Abstract
The PmrA/PmrB regulatory system of Salmonella enterica controls the modification of lipid A with aminoarabinose and phosphoethanolamine. The aminoarabinose modification is required for resistance to the antibiotic polymyxin B, as mutations of the PmrA-activated pbg operon or ugd gene result in strains that lack aminoarabinose in their lipid A molecules and are more susceptible to polymyxin B. Additional PmrA-regulated genes appear to participate in polymyxin B resistance, as pbgP and ugd mutants are not as sensitive to polymyxin B as a pmrA mutant. Moreover, the role that the phosphoethanolamine modification of lipid A plays in the resistance to polymyxin B has remained unknown. Here we address both of these questions by establishing that the PmrA-activated pmrC gene encodes an inner membrane protein that is required for the incorporation of phosphoethanolamine into lipid A and for polymyxin B resistance. The PmrC protein consists of an N-terminal region with five transmembrane domains followed by a large periplasmic region harboring the putative enzymatic domain. A pbgP pmrC double mutant resembled a pmrA mutant both in its lipid A profile and in its susceptibility to polymyxin B, indicating that the PmrA-dependent modification of lipid A with aminoarabinose and phosphoethanolamine is responsible for PmrA-regulated polymyxin B resistance.
Polymyxin B is a cyclic antimicrobial lipopeptide produced by the soil bacterium Paenibacillus polymyxa (33). While the mechanism of killing of polymyxin B is not completely understood, the cationic polymyxin B is believed to bind initially to the anionic surfaces of gram-negative bacteria, in particular to the lipopolysaccharide (LPS) (46). This electrostatic interaction apparently allows polymyxin B to gain access to the bacterial inner membrane, which is its presumed target. Gram-negative bacteria that are resistant to polymyxin B possess mechanisms that modify the LPS by neutralizing its negative charge, which decreases the binding of polymyxin B (30, 37, 45). Strains that exhibit resistance to polymyxin B also display resistance to antimicrobial peptides and proteins from human neutrophils (36).
In Salmonella enterica serovar Typhimurium, polymyxin B resistance is controlled primarily by the PmrA/PmrB regulatory system (35, 44). A polymyxin B-resistant strain that expresses a constitutively active PmrA protein displays increased levels of aminoarabinose and phosphoethanolamine in the lipid A portion of the LPS (20), suggesting that these PmrA-controlled modifications are required for polymyxin B resistance. Consistent with this notion, the PmrA-activated ugd gene and pbg operon (designated pmrF by Gunn et al. [13] and arn by Trent et al. [43]) are necessary for both the biosynthesis and incorporation of aminoarabinose into lipid A (13) and for polymyxin B resistance (12, 13). Yet, pbgP and ugd mutants are not as polymyxin sensitive as a pmrA null mutant (24), indicating that an additional PmrA-regulated gene(s) is required for polymyxin B resistance. pmrA null mutants produce lipid A species that lack aminoarabinose and phosphoethanolamine, whereas strains with a block in the synthesis pathway for aminoarabinose due to mutations in the pbgP operon have increased levels of phosphoethanolamine-modified lipid A (52). While this indicates that the PmrA/PmrB system is absolutely needed for decorating lipid A with aminoarabinose and phosphoethanolamine, the PmrA-regulated determinant(s) responsible for the modification of lipid A with phosphoethanolamine and the role that such a modification plays in polymyxin resistance have remained unknown.
Transcription of PmrA-activated genes is promoted by Fe3+, which is sensed by the sensor protein PmrB (48), and by low levels of Mg2+ in a process that requires the PhoP/PhoQ regulatory system (41) and the PhoP-activated PmrD protein (24). In addition to the increased susceptibility towards polymyxin B (12), pmrA null mutants are hypersusceptible to killing by Fe3+ (2) and mildly attenuated for virulence in mice (15). The PmrA/PmrB system is encoded by the pmrCAB operon and is apparently expressed from both a PmrA-activated promoter upstream of the pmrC gene (47) and a constitutive promoter within the pmrC coding region (14, 41).
In this paper, we demonstrate that the PmrA-activated pmrC gene encodes an inner membrane protein that is required for polymyxin resistance and for the incorporation of phosphoethanolamine into lipid A. We determined that the inactivation of both the pbgP and pmrC genes results in a strain that resembles a pmrA mutant both in its susceptibility to polymyxin B and in its lipid A profile. Our results indicate that the PmrA-regulated incorporation of aminoarabinose and phosphoethanolamine into lipid A is responsible for PmrA-mediated polymyxin B resistance in S. enterica.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used for this study are listed in Table 1. All S. enterica serovar Typhimurium strains used for this study were derived from the wild-type strain 14028s. Phage P22-mediated transductions were performed as described previously (7). Bacteria were grown at 37°C with aeration in Luria-Bertani (LB) broth or in N minimal medium, pH 7.7 or 5.8, supplemented with 0.1% Casamino Acids, 38 mM glycerol, and 10 μM or 10 mM MgCl2 (39). When necessary, antibiotics were added to the following final concentrations: ampicillin, 50 μg/ml; chloramphenicol, 20 μg/ml; kanamycin, 50 μg/ml; and tetracycline, 10 μg/ml. Escherichia coli DH5α (18) was used as a host for the preparation of plasmid DNA.
TABLE 1.
Bacterial strains and plasmids used for this study
| Strain or plasmid | Descriptiona | Reference or source |
|---|---|---|
| Strains | ||
| S. enterica | ||
| 14028s | Wild type | 9 |
| EG13927 | ΔpmrC1::Cmr, partial deletion (1,096 bp of the 1,641-bp pmrC gene) | This study |
| EG14590 | ΔpmrC1.1, Cmr removed from strain EG13927 | This study |
| EG13633 | ΔpmrC2::Cmr, complete deletion of the pmrC open reading frame | This study |
| EG9241 | pbgP1::MudJ | 40 |
| EG14372 | pbgP1::MudJ ΔpmrC1 | This study |
| EG14375 | pbgP1::MudJ ΔpmrC2 | This study |
| EG9492 | pmrA505 zjd::Tn10d-Cmr | 12 |
| EG9868 | pmrA505 pbgP1::MudJ zjd::Tn10d-Cmr | 12 |
| EG14367 | pmrA505 ΔpmrC::Kmrzjd::Tn10d-Cmr | This study |
| EG14368 | pmrA505 ΔpmrC1 zjd::Tn10d-Cmr | This study |
| EG14369 | pmrA505 pbgP1::MudJ ΔpmrC1 zjd::Tn10d-Cmr | |
| EG7139 | pmrA::Cmr | This study |
| EG14286 | phoN::Cmr | 41 |
| EG14656 | EG14590 containing pBAC108L plasmid | This study |
| EG14595 | EG14590 containing ppmrC plasmid | This study |
| EG14592 | EG14590 containing ppmrCFLAG plasmid | This study |
| E. coli | ||
| DH5α | supE44 ΔlacU169 (φ80 lacZΔM15) hsdR17recA1 endA1 gyrA96 thi-1 relA1 | (18) |
| Plasmids | ||
| pBAC108L | Mini-F, Cmr | 38 |
| ppmrC | pBAC108L harboring pmrC coding region with its own promoter | This study |
| ppmrCFLAG | pBAC108L harboring the pmrC coding region with its own promoter and harboring sequences encoding a FLAG epitope sequence at the 3′ end | This study |
| pKD3 | repR6K γ FRT Cmr FRT Apr | 6 |
| pKD4 | repR6K γ FRT Kmr FRT Apr | 6 |
| pKD46 | reppSC101ts paraBAD γ β Exo Apr | 6 |
| pCP20 | reppSC101ts Cmr cI857 λPR Apr | 3 |
| pCE36 | repR6Kγ FRT lacZY this Kmr | 8 |
| pCL1920 | reppSC101ts Spr | 27 |
| pPmrC150 | pCL1920 harboring truncated pmrC150 | This study |
| pPmrC181 | pCL1920 harboring truncated pmrC181 | This study |
| pPmrC295 | pCL1920 harboring truncated pmrC295 | This study |
| pPmrC150lacZ′ | pPmrC150, fusion of pmrC150 and lacZ fragment | This study |
| pPmrC150phoA′ | pPmrC150, fusion of pmrC150 and phoA fragment | This study |
| pPmrC181lacZ′ | pPmrC181, fusion of pmrC181 and lacZ fragment | This study |
| pPmrC181phoA′ | pPmrC181, fusion of pmrC181 and phoA fragment | This study |
| pPmrC295lacZ′ | pPmrC181, fusion of pmrC295 and lacZ fragment | This study |
| pPmrC295phoA′ | pPmrC181, fusion of pmrC295 and phoA fragment | This study |
FRT, FLP recognition target.
Plasmid constructions.
For construction of the single-copy plasmid pBAC108L-pmrC (ppmrC), the pmrC coding and promoter regions were PCR amplified with primers 3109 (5′-GATTGGATCCGTCGCGTTTGTGTATTGCATCTGG-3′) and 2208 (5′-CCCAAGCTTCATTCGCTTAGTCTCCTGCACGG-3′), and 14028s genomic DNA as the template. The amplified PCR fragment was digested with BamHI and HindIII and cloned between the BamHI and HindIII sites of plasmid pBAC108L (38). For the construction of plasmid pBAC108L-pmrCflag (ppmrCFLAG), the pmrC coding region with its own promoter and a FLAG epitope sequence right before the stop codon was PCR amplified with primers 3109 and 3179 (5′-TCAGAAGCTTCACTTGTCATCGTCGTCCTTGTAGTCTTCGCTTAGTCTCCTGCACGGTTG-3′) and 14028s genomic DNA as the template (the DNA sequence encoding the FLAG epitope is underlined). The amplified PCR fragment was digested with BamHI and HindIII and cloned between the BamHI and HindIII sites of plasmid pBAC108L. DNA sequencing verified that the cloned segment had the expected pmrC sequence.
Construction of pmrC mutants.
For the generation of the ΔpmrC1 strain, which harbors a 1,096-bp deletion of the 1,641-bp pmrC gene, a DNA fragment containing a chloramphenicol resistance cassette was PCR amplified with primers 2635 (5′-GCCTGAACATTGCGTTCTACAAGCAGGTACTACAAGACCTGTGTAGGCTGGAGCTGCTTC-3′) and 2636 (5′-GGTGTTGATCAACTGCTCTTGGGAACAGTTCTGAATTTCGCATATGAATATCCTCCTTAG-3′) and plasmid pKD3 (6) as the template, and was used to transform a derivative of strain 14028s as described previously (23). The ΔpmrC1.1 strain, in which the chloramphenicol resistance cassette was removed from the ΔpmrC1 strain by using plasmid pCP20 (3), was used as a host for plasmid pBAC108L, ppmrC, or ppmrCFLAG. Strains ΔpmrC1 and ΔpmrC1.1 exhibited the same lipid A profile and polymyxin B susceptibility.
For construction of the pmrA505 ΔpmrC1.1 strain, a DNA fragment containing a kanamycin resistance cassette was PCR amplified with primers 2807 (5′-GCCTGAACATTGCGTTCTACAAGCAGGTACTACAAGACCT CATATGAATATCCTCCTTAG-3′) and 2808 (5′-GGTGTTGATCAACTGCTCTTGGGAACAGTTCTGAATTTCG GTGTAGGCTGGAGCTGCTTC-3′) and plasmid pKD4 (6) as the template, and was used to transform the pmrA505 strain to generate a pmrA505 ΔpmrC:Kmr strain. The kanamycin resistance cassette was removed from this strain by using plasmid pCP20 (3) to generate the pmrA505 ΔpmrC1.1 strain. To construct the ΔpmrC2 strain, which has a deletion of the entire pmrC coding region, we followed the strategy described above, using primers 2147 (5′-CTTTGTCACGATTAGCGTCACCGAATCGATGGACGCATCAACGTGTAGGCTGGAGCTGCTTC-3′) and 2148 (5′-CCCCTGTAATAATAGCGTGTCGTCTTCAACAATCAGTATCTTCATCATATGAATATCCTCCTTA-3′). The structure of the pmrC region in the generated mutants was confirmed by Southern blot hybridization and/or PCR analysis.
β-Galactosidase assays.
β-Galactosidase assays were performed in duplicate and the activity was determined as described previously (29).
Polymyxin B killing assay.
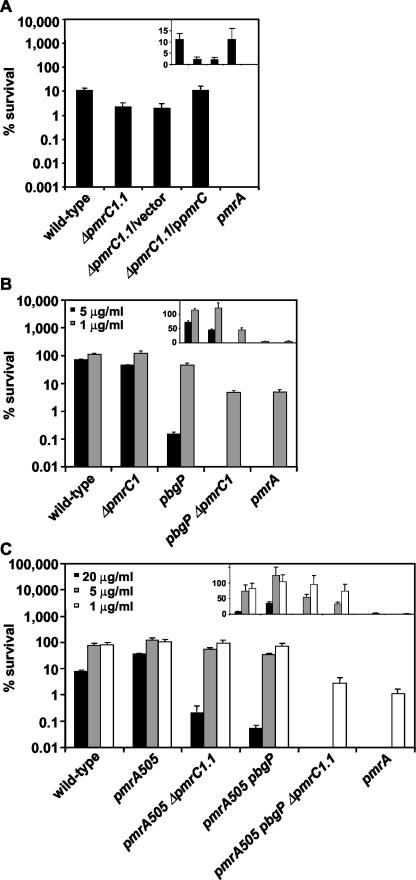
Cells were harvested from an overnight culture grown in N minimal medium at pH 7.7 with 10 mM MgCl2, washed three times with N minimal medium at pH 7.0 without MgCl2, and diluted 1:100 in N minimal medium, pH 5.8, with 10 μM MgCl2. Bacteria were grown for 4 h at 37°C with aeration to an optical density at 600 nm (OD600) of 0.3 to 0.4 and were then diluted 1:100 in LB broth. Fifty microliters of the diluted bacterial culture was mixed with 50 μl of polymyxin B dissolved in a phosphate-buffered saline (PBS) solution and was placed in a 96-well plate (Cell Culture Cluster; Costar). After 1 h of incubation at 37°C with aeration, cultures were serially diluted in PBS and plated onto LB agar plates to determine the number of CFU after an overnight incubation. The percent survival was calculated as follows: (CFU of polymyxin B-treated culture/CFU of PBS-treated culture) × 100 (11). The statistical significance of the polymyxin B susceptibility data was analyzed by a two-tailed Student's t test by using Excel software. The null hypothesis was zero for mean difference comparisons, and P values are reported for this analysis (see Fig. 3).
FIG. 3.
(A) Polymyxin B killing assay of wild-type (14028s), ΔpmrC1.1 (EG14590), ΔpmrC1.1/vector (EG14656), ΔpmrC1.1/ppmrC (EG14595), and pmrA (EG7139) strains grown to logarithmic phase in N-minimal medium, pH 5.8, with 10 μM MgCl2. Polymyxin B was added to a final concentration of 10 μg/ml, and the bacteria were incubated for 1 h at 37°C. The samples were diluted in PBS and plated on LB agar plates to determine the numbers of CFU. Survival values given are relative to those of PBS-treated samples. The ΔpmrC1.1 (EG14590) and ΔpmrC1.1/vector (EG14656) strains were significantly more sensitive to polymyxin B than was the wild-type (14028s) strain (P < 0.01). The complemented strain ΔpmrC1.1/ppmrC (EG14595) was significantly more resistant to polymyxin B than were strains ΔpmrC1.1 (EG14590) and ΔpmrC1.1/vector (EG14656) (P < 0.01). (B) Polymyxin B killing assay of wild-type (14028s), ΔpmrC1 (EG13927), pbgP (EG9241), pbgP ΔpmrC1 (EG14372), and pmrA (EG7139) strains grown and tested as described above, except that polymyxin B was added at final concentrations of 1 and 5 μg/ml. The difference in the polymyxin B (1 μg/ml) susceptibilities of strains pbgP ΔpmrC1 (EG14372) and pmrA (EG7139) was not statistically significant (P = 0.7), indicating that the pbgP and pmrC loci mediate PmrA-controlled polymyxin B resistance. (C) Polymyxin B killing assay of wild-type (14028s), pmrA505 (EG9492), pmrA505 ΔpmrC1.1 (EG14368), pmrA505 pbgP (EG9868), pmrA505 pbgP ΔpmrC1.1 (EG14369), and pmrA (EG7139) strains grown and tested as described for panel A, except that polymyxin B was added at 1, 5, and 20 μg/ml. Note the logarithmic scale (a linear scale is used in the insets) on the y axis. The data correspond to mean values from three independent sets of experiments performed in duplicate. The data demonstrate that the inactivation of the pmrC gene increases the susceptibility of cells to polymyxin B and that a pbgP ΔpmrC1 double mutant exhibits the same level of polymyxin B susceptibility as the pmrA null mutant.
Subcellular localization of PmrC protein.
Inner and outer membranes were prepared as follows. A pmrC strain harboring the ppmrCFLAG plasmid, which carries the pmrC gene with its own promoter and a sequence encoding a FLAG epitope at the 3′ end immediately upstream of the stop codon, was grown overnight in N minimal medium, pH 7.7, with 10 mM MgCl2. The next day, the cells were harvested; washed three times with N minimal medium, pH 7.0, without MgCl2; diluted 1:100 in 200 to 300 ml of N minimal medium, pH 7.7, with 10 μM MgCl2; and grown for 4 h at 37°C with aeration. The cells were then harvested, washed once with PBS, and resuspended in 4 ml of PBS containing sucrose (20%) and lysozyme (100 μg/ml). After being incubated on ice for 30 min, the cells were opened by sonication. A sucrose gradient ultracentrifugation procedure (32, 49) was used, with modifications (www.cmdr.ubc.ca/bobh/methodsall.html), to isolate the inner and outer membranes. Cell debris was removed by centrifugation at 4,000 × g for 15 min, and the whole-cell lysate was loaded on top of a sucrose gradient made with 4 ml each of 60 and 70% sucrose in a Beckman Ultra-Clear centrifuge tube followed by centrifugation in an SW41 rotor at 38,000 rpm for 20 h at 4°C. Bands between 20 and 60% (upper, reddish band) and between 60 and 70% (lower, white band) sucrose, corresponding to the inner and outer membranes, respectively, were collected and dialyzed overnight against PBS. Protein concentrations were determined by a modified Lowry method (1), with bovine serum albumin used as a standard protein. NADH oxidase activity, which was measured as described previously (32), was used as a marker for inner membrane purity. Inner and outer membrane preparations (20 μg of protein each) were run in a sodium dodecyl sulfate (SDS)-10% polyacrylamide gel, transferred onto a nitrocellulose membrane, and developed by using an anti-FLAG M2 monoclonal antibody (Sigma), an anti-mouse immunoglobulin G horseradish peroxidase-linked antibody, and the ECL detection system (Amersham Biosciences).
Construction of genes encoding chimeric PmrC-LacZ and PmrC-PhoA proteins.
A lacZ DNA fragment missing nine codons at the 5′ end (lacZ′) (26) was PCR amplified by using E. coli MG1655 genomic DNA as a template and the following primer pairs: 4140 (5′-GATCCCTAGGGCCGTCGTTTTACAACGTCGTGAC-3′) and 4141 (5′-CCGGAAGCTTTTATTTTTGACACCAGACCAACTG-3′), introducing AvrII (CCTAGG) and HindIII (AAGCTT) restriction sites, respectively; or 4142 (5′-GATCGCTAGCGCCGTCGTTTTACAACGTCGTGAC-3′) and 4141, introducing NheI (GCTAGC) and HindIII (AAGCTT) restriction sites, respectively. A phoA gene segment missing 13 codons at the 5′ end (phoA′) (21) was PCR amplified by using E. coli MG1655 genomic DNA as a template and the following primer pairs: 4143 (5′-GATCCCTAGGCTGTTTACCCCTGTGACAAAAGCC-3′) and 4144 (5′-GGGCAAGCTTTTATTTCAGCCCCAGAGCGGCTTT-3′), introducing AvrII (CCTAGG) and HindIII (AAGCTT) restriction sites, respectively; or 4145 (5′-GATCGCTAGCCTGTTTACCCCTGTGACAAAAGCC-3′) and 4144, introducing NheI (GCTAGC) and HindIII (AAGCTT) restriction sites, respectively. (Restriction sites in the primers are underlined.)
DNA fragments encoding the truncated PmrC proteins PmrC1-150, PmrC1-181, and PmrC1-295 were PCR amplified by using 14028s genomic DNA as a template and the following pair of primers: 3109 (5′-GATTGGATCCGTCGCGTTTGTGTATTGCATCTGG-3′) and 4146 (5′-GATCCTGCAGCCTAGGCGTCGCCGGACGGATTTTGACCCA-3′) for PmrC1-150, 3109 and 4147 (5′-GATCCTGCAGGCTAGCGTAATCTTTATAGAAAAAGGCGGC-3′) for PmrC1-181, and 3109 and 4148 (5′-GATCCTGCAGCCTAGGCATATCAGAAAACATGCAGGGAAC-3′) for PmrC1-295 (the following restriction sites in the primers are underlined: AvrII [CCTAGG], BamHI [GGATCC], NheI [GCTAGC], and PstI [CTGCAG]). The PCR-amplified DNA fragments were first digested with BamHI and PstI and cloned between the BamHI and PstI sites of plasmid pCL1920 (27) to generate plasmids pPmrC150, pPmrC181, and pPmrC295. The fragments harboring the lacZ′ and phoA′ genes digested with AvrII and HindIII were cloned between the AvrII and HindIII sites of plasmids pPmrC150 and pPmrC295 to generate plasmids pPmrC150-lacZ′, pPmrC150-phoA′, pPmrC295-lacZ′, and pPmrC295-phoA′. The fragments digested with NheI and HindIII were cloned between the NheI and HindIII sites of plasmid pPmrC181 to generate plasmids pPmrC181-lacZ′ and pPmrC181-phoA′. In plasmids pPmrC150-lacZ′ and pPmrC150-phoA′, the lacZ′ and phoA′ genes were fused in frame to pmrC right after the sequence encoding the fourth predicted transmembrane domain. In plasmids pPmrC181-lacZ′ and pPmrC181-phoA′, the lacZ′ and phoA′ genes were fused in frame to pmrC right after the sequence encoding the fifth predicted transmembrane domain. In plasmids pPmrC295-lacZ′ and pPmrC295-phoA′, the lacZ′ and phoA′ genes were fused in frame to pmrC right after the sequence encoding the sixth predicted transmembrane domain. These plasmids were transformed into a Salmonella strain with a deletion of the phoN gene, which was constructed as described previously (6), with plasmid pKD3 as the template and with primers 2935 (5′-GGATTACATCTGTTTATTATTGCCTGATCCGGAGTGAGTCTTTGTGTAGGCTGGAGCTGCTTC-3′) and 2936 (5′-GTTTGGGGTGATCTTCTTTACTCAATAAATTATTTTTGTCGTCATATGAATATCCTCCTTA-3′). The production of alkaline phosphatase by strains expressing PmrC-PhoA proteins was determined on LB agar plates containing 5-bromo-4-chloro-3-indolylphosphate (XP; 40 μg/ml). The production of β-galactosidase by strains expressing PmrC-LacZ proteins was determined on LB agar plates containing 5-bromo-4-chloro-3-indolyl-β-d-galactoside (X-Gal; 40 μg/ml).
Preparation of lipid A samples for MALDI-TOF mass spectrometry analysis.
Lipid A samples were prepared as described previously (50), with a slight modification: cells were harvested from overnight cultures grown in N minimal medium, pH 7.7, with 10 mM MgCl2; washed three times with N minimal medium, pH 7.0, without MgCl2; and diluted 1:100 in N minimal medium, pH 5.8, with 10 μM MgCl2. After growth for 4 h at 37°C with aeration, the cells were harvested, washed once with PBS, and resuspended in 300 μl of Tri-Reagent (Molecular Research Center) for the amount of cells harvested from 30 ml of culture at an OD600 of ∼0.4. After an incubation for 20 min at room temperature, 30 μl of chloroform was added, and the samples were vortexed vigorously and incubated for 15 min at room temperature. The phases were separated by centrifugation at 12,000 × g for 10 min, and the upper phase was transferred to a new tube. One hundred microliters of water was added to the lower phase, vortexed, incubated for 15 min, and centrifuged at 12,000 × g for 10 min. The upper phase was combined with the upper phase recovered as described above. This extraction was performed twice. The combined upper phases were dried in a speed-vac apparatus (model RC10.22; Jouan, Winchester, Va.) and dissolved in 500 μl of hydrolysis buffer, pH 4.5, containing 12.5 mM sodium acetate and 1% SDS. For the release of lipid A from the LPS, samples were boiled for 1 h at 100°C, dried in a speed-vac, and resuspended in a mixture of 100 μl of water and 500 μl of acidified ethanol (made by mixing 100 μl of 4 M HCl with 20 ml of 95% ethanol). The pellet was harvested by centrifugation at 2,060 × g for 10 min, washed with 500 μl of 95% ethanol, and centrifuged again at 2,060 × g for 10 min. The washing steps were repeated to completely remove SDS. The pellet was dried at room temperature for 5 min, and lipid A was dissolved by the addition of 100 μl of chloroform and methanol (3:1) and was used for matrix-assisted laser desorption-ionization time-of-flight (MALDI-TOF) mass spectrometry analysis. MALDI-TOF mass spectrometry analyses of lipid A were performed with the negative-ion mode of a Voyager DE STR mass spectrometer (PerSeptive Biosystems, Framingham, Mass.) equipped with a 337-nm nitrogen laser with delayed extraction. Analyses were carried out in the reflector mode at a mass range of m/z 1,500 to 3,000, with an accelerating voltage of 20 kV and a delay time of 300 ns. The instrument was externally calibrated. A low-mass gate value of m/z 500 was selected to avoid saturation of the detector. 2,5-Dihydroxybenzoic acid at 10 μg/μl in 70% acetonitrile-0.1% trifluoroacetic acid was used as a matrix. The final mass spectra were obtained from an average of 5 to 10 spectra, and each spectrum was a collection from 200 laser shots.
RESULTS
The Salmonella PmrC protein exhibits sequence identity with Neisseria proteins implicated in the incorporation of phosphoethanolamine into LPS.
To identify Salmonella genes responsible for the phosphoethanolamine modification of lipid A, we conducted a BLAST search of the Salmonella genome by using as the query the amino acid sequence of the Neisseria meningitidis Lpt-3 protein, which had been implicated in the phosphoethanolamine modification of the heptose residue in the core oligosaccharide portion of the LPS (28). We recovered four open reading frames (PmrC, YbiP, YhjW, and YijP) (Table 2) and decided to focus on the PmrC protein because it is encoded in the PmrA-dependent pmrCAB operon (41) and because we were interested in phosphoethanolamine modifications that are regulated by PmrA. We then used the amino acid sequence of the PmrC protein to query the Neisseria genome and obtained three genes: the expected NMB2010 gene (lpt-3), NMB0415, which appears to be a pseudogene, and NMB1638 (lptA), which has been shown to be required for the incorporation of phosphoethanolamine into the lipid A moiety of the LPS (5). The Salmonella PmrC protein exhibited the highest identity with the NMB1638 gene product (42% identity and 60% similarity). However, the regions of sequence identity and similarity were not evenly distributed: these proteins were 48% identical (65% similar) in the C-terminal 340 residues but only 30% identical (53% similar) in the N-terminal 176 amino acids. This analysis suggested that the PmrA-regulated pmrC gene might be involved in the phosphoethanolamine modification of the LPS.
TABLE 2.
Salmonella open reading frames exhibiting sequence similarity to the lpt-3 gene product of N. meningitidis MC58
| Locus | Gene name | % Amino acid identity (no. with identity/total) | % Amino acid similarity (no. with similarity total) |
|---|---|---|---|
| STM3635 | yhjW | 24 (137/562) | 41 (236/562) |
| STM4293 | pmrC | 25 (71/279) | 43 (122/279) |
| STM4118 | yijP | 23 (64/272) | 38 (104/272) |
| STM0834 | ybiP | 23 (61/260) | 41 (107/260) |
Construction of a nonpolar pmrC mutant.
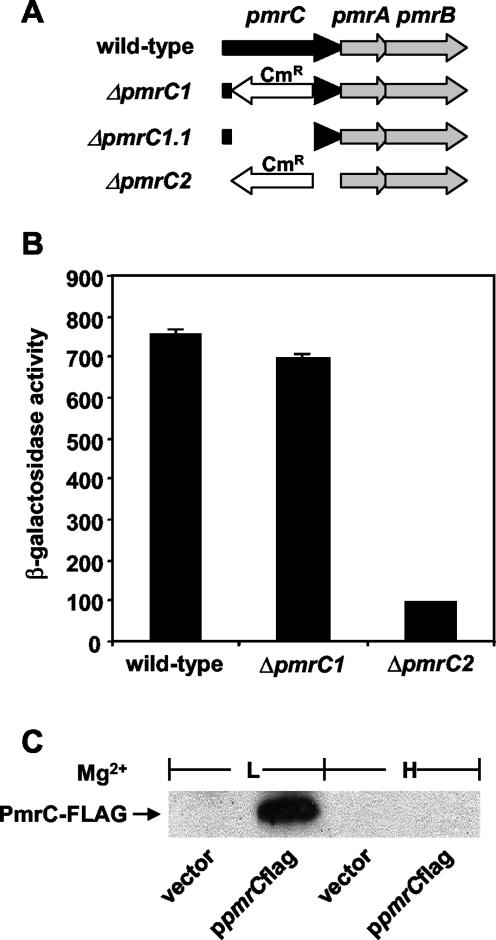
To examine the function of the pmrC gene, we constructed a nonpolar pmrC mutant that lacked 1,096 of the 1,641 bp of the pmrC gene (Fig. 1A). This mutant retained 185 bp at the 5′ end of the pmrC coding region as well as the last 360 bp of the pmrC open reading frame. The latter region contains a putative promoter that apparently directs the constitutive expression of the downstream pmrA and pmrB genes (13, 41) (Fig. 1A). The generated mutation (designated ΔpmrC1) was not polar on the pmrA and pmrB genes because the same levels of transcription of the PmrA-activated pbgP gene were displayed by isogenic wild-type and ΔpmrC1 strains (Fig. 1B). Moreover, a PmrC-FLAG protein that was expressed from the pmrC promoter carried by a single-copy-number plasmid exhibited normal regulation in the ΔpmrC1.1 mutant: the protein was produced when bacteria were grown in low, but not high, levels of Mg2+ (Fig. 1C). The behavior of the ΔpmrC1 mutant contrasted with that exhibited by a strain with a deletion of the complete pmrC coding region (ΔpmrC2) (Fig. 1A), which showed levels of pbgP transcription that were 10 times lower than those displayed by the wild-type strain (Fig. 1B). These results support the notion that there is a promoter within the pmrC coding region that promotes the transcription of the downstream pmrA and pmrB genes. Furthermore, they indicate that the generated ΔpmrC1 and ΔpmrC1.1 mutations do not affect the expression of the pmrA and pmrB genes, and this allowed us to examine the phenotypes resulting from the absence of a functional pmrC gene.
FIG. 1.
(A) Schematic representation of the pmrCAB operon in wild-type Salmonella and in mutants with a partial (ΔpmrC1 and ΔpmrC1.1) or complete (ΔpmrC2) deletion of the pmrC open reading frame. (B) β-Galactosidase activity (in Miller units) expressed by strains harboring a chromosomal lac transcriptional fusion to the PmrA-activated pbgP gene that were grown logarithmically in N-minimal medium, pH 5.8, with 10 μM MgCl2. Transcription was investigated in wild-type (14028s), ΔpmrC1 (EG13927), and ΔpmrC2 (EG13633) genetic backgrounds. Data correspond to mean values from three independent sets of experiments performed in duplicate. Transcription of the PmrA-activated pbgP gene was similar in the wild-type and ΔpmrC1 strains, but it was decreased in the ΔpmrC2 mutant. (C) Western blot analysis of cell extracts prepared from the ΔpmrC1.1 mutant (EG14592) containing the ppmrCFLAG plasmid, which expresses the pmrCflag gene from its own promoter, after logarithmic growth in N-minimal medium, pH 7.7, with 10 μM (L) or 10 mM (H) MgCl2. The total protein from equal amounts of bacterial cells, as adjusted by the OD600, was run in an SDS-10% polyacrylamide gel, transferred onto a nitrocellulose membrane, and developed by using anti-FLAG antibodies. The ΔpmrC1.1 mutant displays normal PmrA regulation, as the PmrC-FLAG protein is produced by bacteria grown in a low Mg2+ concentration but is not detected when bacteria are grown in a high Mg2+ concentration.
Mutation of the pmrC gene results in lipid A that lacks phosphoethanolamine.
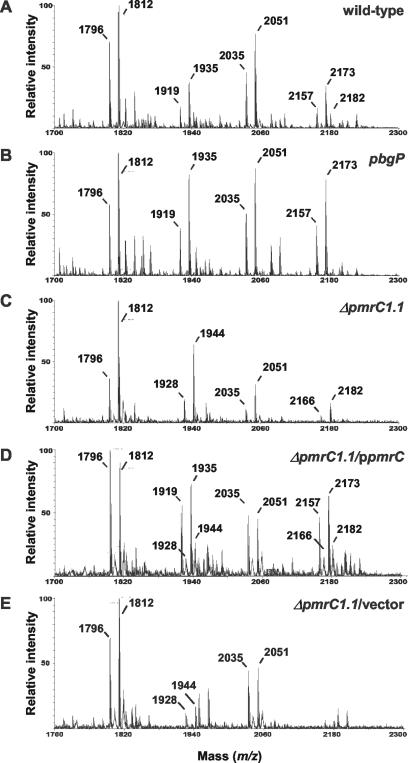
To examine whether the pmrC gene is necessary for the incorporation of phosphoethanolamine into lipid A, we used negative-ion-mode MALDI-TOF mass spectrometry to analyze the lipid A species from wild-type pbgP, ΔpmrC1.1, and pmrA strains and strains grown at a low pH and with a low level of Mg2+, which are conditions that promote the transcription of PmrA-activated genes (41). Because the chemical structures and m/z values for most of the lipid A species in S. enterica had been previously assigned (16, 51-53), we focused on the differences in lipid A profiles between wild-type and mutant strains, putting particular emphasis on the representative molecular ions ([M − H]−) of lipid A species modified with phosphoethanolamine or aminoarabinose, which are governed by the PmrA/PmrB system.
A molecular ion ([M − H]−) at m/z 1,796 was considered to be the prototype lipid A, a hexa-acylated lipid A 1,4′-bisphosphate (i.e., diphosphorylated lipid A) (Fig. 2A). The m/z values corresponding to phosphoethanolamine-modified lipid A molecular ions ([M − H]−) are as follows: m/z 1,919, a diphosphorylated lipid A (m/z 1,796) bearing a phosphoethanolamine of 124 average mass units (amu) at the 1 or 4′ phosphate of lipid A; m/z 1,935, a hydroxylated form of m/z 1,919; m/z 2,157, a palmitoylated form of m/z 1,919; and m/z 2,173, a palmitoylated form of m/z 1,935. The molecular ions ([M − H]−) of lipid A species modified with aminoarabinose are as follows: m/z 1,928, a diphosphorylated lipid A (m/z 1,796) bearing an aminoarabinose (132 amu) at the 1 or 4′ phosphate of lipid A; m/z 1,944, a hydroxylated form of m/z 1,928; m/z 2,166, a palmitoylated form of m/z 1,928; and m/z 2,182, a hydroxylated form of m/z 2,166. m/z 1,812 represents a hydroxylated form of the prototype lipid A (m/z 1797), and m/z 2,035 and 2,051 represent a diphosphorylated lipid A molecular ion ([M − H]−) bearing a palmitate group and a hydroxyl group, respectively (Fig. 2A). The latter modifications are known to be regulated by the PhoP/PhoQ system (10, 16, 17), which is activated under the low-Mg2+ conditions used to grow the organisms (39), and were used as internal controls for our lipid A analyses.
FIG. 2.
Lipid A species profiles from wild-type (14028s) (A), pbgP (EG9241) (B), ΔpmrC1.1 (EG14590) (C), ΔpmrC1.1/ppmrC (EG14595) (D), and ΔpmrC1.1/vector (EG14656) (E) strains grown to logarithmic phase in N-minimal medium, pH 5.8, with 10 μM MgCl2, and analyzed by negative-ion-mode MALDI-TOF mass spectrometry. These profiles show that the pmrC mutant lacks lipid A species modified with phosphoethanolamine.
The ΔpmrC1.1 mutant lacked peaks at m/z 1,919, 1,935, 2,157, and 2,173, which correspond to phosphoethanolamine-modified lipid A species (Fig. 2C). On the other hand, this mutant retained molecular ions corresponding to lipid A species modified with aminoarabinose at m/z 1,928, 1,944, 2,166, and 2,182 (Fig. 2C), which, as expected (14), were absent from the pbgP mutant (Fig. 2B). The lipid A profile of the ΔpmrC1.1 mutant was solely due to the absence of the pmrC gene function, as the phosphoethanolamine-modified lipid A molecular ions (peaks at m/z 1,919, 1,935, 2,157, and 2,173) were present in the lipid A species of a ΔpmrC1.1 strain harboring a plasmid with a wild-type copy of the pmrC gene (Fig. 2D), but not in a ΔpmrC1.1 strain with a vector control (Fig. 2E). These results demonstrate that the pmrC gene is required for the incorporation of phosphoethanolamine into lipid A.
The pmrC gene is required for resistance to polymyxin B.
We determined that the ΔpmrC1.1 mutant was three- to fivefold more sensitive to polymyxin B than was the wild-type strain (Fig. 3A). This phenotype was due to the lack of the pmrC gene function, as a plasmid carrying a wild-type copy of the pmrC gene restored wild-type levels of polymyxin B resistance to the ΔpmrC1.1 mutant (Fig. 3A). Moreover, the ΔpmrC1.1 mutation decreased polymyxin B resistance even in the polymyxin-resistant pmrA505 strain (Fig. 3C), which expresses PmrA-regulated genes even under noninducing conditions (24). Because the ΔpmrC1.1 strain lacked phosphoethanolamine but retained aminoarabinose in lipid A (Fig. 2C), these results demonstrate that the ability to modify lipid A with phosphoethanolamine is necessary for polymyxin B resistance.
A mutant defective in both the pbgP and pmrC genes has the same lipid A profile and susceptibility to polymyxin B as a pmrA null mutant.
When grown under low-Mg2+ and mildly acidic conditions, mutants defective in the pbgP or pmrC genes are more sensitive to polymyxin B than the wild-type strain but are not quite as sensitive as a pmrA null mutant (Fig. 3B) (24). On the other hand, a pbgP ΔpmrC1 double mutant displayed the same level of polymyxin B susceptibility as a pmrA null mutant (Fig. 3B). Consistent with this result, the inactivation of both the pmrC and pbgP genes in the polymyxin B-resistant pmrA505 genetic background reduced polymyxin B resistance to the levels of the pmrA null mutant (Fig. 3C). These results indicate that the pbg operon and the pmrC genes are solely responsible for PmrA-regulated polymyxin B resistance. (This is in addition to the ugd gene, which exhibits a similar susceptibility phenotype as the pbgP mutant, consistent with these loci encoding proteins mediating the biosynthesis of aminoarabinose.)
To further explore the association between polymyxin B resistance and lipid A modifications, we examined the lipid A profiles of pmrA, pbgP ΔpmrC1, pmrA505, and pmrA505 pbgP ΔpmrC1.1 strains. The lipid A from the pmrA mutant lacked molecular ions ([M − H]−) corresponding to those modified with either phosphoethanolamine (peaks at m/z 1,919, 1,935, 2,153, and 2,173) or aminoarabinose (peaks at m/z 1,928 1,944, 2,166, and 2,182) (Fig. 4D), which was consistent with previous reports (52). Likewise, inactivation of both the pbgP and pmrC genes in either a pmrA+ (Fig. 4C) or pmrA505 (Fig. 4B) background resulted in a strain with the same lipid A profile as that exhibited by the pmrA null mutant (Fig. 4D), which lacks the modifications displayed by the pmrA505 strain (Fig. 4A). Taken together with the results of the polymyxin susceptibility assays (Fig. 3), this analysis indicates that PmrA-controlled polymyxin B resistance is mediated by the aminoarabinose and phosphoethanolamine modifications of lipid A.
FIG. 4.
Lipid A species profiles for the pmrA505 (EG9492) (A), pmrA505 pbgP ΔpmrC1.1 (EG14369) (B), pbgP ΔpmrC1 (EG14372) (C), and pmrA (EG7139) (D) strains grown to logarithmic phase in N-minimal medium, pH 5.8, with 10 μM MgCl2, and analyzed by negative-ion-mode MALDI-TOF mass spectrometry. These profiles show that the pbgP ΔpmrC1 and pmrA505 pbgP ΔpmrC1.1 mutants have the same lipid A profile as the pmrA null mutant.
The pmrC gene is dispensable for resistance to Fe3+.
The pmrA mutant exhibits hypersusceptibility to killing by Fe3+, but the targets of PmrA regulation that are responsible for Fe3+ resistance have remained unknown (48). Thus, we tested the ΔpmrC1 and pbgP ΔpmrC1 mutants for Fe3+ sensitivity and found that they retained wild-type levels of resistance to Fe3+ (data not shown), suggesting that the pmrC gene is not required for this property.
PmrC is an inner membrane protein with a large periplasmic domain.
The PSORT-B subcellular localization program (www.psort.org/psortb/index.html) predicted an inner membrane location for the PmrC protein. Thus, to examine the subcellular location of the PmrC protein, we conducted a Western blot analysis of inner and outer membrane preparations from a ΔpmrC1.1 derivative expressing a C-terminal FLAG-tagged PmrC protein from the pmrC promoter. The PmrC protein localized to the inner membrane (Fig. 5A), which makes physiological sense because that is where the largest pool of phosphatidylethanolamine in the bacterial cell is located, and phosphatidylethanolamine is the donor of phosphoethanolamine in E. coli (19) and Salmonella (Yixin Shi and Eduardo A. Groisman, unpublished results).
FIG. 5.
(A) Western blot analysis of inner and outer membranes prepared from the ΔpmrC1.1 strain containing either the pBAC108L vector (EG14656) or the ppmrCFLAG plasmid (EG14592), which carries a pmrC gene directed by its own promoter and expresses a PmrC protein tagged with a FLAG epitope at its C terminus. Bacteria were grown to the logarithmic phase in N-minimal medium, pH 7.7, with 10 μM MgCl2. Inner and outer membranes were prepared by sucrose density gradient centrifugation. Twenty micrograms of protein from the inner and outer membranes was boiled for 10 min, run in an SDS-10% polyacrylamide gel, transferred onto a nitrocellulose membrane, and developed by using anti-FLAG antibodies. To examine the purity of the membrane preparations, we determined the NADH oxidase activity by measuring the oxidation of NADH at 340 nm, and these values are expressed as follows: 100 × μmol of substrate oxidized/min/mg of protein. The analysis demonstrates that the PmrC protein localizes to the inner membrane. (B) Kyte-Doolittle hydropathy plot (25) of the PmrC protein generated by DNA Strider 1.3 software. (C) The left panel shows the predicted topology of the PmrC protein. The numbers correspond to the positions in the PmrC protein at which in-frame fusions were generated to the PhoA and LacZ proteins. The right panel shows alkaline phosphatase and β-galactosidase activities displayed by the phoN strain (EG14286) harboring plasmids pPmrC150-lacZ′, pPmrC150-phoA′, pPmrC181-lacZ′, pPmrC181-phoA′, pPmrC295-lacZ′, and pPmrC295-phoA′ when streaked onto LB agar plates containing either XP (40 μg/ml) or X-Gal (40 μg/ml). These data suggest that the C-terminal region (amino acids 177 to 547) of PmrC localizes to the periplasm.
An analysis of the PmrC protein by the TMpred program (www.ch.embnet.org/software/TMPRED_form.html) suggested the presence of five to six transmembrane domains in the N-terminal region that could mediate membrane association (Fig. 5B) and of a C-terminal region that could be responsible for the predicted phosphoethanolamine transferase activity. The five-transmembrane-domain model predicts that amino acids 1 to 176 mediate membrane association and that the C-terminal 371 amino acids are located in the periplasm. On the other hand, the six-transmembrane-domain model predicts that amino acids 1 to 291 mediate membrane association and that the remaining C-terminal region of the PmrC protein is in the cytoplasm.
To investigate the topology of the PmrC protein, we evaluated the β-galactosidase and alkaline phosphatase activities of a Salmonella strain with a deletion of the phoN gene and harboring plasmids with in-frame lacZ or phoA fusions to the 3′ end of the pmrC gene truncated at different positions. These fusions were predicted to generate chimeric proteins with LacZ or PhoA immediately after the predicted fourth, fifth, and sixth transmembrane domains of PmrC (Fig. 5C). (The use a phoN mutant facilitated the detection of alkaline phosphatase activity, which can be obscured by the phoN-encoded nonspecific acid phosphatase.) We detected alkaline phosphatase activity in the strains expressing the PhoA chimera harboring the N-terminal 181 and 295 residues of PmrC but not in that expressing a chimera harboring the N-terminal 150 residues (Fig. 5C). Consistent with these results, the strains expressing the LacZ chimera harboring the N-terminal 181 and 295 residues of PmrC produced no β-galactosidase activity, whereas the strain with LacZ fused to the N-terminal 150 residues did (Fig. 5C). These results suggest that the PmrC protein harbors five transmembrane domains that are followed by a large periplasmic region.
DISCUSSION
The PmrA/PmrB two-component regulatory system has been implicated in the modification of the 1 and 4′ positions of lipid A with aminoarabinose and phosphoethanolamine (52). The synthesis of aminoarabinose is mediated by the PmrA-activated ugd gene and pbgP operon (43), which are necessary for resistance to polymyxin B. We have now established that the PmrA-activated pmrC gene is necessary for the phosphoethanolamine modification of lipid A (Fig. 2) and for resistance to polymyxin B (Fig. 3).
The PmrC protein exhibits sequence identity with two Neisseria proteins that are implicated in the incorporation of phosphoethanolamine into lipid A and the core region of the LPS (5, 28). There is a higher degree of sequence identity between the Salmonella PmrC and Neisseria LptA proteins in the C-terminal region, possibly reflecting the fact that both of these proteins are necessary for the modification of lipid A with phosphoethanolamine (Fig. 2) (5). A search of the conserved domain database (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi), using the C-terminal region (amino acids 177 to 547) of PmrC as a query, retrieved the catalytic domains of the phosphoglycerol transferase and sulfatase families. This makes sense because phosphoglycerol transferase uses phosphatidylglycerol as a donor of phosphoglycerol in E. coli (22) and because phosphatidylglycerol and phosphatidylethanolamine are structurally similar. Moreover, a sulfatase catalyzes the hydrolysis of a sulfate group, which is similar in size to a phosphate group (31P versus 32S). Interestingly, the sulfatase is closely related to the sulfotransferases of mycobacteria in terms of substrate binding, i.e., binding of a sulfate group (31), which incorporate a sulfate group into the glycopeptidolipid (4), the equivalent of the LPS in gram-negative bacteria. Cumulatively, our results suggest that PmrC is a phosphoethanolamine transferase that uses phosphatidylethanolamine as a donor of phosphoethanolamine.
The PmrC protein localizes to the inner membrane (Fig. 5A) and appears to have two distinct domains. The N-terminal 176-amino-acid domain harbors several stretches of hydrophobic amino acids that may constitute transmembrane segments (Fig. 5B) and likely mediates the membrane association of the PmrC protein. The alkaline phosphatase and β-galactosidase activities displayed by strains expressing different chimeric PmrC proteins suggest that the C-terminal 370-amino-acid domain is present in the periplasmic side of the inner membrane (Fig. 5C). The location and topology of the PmrC protein would allow it to catalyze the incorporation of phosphoethanolamine into lipid A by using phosphatidylethanolamine (19), the most abundant phospholipid in E. coli (34) and Salmonella (Shi and Groisman, unpublished results), as a substrate.
Mutants in the regulatory protein PmrA that are resistant to polymyxin B exhibit increased levels of aminoarabinose and phosphoethanolamine in lipid A (20). We have now established that both of these PmrA-controlled modifications are required for polymyxin B resistance, as a pbgP ΔpmrC1 double mutant is as susceptible to polymyxin B as a pmrA null mutant (Fig. 3B) and has a lipid A profile that is identical to that of a pmrA null mutant, lacking both aminoarabinose and phosphoethanolamine (Fig. 4). This is true even when the pbgP and pmrC genes are mutated in the hyperactive pmrA505 genetic background (Fig. 3C and 4). While the pmrA null mutant is ∼10,000-fold more susceptible to polymyxin B than the wild-type strain, we were surprised to find that this is more than the sum of the susceptibilities displayed by mutants defective in pbgP or pmrC (Fig. 3B). This suggests that when Salmonella lacks the ability to perform a particular type of lipid A modification, a different type of modification may be enhanced. Indeed, phosphoethanolamine-modified lipid A accumulates to higher levels in a pbgP (pmrF) mutant of E. coli than in the wild-type strain (52). Taken together, these results establish that the PmrA-controlled phosphoethanolamine modification of lipid A is essential for full resistance to polymyxin B.
It has been hypothesized that two promoters mediate the transcription of the pmrA and pmrB genes: a PmrA-activated promoter located upstream of the pmrC gene in the pmrCAB operon and a constitutive promoter located within the pmrC open reading frame. Whereas the PmrA-regulated promoter has been defined by S1 mapping experiments (47), evidence for the constitutive promoter is based on the ability of a 346-bp fragment from the pmrC coding region to promote transcription from a plasmid-linked promoterless reporter gene (14) and the fact that pmrC-lac fusions generated with the MudJ transposon near the 3′ end, but within the pmrC coding region, exhibit normal PmrA-dependent transcription (41). We have now provided genetic evidence for the presence of a promoter within the pmrC gene by establishing that the deletion of the complete pmrC open reading frame abolished PmrA-mediated transcription, whereas a strain retaining 360 bp at the 3′ of the pmrC gene exhibited normal PmrA-controlled transcription (Fig. 1B). As described for the PhoP/PhoQ two-component regulatory system (42), this constitutive promoter may provide the basal levels of PmrA and PmrB proteins that are required in order to respond to environmental changes.
Finally, the availability of strains that are specifically defective in the phosphoethanolamine modification of lipid A makes it possible to examine the role that this modification plays in resistance to other antimicrobial peptides and in potential interference with signaling by host cells.
Acknowledgments
We thank Yixin Shi for excellent discussions. We are grateful to Russell W. Carlson and Anup Datta for advice regarding the isolation of lipid A.
This work was supported in part by grant AI42236 from the National Institute of Health to E.A.G., who is an investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Bensadoun, A., and D. Weinstein. 1976. Assay of proteins in the presence of interfering materials. Anal. Biochem. 70:241-250. [DOI] [PubMed] [Google Scholar]
- 2.Chamnongpol, S., W. Dodson, M. J. Cromie, Z. L. Harris, and E. A. Groisman. 2002. Fe(III)-mediated cellular toxicity. Mol. Microbiol. 45:711-719. [DOI] [PubMed] [Google Scholar]
- 3.Cherepanov, P. P., and W. Wackernagel. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9-14. [DOI] [PubMed] [Google Scholar]
- 4.Converse, S. E., J. D. Mougous, M. D. Leavell, J. A. Leary, C. R. Bertozzi, and J. S. Cox. 2003. MmpL8 is required for sulfolipid-1 biosynthesis and Mycobacterium tuberculosis virulence. Proc. Natl. Acad. Sci. USA 100:6121-6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cox, A. D., J. C. Wright, J. Li, D. W. Hood, E. R. Moxon, and J. C. Richards. 2003. Phosphorylation of the lipid A region of meningococcal lipopolysaccharide: identification of a family of transferases that add phosphoethanolamine to lipopolysaccharide. J. Bacteriol. 185:3270-3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davis, R. W., D. Bolstein, and J. R. Roth. 1980. Advanced bacterial genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 8.Ellermeier, C. D., A. Janakiraman, and J. M. Slauch. 2002. Construction of targeted single copy lac fusions using lambda Red and FLP-mediated site-specific recombination in bacteria. Gene 290:153-161. [DOI] [PubMed] [Google Scholar]
- 9.Fields, P. I., E. A. Groisman, and F. Heffron. 1989. A Salmonella locus that controls resistance to microbicidal proteins from phagocytic cells. Science 243:1059-1062. [DOI] [PubMed] [Google Scholar]
- 10.Gibbons, H. S., S. Lin, R. J. Cotter, and C. R. Raetz. 2000. Oxygen requirement for the biosynthesis of the S-2-hydroxymyristate moiety in Salmonella typhimurium lipid A. Function of LpxO, a new Fe2+/alpha-ketoglutarate-dependent dioxygenase homologue. J. Biol. Chem. 275:32940-32949. [DOI] [PubMed] [Google Scholar]
- 11.Groisman, E. A., F. Heffron, and F. Solomon. 1992. Molecular genetic analysis of the Escherichia coli phoP locus. J. Bacteriol. 174:486-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Groisman, E. A., J. Kayser, and F. C. Soncini. 1997. Regulation of polymyxin resistance and adaptation to low-Mg2+ environments. J. Bacteriol. 179:7040-7045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gunn, J. S., K. B. Lim, J. Krueger, K. Kim, L. Guo, M. Hackett, and S. I. Miller. 1998. PmrA-PmrB-regulated genes necessary for 4-aminoarabinose lipid A modification and polymyxin resistance. Mol. Microbiol. 27:1171-1182. [DOI] [PubMed] [Google Scholar]
- 14.Gunn, J. S., and S. I. Miller. 1996. PhoP-PhoQ activates transcription of pmrAB, encoding a two-component regulatory system involved in Salmonella typhimurium antimicrobial peptide resistance. J. Bacteriol. 178:6857-6864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gunn, J. S., S. S. Ryan, J. C. Van Velkinburgh, R. K. Ernst, and S. I. Miller. 2000. Genetic and functional analysis of a PmrA-PmrB-regulated locus necessary for lipopolysaccharide modification, antimicrobial peptide resistance, and oral virulence of Salmonella enterica serovar Typhimurium. Infect. Immun. 68:6139-6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo, L., K. B. Lim, J. S. Gunn, B. Bainbridge, R. P. Darveau, M. Hackett, and S. I. Miller. 1997. Regulation of lipid A modifications by Salmonella typhimurium virulence genes phoP-phoQ. Science 276:250-253. [DOI] [PubMed] [Google Scholar]
- 17.Guo, L., K. B. Lim, C. M. Poduje, M. Daniel, J. S. Gunn, M. Hackett, and S. I. Miller. 1998. Lipid A acylation and bacterial resistance against vertebrate antimicrobial peptides. Cell 95:189-198. [DOI] [PubMed] [Google Scholar]
- 18.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 19.Hasin, M., and E. P. Kennedy. 1982. Role of phosphatidylethanolamine in the biosynthesis of pyrophosphoethanolamine residues in the lipopolysaccharide of Escherichia coli. J. Biol. Chem. 257:12475-12477. [PubMed] [Google Scholar]
- 20.Helander, I. M., I. Kilpelainen, and M. Vaara. 1994. Increased substitution of phosphate groups in lipopolysaccharides and lipid A of the polymyxin-resistant pmrA mutants of Salmonella typhimurium: a 31P-NMR study. Mol. Microbiol. 11:481-487. [DOI] [PubMed] [Google Scholar]
- 21.Hoffman, C. S., and A. Wright. 1985. Fusions of secreted proteins to alkaline phosphatase: an approach for studying protein secretion. Proc. Natl. Acad. Sci. USA 82:5107-5111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jackson, B. J., and E. P. Kennedy. 1983. The biosynthesis of membrane-derived oligosaccharides. A membrane-bound phosphoglycerol transferase. J. Biol. Chem. 258:2394-2398. [PubMed] [Google Scholar]
- 23.Kato, A., T. Latifi, and E. A. Groisman. 2003. Closing the loop: the PmrA/PmrB two-component system negatively controls expression of its posttranscriptional activator PmrD. Proc. Natl. Acad. Sci. USA 100:4706-4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kox, L. F., M. M. Wosten, and E. A. Groisman. 2000. A small protein that mediates the activation of a two-component system by another two-component system. EMBO J. 19:1861-1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kyte, J., and R. F. Doolittle. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157:105-132. [DOI] [PubMed] [Google Scholar]
- 26.Lawley, B., and A. J. Pittard. 1994. Regulation of aroL expression by TyrR protein and Trp repressor in Escherichia coli K-12. J. Bacteriol. 176:6921-6930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lerner, C. G., and M. Inouye. 1990. Low copy number plasmids for regulated low-level expression of cloned genes in Escherichia coli with blue/white insert screening capability. Nucleic Acids Res. 18:4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mackinnon, F. G., A. D. Cox, J. S. Plested, C. M. Tang, K. Makepeace, P. A. Coull, J. C. Wright, R. Chalmers, D. W. Hood, J. C. Richards, and E. R. Moxon. 2002. Identification of a gene (lpt-3) required for the addition of phosphoethanolamine to the lipopolysaccharide inner core of Neisseria meningitidis and its role in mediating susceptibility to bactericidal killing and opsonophagocytosis. Mol. Microbiol. 43:931-943. [DOI] [PubMed] [Google Scholar]
- 29.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 30.Moskowitz, S. M., R. K. Ernst, and S. I. Miller. 2004. PmrAB, a two-component regulatory system of Pseudomonas aeruginosa that modulates resistance to cationic antimicrobial peptides and addition of aminoarabinose to lipid A. J. Bacteriol. 186:575-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mougous, J. D., R. E. Green, S. J. Williams, S. E. Brenner, and C. R. Bertozzi. 2002. Sulfotransferases and sulfatases in mycobacteria. Chem. Biol. 9:767-776. [DOI] [PubMed] [Google Scholar]
- 32.Orndorff, P. E., and M. Dworkin. 1980. Separation and properties of the cytoplasmic and outer membranes of vegetative cells of Myxococcus xanthus. J. Bacteriol. 141:914-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paulus, H., and E. Gray. 1964. The biosynthesis of polymyxin B by growing cultures of Bacillus polymyxa. J. Biol. Chem. 239:865-871. [PubMed] [Google Scholar]
- 34.Pluschke, G., and P. Overath. 1981. Function of phospholipids in Escherichia coli. Influence of changes in polar head group composition on the lipid phase transition and characterization of a mutant containing only saturated phospholipid acyl chains. J. Biol. Chem. 256:3207-3212. [PubMed] [Google Scholar]
- 35.Roland, K. L., L. E. Martin, C. R. Esther, and J. K. Spitznagel. 1993. Spontaneous pmrA mutants of Salmonella typhimurium LT2 define a new two-component regulatory system with a possible role in virulence. J. Bacteriol. 175:4154-4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shafer, W. M., L. E. Martin, and J. K. Spitznagel. 1984. Cationic antimicrobial proteins isolated from human neutrophil granulocytes in the presence of diisopropyl fluorophosphate. Infect. Immun. 45:29-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shimomura, H., M. Matsuura, S. Saito, Y. Hirai, Y. Isshiki, and K. Kawahara. 2003. Unusual interaction of a lipopolysaccharide isolated from Burkholderia cepacia with polymyxin B. Infect. Immun. 71:5225-5230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shizuya, H., B. Birren, U. J. Kim, V. Mancino, T. Slepak, Y. Tachiiri, and M. Simon. 1992. Cloning and stable maintenance of 300-kilobase-pair fragments of human DNA in Escherichia coli using an F-factor-based vector. Proc. Natl. Acad. Sci. USA 89:8794-8797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Snavely, M. D., C. G. Miller, and M. E. Maguire. 1991. The mgtB Mg2+ transport locus of Salmonella typhimurium encodes a P-type ATPase. J. Biol. Chem. 266:815-823. [PubMed] [Google Scholar]
- 40.Soncini, F. C., E. Garcia Vescovi, F. Solomon, and E. A. Groisman. 1996. Molecular basis of the magnesium deprivation response in Salmonella typhimurium: identification of PhoP-regulated genes. J. Bacteriol. 178:5092-5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Soncini, F. C., and E. A. Groisman. 1996. Two-component regulatory systems can interact to process multiple environmental signals. J. Bacteriol. 178:6796-6801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Soncini, F. C., E. G. Vescovi, and E. A. Groisman. 1995. Transcriptional autoregulation of the Salmonella typhimurium phoPQ operon. J. Bacteriol. 177:4364-4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Trent, M. S., A. A. Ribeiro, S. Lin, R. J. Cotter, and C. R. Raetz. 2001. An inner membrane enzyme in Salmonella and Escherichia coli that transfers 4-amino-4-deoxy-l-arabinose to lipid A: induction on polymyxin-resistant mutants and role of a novel lipid-linked donor. J. Biol. Chem. 276:43122-43131. [DOI] [PubMed] [Google Scholar]
- 44.Vaara, M., T. Vaara, M. Jensen, I. Helander, M. Nurminen, E. T. Rietschel, and P. H. Makela. 1981. Characterization of the lipopolysaccharide from the polymyxin-resistant pmrA mutants of Salmonella typhimurium. FEBS Lett. 129:145-149. [DOI] [PubMed] [Google Scholar]
- 45.Vaara, M., T. Vaara, and M. Sarvas. 1979. Decreased binding of polymyxin by polymyxin-resistant mutants of Salmonella typhimurium. J. Bacteriol. 139:664-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vaara, M., and P. Viljanen. 1985. Binding of polymyxin B nonapeptide to gram-negative bacteria. Antimicrob. Agents Chemother. 27:548-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wosten, M. M., and E. A. Groisman. 1999. Molecular characterization of the PmrA regulon. J. Biol. Chem. 274:27185-27190. [DOI] [PubMed] [Google Scholar]
- 48.Wosten, M. M., L. F. Kox, S. Chamnongpol, F. C. Soncini, and E. A. Groisman. 2000. A signal transduction system that responds to extracellular iron. Cell 103:113-125. [DOI] [PubMed] [Google Scholar]
- 49.Yamato, I., Y. Anraku, and K. Hirosawa. 1975. Cytoplasmic membrane vesicles of Escherichia coli. A simple method for preparing the cytoplasmic and outer membranes. J. Biochem. (Tokyo) 77:705-718. [DOI] [PubMed] [Google Scholar]
- 50.Yi, E. C., and M. Hackett. 2000. Rapid isolation method for lipopolysaccharide and lipid A from gram-negative bacteria. Analyst 125:651-656. [DOI] [PubMed] [Google Scholar]
- 51.Zhou, Z., S. Lin, R. J. Cotter, and C. R. Raetz. 1999. Lipid A modifications characteristic of Salmonella typhimurium are induced by NH4VO3 in Escherichia coli K12. Detection of 4-amino-4-deoxy-l-arabinose, phosphoethanolamine and palmitate. J. Biol. Chem. 274:18503-18514. [DOI] [PubMed] [Google Scholar]
- 52.Zhou, Z., A. A. Ribeiro, S. Lin, R. J. Cotter, S. I. Miller, and C. R. Raetz. 2001. Lipid A modifications in polymyxin-resistant Salmonella typhimurium: PMRA-dependent 4-amino-4-deoxy-l-arabinose, and phosphoethanolamine incorporation. J. Biol. Chem. 276:43111-43121. [DOI] [PubMed] [Google Scholar]
- 53.Zhou, Z., A. A. Ribeiro, and C. R. Raetz. 2000. High-resolution NMR spectroscopy of lipid A molecules containing 4-amino-4-deoxy-l-arabinose and phosphoethanolamine substituents. Different attachment sites on lipid A molecules from NH4VO3-treated Escherichia coli versus kdsA mutants of Salmonella typhimurium. J. Biol. Chem. 275:13542-13551. [DOI] [PubMed] [Google Scholar]