated several times for recall and precision. Recall has been evaluated to approximate 0.60.17,28 In previous work identifying drug–drug interactions using semantic predications,14 we undertook a formal linguistic evaluation for three predication types: gene–drug, drug–gene, and gene– function. The overall precision was 0.60 and varied slightly for each type (0.61 for drug–gene, 0.65 for gene–drug, and 0.54 for gene–function).
Identification of known prostate cancer targets
Our results are limited in several ways. One is due to a physician having manually reviewed a relatively small, randomized subset of candidates. Through this process, we were able to identify drug–gene and gene–cancer pairs (eg, tanshinone II A INHIBITS AR, AR ASSOCIATED_WITH MNP) by looking for specific known targets (prostate cancer-specific androgen receptor and androgen synthesis pathways).
However, many complete pairs still did not appear in our filtered set; typically, only the drug–gene predication occurred (or less commonly we found only the gene–cancer relationship). There are two major reasons for these missed relationships, both due to decisions made when post-processing the extracted predications.
SemRep is not always able to resolve ambiguous gene/protein names, for example, Steroid 17-alpha-monooxygenase versus CYP17A1. In such cases, both concepts are included in the predication in the database. For this study, we eliminated these predications from further processing. Since this step significantly reduced the size of our results, disambiguation of such cases needs to be pursued in future work.
Another post-processing step that reduced results was keeping only specific drugs and genes, while removing relationships in which one of the arguments was a class of drugs (eg, anthracyclines or estrogen antagonists) or proteins (eg, HSP90 heat-shock proteins). Results containing drug classes would likely be nearly as useful as specific compounds. On the other hand, including specific drug–gene and gene–cancer relationships along with gene families would increase recall and provide more candidates but would also significantly increase noise and decrease precision.
Limitations and future work
One limitation to this work is that we depend on previous evaluations of SemRep predications and these evaluations did not include all of our predication types, specifically gene–gene or gene–cancer predications. Although these types are similar to those included in evaluations and relatively consistent within other similar types, an evaluation on these specific predication types may provide additional validation of our methodology.
Our Step 4, physician selection, limits the number of potential pathways analyzed because, instead of equal consideration of each and every predication, selection is somewhat limited to a human-readable amount of component predications and subject to individual bias. Machine learning or similar predictive techniques may be able to simulate selection process given prior selections as training data. This in turn may increase the amount of candidates that may be considered computationally and reduce the amount that needs to be considered by humans as a last step.
An essential part of this physician selection was distinguishing whether the cancer genes within the predications were likely to have a “driver” or “passenger” role. This need arose in part from the underspecified nature of SemRep predications, especially in the case of the predicate ASSOCIATED_WITH. Because this relationship can either indicate a promoting or decreasing effect, further clarification was gathered from the source text.
One concern that may be significant in our approach is that the compounds extracted by SemRep are from the 2006 version of the UMLS to avoid increased ambiguity in the 2012 version, and so we are not able to consider potential drugs that were added to the newer version. Even the 2012 version may leave out a considerable amount of potential drugs and using another source for chemical compounds might increase the number of drug–gene assertions extracted.
Just as this approach is an extension of our previous discovery of potential drug–drug interactions, it too can be easily extended to consider other cancers as well as different diseases, conditions, and syndromes. In addition, more levels of gene–gene interactions can be added, extending the schemas to Drug→Gene1→Gene2→ Gene3→Cancer, Drug→Gene1→Gene2 →Gene3→Gene4→Cancer, etc. The gene position could also be substituted with an established biochemical pathway using predications that assert that a gene interacts with a given pathway and that pathway is associated with cancer. This would allow a broadening of the search and produce a greater number of candidate drugs.
Conclusion
We present a method to identify potential prostate cancer drugs that takes advantage of the wealth of biomedical literature knowledge contained in the MEDLINE database. In our study, we identified 18 potential prostate cancer drugs that have not previously been used for prostate cancer. Our methodology was also able to identify three substances that have already been used in prostate cancer treatment.
Figure 3.
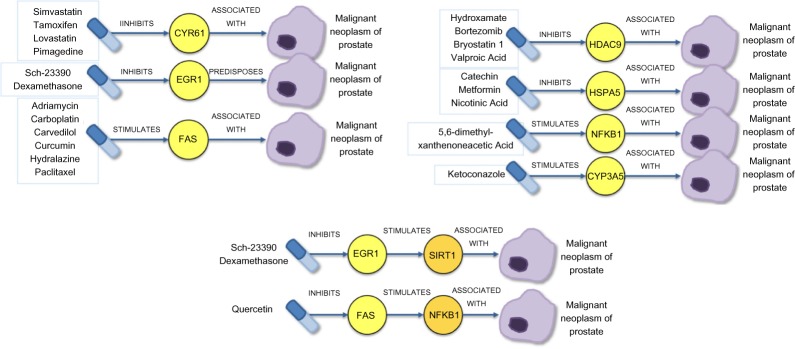
The resulting drug candidates and their mechanism of action in treating prostate cancer are represented schematically.
Footnotes
Author Contributions
Conceived the concepts: RZ, MJC. Analyzed the data: RZ, MJC. Wrote the first draft of the manuscript: RZ, MJC. Contributed to the writing of the manuscript: RZ, MJC, MF, HK, TCR, SP, GBM. Agree with manuscript results and conclusions: RZ, MJC, MF, HK, TCR, SP, GBM. Jointly developed the structure and arguments for the paper: RZ, MJC, MF, HK, TCR, SP, GBM. Made critical revisions and approved final version: RZ, MJC, MF, HK, TCR, SP, GBM. All authors reviewed and approved of the final manuscript.
ACADEMIC EDITOR: JT Efird, Editor in Chief
FUNDING: This research was supported in part by the Intramural Research Program of the NIH, National Library of Medicine. This research was supported by the Agency for Healthcare Research & Quality grant (#1R01HS022085–01) (GMM) and University of Minnesota Clinical and Translational Science Award (#8UL1TR000114–02) (Blazer). The authors confirm that the funder had no influence over the study design, content of the article, or selection of this journal.
COMPETING INTERESTS: Authors disclose no potential conflicts of interest.
This paper was subject to independent, expert peer review by a minimum of two blind peer reviewers. All editorial decisions were made by the independent academic editor. All authors have provided signed confirmation of their compliance with ethical and legal obligations including (but not limited to) use of any copyrighted material, compliance with ICMJE authorship and competing interests disclosure guidelines and, where applicable, compliance with legal and ethical guidelines on human and animal research participants. Provenance: the authors were invited to submit this paper.
REFERENCES
- 1.American Cancer Society 2014. Available at http://www.cancer.org/research/cancer-factsstatistics/cancerfactsfigures.
- 2.Prostate Cancer Treatment. 2014. Available at http://www.cancer.gov/cancertopics/treatment/prostate.
- 3.Potosky AL, Davis WW, Hoffman RM, et al. Five-year outcomes after prostatectomy or radiotherapy for prostate cancer: the prostate cancer outcomes study. J Natl Cancer Inst. 2004;96(18):1358–67. doi: 10.1093/jnci/djh259. [DOI] [PubMed] [Google Scholar]
- 4.Bishoff JT, Motley G, Optenberg SA, et al. Incidence of fecal and urinary incontinence following radical perineal and retropubic prostatectomy in a national population. J Urol. 1998;160(2):454–8. [PubMed] [Google Scholar]
- 5.Fowler FJ, Jr, McNaughton Collins M, Walker Corkery E, Elliott DB, Barry MJ. The impact of androgen deprivation on quality of life after radical prostatectomy for prostate carcinoma. Cancer. 2002;95(2):287–95. doi: 10.1002/cncr.10656. [DOI] [PubMed] [Google Scholar]
- 6.Vernon JA, Golec JH, Dimasi JA. Drug development costs when financial risk is measured using the Fama-French three-factor model. Health Econ. 2010;19(8):1002–5. doi: 10.1002/hec.1538. [DOI] [PubMed] [Google Scholar]
- 7.Kilicoglu H, Shin D, Fiszman M, Rosemblat G, Rindflesch TC. SemMedDB: a PubMed-scale repository of biomedical semantic predications. Bioinformatics. 2012;28(23):3158–60. doi: 10.1093/bioinformatics/bts591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rindflesch TC, Fiszman M. The interaction of domain knowledge and linguistic structure in natural language processing: interpreting hypernymic propositions in biomedical text. J Biomed Inform. 2003;36(6):462–77. doi: 10.1016/j.jbi.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 9.Bodenreider O. The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic Acids Res. 2004;32(Database issue):D267–70. doi: 10.1093/nar/gkh061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McCray AT, Srinivasan S, Browne AC. Lexical methods for managing variation in biomedical terminologies. Proc Annu Symp Comput Appl Med Care. 1994:235–9. [PMC free article] [PubMed] [Google Scholar]
- 11.Smith L, Rindflesch T, Wilbur WJ. MedPost: a part-of-speech tagger for bio-Medical text. Bioinformatics. 2004;20(14):2320–1. doi: 10.1093/bioinformatics/bth227. [DOI] [PubMed] [Google Scholar]
- 12.Aronson AR. Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program. AMIA Annu Symp Proc. 2001:17–21. [PMC free article] [PubMed] [Google Scholar]
- 13.Ahlers CB, Fiszman M, Demner-Fushman D, Lang FM, Rindflesch TC. Extracting semantic predications from Medline citations for pharmacogenomics. Pac Symp Biocomput. 2007:209–20. [PubMed] [Google Scholar]
- 14.Zhang R, Cairelli MJ, Fiszman M, et al. Using semantic predications to uncover drug-drug interactions in clinical data. J Biomed Inform. 2014;49:134–47. doi: 10.1016/j.jbi.2014.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hristovski D, Friedman C, Rindflesch TC, Peterlin B. Exploiting semantic relations for literature-based discovery. AMIA Annu Symp Proc. 2006:349–53. [PMC free article] [PubMed] [Google Scholar]
- 16.Hristovski D, Friedman C, Rindflesch TC, Peterlin B. Literature-based discovery using natural language processing. In: Bruza P, Weeber M, editors. Literature-based discovery. Berlin: Springer-Verlag; pp. 153–72. [Google Scholar]
- 17.Ahlers CB, Hristovski D, Kilicoglu H, Rindflesch TC. Using the literature-based discovery paradigm to investigate drug mechanisms. AMIA Annu Symp Proc. 2007;2007:6–10. [PMC free article] [PubMed] [Google Scholar]
- 18.Cohen T, Widdows D, Schvaneveldt RW, Davies P, Rindflesch TC. Discovering discovery patterns with predication-based semantic indexing. J Biomed Inform. 2012;45(6):1049–65. doi: 10.1016/j.jbi.2012.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chun HW, Tsuruoka Y, Kim JD, et al. Automatic recognition of topic-classified relations between prostate cancer and genes using MEDLINE abstracts. BMC Bioinformatics. 2006;7(suppl 3):S4. doi: 10.1186/1471-2105-7-S3-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Epstein RJ. Unblocking blockbusters: using Boolean text-mining to optimise clinical trial design and timeline for novel anticancer drugs. Cancer Inform. 2009;7:231–8. doi: 10.4137/cin.s2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deng X, Geng H, Bastola DR, Ali HH. Link test–a statistical method for finding prostate cancer biomarkers. Comput Biol Chem. 2006;30(6):425–33. doi: 10.1016/j.compbiolchem.2006.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu J, Huang G, Li HP, et al. Prediction of cancer drugs by chemical-chemical interactions. PLoS One. 2014;9(2):e87791. doi: 10.1371/journal.pone.0087791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hong JY, Nam EM, Lee J, et al. Randomized double-blinded, placebo-controlled phase II trial of simvastatin and gemcitabine in advanced pancreatic cancer patients. Cancer Chemother Pharmacol. 2014;73(1):125–30. doi: 10.1007/s00280-013-2328-1. [DOI] [PubMed] [Google Scholar]
- 24.Ahmed TA, Hayslip J, Leggas M. Pharmacokinetics of high-dose simvastatin in refractory and relapsed chronic lymphocytic leukemia patients. Cancer Chemother Pharmacol. 2013;72(6):1369–74. doi: 10.1007/s00280-013-2326-3. [DOI] [PubMed] [Google Scholar]
- 25.Han JY, Lim KY, Yu SY, Yun T, Kim HT, Lee JS. A phase 2 study of irinotecan, cisplatin, and simvastatin for untreated extensive-disease small cell lung cancer. Cancer. 2011;117(10):2178–85. doi: 10.1002/cncr.25790. [DOI] [PubMed] [Google Scholar]
- 26.Lissoni P, Vigano P, Vaghi M, et al. A phase II study of tamoxifen in hormone-resistant metastatic prostate cancer: possible relation with prolactin secretion. Anticancer Res. 2005;25(5):3597–9. [PubMed] [Google Scholar]
- 27.Petrioli R, Fiaschi AI, Francini E, Pascucci A, Francini G. The role of doxorubicin and epirubicin in the treatment of patients with metastatic hormone-refractory prostate cancer. Cancer Treat Rev. 2008;34(8):710–8. doi: 10.1016/j.ctrv.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 28.Kilicoglu H, Fiszman M, Rosemblat G, Marimpietri S, Rindflesch TC. Arguments of nominals in semantic interpretation of biomedical text. Proc BioNLP Workshop. 2010:46–54. [Google Scholar]


