Summary
RECQL4, a RecQ helicase deficient in Rothmund-Thomsons syndrome, is a multifunctional participant of DNA metabolism. Here, we show that RECQL4 interacts with the Ku complex of DNA-PK and participates in DNA repair via NHEJ.
Abstract
RECQL4, a member of the RecQ helicase family, is a multifunctional participant in DNA metabolism. RECQL4 protein participates in several functions both in the nucleus and in the cytoplasm of the cell, and mutations in human RECQL4 are associated with three genetic disorders: Rothmund-Thomson, RAPADILINO and Baller-Gerold syndromes. We previously reported that RECQL4 is recruited to laser-induced DNA double-strand breaks (DSB). Here, we have characterized the functional roles of RECQL4 in the non-homologous end joining (NHEJ) pathway of DSB repair. In an in vitro NHEJ assay that depends on the activity of DNA-dependent protein kinase (DNA-PK), extracts from RECQL4 knockdown cells display reduced end-joining activity on DNA substrates with cohesive and non-cohesive ends. Depletion of RECQL4 also reduced the end joining activity on a GFP reporter plasmid in vivo. Knockdown of RECQL4 increased the sensitivity of cells to γ-irradiation and resulted in accumulation of 53BP1 foci after irradiation, indicating defects in the processing of DSB. We find that RECQL4 interacts with the Ku70/Ku80 heterodimer, part of the DNA-PK complex, via its N-terminal domain. Further, RECQL4 stimulates higher order DNA binding of Ku70/Ku80 to a blunt end DNA substrate. Taken together, these results implicate that RECQL4 participates in the NHEJ pathway of DSB repair via a functional interaction with the Ku70/Ku80 complex. This is the first study to provide both in vitro and in vivo evidence for a role of a RecQ helicase in NHEJ.
Introduction
The RecQ family of DNA helicases consists of evolutionarily conserved proteins with Escherichia coli’s RecQ as the prototypical member. Human RecQ helicases, RECQL1, Bloom (BLM), Werner (WRN), RECQL4 and RECQL5, participate in several cellular functions including DNA replication, DNA repair, transcription, genome organization, mitochondrial maintenance, senescence and apoptosis (1–3). RECQL4, expressed in many human cell types and tissues, consists of 1208 amino acids (aa) with a characteristic RecQ helicase domain in the center. Like the other human RecQ helicases, RECQL4 is a DNA-dependent adenosine triphosphatase (ATPase) and 3′–5′ helicase enzyme. In contrast to WRN and BLM, RECQL4 shows more robust strand annealing activity than helicase activity (4). Interestingly, the N- and C-terminal regions of RECQL4 contain distinct sequences not present in the other RecQ family members. The N-terminal region of RECQL4 contains an Sld2-like domain (3,4) as well as nuclear and mitochondrial targeting sequences rendering the protein able to perform functions in the nucleus as well as in mitochondria. RECQL4 lacks the HRDC domain, present in WRN and BLM, and a recent bioinformatics study suggests the presence of a RQC domain in RECQL4 (5).
Mutations in three of the RecQ helicases, BLM, WRN and RECQL4, are associated with heritable diseases. The RECQL4 mutation spectrum disorders include Rothmund-Thomson (RTS), RAPADILINO (RAPA) and Baller-Gerold (BGS) syndromes (6). The common features among these diseases are short stature, skeletal abnormalities and radial ray defects. Interestingly, patients with either RTS or RAPA are at elevated risks for osteosarcomas and lymphomas (6,7). Increased expression of RECQL4 has been found to be associated with carcinogenesis in metastatic prostate (8), sporadic osteosarcoma (9), primary colorectal (10), breast (11) and cervical (12) cancers. Thus, there is significant interest in RECQL4 as both a tumor suppressor and a marker of tumor progression.
Cells derived from RTS patients show chromosomal abnormalities such as trisomy, aneuploidy and chromosomal rearrangements and defects in DNA repair (3,13,14). Defects in DNA repair processes are associated with chromosomal abnormalities and carcinogenesis (15,16). Recently, we reported that the helicase and ATPase activities of RECQL4 are compromised in patients with RECQL4-associated diseases (17,18). Interestingly, the trend is that RECQL4 patient mutations, which cause protein instability segregate with RTS, whereas those mutations which cause reduced helicase activity are associated with RAPA.
Double-strand breaks (DSB) are among the most potent and deleterious forms of cellular DNA damage causing mutagenic and developmental defects, gross chromosomal rearrangements, cell death and malignancy (19). In higher eukaryotes, DSB are repaired by two distinct pathways: homologous recombination (HR) and non-homologous end joining (NHEJ). The HR pathway is a high fidelity DNA repair mechanism, which requires homologous sequences primarily from the sister chromatids and preferentially occurs during the S/G2 phase of the cell cycle. In contrast, the NHEJ pathway is an error prone mechanism where two ends of the DSB are joined via a process that is largely independent of terminal DNA sequence homology (20,21). In spite of its error prone nature, NHEJ is the predominant form of DSB repair in human somatic cells. The essential factors involved in NHEJ are the DNA-dependent protein kinase (DNA-PK) complex, which include the catalytic subunit of DNA-PK (DNA-PKcs), the Ku70/Ku80 heterodimer and the XRCC4/Ligase IV (X4L4) complex. Cells deficient in any of these proteins are defective in DSB repair. NHEJ is initiated by the binding of the Ku70/Ku80 heterodimer to the DSB ends. Live cell imaging following laser micro-irradiation indicates that DNA-PKcs, Cernunnos (XRCC4 like factor, XLF) and the preassembled X4L4 are recruited to Ku-bound DSB (22). Autophosphorylation of DNA-PKcs mediates a conformational change required for the activation and recruitment of end processing enzymes such as the Artemis and the members of the X family of DNA polymerases. Finally ligation of the processed DSB requires the X4L4 complex which is stimulated in vitro by XLF (21,23).
The roles of all human RecQ helicases, i.e. WRN, BLM, RECQL1, RECQL4 and RECQL5 have been studied in HR (24,25), but much less is known about their roles in NHEJ. The studies so far have largely been focused on WRN and to a lesser extent on RECQL4 (26–30). Previous reports suggested that RECQL4 is involved in DSB repair (13,14,31), but its role in human NHEJ has not been investigated. Our live cell microscopy studies indicated that RECQL4 is recruited early to the laser-induced DSB and remains for a shorter duration than WRN and BLM. Further, the recruitment of RECQL4 to DNA damage sites was independent of the functions of ataxia telangiectasia mutated (ATM) (14), a regulator of the HR pathway.
Based on the above observations we speculated that, in humans, RECQL4 might play an important role in NHEJ. We therefore investigated the functional roles of RECQL4 in NHEJ. We report here that RECQL4-deficient cells show increased sensitivity to γ-irradiation and accumulate higher levels of 53BP1 foci after γ-irradiation compared with scramble control cells, suggesting a defect in DSB processing. We also show that RECQL4 functionally interacts with the Ku70/Ku80 heterodimer via its N-terminus, but not with other components (Artemis, Ligase IV and XLF) of the NHEJ pathway. Moreover, total end-joining activity was significantly reduced in RECQL4-deficient cells compared with scrambled control cells using an in vitro plasmid based NHEJ assay and an in vivo GFP reporter plasmid based end-joining assay. Interestingly, RECQL4 also stimulated higher order binding of the Ku70/Ku80 complex to a blunt end DNA substrate. These findings suggest the involvement of RECQL4 in NHEJ via its functional interaction with the Ku70/Ku80 complex.
Materials and methods
Cell lines
HeLa, U2OS and GM637 cell lines, used in this study, were cultured in Dulbecco’s modified Eagle’s medium (Life Technologies) or Minimum essential medium (Life Technologies) supplemented with growth factors as before (14).
Knockdown of RECQL4
Endogenous RECQL4 was knocked down using small interfering RNA (siRNA) or short hairpin RNA (shRNA). RECQL4 knockdown (RECQL4 KD) and scrambled control cells were generated in HeLa and U2OS cell lines using Mission shRNA lentiviral construct TRCN0000051169 (5′-CCTCGATTCCATTATCATTTA-3′) for RECQL4 KD (Sigma–Aldrich) as described before (32). siRNA-mediated knockdown experiments were performed in GM637 cell line using ON-TARGETplus control (C) and RECQL4 siRNA (RQ4: 5′-CAA UACAGCUUACCGUACA-3′, RQ4-3a: 5′-GCTC CAAAA TGCAG AAT AA-3′, RQ4-3b: 5′-ACTGAGGACCTGGGCAAAA-3′) and INTERFERin (Polyplus transfection) transfection reagent.
Colonogenic survival
HeLa scrambled and RECQL4 KD cells were seeded in triplicate at a cell density of 2500 and 4000 cells per well, respectively, in six-well Petri dishes. The following day, cells were γ-irradiated with 0, 1, 2, 3 and 4 Gy and allowed to grow in normal growth conditions for 13 days and the colonogenic survival was measure as described previously (14).
Immunoprecipitation and immunoblotting
Co-immunoprecipitation assays were carried out as described previously (32) using HeLa cell extracts or recombinant RECQL4 (10 pmol) and Ku70/Ku80 heterodimer (20 pmol) proteins. Antibodies used for immunoprecipitation and immunoblotting were rabbit anti-RECQL4 (32), anti-Ku70 (ab53129; Abcam), anti-Ku80 (sc9034; Santa Cruz Biotechnology) and normal rabbit IgG (sc2027; Santa Cruz Biotechnology), anti-ATM (ab32420; Abcam), anti-phosphoATM (S1981) (ab32420), anti-NBS1 (611871, BD Biosciences), anti-phophoNBS1 (S343) (05-663, Upstate), antiChk2 (Sc-9064, Santa Cruz), anti-phosphoChk2 (T387) (2668, Cell Signaling). For overexpression of RECQL4, HeLa cells were transfected with pcDNA 3.1 containing 3X-FLAG tagged full length RECQL4 (1–1208) or N-terminal RECQL4 (1–447) or helicase domain of RECQL4 (447–830) or C-terminal RECQL4 (830–1208) and jetPRIME transfection reagent (Polyplus transfection).
End-joining extracts
Cell extract preparation was carried out as described previously (33–35). Briefly, exponentially growing cells were lysed through three freeze/thaw cycles in lysis buffer [25 mM Tris-HCl (pH 7.5), 333 mM KCl, 1.5 mM ethylenediaminetetraacetic acid (EDTA) and 4 mM dithiothreitol (DTT)], containing protease inhibitor cocktail (Roche Diagnostics) and phosphatase inhibitor cocktails I and II (Sigma–Aldrich) or by Dounce homogenizer as described previously (35). Lysates were incubated at 4°C for 20min, cleared by centrifugation, and dialyzed against dialysis buffer [20 mM Tris-HCl (pH 8.0), 100 mM potassium acetate, 20% glycerol, 0.5 mM EDTA pH 8.0, 1 mM DTT]. Protein concentration was determined by Bradford assay (Bio-Rad) and end-joining extracts were snap-freezed and stored at −80°C until use.
In vitro end joining
The assays were performed with 5 or 10 ng of 2.6 kb cohesive and 5.7 kb non-cohesive DNA substrates generated from pUC18, pKS and pSingle-tTS-shRNA plasmids (35) and end-joining extracts (20 or 40 μg) for 1–3 h at 25°C in 10 or 20 μl reaction containing 50 mM Triethanolamine (pH 8.0), 0.5 mM magnesium acetate, 1 mM DTT, 0.1 mg/ml bovine serum albumin (BSA), 60 mM potassium acetate, 1 mM adenosine triphosphate (ATP) or [20 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid (pH 7.5), 10 mM MgCl2, 80 mM KCl, 1 mM DTT, 1 mM ATP and 50 μM deoxyribonucleotide triphosphates]. The reaction was stopped with 50 mM EDTA and 80 μg/ml RNaseA at 37°C, and deproteinized by incubating with proteinase K (2 mg/ml) for 60 min at 37°C. Ligation products were separated in a 0.7% agarose gel and stained with SYBR-Gold (Invitrogen). Fluorescence was detected with Molecular Imager Gel Doc XR system (Bio-Rad) and DNA bands were analyzed with the ImageJ software (version 1.4). Cohesive substrates were derived by using BamHI or SalI or EcoRI (New England Biologicals) restriction enzymes, and non-cohesive substrate was derived by using BstXI (New England Biologicals).
In vivo NHEJ
The scrambled and RECQL4 KD cells (5 × 105) were transfected with 50 ng of pEGFP-Pem1-Ad2 reporter plasmid (35,36) digested with HindIII or I-SceI (New England Biologicals) and 25 ng of pDsRed-Express-C1 (Clontech) using jetPRIME. The cells were maintained in drug-free Dulbecco’s modified Eagle’s medium containing 10% fetal bovine serum at 37°C. Twenty-four-hour post transfection, cells were harvested and washed with ice cold phosphate-buffered saline by centrifugation at 4°C and 500g for 5 min. The cells were resuspended in 0.5 ml phosphate-buffered saline, and assayed for the expression of enhanced green fluorescent protein (EGFP) and DsRed by flow cytometry (Accuri C6; BD Biosciences).
DNA mobility shift
A pair of 80 nt long oligonucleotides G80 and C80 (37) were used to generate blunt end duplex DNA substrate GC80. The G80 oligonucleotide was 5′ end labeled with γ-32P-ATP (PerkinElmer Life Sciences) and then annealed to the C80. For DNA binding, 0.5 nM of labeled GC80 substrate was incubated with either Ku complex alone or Ku complex in the presence of full length RECQL4 or N-terminal 1–492 aa (NTD492) or N-terminal 1–240 (NTD240) or WRN in a buffer containing 30 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid (pH 7.3), 40 mM KCl, 8 mM MgCl2, 1 mM DTT, 100 ng/ml BSA and 5% glycerol at room temperature for 30 min. Incubated samples were electrophoresed on a 5% polyacrylamide gel (29:1 cross-linking ratio) and exposed to PhosphorImager plate (GE Healthcare). DNA mobility shift results were analyzed using Typhoon Fluorimager and ImageQuant version 5.2 (GE Healthcare).
Helicase activity
Helicase reactions were performed as described previously (32). Briefly, the reactions were carried out in 10 μl reaction volume in a buffer containing 20 mM Tris-HCl (pH 7.4), 20 mM NaCl, 25 mM KCl, 1 mM DTT, 5% glycerol, 2.5 mM MgCl2, 2.5 mM ATP and 100 μg/ml BSA. Indicated amounts of proteins were incubated with 0.5 nM of substrate, and incubated at 37ºC for 30 min. The reactions were stopped by 3X stop dye (50 mM EDTA, 40% glycerol, 0.9% sodium dodecyl sulfate, 0.05% bromophenol blue and 0.05% xylene cyanol). The products were separated by electrophoresis on 8% native polyacrylamide gels, exposed to a PhosphorImager plate and analyzed by ImageQuant version 5.2.
ATPase activity
ATPase assays were performed as described previously (17,38). Briefly, indicated amount of proteins were incubated in reaction buffer [30 mM Tris-HCl (pH 7.4), 50 mM KCl, 5 mM MgCl2, 11% glycerol, 0.1 mg/ml BSA, 1 mM DTT, 50 μM (cold) ATP, 10 μM 10 μCi/μl (γ-32P) ATP] with 100 nM 60 nt oligonucleotide for 1 h at 37°C. About 167 mM of EDTA was added to stop reactions before separating samples by thin-layer chromatography on Baker-flex Cellulose PEI sheets (J.T. Baker). The PEI sheet was exposed to a PhosphorImager plate, and the results were analyzed and quantified using ImageQuant version 5.2.
Results
RECQL4-deficient cells show end-joining defects in vitro
NHEJ, the predominant DSB repair pathway, is active throughout the cell cycle and repairs DSB during S/G2 phases when HR, which needs sister chromatid is absent (21). Expression of RECQL4 is observed during all phases of the cell cycle with higher amounts in the S-phase (39). A previous report using the Xenopus model system reported that RECQL4 functions in DSB repair along with the Ku complex (13). The Ku heterodimer proteins recognize DSB and initiate NHEJ, which depends on the kinase activity of DNA-PKcs (21). We previously documented ATM-independent recruitment of RECQL4 to microirradiation-induced DNA damage sites (14). Based on these observations we hypothesized that RECQL4 may play a role in NHEJ and its deficiency would result in decreased DNA end joining activity.
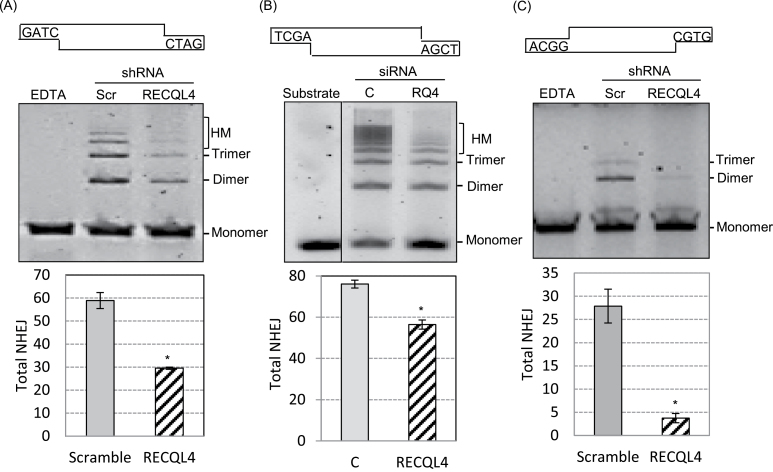
In vitro NHEJ assays have been widely used to assess the role of various proteins of interest in NHEJ (40). In these assays, a linearized plasmid is incubated with total cellular or nuclear extract that generates high molecular weight DNA oligomers via NHEJ. To examine the role of RECQL4 in NHEJ in vitro, we performed NHEJ assays (33,35) using whole cell extracts prepared from HeLa, U2OS and GM637 cell lines depleted for RECQL4. Linearized pUC18 (Figure 1A and B) or pKS plasmids (Supplementary Figure 1, available at Carcinogenesis Online) with BamHI or SalI or EcoRI were used as DNA substrates with cohesive ends. Resolution of NHEJ reaction products on agarose gels revealed the presence of multiple bands generated from ligation of input substrates. The results showed that the extracts from RECQL4 KD U2OS cells had significantly reduced (50–30%) total end-joining activity compared with extracts from scrambled control cell extracts (Figure 1A, compare lanes 2 and 3; Supplementary Figure 1, available at Carcinogenesis Online). The effect was more significant for higher multimers (trimers and above). Further, we tested the effect of knockdown of RECQL4 in primary human fibroblasts, GM637, on NHEJ efficiency in vitro. Using a SalI linearized DNA substrate, which contains cohesive DNA ends, we found that depletion of RECQL4 reduced the end-joining activity by ~20% compared with control cell extracts (Figure 1B). Under similar experimental conditions, we also tested normal and RECQL4-depleted HeLa cell extracts for end-joining activity using DNA substrates containing compatible ends and found similar results (data not shown).
Fig. 1.
RECQL4 deficiency reduces in vitro NHEJ. (A) In vitro NHEJ assay on cohesive-end containing DNA substrate with cellular extracts prepared from scrambled and RECQL4 KD U2OS cells. The end-joining assays were performed with BamHI-linearized pUC18 DNA substrate. The products were resolved by agarose gel electrophoresis and quantified with the ImageJ software. Bar graph below the gel shows the total end joining from three independent experiments. Error bars represent the standard deviation (±). (B) In vitro NHEJ with siRNA transfected GM637 cell extracts. pUC18 linearized with SalI served as DNA substrate. Quantification of the NHEJ results shown in the bar graph, below the gel, was performed as in panel A. (C) In vitro NHEJ assay with non-cohesive DNA substrate. The end-joining assays were performed with BstXI-linearized 5.7 kb non-cohesive end DNA substrate generated from pSingle-tTS-shRNA plasmid, and cell extracts as in panel A. DNA substrates, with end sequence, used in the NHEJ reactions are shown on top of each gel. EDTA, reaction in the presence of EDTA; Scr, scramble; C, ONTARGETplus control; RQ4, RECQL4. *P < 0.05.
Next, we examined if different nucleotide sequences at the DNA ends affected the total NHEJ activity of RECQL4-deficient cells. NHEJ assays were done with plasmid DNA containing non-cohesive ends, which require end processing activities, and with cellular extracts prepared from scrambled or RECQL4 KD U2OS cells. Non-cohesive DNA substrates, 5.7 kb, with incompatible 3′ protruding ends were prepared by digesting the pSingle-tTS-shRNA with BstXI restriction enzyme (35). Analysis of end-joined products revealed that knockdown of RECQL4 reduced the NHEJ activity on the substrates with non-cohesive ends. Extracts from RECQL4 KD cells showed ~7-fold reduction in end-joining activity (Figure 1C). Taken together, these results suggest that the depletion of RECQL4 results in significant loss of NHEJ activity in vitro.
RECQL4 knockdown decreases NHEJ efficiency in vivo
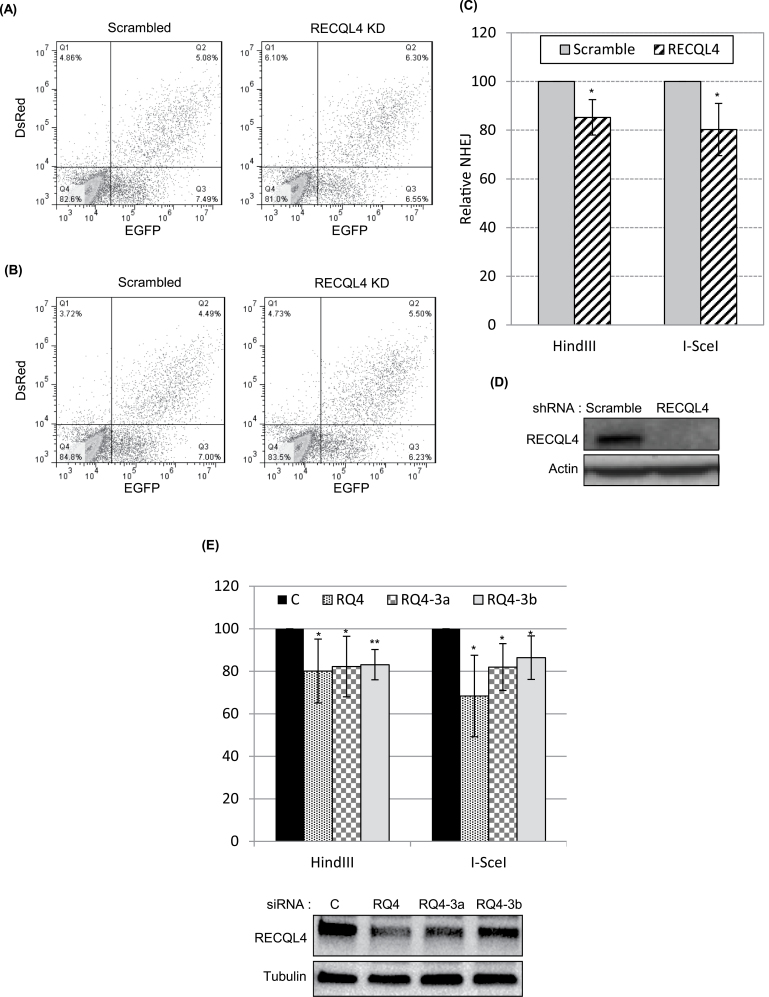
To strengthen our in vitro NHEJ observations in RECQL4 knockdown cell extracts, we used a plasmid based reporter assay for measuring NHEJ in vivo (36). The reporter plasmid, pEGFP-Pem1-Ad2 containing a disrupted EGFP, was linearized with HindIII or I-SceI to generate cohesive and non-cohesive DNA ends, respectively. Expression of EGFP from the reporter plasmid requires end joining of linearized plasmid by NHEJ (35,36). U2OS cells expressing scrambled and RECQL4 shRNA were transfected with pEGFP-Pem1-Ad2 reporter plasmid containing cohesive or non-cohesive ends, and concomitantly with a control reporter plasmid expressing red fluorescence (DsRed). NHEJ-dependent expression of EGFP was detected by fluorescence-activated cell sorter (FACS) analysis, and the data were normalized to DsRed. Our results showed that RECQL4 KD U2OS cells had decreased NHEJ activity compared with scrambled control cells using both cohesive (Figure 2A and C) and non-cohesive end-containing plasmids (Figure 2B and C). Normalized expression of EGFP from multiple experiments showed that DNA end-joining efficiency in RECQL4KD cells was reduced by ~18–20% when compared with control cell. Western blots showed almost complete reduction in the RECQL4 (95%) protein levels in RECQL4 KD cells (Figure 2D). Next we tested the in vivo NHEJ efficiency in GM0637 human normal fibroblasts transiently transfected with three different siRNAs against RECQL4 (Figure 2E). Two of the three siRNA (RQ4-3a and RQ4-3b) were designed to target the 3′ UTR region of RECQL4 mRNA. Knockdown of RECQL4 by the three siRNAs significantly reduced the efficiency of NHEJ when compared with control cells. RECQL4 knockdown efficiency was determined by immunoblotting (Figure 2E). Taken together, the results obtained from in vitro and in vivo NHEJ assays support the role of RECQL4 in DNA repair via the NHEJ pathway.
Fig. 2.
RECQL4 knockdown decreases in vivo NHEJ efficiency. (A and B) In vivo NHEJ assays with cohesive and non-cohesive end containing DNA substrates, respectively. U2OS scrambled and RECQL4 KD cells were transfected with either HindIII digested (cohesive ends) or I-SceI digested (non-cohesive ends) linearized pEGFP-Pem1-Ad2 reporter plasmid. Twenty-four-hour post transfection, the cells were harvested, fixed and, analyzed by flow cytometry for EGFP and DsRed positive cells. Efficiency of NHEJ is calculated by comparing total EGFP positive (quartile 2 and 3) with total DsRed positive (quartile 1 and 2) cells. (C) Bar graph represents the quantification of EGFP-positive cells normalized to DsRed-positive cells from three independent experiments. The error bars represent the standard deviation. (D) Immunoblot showing the levels of RECQL4 in U2OS cells transduced with lentivirus containing scrambled and RECQL4 shRNA. (E) Bar graph showing in vivo NHEJ in GM0637 cells. Three independent NHEJ experiments were performed and the results were quantified as in panel C. Immunoblots represent protein levels in siRNA transfected cells. *P < 0.05, **P < 0.01.
RECQL4 KD cells are sensitive to γ-irradiation and accumulate 53BP1 foci
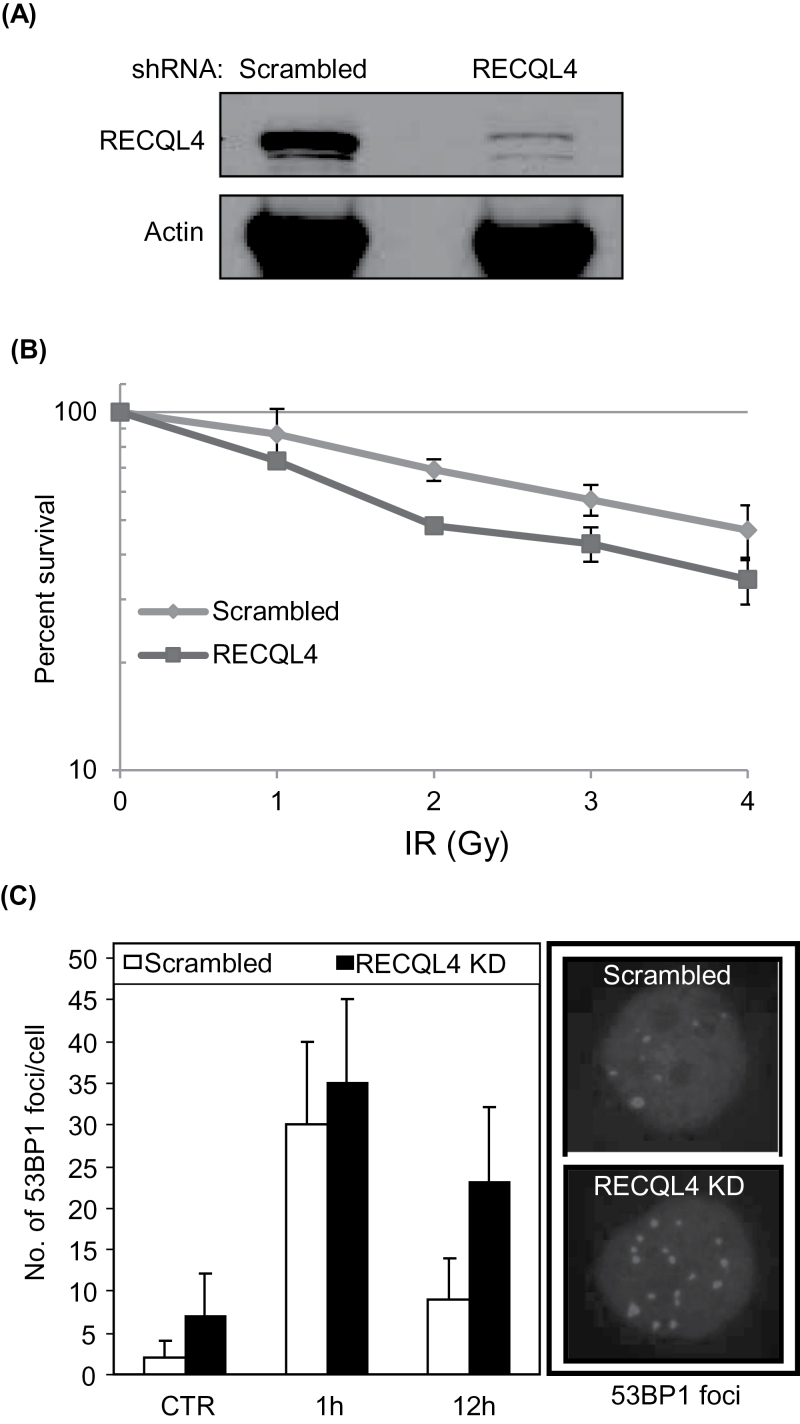
To study the effects of RECQL4 knockdown on the sensitivity of the cells to DNA damage induced by ionizing radiation, HeLa cells were infected with lentivirus carrying shRNA targeting the RECQL4 transcripts (32). Western analysis of knockdown cells indicates more than 90% reduction in RECQL4 expression (Figure 3A). Knockdown cells were also analyzed for the checkpoint proteins ATM, Chek2 and NBS1 (Supplementary Figure 2, available at Carcinogenesis Online). Quantitation of immunoblots showed an increase in the activated forms of these tested proteins in RECQL4 knockdown cells. To examine the survival efficiency of HeLa scrambled and RECQL4 KD cells after γ-irradiation, the cells were treated with increasing doses of γ-radiation and survival efficiency was measured after 13 days post irradiation by colony forming assay. The results showed that the RECQL4 cells were more sensitive to γ-irradiation than scrambled control cells (Figure 3B). At each radiation dose tested, RECQL4 KD cells displayed ~20% lower survival compared with control cells indicating the importance of RECQL4 in DNA repair.
Fig. 3.
RECQL4 depleted cells are moderately sensitive to γ-irradiation and show increased accumulation of 53BP1 foci. (A) Western blots showing the RECQL4 protein levels in scrambled and RECQL4 KD HeLa cells. (B) Survival efficiencies of HeLa scrambled and RECQL4 KD cells after different doses of γ-irradiation. The experiments were performed in triplicate and the error bars represent the standard deviation. (C) Graph showing the average number of 53BP1 foci/cell in HeLa scrambled and RECQL4 KD cells. Cells irradiated with 2 Gy of IR were allowed to recover for 1 and 12 h under normal growth conditions. Controls, CTR, represent un-irradiated cells. Representative images of 53BP1 foci in scrambled and RECQL4 KD cells at 12 h after irradiation are shown on right.
Next to determine the DNA repair efficiency, scrambled and RECQL4 KD cells were γ-irradiated with 2 Gy and tested for the presence of DSB foci after 12 h of recovery under normal growth conditions. Since 53BP1 foci are widely used as markers for DSBs in cells (41), irradiated and unirradiated cells were immunostained with anti-53BP1 antibody and examined by confocal laser scanning microscopy. The cells accumulated 53BP1 foci immediately after irradiation, and a significantly higher number of foci (~2.5-fold) remained in the γ-irradiated RECQL4 KD cells compared with scrambled cells after 12 h, (Figure 3C). Examination of unirradiated cells indicated that RECQL4 KD cells contained ~7 foci per cell (Figure 3C), ~3-fold higher than control cells, which might be generated in response to endogenous DNA damaging agents. These results suggest that DSB repair kinetics is delayed in RECQL4 KD cells compared with scrambled control cells, hence suggesting the importance of RECQL4 in DSB repair. In order to characterize the involvement of RECQL4 in DSB repair, we next examined its interaction with the proteins involved in NHEJ.
RECQL4 interacts with the Ku70/Ku80 heterodimer
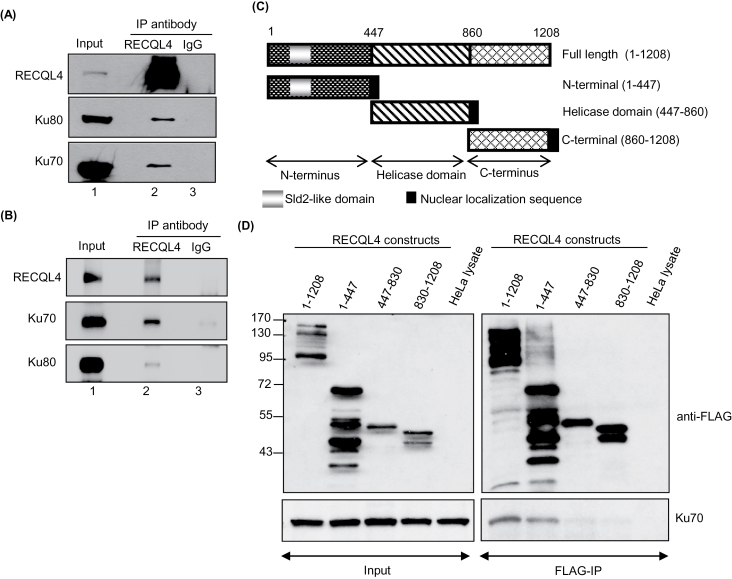
To investigate the role for RECQL4 in NHEJ in human cells, we asked if RECQL4 interacted with the key proteins of NHEJ using co-immunoprecipitation experiments in HeLa cell extracts. The immunoprecipitation experiments were performed in the presence of ethidium bromide (EtBr) to detect nucleic acid-independent protein–protein interactions (42). Immunoblotting demonstrated that endogenous RECQL4 specifically interacts with Ku70 and Ku80 (Figure 4A) but not with Artemis, Ligase IV, or XLF under the tested conditions (Supplementary Figure 3A, available at Carcinogenesis Online). Reciprocally, RECQL4 co-immunoprecipated with Ku proteins (Supplementary Figure 3B, available at Carcinogenesis Online) but the interaction of RECQL4 with DNA-PKcs was inconsistent (data not shown). These results suggest that RECQL4 and the Ku70/Ku80 heterodimer are in complex in vivo (Figure 4A). To rule out the possibility that other proteins mediated this interaction, we carried out the immunoprecipitation experiments using purified RECQL4 and Ku70/Ku80 heterodimer proteins. The results suggest that there is a direct physical interaction between RECQL4 and the Ku70/Ku80 (Figure 4B). Next, to map the interaction between RECQL4 and Ku, HeLa cells were transiently transfected with plasmids containing different domains of RECQL4 (Figure 4C) tagged with a FLAG sequence at the C-terminus. Immunoblotting of the cell lysates with FLAG antibody from cells expressing RECQL4 constructs revealed the presence of multiple bands along with the expected RECQL4 protein bands (Figure 4D, input lanes) which could be degradation products of the FLAG-RECQL4 protein. Immunoprecipitation with FLAG-M2 beads pulled down the ectopically expressed full length RECQL4 (1–1208 aa), N-terminal RECQL4 (1–447 aa), helicase domain (447–830) and C-terminal RECQL4. Examination of IP-fractions revealed that Ku70 efficiently immunoprecipitates with the full length RECQL4 and the N-terminal domain of RECQL4 (Figure 4D), indicating the importance of N-terminal domain of RECQL4 for the formation of RECQL4-Ku complex. Taken together, these results suggest that the RECQL4 and the Ku70/Ku80 heterodimer complex physically interact in vivo.
Fig. 4.
RECQL4 interacts with Ku70/Ku80 heterodimer complex.
(A) Endogenous RECQL4 co-immunoprecipitates endogenous Ku70/Ku80 heterodimer. Immunoprecipitation was performed with RECQL4 (lane 2) and normal IgG (lane 3) antibodies, and RECQL4, Ku70 and Ku80 were detected by immunoblotting. (B) Purified RECQL4 and Ku70/Ku80 proteins interact in vitro. Immunoprecipitation and immunoblotting were performed as in panel A. (C) Diagram of RECQL4 constructs used for mapping RECQL4–Ku interactions. (D) N-terminal region of RECQL4 mediates the RECQL4–Ku interaction. Immunoprecipitation was performed with FLAG-M2 beads using HeLa cells expressing FLAG tagged full length RECQL4 (1–1208), N-terminal RECQL4 (1–447), Helicase domain of RECQL4 (447–830) and C-terminal RECQL4 (830–1208). RECQL4 and Ku70 were detected with anti-FLAG and anti-Ku70 antibodies. HeLa cell lysate served as control for IP.
Ku70/Ku80 inhibits the helicase activity of RECQL4 without affecting its ATPase activity
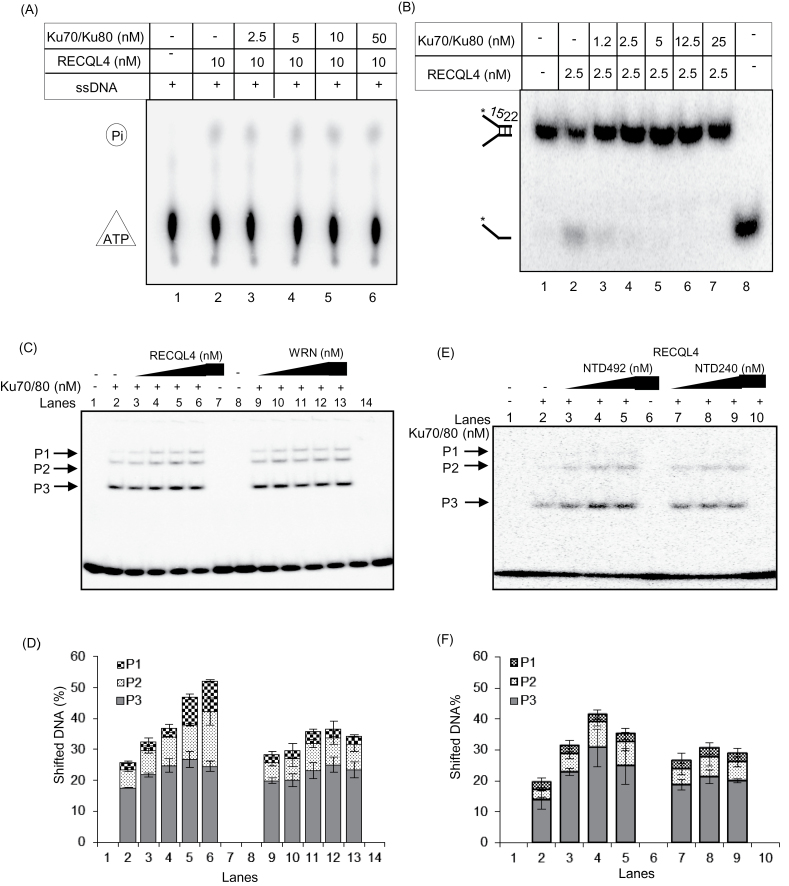
RECQL4 exhibits ATPase and helicase activities, hallmark features of the RecQ helicases (38,43). Therefore, we investigated if the Ku70/Ku80 complex would affect these activities. The ATPase activity was measured using a 60 mer ssDNA oligonucleotide, in the absence or the presence of Ku70/Ku80 heterodimer. The results showed that the Ku70/Ku80 complex does not affect the ATPase activity of RECQL4, in vitro (Figure 5A). We then examined the helicase activity of RECQL4 in the absence or presence of the Ku70/Ku80 heterodimer. RECQL4 has unwinding activity on very short DNA duplexes (38), and we used a short-forked DNA substrate with a 22 mer duplex region in this study. Our results showed that the Ku70/Ku80 heterodimer inhibits the helicase activity of RECQL4 (Figure 5B, lanes 3–7). This inhibition may be due to the strong competitive binding of the Ku70/Ku80 heterodimer to the duplex DNA substrate, thus making the substrate inaccessible to RECQL4.
Fig. 5.
Mutual regulation of RECQL4 and Ku70/Ku80. (A) ATPase activity of RECQL4 in the absence or the presence of Ku70/Ku80 heterodimer. The ATPase assay was performed in the presence of 60 mer ssDNA and indicated proteins. (B) Helicase activity of RECQL4 in the absence or the presence of Ku70/Ku80 heterodimer. The helicase assay was performed with RECQL4 protein on 22 mer forked duplex DNA substrate in the absence (lane 2) or the presence of increasing concentrations of Ku70/Ku80 heterodimer (lanes 3–7). Lane 1, labeled substrate; lane 8, denatured substrate. (C) RECQL4 enhances higher order binding of Ku70/Ku80 complex to the blunt end dsDNA substrate. The Ku70/Ku80 heterodimer (2.5 nM) was incubated with radio-labeled 80 nt double-stranded oligonucleotide, in the absence or the presence of increasing concentrations (2.5, 5, 10 and 20 nM) of either RECQL4 (lanes 3–6) or WRN protein (lanes 10–13). The mobility shift of Ku70/Ku80 binding to DNA substrate alone is shown (lanes 2 and 9). The mobility shifts by RECQL4 and WRN binding to DNA substrate are shown in lanes 7 and 14, respectively. The different DNA binding complexes formed by Ku70/Ku80 heterodimer is marked as P1, P2 and P3. (D) Graph showing the quantification of various protein-DNA complexes observed in electrophoretic mobility shift assay (EMSA) shown in panel C. The error bars represent the standard deviation of three independent experiments. (E) N-terminal domain of RECQL4 stimulates in vitro DNA binding of Ku. EMSA was performed as in Panel C with 2.5, 5 and 20 nM of NTD492 and NTD240. NTD492, N-terminal domain 1–492; NTD240, N-terminal domain 1–240. (F) Graph showing P1, P2 and P3 complexes. Error bars represent standard deviation of three independent experiments.
RECQL4 enhances higher order binding of the Ku70/Ku80 complex to a blunt end double-stranded DNA substrate
The initial step of NHEJ is the binding of the Ku70/Ku80 heterodimer to the ends of the broken DNA, followed by the recruitment of DNA-PKcs, forming the DNA-PK complex, and downstream processing. Upon binding to DNA, the Ku70/Ku80 heterodimer forms higher order protein-DNA complexes, which migrate distinctly in an electrophoretic mobility shift assay (44). We investigated if RECQL4 had any effect on the DNA binding ability of the Ku70/Ku80 heterodimer. Electrophoretic mobility shift assay was performed with the Ku70/Ku80 heterodimer protein using an 80 nt blunt end dsDNA substrate in the absence or presence of RECQL4 or WRN proteins. WRN was used in the assay because we previously reported an interaction between Ku and WRN (45) and because WRN plays a role in the DNA repair processes. The Ku70/Ku80 heterodimer itself formed three different protein-DNA complexes with the substrate, marked as P1, P2 and P3 (Figure 5C, lanes 2 and 9). In the presence of RECQL4, the association of the Ku70/Ku80 heterodimer in these complexes was higher (Figure 5C and D, lanes 3–6). The total DNA binding of Ku70/Ku80 in all three complexes taken together (combined P1, P2 and P3) was enhanced up to ~2-fold in the presence of 8:1 molar ratio of RECQL4 to Ku70/Ku80 (Figure 5C and D, compare lane 6 with lane 2). The most significant stimulatory effect was observed with higher order complexes P1 and P2 (Figure 5D, compare P1 and P2, lane 2 and lane 6), whereas levels of P3 remained similar. For P1 and P2 complexes, the stimulatory effect was approximately 5.5- and 4-fold, respectively (Figure 5D, compare lane 2 with lane 6). However, RECQL4 alone could not form any detectable protein-DNA complexes under our experimental conditions, probably because of its weak ability to bind blunt ended dsDNA (Figure 5C, lane 7). Since WRN also interacts with Ku70/Ku80, we asked if WRN had a similar effect on the DNA binding ability of the Ku complex. However, WRN did not significantly affect the higher order binding ability of the Ku70/Ku80 heterodimer to dsDNA (Figure 5C and D, lanes 9–13).
Our co-immunoprecipitation results shown in Figure 4D indicate that N-terminal region of RECQL4 mediates the interaction between RECQL4 and the Ku70/Ku80 complex. To test the stimulatory effect of the N-terminal domain of RECQL4 on the formation of Ku-DNA complexes, electrophoretic mobility shift assay was performed with two fragments of the N-terminal domain (NTD492 and NTD240) (Figure 5E). Formation of slow migrating P2 and P3, Ku-DNA complexes, were stimulated in the presence of the complete N-terminal domain (NTD492) (Figure 5E and F) similarly to the full length RECQL4 (Figure 5C). The intensity of P2 and P3 increased with increasing amounts of NTD492. This stimulation was not observed when Ku and DNA were co-incubated with the N-terminal domain contained 1–240 aa (NTD240). Together, these results suggest that RECQL4 specifically stimulates the DNA binding activity of the Ku70/Ku80 heterodimer which might have an effect on NHEJ.
Discussion
RecQ helicases play important roles in many aspects of DNA metabolism including DNA repair (1,46,47). Studies have shown that RecQ helicases have critical functions at various steps of DSB repair (25). In this study, we sought to investigate the roles of one of the human RecQ helicases, RECQL4, in the NHEJ pathway of DSB repair. Like WRN, BLM and RECQL5, RECQL4 accumulates at the sites of DSB, but has distinct dissociation kinetics compared with the other RecQ helicases (14,48–50). WRN interacts with various NHEJ factors including Ku70/Ku80, DNA-PKcs and the X4L4 complex, which in turn modulate WRN’s exonuclease and helicase functions that are important for NHEJ (26,28,29,48,51). One study reported that RECQL1 is in complex with the Ku heterodimer (30). Thus far studies that demonstrate roles for RecQ helicases in NHEJ have only used in vitro systems; however, here we have studied the functions of RECQL4 in NHEJ both in vitro and in vivo and provide cellular and biochemical support for a role of RECQL4 in NHEJ.
Depletion of RECQL4 reduced the end-joining activity, both in vitro and in vivo. RECQL4 was actively involved in the resolution of non-ligatable DNA end substrates as well as cohesive substrates (Figure 1). Substrates with cohesive ends can be readily joined by simple ligation as contrasted to non-cohesive substrates, which need processing of the ends by nucleases and polymerases before ligation. Both exogenous and endogenous DNA damages generate a variety of broken DNA ends at the DSB, which require processing by nucleases like Artemis and the X-family of DNA polymerases (21). Knockdown of RECQL4 displayed greater defect on repair of non-cohesive DNA substrates suggesting that RECQL4 deficiency inhibits the end processing activities of NHEJ. Structural and functional evidence suggest that WRN also participates in DNA end processing using its unique exonuclease domain (27,29,52,53). We previously reported that WRN, BLM and RECQL4 are recruited to DSBs induced by micro-irradiation (14). RECQL4 enhances the binding of polymerase γ to the DNA and potentiate the exonuclease and polymerase activities of the polymerase (54). It is possible that RECQL4 might be acting in association with other RecQ helicases or with DNA repair DNA polymerases to promote processing of non-ligatable DSBs, and its deficiency leads to an accumulation of unresolved non-ligatable DNA. Likewise, in vivo NHEJ assays in RECQL4 KD cells showed a significant reduction in the NHEJ activity. However, the reduction was not as marked as in the in vitro NHEJ. This could be due to the different assay system or to the cellular environment where various factors might compensate for the loss of RECQL4.
A previous study with Xenopus egg extracts found an indirect interaction between RECQL4 and Ku70 on DSB containing chromatin using chromatin-immunoprecipitation experiments (13). Further, results from co-IP experiments in the absence of EcoRI, which induce DSB on sperm DNA, implied a lack of detectable direct interaction between RECQL4 and the Ku complex. Based on their results, the authors suggested that RECQL4 loads adjacent to the Ku complex on damaged chromatin (13). Our co-IP data indicate the presence of a direct physical interaction between human RECQL4 and the Ku heterodimer. These proteins co-immunoprecipitate with one another and the interaction is not dependent on DNA, suggesting a preformed protein complex, before binding to DNA. The observed differences in RECQL4–Ku interactions in human and Xenopus systems might be a result of several factors including the species differences in the RECQL4 itself. Comparison of human- and Xenopus-RECQL4 protein sequence indicates a significant difference as the proteins are only 52% identical in amino acid composition. Notably, Xenopus RECQL4 contains an additional ~300 aa. Domain mapping studies provided in the present study indicate the importance of the N-terminus of RECQL4 in mediating the interaction with Ku, and combined with our previous results on N-terminus region (363–493 aa)-dependent recruitment of RECQL4 to DSB (14), provide insights into the roles of RECQL4 in NHEJ. The N-terminus of RECQL4 is functionally important in mediating various protein interactions with MCM10, BLM, p300 and others (3). Unlike WRN, which interacts with several NHEJ proteins, RECQL4 is associated specifically with the Ku70/Ku80 heterodimer and not with Artemis, Ligase IV, or XLF. Thus RECQL4 may be involved in the initial steps of DSB repair, supporting our previous finding that RECQL4 stays at the DSB sites for a very short time period (14). We propose that RECQL4 binds to DNA adjacent to the broken ends, facilitating the higher order co-operative binding of Ku70/Ku80 heterodimer at the DNA ends. Ku70/Ku80 DNA-end binding is then followed by the downstream steps of NHEJ, that is, assembly of the DNA-PKcs subunit and X4L4 ligation complex. This hypothesis is endorsed by our biochemical observations that RECQL4 enhances higher order binding of the Ku70/Ku80 heterodimer to DNA (Figure 5D and E). Alternatively, RECQL4 might destabilize DNA ends locally by its helicase activity and help in the recruitment and formation of a higher order DNA repair complex assembly at, or near, the broken ends. Based on our results, we propose a model for the role of RECQL4 in NHEJ (Supplementary Figure 4, available at Carcinogenesis Online). Interaction of RECQL4 with Ku70/Ku80, in the vicinity of DSB generated by IR, enhances the association and stabilization of the Ku70/Ku80 complex on the DNA to initiate an efficient repair pathway. RECQL4 might locally unwind and destabilize the DSB ends, allowing access to other DNA repair proteins and facilitating the establishment of the repair complex at the DSB. Inhibition of RECQL4’s helicase activity by Ku may favor reduction of unwanted extensive unwinding of DNA ends. In the later steps, on DSB with non-cohesive ends, RECQL4 and Ku70/Ku80 might co-ordinate with nucleases like Artemis or WRN and with polymerases to mediate the DNA end processing to generate ligatable ends. Evidence provided here suggests that RECQL4 would act as an accessory factor, rather than an essential factor, by interacting with the Ku70/Ku80 complex, and play a role in the initial steps of NHEJ.
Defects in DNA repair result in chromosome fusions, chromosome aberrations, translocations and mitotic failure promoting cancer (55–57). Dysregulation of RECQL4 has been observed in a variety of tumors (8,10–12) and patients with RTS and RAPA conditions, caused by mutation in RECQL4, show high incidence of chromosome aberrations, and are at elevated risk for osteosarcomas and lymphomas (6,7,58). Furthermore, altered expression levels of RECQL4 and mutations in RECQL4 are being identified in a variety of tumor types. Thus, there is significant interest in RECQL4 as a tumor suppressor and cancer biomarker. Previously, we described RECQL4’s role in base excision repair (59) and recruitment to DSBs but its role in DSB repair was unclear. Here we show that RECQL4 participates in NHEJ via its interaction with Ku, and its deficiency results in defects in DNA repair which could have direct and indirect effects on the development of tumors observed in patients with mutated RECQL4.
Supplementary material
Supplementary Figures 1–4 can be found at http://carcin.oxfordjournals.org/
Funding
Intramural Program of the National Institute on Aging (ZIA AG000726), National Institutes of Health, USA.
Supplementary Material
Acknowledgements
We would like to thank Drs Manikandan Paramasivam and Venkateswarlu Popuri for critical reading of the manuscript. We thank Dr Michael B.Mathews, Rutgers New Jersey Medical School and Dr Vera Gorbunova of University of Rochester for providing plasmids for NHEJ assays. We thank Dr Dik van Gent, Erasmus MC, the Netherlands, for Ku constructs.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations:
- ATM
ataxia telangiectasia mutated
- ATP
adenosine triphosphate
- ATPase
adenosine triphosphatase
- BGS
Baller-Gerold syndrome
- BLM
Bloom
- BSA
bovine serum albumin
- DNA-PK
DNA-dependent protein kinase
- DSB
double-strand breaks
- DTT
dithiothreitol
- EDTA
ethylenediaminetetraacetic acid
- EGFP
enhanced green fluorescent protein
- HR
homologous recombination
- NHEJ
non-homologous end joining
- RAPA
RAPADILINO syndrome
- RTS
Rothmund-Thomson syndrome
- shRNA
short hairpin RNA
- siRNA
small interfering RNA
- WRN
Werner
- XLF
XRCC4 like factor.
References
- 1. Bohr V.A. (2008). Rising from the RecQ-age: the role of human RecQ helicases in genome maintenance. Trends Biochem. Sci., 33, 609–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chu W.K., et al. (2009). RecQ helicases: multifunctional genome caretakers. Nat. Rev. Cancer, 9, 644–654 [DOI] [PubMed] [Google Scholar]
- 3. Croteau D.L., et al. (2012). RECQL4 in genomic instability and aging. Trends Genet., 28, 624–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Xu X., et al. (2009). Dual DNA unwinding activities of the Rothmund-Thomson syndrome protein, RECQ4. EMBO J., 28, 568–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Marino F., et al. (2013). Bioinformatic analysis of RecQ4 helicases reveals the presence of a RQC domain and a Zn knuckle. Biophys. Chem., 177–178, 34–39 [DOI] [PubMed] [Google Scholar]
- 6. Siitonen H.A., et al. (2009). The mutation spectrum in RECQL4 diseases. Eur. J. Hum. Genet., 17, 151–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Simon T., et al. (2010). Multiple malignant diseases in a patient with Rothmund-Thomson syndrome with RECQL4 mutations: case report and literature review. Am. J. Med. Genet. A, 152A, 1575–1579 [DOI] [PubMed] [Google Scholar]
- 8. Su Y., et al. (2010). Human RecQL4 helicase plays critical roles in prostate carcinogenesis. Cancer Res., 70, 9207–9217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Maire G., et al. (2009). Recurrent RECQL4 imbalance and increased gene expression levels are associated with structural chromosomal instability in sporadic osteosarcoma. Neoplasia, 11, 260–268, 3p following 268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lao V.V., et al. (2013). Altered RECQ helicase expression in sporadic primary colorectal cancers. Transl. Oncol., 6, 458–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Santarpia L., et al. (2013). DNA repair gene patterns as prognostic and predictive factors in molecular breast cancer subtypes. Oncologist, 18, 1063–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Das M., et al. (2013). Over expression of minichromosome maintenance genes is clinically correlated to cervical carcinogenesis. PLoS ONE, 8, e69607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kumata Y., et al. (2007). Possible involvement of RecQL4 in the repair of double-strand DNA breaks in Xenopus egg extracts. Biochim. Biophys. Acta, 1773, 556–564 [DOI] [PubMed] [Google Scholar]
- 14. Singh D.K., et al. (2010). The involvement of human RECQL4 in DNA double-strand break repair. Aging Cell, 9, 358–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Moraes M.C., et al. (2012). DNA repair mechanisms protect our genome from carcinogenesis. Front. Biosci. (Landmark Ed)., 17, 1362–1388 [DOI] [PubMed] [Google Scholar]
- 16. Moynahan M.E., et al. (2010). Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat. Rev. Mol. Cell Biol., 11, 196–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jensen M.B., et al. (2012). The helicase and ATPase activities of RECQL4 are compromised by mutations reported in three human patients. Aging (Albany NY)., 4, 790–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Croteau D.L., et al. (2012). RAPADILINO RECQL4 mutant protein lacks helicase and ATPase activity. Biochim. Biophys. Acta, 1822, 1727–1734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jackson S.P., et al. (2009). The DNA-damage response in human biology and disease. Nature, 461, 1071–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Krejci L., et al. (2012). Homologous recombination and its regulation. Nucleic Acids Res., 40, 5795–5818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lieber M.R. (2010). The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem., 79, 181–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yano K., et al. (2008). Live cell imaging of XLF and XRCC4 reveals a novel view of protein assembly in the non-homologous end-joining pathway. Cell Cycle, 7, 1321–1325 [DOI] [PubMed] [Google Scholar]
- 23. Dobbs T.A., et al. (2010). A structural model for regulation of NHEJ by DNA-PKcs autophosphorylation. DNA Repair (Amst)., 9, 1307–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Singh D.K., et al. (2009). Roles of RECQ helicases in recombination based DNA repair, genomic stability and aging. Biogerontology, 10, 235–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Singh D.K., et al. (2012). RecQ helicases in DNA double strand break repair and telomere maintenance. Mutat. Res., 736, 15–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cooper M.P., et al. (2000). Ku complex interacts with and stimulates the Werner protein. Genes Dev., 14, 907–912 [PMC free article] [PubMed] [Google Scholar]
- 27. Li B., et al. (2001). Requirements for the nucleolytic processing of DNA ends by the Werner syndrome protein–Ku70/80 complex. J. Biol. Chem., 276, 9896–9902 [DOI] [PubMed] [Google Scholar]
- 28. Karmakar P., et al. (2002). Werner protein is a target of DNA-dependent protein kinase in vivo and in vitro, and its catalytic activities are regulated by phosphorylation. J. Biol. Chem., 277, 18291–18302 [DOI] [PubMed] [Google Scholar]
- 29. Kusumoto R., et al. (2008). Werner protein cooperates with the XRCC4-DNA ligase IV complex in end-processing. Biochemistry, 47, 7548–7556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Parvathaneni S., et al. (2013). Human RECQ1 interacts with Ku70/80 and modulates DNA end-joining of double-strand breaks. PLoS ONE, 8, e62481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Petkovic M., et al. (2005). The human Rothmund-Thomson syndrome gene product, RECQL4, localizes to distinct nuclear foci that coincide with proteins involved in the maintenance of genome stability. J. Cell Sci., 118(Pt 18), 4261–4269 [DOI] [PubMed] [Google Scholar]
- 32. Singh D.K., et al. (2012). The human RecQ helicases BLM and RECQL4 cooperate to preserve genome stability. Nucleic Acids Res., 40, 6632–6648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang H., et al. (2003). Biochemical evidence for Ku-independent backup pathways of NHEJ. Nucleic Acids Res., 31, 5377–5388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Buck D., et al. (2006). Cernunnos, a novel nonhomologous end-joining factor, is mutated in human immunodeficiency with microcephaly. Cell, 124, 287–299 [DOI] [PubMed] [Google Scholar]
- 35. Shamanna R.A., et al. (2011). The NF90/NF45 complex participates in DNA break repair via nonhomologous end joining. Mol. Cell. Biol., 31, 4832–4843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Seluanov A., et al. (2004). DNA end joining becomes less efficient and more error-prone during cellular senescence. Proc. Natl Acad. Sci. USA, 101, 7624–7629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Machwe A., et al. (2005). RecQ family members combine strand pairing and unwinding activities to catalyze strand exchange. J. Biol. Chem., 280, 23397–23407 [DOI] [PubMed] [Google Scholar]
- 38. Rossi M.L., et al. (2010). Conserved helicase domain of human RecQ4 is required for strand annealing-independent DNA unwinding. DNA Repair (Amst)., 9, 796–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kitao S., et al. (1998). Cloning of two new human helicase genes of the RecQ family: biological significance of multiple species in higher eukaryotes. Genomics, 54, 443–452 [DOI] [PubMed] [Google Scholar]
- 40. Pastwa E., et al. (2009). In vitro non-homologous DNA end joining assays—the 20th anniversary. Int. J. Biochem. Cell Biol., 41, 1254–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schultz L.B., et al. (2000). p53 binding protein 1 (53BP1) is an early participant in the cellular response to DNA double-strand breaks. J. Cell Biol., 151, 1381–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lai J.S., et al. (1992). Ethidium bromide provides a simple tool for identifying genuine DNA-independent protein associations. Proc. Natl Acad. Sci. USA, 89, 6958–6962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Macris M.A., et al. (2006). Biochemical characterization of the RECQ4 protein, mutated in Rothmund-Thomson syndrome. DNA Repair (Amst)., 5, 172–180 [DOI] [PubMed] [Google Scholar]
- 44. Wang J., et al. (1998). Identification of two domains of the p70 Ku protein mediating dimerization with p80 and DNA binding. J. Biol. Chem., 273, 842–848 [DOI] [PubMed] [Google Scholar]
- 45. Karmakar P., et al. (2002). Ku heterodimer binds to both ends of the Werner protein and functional interaction occurs at the Werner N-terminus. Nucleic Acids Res., 30, 3583–3591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bernstein K.A., et al. (2010). The RecQ DNA helicases in DNA repair. Annu. Rev. Genet., 44, 393–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Larsen N.B., et al. (2013). RecQ Helicases: conserved guardians of genomic integrity. Adv. Exp. Med. Biol., 767, 161–184 [DOI] [PubMed] [Google Scholar]
- 48. Lan L., et al. (2005). Accumulation of Werner protein at DNA double-strand breaks in human cells. J. Cell Sci., 118(Pt 18), 4153–4162 [DOI] [PubMed] [Google Scholar]
- 49. Karmakar P., et al. (2006). BLM is an early responder to DNA double-strand breaks. Biochem. Biophys. Res. Commun., 348, 62–69 [DOI] [PubMed] [Google Scholar]
- 50. Popuri V., et al. (2012). Recruitment and retention dynamics of RECQL5 at DNA double strand break sites. DNA Repair (Amst)., 11, 624–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Li B., et al. (2000). Functional interaction between Ku and the Werner syndrome protein in DNA end processing. J. Biol. Chem., 275, 28349–28352 [DOI] [PubMed] [Google Scholar]
- 52. Perry J.J., et al. (2006). WRN exonuclease structure and molecular mechanism imply an editing role in DNA end processing. Nat. Struct. Mol. Biol., 13, 414–422 [DOI] [PubMed] [Google Scholar]
- 53. Orren D.K., et al. (2001). A functional interaction of Ku with Werner exonuclease facilitates digestion of damaged DNA. Nucleic Acids Res., 29, 1926–1934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Gupta S., et al. (2014). RECQL4 and p53 potentiate the activity of polymerase γ and maintain the integrity of the human mitochondrial genome. Carcinogenesis, 35, 34–45 [DOI] [PubMed] [Google Scholar]
- 55. Difilippantonio M.J., et al. (2000). DNA repair protein Ku80 suppresses chromosomal aberrations and malignant transformation. Nature, 404, 510–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hayashi M.T., et al. (2013). DNA damage associated with mitosis and cytokinesis failure. Oncogene, 32, 4593–4601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Bunting S.F., et al. (2013). End-joining, translocations and cancer. Nat. Rev. Cancer, 13, 443–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Larizza L., et al. (2006). Rothmund-Thomson syndrome and RECQL4 defect: splitting and lumping. Cancer Lett., 232, 107–120 [DOI] [PubMed] [Google Scholar]
- 59. Schurman S.H., et al. (2009). Direct and indirect roles of RECQL4 in modulating base excision repair capacity. Hum. Mol. Genet., 18, 3470–3483 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.