Summary
The first study to demonstrate HS to affect GEP binding on the cell surface.
Abstract
Granulin-epithelin precursor (GEP) is a pluripotent secretory growth factor which promotes cancer progression in a number of human cancers. However, how cancer cells interact with GEP remains unknown. In this study, we aimed to identify the cell surface-binding partner of GEP on liver cancer cells. Human recombinant GEP (rGEP) was expressed and purified to homogeneity. The rGEP was shown to trigger phosphorylation of AKT and ERK1/2 in liver cancer cells. We demonstrated cell surface attachment of rGEP, which was blocked by prebinding of platelet-derived growth factor-AA, platelet-derived growth factor-BB and fibroblast growth factor-2. Therefore, heparan sulfate (HS) had been reasoned as the binding partner of rGEP. Heparinase digestion validated the role of HS on supporting the attachment. The heparin-binding domain of GEP was mapped to RRH(555-557) in the C-terminal region. Suppression of the HS polymerase exostosin-1 reduced the rGEP binding and rGEP-mediated signaling transduction. Suppression of a specific HS proteoglycan, glypican-3, also showed a partial reduction of rGEP binding and an inhibition on rGEP-mediated activation of AKT. Furthermore, glypican-3 was shown to correlate with the expressions of GEP in clinical samples (Spearman’s ρ = 0.363, P = 0.001). This study identified HS, partly through glypican-3, as a novel binding partner of GEP on the surface of liver cancer cells.
Introduction
Hepatocellular carcinoma (HCC) is a malignant neoplasm of hepatocytes and it accounts for >80% of primary liver cancers (1). This disease ranks as the fifth most common cancer worldwide and is the third leading cancer killer. Because liver cancer is frequently diagnosed at an advanced stage, only ~30–40% of patients are eligible for curative treatment, which includes surgical resection, transplantation or percutaneous ablation (2). Other patients have to receive systemic chemotherapies but the response rates are <20% (3). Moreover, even after surgical resection, the rate of recurrence is relatively high at ~60% (4). Consequently, the 5 year survival rates of HCC patients are <15% in most countries (5). As a major global health problem, the current technologies in early diagnosis and prognosis of HCC are unsatisfactory. A better understanding of the molecular mechanisms of the disease is essential for developing diagnostic method and for seeking alternative or supportive therapies to manage liver malignancy.
Granulin-epithelin precursor (GEP, also named progranulin, acrogranin or PC-derived growth factor) is a secretory growth factor that constitutes of seven and half cysteine rich granulin subunits. While granulin subunits are involved in regulating inflammation (6), its precursor, GEP, plays a wide range of biological roles. It was demonstrated to regulate cancer progression (7), fetal development (8) and tissue repair (9). Besides, mutation at the gene of GEP affects neuron survival and causes frontotemporal dementia (10,11). We previously showed that GEP overexpression was found in >70% of human HCC (12). Besides, GEP was demonstrated as a hepatic oncofetal protein which regulated embryonic stem cell-related signaling molecules (13). Functional studies showed that GEP controlled cell proliferation, invasion, tumorigenicity and chemoresistance of HCC (12,14). In animal model, anti-GEP monoclonal antibody (mAb) was demonstrated to inhibit the growth of established HCC, providing evidences that GEP is a potential therapeutic target for HCC (15). Therefore, it is urgently needed to clarify the role of GEP in HCC by characterizing the downstream mechanisms of GEP overexpression.
Although the signaling transduction mediated by extracellular recombinant GEP (rGEP) has not been characterized in liver cancer cells, studies of other cancer cell types suggested its paracrine effect at triggering several signaling pathways. They include the ERK pathway (16–19), the PI3K cascade (16,18,19) and the focal adhesion signaling cascade (16,17). These rGEP-triggered activations are expected to be regulated by cell surface receptors. Cell surface-binding partners of GEP have not yet been identified from cancer cells but from neuronal cell and chondrocyte as sortilin-1 (20) and tumor necrosis factor (TNF) receptors (21), respectively. Interactions between these molecules and GEP were not shown to regulate cell signaling pathways, although sortilin-1 was shown to mediate endocytosis of extracellular GEP (20). On the other hand, in the presence of CpG-ONDs, proteolytic fragments of GEP interact with toll-like receptor 9 in the endolysosomal compartments of toll-like receptor 9-expressing RAW macrophages (22).
In this study, purified rGEP was assayed for its paracrine effect and physical attachment in liver cancer cells. The signaling pathways activated by exogenous rGEP were determined in liver cancer cell lines. The binding of rGEP on the cell surface of liver cancer cells was demonstrated in flow cytometry and characterized by competitive assays. Using heparinase digestion, we concluded that the interaction between GEP and cell surface is mediated by heparan sulfate (HS). The heparin-binding domain of GEP was mapped as in the C-terminal region of the protein, mainly contributed by the RRH(555-557) residues. A HS polymerase, exostosin-1 (EXT1), and one of the HS proteoglycans (HSPGs), glypican-3 (GPC3), were suppressed to validate the role of HS in GEP binding and signaling transduction. Transcript levels of GEP and GPC3 were shown to correlate in clinical samples. This study showed that HS as a novel cell surface-binding partner of GEP, implying a potential role of HS in HCC tumorigenesis.
Materials and methods
Cell lines
Two human liver cancer cell lines, Hep3B and HepG2 (American Type Culture Collection, ATCC, Manassas, VA), were grown in advanced minimum essential medium (AMEM) containing 10% fetal bovine serum (FBS) and l-glutamine supplement at 37°C in 5% CO2. GEP-suppressed Hep3B (Hep3B-sh1) was generated previously (12) and was maintained in 0.4mg/ml G418 in complete medium. Selection drug was withdrawn during assay. The African green monkey cell line COS-1 (ATCC) was cultured in high glucose Dulbecco’s modified Eagle’s medium with 10% FBS at 37°C in 5% CO2. All cell lines were authenticated and characterized by the company. The cells were expanded immediately and multiple aliquots were cryopreserved. Cells were used within 6 months of resuscitation.
Antibodies used for flow cytometry and immunoblotting
Anti-GEP mAb generated against the GEP carboxyl-terminus (15) (Versitech, Hong Kong) was used in rGEP binding neutralization. This antibody was conjugated with fluorescein isothiocyanate (FITC) (ThermoFisher) and was used to detect endogenous GEP in flow cytometry. Anti-HS mAb (10E4 epitope; USBiological), mouse IgM isotypic control (eBiosciences), anti-mouse IgG and anti-rabbit IgG secondary antibodies (Dako, Carpinteria, CA) and FITC-anti-mouse IgG/IgM (BD Biosciences) were purchased. Alexa Fluor® 647 ERK1/2 (pT202/pY204), Alexa Fluor® 647 AKT (pS473) and Alexa Fluor® 647 mouse IgG1κ isotype control were purchased from BD Biosciences. Anti-His(C-term)-FITC antibody and unconjugated anti-His (C-term) antibody are from Life Technologies. Anti-GPC3 antibody (1G12) was obtained from Santa Cruz for detection in immunoblotting while anti-GPC3 antibody from R&D Systems was used in flow cytometry.
Recombinant growth factors used in competitive binding assay
Recombinant human growth factors were used for competitive binding assay. Hepatocyte growth factor (HGF), epidermal growth factor (EGF) and TNFα were purchased from Sigma-Aldrich. Vascular endothelial growth factor-165 (VEGF-165) is from R&D Systems. Platelet-derived growth factor-AA (PDGF-AA) and PDGF-BB were purchased from Cell Signaling Technology. Fibroblast growth factor-1 (FGF-1) and FGF-2 were purchased from Life Technologies.
rGEP and GEP derivatives
The coding sequences of human GEP (aa 14–593), granulin A (aa 281–336), granulin B (aa 206–261), granulin E (aa 518–573) and four GEP truncated mutants, N492 (aa 14–492), C101 (aa 493–593), C77 (aa 517–593) and C51 (aa 543–593), were cloned upstream of a HIS-tag in an expression vector pSecTag2/Hygro (Life Technologies) through HindIII and BamHI restriction sites. Three GEP derivatives with the alanine mutations at RRH(555-557)→3A, RR(574-575)→2A, or both of them (5A), were generated by site directed mutagenesis and were cloned to pSecTag2/Hygro through the same restriction sites. The plasmids were transfected to COS-1 using Lipofectamine (Life Technologies) according to the manufacturer’s instructions. The transfected cells were cultured under the selection of 150 µg/ml hygromycin (Life Technologies). After selection, the cells were grown in 1% FBS and the secreted recombinant proteins were collected from culture medium. Recombinant proteins were purified by nickel gel chromatography and quantified by DC protein assay (Bio-Rad Laboratories).
rGEP cross-linking
Purified recombinant protein was incubated with different amounts of heparin at room temperature for 20 min and was cross-linked by incubation with 1mM disuccinimidyl suberate (DSS, Pierce) at room temperature for another 20 min. The protein was then denatured and separated in sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE). The protein was detected by anti-His antibody in immunoblotting.
SDS–PAGE and immunoblotting
Indicated amounts of proteins were separated in SDS–PAGE gel in denatured condition. Proteins were electro-transferred onto polyvinylidene fluoride membranes. The membrane was blocked by 5% skim milk and subsequently incubated overnight at 4°C with corresponding primary antibodies. Detection was performed using horseradish peroxidase-labeled secondary antibodies with enhanced chemiluminescence (GE healthcare Life Sciences). For deglycosylation, purified recombinant protein was digested with PNGase F (New England Biolabs) at 37°C for 1 h after denaturation to remove the N-linked glycan. Proteins were applied to the SDS–PAGE after denaturing.
Cell surface binding assay
Cells were detached by 5mM ethylenediaminetetraacetic acid (EDTA) in phosphate-buffered saline (PBS) after culturing in complete medium. After washing with blocking buffer (1% bovine serum albumin, 0.03% NaN3 in PBS), detached cells were incubated with purified rGEP, the truncated mutants or the derivatives on ice for 40 min. The recombinant proteins bound on the cell surface were detected by anti-His(C-term)-FITC antibody and measured by flow cytometry. In the competitive binding assay, cells were incubated with various recombinant growth factors on ice for 40 min and washed before the incubation of rGEP protein. In heparin displacement binding assay, cells were incubated with rGEP, and then with indicated amount of heparin on ice for 15 min, followed by anti-His antibody detection.
Cell signaling assay
Cells were seeded and cultured in complete medium for 24 h and were starved in plain minimal essential medium (MEM) for 48 h. If transfection involved, transfection mix was changed to complete medium at 24 h posttransfection. After 24 h, the cells were starved in plain MEM for 48 h. Cells were incubated with different concentrations of rGEP or EGF for 5 min at 37°C. After detachment by trypsin–EDTA, ~3×105 cells were fixed in 4% formaldehyde at room temperature for 10 min. The fixed cells were permeabilized by 70% methanol/30% PBS on ice for 20 min. After washing with blocking buffer (3% FBS in PBS), cells were stained with Fluor® 647 AKT (pS473) and Alexa Fluor® 647 ERK1/2 (pT202/pY204) antibodies and then analyzed in FACSCalibur flow cytometer (BD Biosciences). For inhibition experiment, cells were incubated with wortmannin (100nM) or U0126 (10 µM) for 1 h at 37°C prior to rGEP stimulation.
Confocal imaging
Cells were seeded on a lysine coated slide and cultured in complete medium. For determining the attachment of exogenous rGEP on cell surface, cells were treated with 10mM NaN3 and 2mM NaF for 10 min at 37°C. Cells were then blocked by 1% bovine serum albumin in PBS and incubated with rGEP at 4°C. The cells were incubated with specific antibody against GEP (Santa Cruz) and then second antibody. Afterwards, the membrane was stained by Texas Red®-X conjugated WGA (Life Technologies). The cells were fixed by 4% formaldehyde and permeabilized by 0.25% saponin. Nuclei were stained by 4′,6-diamidino-2-phenylindole before mounting. Confocal images were captured from Carl Zeiss LSM 700.
Depletion of cell surface HS
Cells cultured in six-well plate were treated with heparinase III (Sigma-Aldrich) or ABC chondroitinase (Sigma-Aldrich) at 37°C for 1 h in lyase buffer (0.5mM MgCl2, 1mM CaCl2, 0.1% glucose, 0.5% bovine serum albumin in PBS, pH 7.2). The treated cells then proceeded to HS detection using HS mAb followed by FITC goat anti-mouse IgG/IgM. Mouse IgM was used as isotypic control. Treated cells were also subjected to rGEP binding assay and cell surface GEP detection.
Heparin sepharose chromatography
Conditioned media containing the rGEP protein, its derivatives, truncated mutants and granulin subunits were collected and 3ml of each was directly applied to 0.1ml heparin sepharose (GE healthcare Life Sciences), which has been equilibrated with Tris buffer (10mM Tris, pH7). After 30 min incubation in room temperature with gentle rotation, sepharose was washed twice with the Tris buffer. Proteins were then eluted with the Tris buffer supplementing with a stepwise gradient of increasing NaCl from 0.125 to 2M. All the washes and eluates were analyzed by SDS–PAGE and immunoblotting followed by anti-His antibody detection.
Clinical samples
Patients underwent curative partial hepatectomy for liver cancer between October 2002 and September 2005 at Queen Mary Hospital, Hong Kong, were recruited with written inform consent. The study protocol was approved by the Institutional Review Board of the University of Hong Kong/Hospital Authority Hong Kong West Cluster. Total RNA was extracted from snap frozen tissue specimens for mRNA expression study using real-time quantitative PCR (12,14). The GPC3 data presented were new data, whereas part of the GEP data had been extracted from the previous reported cohort (14) and the current study had increased the sample size to paralleled datasets on GEP and GPC3.
Suppression of GPC3 and EXT1
The expressions of GPC3 and EXT1 in Hep3B and HepG2 cells were suppressed by transfection of specific siRNAs (Life Technologies). Reverse transfection using Lipofectamine™ 2000 (Life Technologies) was adopted to introduce the siRNA as described (14). Briefly, 5 µg/ml lipofectamine and 300nM siRNA were mixed in 1ml plain medium in each well of a six-well plate. After 15 min incubation, 2 × 105 cells in complete medium were added to the transfection mixture. Cells were then incubated at 37°C for 48 h and harvested for immunoblotting and flow cytometric analyses. Cells were also harvested for obtaining the first strand cNDA by the Cells-to-CT kit (Life Technologies) according to the manufacturer’s instructions.
Real-time quantitative reverse transcription–PCR
The primers and probes used for GPC3 and 18S are Pre-designed TaqMan Assay Reagents purchased from Life Technologies. The primers and probes for GEP were described previously (1). The delta Ct between GPC3 and 18S as well as GEP and 18S were used for comparison of mRNA level in different treatments and in different clinical specimens. The mRNA expression was presented as the relative fold change.
Statistical analyses
Continuous variables were assessed by Spearman correlation, comparison between groups by Student’s t-test or one-way analysis of variance as appropriate and described in the text. P values <0.05 were considered significant. Statistical analyses were performed by SPSS version 16 (SPSS, Chicago, IL).
Results
Expression of rGEP and its biological functions
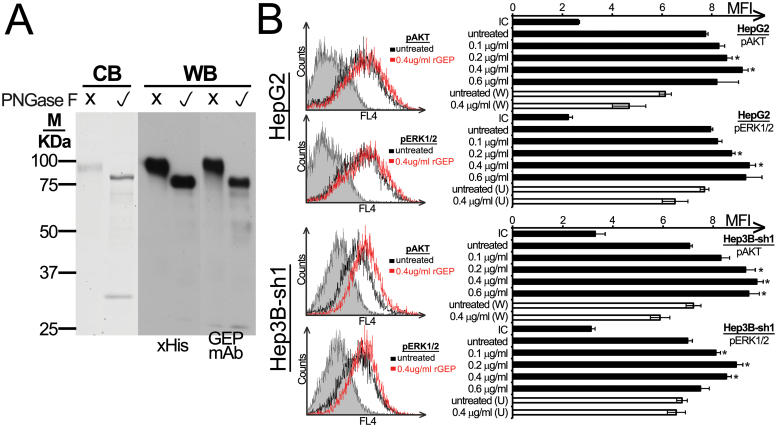
rGEP fused with C-terminal HIS-tag was expressed from a stable clone of pSecTag2/Hygro-rGEP transfected COS-1. Secreted rGEP was collected from the medium and purified to ho mogeneity. Molecular weights of rGEP were estimated to ~90 and 70 kDa, respectively, before and after the cleavage of the N-glycan (Figure 1A; Supplementary Figure S1, available at Carcinogenesis Online). A range of 0.1–0.4 µg/ml rGEP was found to trigger ph osphorylation of AKT and ERK1/2 in HepG2 and Hep3B-sh1 cells dose dependently (Figure 1B; Supplementary Figure S2, available at Carcinogenesis Online). These activations could be counteracted by pretreating the cells with PI3K inhibitor wortmannin and MEK inhibitor U0126 (Figure 1B). The activation of intracellular signaling pathways implicated that the rGEP is biologically functional and these signaling pathways maybe regulated through cell surface-binding partner of GEP.
Fig. 1.
Purity and signaling transduction of rGEP. (A) Purified rGEP was either untreated or deglycosylated by PNGase F and was analyzed in SDS–PAGE. Left panel shows the visualization of Coomassie blue staining (CB), where PNGase F was found at ~30kDa. The middle and right panels show the western blot (WB) analysis after detection of anti-His antibody and GEP mAb, respectively. One microgram of rGEP was used for each lane in Coomassie blue staining, whereas 10ng was used for western blot. (B) HepG2 and the GEP-suppressed Hep3B-sh1 were FBS-starved for 48h and were incubated with rGEP for 5min, followed by trypsin-EDTA detachment, formaldehyde fixation and 70% methanol permeabilization. Specific antibodies against pAKT and pERK1/2 were used for detection. If inhibitors were involved, 100nM wortmannin (W) or 10 µM U0126 (U) was added to the cells 1h in prior to the assay and was withdrawn before rGEP incubation. Representative histograms with isotypic controls (filled area), untreated samples (black line) and cells treated with 0.4 µg/ml rGEP (red line) are shown. The geometric mean fluorescent intensity (MFI) of each sample is shown in the graph. In parallel with rGEP, EGF was used as a positive control for the activation of signaling pathways (data not shown). Asterisks represent significant differences from the control without adding rGEP at 95% level according to Student’s t-test.
rGEP attaches to liver cancer cell surface
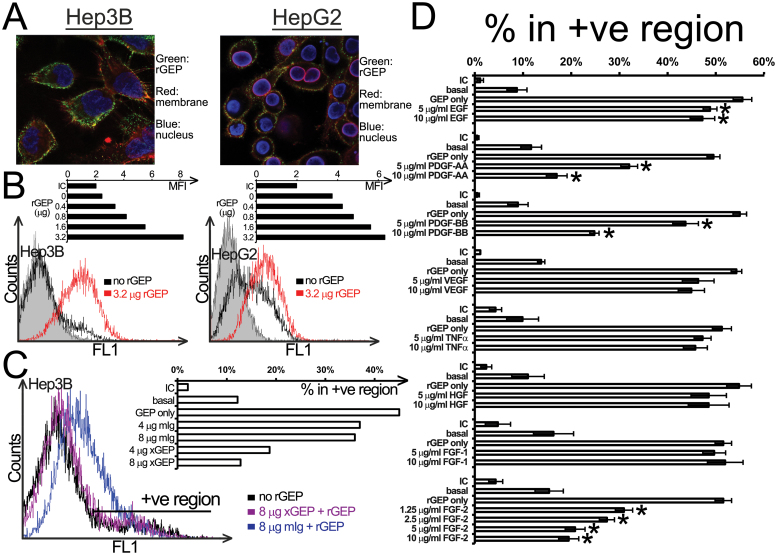
After demonstrating the exogenous rGEP could trigger intracellular signaling pathways in liver cancer cell lines, we reasoned that rGEP might interact with specific binding partner(s) on the cell surface. Therefore, purified rGEP was added to Hep3B and HepG2 and cell surface binding was detected by GEP antibody (Santa Cruz). The confocal images of the cells showed rGEP (green) localized on plasma membrane (red) (Figure 2A). Besides, detached HCC cells were incubated with purified rGEP at 4°C to quantify the rGEP binding by flow cytometry. In Figure 2B, rGEP was demonstrated to bind to both Hep3B and HepG2 from 0.4 to 3.2 µg without achieving plateau. Similar binding effects were demonstrated in Hep3B-sh1 and Huh-7 cells (data not shown). The binding of rGEP could be neutralized by preincubating the protein with GEP mAb, confirming the binding is specific (Figure 2C).
Fig. 2.
Binding properties of rGEP on liver cancer cell lines. (A) Confocal images of rGEP binding on HCC cell surface. Hep3B and HepG2 were incubated with rGEP, specific GEP antibody, anti-rabbit second antibody and Texas Red-conjugated WGA. The cells were then fixed, permeabilized and stained with 4′,6-diamidino-2-phenylindole. (B) Cell surface-binding capacity of Hep3B (left panel) and HepG2 (right panel) for rGEP. Detached cells were incubated with different amounts (0.4–3.2 µg) of rGEP at 4°C and the cell surface binding of rGEP was detected by FITC-anti-His antibody. The histograms show fluorescent intensity of cells and the shaded peaks are isotypic controls. The bar charts show the geometric mean fluorescent intensity (MFI) of different samples. (C) Neutralization of rGEP binding by GEP mAb (×GEP). GEP mAb or mouse IgG were premixed with 0.8 µg rGEP. This rGEP-antibody mixture was added to EDTA-detached Hep3B for binding. The histogram shows the fluorescent intensity of cells bound with pretreated rGEP. The bar chart shows the percentage of cells in the positive region in different samples. Untreated cells detected by isotypic control antibody (IC) and anti-His antibody (basal) are also shown. (D) Competitive binding assay of rGEP. Different growth factors were preincubated with detached Hep3B at 4°C, followed by the binding of 0.8 µg rGEP, detection of anti-His antibody and quantification in flow cytometry. The bar chart shows the percentage of cells in the positive region where the IC and the anti-His basal levels are also shown. The percentages shown are the means and standard deviations obtained from three independent assays. Asterisks represent significant differences from the ‘rGEP only’ at 95% level according to Student’s t-test.
In order to characterize the interaction between rGEP and the cell surface-binding partner(s), eight recombinant human growth factors were used to compete the binding with rGEP. Among which, preincubation of PDGF-AA, PDGF-BB and FGF-2 were found to significantly reduce the binding of rGEP, with the strongest effect exerted by FGF-2 (Figure 2D). Similar results were demonstrated in another HCC cell line Huh-7 (data not shown). These results implied GEP might share common binding partner with these three growth factors.
Cell surface HS is essential for GEP binding
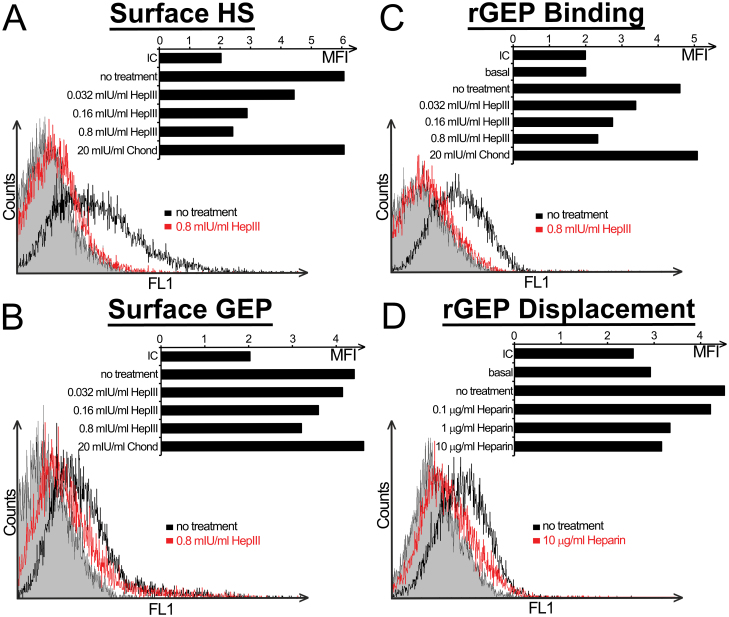
Because PDGF-AA, PDGF-BB and FGF-2 have been shown to interact with cell surface HS, we speculated it as a potential binding partner of rGEP. Different amounts of heparinase III were used to cleave HS from cell surface of liver cancer cell lines to determine its role in the binding of rGEP. Depletion of cell surface HS by the treatment of heparinase III was confirmed in flow cytometry (Figure 3A) and this depletion dramatically reduced the rGEP binding capacity of the cell line (Figure 3B) as well as the expression level of GEP on the cell surface (Figure 3C), implicating a direct interaction between HS and GEP on the HCC cell surface. On the other hand, heparin, a highly sulfated structural analogues of HS (23), was used to displace the bound rGEP from cell surface HS. In Figure 3D, heparin displaces the bound rGEP from the cell surface in a dose-dependent manner. Therefore, these results implicated that the presence of HS is essential for cell surface binding of both rGEP and endogenous secretory GEP.
Fig. 3.
GEP interacts with HS on cell surface of liver cancer cell line. (A–C) Hep3B cells were incubated with the indicated enzymes in lyase buffer at 37°C for an hour. Cells were detached by EDTA and collected for antibody detection and rGEP binding. The bar charts show the geometric mean fluorescent intensity (MFI) of each sample, whereas ‘basal’ represents untreated cells detected by anti-His without the addition of rGEP. Because the basal MFI between treated cells and untreated cells are similar, only that of the untreated cells is shown. The histograms show the peaks of untreated cells and cells treated with 0.8 mIU/ml HepIII, whereas shaded peak represents isotypic control (IC). (A) Cell surface HS of the detached cells was detected by HS mAb. (B) The treated cells were detected by FITC-conjugated GEP mAb to assess the change of cell surface expression of GEP. (C) The detached cells were incubated with 0.8 µg rGEP at 4°C to determine their rGEP binding capacity. The bound rGEP was then detected by anti-His antibody. (D) Hep3B cells were detached and incubated with 0.8 µg rGEP at 4°C. The cells were then incubated with the indicated amounts of heparin on ice for 15min and then washed with blocking buffer for twice while the residual bound rGEP was detected by anti-His antibody. In the histogram, only the IC, untreated cells and the sample washed by 10 µg/ml heparin are shown. Chond: chondroitinase ABC; HepIII: heparinase III; IU: international unit.
Mapping of heparin-binding domain of GEP
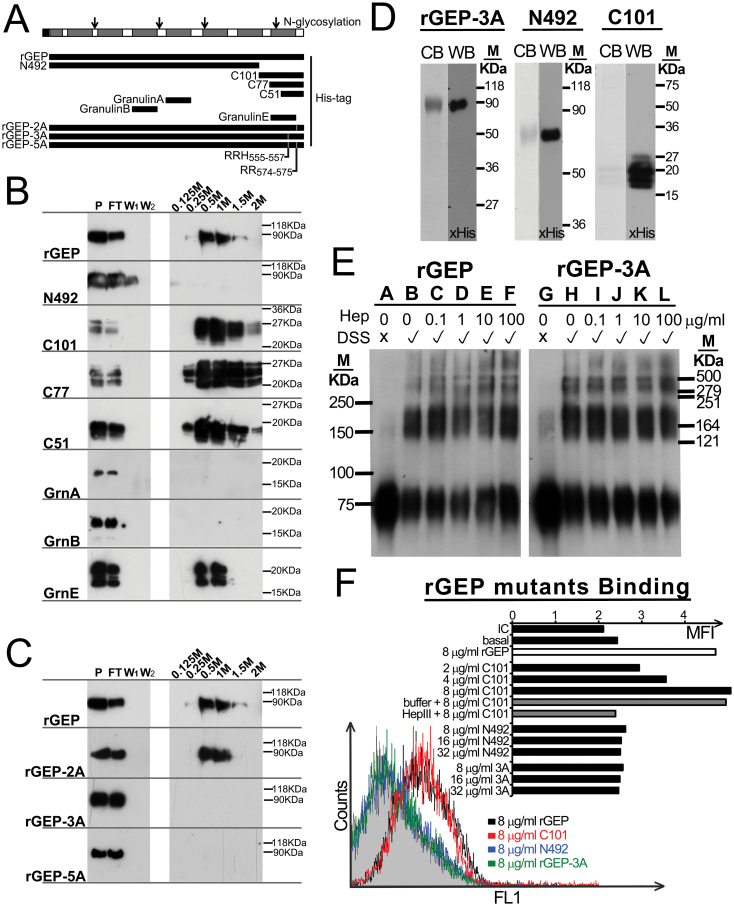
Previous study on heparin-binding domain (24) indicated its constitution of basic residues such as Lys, Arg and His. The C-terminal region of GEP is relatively rich of these basic residues and therefore four truncated mutants of GEP either lacking the C-terminus (N492) or covering different lengths of the C-terminus (C101, C77 and C51) were expressed from stable clones of transfected COS-1 cells (Figure 4A). Heparin sepharose chromatography showed that rGEP, C101, C77 and C51 could bind to the heparin and eluted with 0.5 M sodium chloride, whereas N492 did not bind to the heparin sepharose specifically (Figure 4B). This result showed a direct interaction between heparin and rGEP and implicated the heparin-binding domain of GEP located at the C-terminal 51 residues. From these 51 residues, two regions RRH(555-557) and RR(574-575) were suspected as the amino acids contributing to the binding. Therefore, three rGEP derivatives, rGEP-2A, rGEP-3A and rGEP-5A, were generated by alanine mutation. Our results showed that the alanine mutation of RRH(555-557) alone efficiently abolished the rGEP binding to heparin column, whereas the mutation of RR(574-575) showed no effect (Figure 4C). This indicated the RRH(555-557) is the determining domain in GEP for heparin binding. On the other hand, among the three tested granulin subunits only granulin E bind to the heparin column, implicating a different binding property of the full length GEP from most of the granulin subunits (Figure 4B).
Fig. 4.
Mapping of heparin-binding domain in GEP and the contribution of the heparin-binding domain in HCC cell surface binding. (A) Schematic diagram of GEP protein, rGEP and derivatives and three granulin subunits. The seven and half granulins are shown as grey boxes in the GEP protein. Open arrows represent the N-glycosylation sites identified previously. The locations of rGEP, the deletion mutants (N492, C101, C77 and C51), the derivatives (rGEP-2A, rGEP-3A and rGEP-5A) and three granulin proteins corresponding to the GEP protein are shown. (B and C) Immunoblotting after heparin sepharose chromatography. The rGEP, deletion mutants, derivatives and granulin proteins were collected from media of the transfected COS-1. These media (P) were applied to heparin sepharose for incubation. The flow through (FT) was collected after 30min incubation. The sepharose was washed by Tris buffer for twice (W1 and W2). Elution buffer containing 0.125–2M NaCl was applied sequentially to the sepharose. The recombinant proteins were then detected by anti-His antibody in the immunoblotting. (D) Purified rGEP-3A and rGEP derivatives N492 and C101 were analyzed in SDS–PAGE. Protein was stained by Coomassie blue (CB) or detected by anti-His antibody in western blot (WB). (E) Polymerization of rGEP is independent of heparin and heparin-binding domain. Different amounts of heparin (0–100 µg/ml) were incubated with the recombinant protein at room temperature with or without DSS cross-linking. The polymerization status of rGEP (lanes A–F) and rGEP-3A (lanes G-L) was assessed by SDS–PAGE analysis and immunoblotting detected by anti-His antibody. (F) Indicated amounts of purified rGEP, rGEP-3A, N492 and C101 were incubated with detached Hep3B to compare their binding ability. The binding was then detected by FITC-anti-His antibody. In the case of HS depletion, cells were incubated with heparinase III in lyase buffer at 37°C for an hour before cell detachment. The histogram shows the fluorescent signal of the cells when incubate with 0.8 µg rGEP, N492 and C101. The shaded area represents isotypic control. The bar chart shows the geometric mean fluorescent intensity (MFI) of the cells when incubated with different amount of proteins.
C-terminal region of GEP is essential for HS binding
The GEP mutants rGEP-3A, N492 and C101 were purified (Figure 4D). Both wild-type rGEP and rGEP-3A were assayed for their polymerization. From Figure 4E, both of them showed polymeric structures although portions of them remain their monomeric structures. This result concurs with a recent finding of plasma GEP and rGEP being a homodimer (25). Because oligomerization of HGF is mediated by fragmented HS or heparin (26), polymerization of rGEP was also investigated. However, the tendency of polymeric structures of wild-type and mutant GEP is the same and the addition of heparin showed no effect in the polymer formation. Therefore, the heparin-binding domain and heparin may not involve in the polymeric structure of GEP.
Purified rGEP-3A, N492 and C101 were analyzed for their cell surface-binding ability on Hep3B cells. Flow cytometry showed C101 bound on the cell surface of Hep3B, whereas rGEP-3A and N492 did not. When the HS was depleted by heparinase III from Hep3B, the binding of C101 reduced (Figure 4F). This result implicated the C-terminal region of GEP, contributed by RRH(555-557), is responsible for cell surface binding and this binding is mediated by HS.
Correlation of GEP and GPC3 in clinical specimens
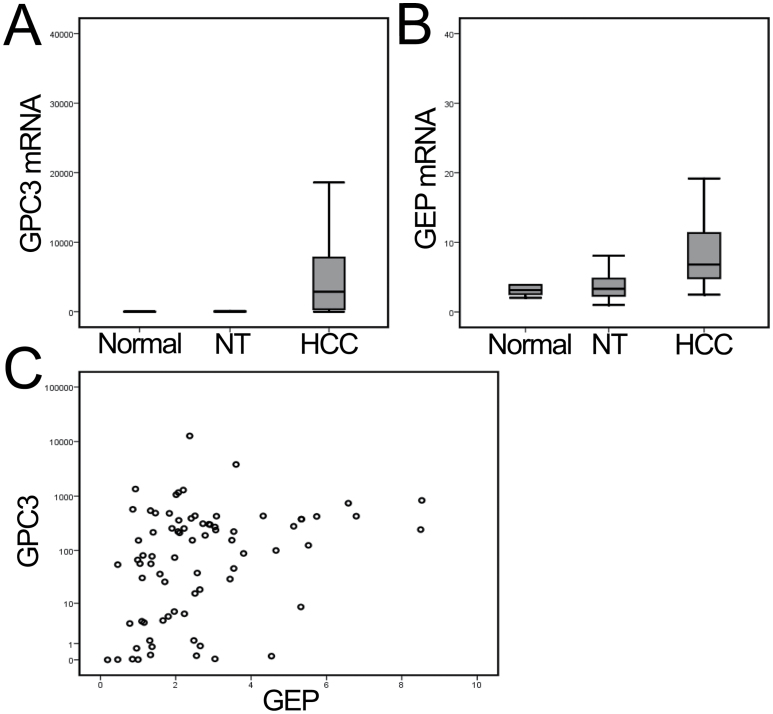
GPC3 is one of the HSPGs located on the cell surface and was found upregulated in HCC tumors (27). The mRNA expression levels of GPC3 were investigated in 77 pairs of HCC and adjacent non-tumor liver tissues, as well as 10 normal liver tissues. Elevated GPC3 transcript levels were observed in tumor compared with non-tumor tissues and normal liver tissues (P < 0.001) (Figure 5A). The GEP and GPC3 levels were significantly correlated (n = 77, Spearman’s ρ correlation coefficient = 0.363, P = 0.001) (Figure 5B), corroborating their biological functions as binding partners.
Fig. 5.
Overexpression of GPC3 and GEP mRNA levels in liver cancer. (A and B) Elevated expressions of GPC3 and GEP were observed in liver cancer (HCC) compared with the paralleled adjacent non-tumor liver tissues (NT) and with normal liver tissue from liver donors (P < 0.001, by one-way analysis of variance). (C) GPC3 and GEP transcript levels (HCC versus NT ratio) were significantly correlated (Spearman’s ρ correlation coefficient = 0.363, P = 0.001).
Role of HSPGs on rGEP binding and signaling induction
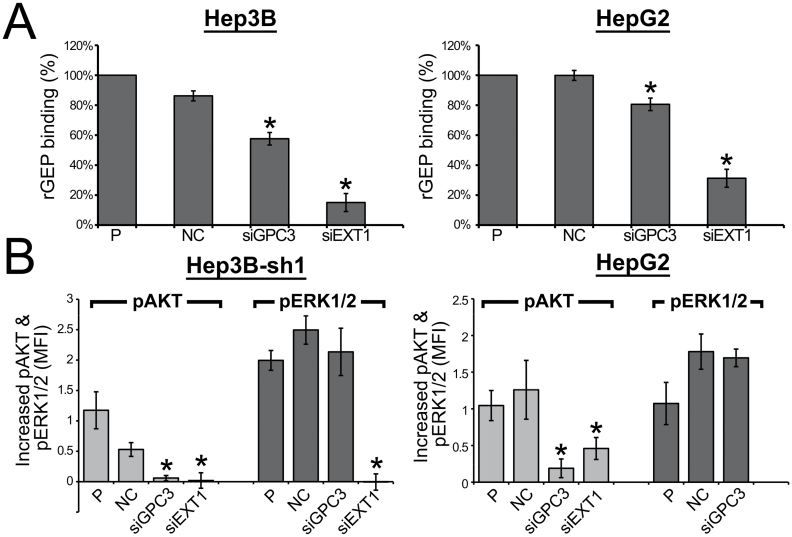
The polymerase of HS, EXT1, and a liver cancer marker, GPC3, were suppressed in two liver cancer cell lines to determine the contribution of HS on rGEP binding and rGEP-mediated signaling transduction. The suppression and binding properties of parental Hep3B and Hep3B-sh1 are very similar and so only that of parental Hep3B were shown. After transfection of specific siRNA, mRNA level of GPC3 of the two cell lines was shown to decrease (Supplementary Figure S3A, available at Carcinogenesis Online). The reduction of protein level of GPC3 was demonstrated in immunoblotting (Supplementary Figure S3B, available at Carcinogenesis Online) and flow cytometry (Supplementary Figure S3C, available at Carcinogenesis Online). Suppression of EXT1 mRNA level and decrease of cell surface HS after EXT1 siRNA transfection are shown in Supplementary Figure S4, available at Carcinogenesis Online. These GPC3- and EXT1-depleted cells were demonstrated to have reduced capacity for rGEP binding (Figure 6A). Notably, the reductions of binding on the GPC3-suppressed cells of ~20–30% compared with the negative control (NC) indicated that other cell surface HSPGs might also interact with GEP. The residual binding of rGEP on EXT1-suppressed cells may reflect the presence of surface HS before knockdown took place while turnover rate of surface HSPGs varies. The decreased rGEP binding shown in NC-treated Hep3B maybe due to cell line-specific effect of lipofectamine, which decrease the amount of HS (Supplementary Figures S3 and S4, available at Carcinogenesis Online). Suppression of EXT1 in Hep3B-sh1 inhibited the rGEP-mediated activation of AKT and ERK1/2, whereas suppression of GPC3 inhibited the AKT activation (Figure 6B). These results suggested that GEP-HS interaction on HCC cell surface plays a determining role on rGEP-mediated signaling transduction in HCC, whereas GPC3 may mainly contribute to rGEP-mediated activation of AKT. In general, siRNA transfection did not affect the basal pAKT or pERK1/2 levels. However, in the transfection of siEXT1 to HepG2 cells, upregulation of pERK1/2 was observed compared with siRNA NC and parental (P) cells in the absence of rGEP (data not shown). This might indicate additional roles of EXT1 in HepG2 MAPK signaling cassette and interfere with rGEP-mediated signaling transduction. Therefore, rGEP-mediated ERK1/2 activation in siEXT1 HepG2 was not shown in Figure 6B.
Fig. 6.
rGEP binding and rGEP-mediated signaling transduction in HCC cells after suppression of GPC3 and EXT1. Cells were transfected with NC siRNA and siRNAs targeting GPC3 and EXT1 (HSS104149 and HSS103435, respectively, from Life Technologies). (A) The cells were detached at 48h posttransfection for rGEP binding assay. The detached cells were incubated with 3.2 µg rGEP, followed by detection of anti-His antibody and analysis in flow cytometry. The chart shows the relative percentage of binding by comparing the MFI of each transfectant to the untreated cells (P). The means and the standard deviations were obtained from three independent assays. Asterisks represent significant differences from the ‘NC’ at 95% level according to Student’s t-test. The binding properties of Hep3B and Hep3B-sh1 are the same. (B) Suppression of GPC3 and EXT1 in HCC cell reduced the rGEP-mediated signaling transduction. Two cell lines (HepG2 and GEP-suppressed Hep3B) were transfected with the siRNAs and were treated with or without 0.2 µg/ml rGEP for 5min. Cells were detached, fixed by formaldehyde, permeabilized by 70% methanol and stained with fluorescein-conjugated specific antibody against pAKT and pERK1/2. The graphs show the difference of MFI with and without rGEP induction, reflecting the increased amount of pAKT and pERK1/2 upon rGEP induction in different siRNA treated cells. Asterisks represent significant differences from the ‘NC’ at 95% level according to Student’s t-test.
Discussion
Previously, the autocrine effects of GEP in cancer progression and chemoresistance have been demonstrated by overexpression and suppression of GEP in HCC cells (12,14). However, the direct downstream mechanisms mediated by GEP have not been characterized yet. This study aimed to characterize how liver cancer cells interact with GEP from an extracellular environment, attributed to the secretory nature of GEP. Therefore, rGEP was expressed from mammalian cells and purified to homogeneity. In the first part of the study, this external source of GEP was demonstrated to mediate phosphorylation of ERK1/2 and Akt (Figure 1B; Supplementary Figure S2, available at Carcinogenesis Online). This observation echoes our previous finding that GEP mAb reduced tumor cell proliferation via the ERK1/2 and PI3K pathways (15). The rGEP-mediated activations of these two pathways have not been reported in HCC but in other cancer models (16–19).
We showed that the rGEP could bind on the plasma membrane of liver cancer cells in confocal imaging (Figure 2A). This binding was shown under a relatively native physiological condition where the cells were attached on slides. This cell surface binding was also demonstrated dose dependently by flow cytometry. However, we did not observe a plateau even at the highest concentration of rGEP (3.2 µg), thereby indicating abundant binding sites for GEP on the cell surface (Figure 2B). The neutralization of the rGEP binding by GEP mAb demonstrated the binding is specific. The immunogen of this mAb is a peptide located at the C-terminal region of GEP covering the RRH(555-557) (15), suggesting the neutralizing effect is due to the blocking of the heparin-binding domain of rGEP. The competitive binding assay showed FGF-2, PDGF-AA and PDGF-BB, but not the other tested growth factors, could block the binding of rGEP (Figure 2D). Because HS is the only cell surface component interacts with these three growth factors, we reasoned that the cell surface binding of rGEP might be mediated by HS. To the best of our knowledge, characterization of rGEP binding on cell surface using competitive binding assay has not performed before. Nevertheless, similar study has been performed on the epithelin-1 subunit from rat GEP. Culouscou and coworkers showed that none of the tested growth factors, including FGF-2, inhibits the binding of epithelin-1 onto a human cell line (28), implying rat epithelin-1 may not interact with cell surface HS. This finding coincides with our results that the equivalent human granulin A does not interact with heparin (Figure 4B).
In the competitive binding assay (Figure 2D), only PDGF-AA, PDGF-BB and FGF-2 showed blocking effect to rGEP. However, many of the tested growth factors, including PDGF-AA (29), PDGF-BB (30), FGF-2 (31), HGF (32), VEGF-165 (33), TNFα (34) and FGF-1 (35), were shown to bind to HS. The selective blocking of rGEP by PDGF-AA, PDGF-BB and FGF-2 might be partly due to their different preference of polysaccharide sequences of HS. The polysaccharide chains are modified by various enzymes for sulfation, epimerization and N-acetylation, resulting in different sequence specificities (36). Differential preferences of HS polysaccharides have been observed among HS-binding growth factors (29,37–39). For instance, FGF-2 was shown to prefer 2-O-sulfate group than 6-O-sulfate (37,39). While for PDGF-AA, all the N-, 2-O- and 6-O-sulfate groups were involved in the interaction (29). Therefore, the blocking effect of these three specific growth factors in our study may indicate GEP shares the similar saccharide preference. On the other hand, the weak heparin binding affinity of VEGF-165 of ~10-fold less than FGF-2 in neutral pH (40) could also contribute to the weak blocking ability. Therefore, the blocking effect of PDGF-AA, PDGF-BB and FGF-2 in our study might indicate that these growth factors show stronger HS affinity than GEP.
Because the histamine residues may contribute to the heparin-binding domain (24) and previous report showed that His-tag may provide a positive effect on HS binding (41), purified rGEP with DDDDK-tag (Abcam) was assayed for cell surface binding and heparin sepharose chromatography and showed a very similar result as our rGEP (Supplementary Figure S1, available at Carcinogenesis Online). It is noted that only a small proportion of histamine shows positive charge in neutral pH. Therefore, although the His-tag may affect the HS binding in acidic intracellular compartments, it may not contribute to the specific HS binding of GEP on the cell surface. The heparin-binding domain was mapped to the RRH(555-557) at the C-terminal region of GEP, implying that HS only binds to the full length GEP but not most of the granulin subunits generated from posttranslational proteolytic cleavage. We showed the recombinant proteins of granulin A and B, which have been isolated from human blood (42), were independent from heparin binding (Figure 4B). This differential binding property between GEP and the subunits may contribute to their different and sometimes opposite biological roles observed from other studies (6,43). Nevertheless, the recombinant putative granulin E, which contains the RRH(555-557), was shown to interact with heparin (Figure 4B).
Although we demonstrated that GEP interacted with HS in liver cancer model, HS from other cell types or tissues might not bind GEP due to the HS sequence specificity. In fact, previous studies have indicated that liver HS is more heavily sulfated than HS from other organs (44), implying liver HS is more negatively charged to provide strong attachment to the ligands. Structural analysis of liver HS revealed it with a considerable proportion of highly sulfated heparin-like structure asymmetrically concentrated to the distal part of the polysaccharides (45). Therefore, interaction between extracellular GEP and cell surface HS may be cell type specific. Nevertheless, because HS binding plays an important role in increasing the local concentration of ligands to specific functional compartment on the cell surface, the assisting role of HS in documented GEP interactions on the cell surface, such as sortilin-1 and TNFα binding (20,21), will deserve further investigation.
HS are O-linked polysaccharide chains covalently attached to the core proteins forming HSPGs. Syndecans and GPCs are the two main families of HSPG that could be found on the cell surface (46). GPC3 was shown to be overexpressed in HCC tumor and modulating activities of growth factors including FGF-2 and BMP-7 (27,47). Our demonstration of reduced rGEP binding on GPC3-suppressed HCC cells indicated GPC3 as potential binding partner of GEP (Figure 6A). The associations of GEP and GPC3 in HCCs further supported their functional relationship (Figure 5). These findings implied a potential involvement of GPC3 in GEP regulated tumorigenesis in HCC.
Generally, biological significance of the interaction between growth factors and HS is to provide an extracellular storage of the growth factors and protect them from proteolysis. In particular, HS regulates the signaling pathways of many HS-binding growth factors by presenting the ligand to the receptor and forming complex with the ligand-receptor. Examples include FGF and FGF receptor, HGF and c-MET, VEGF-165 and VEGF receptor, PDGF-BB and PDGF receptor-α and sonic hedgehog and HSPGs [reviewed in (48)]. Although the primary receptor of GEP has not been identified, ligand binding to cell surface HS may yield a higher local ligand concentration to activate signaling receptor (36). In this study, suppression of a HS polymerase EXT1 and a specific HSPG GPC3 could inhibit the rGEP-mediated cell surface binding and signaling transduction (Figure 6), suggesting an important role of HS for GEP-receptor interaction. Knockdown of GPC3 has been reported to inhibit AKT activation in Hep3B but not in other tested HCC cell lines including HepG2, Huh-7 and Huh-4 (49). Coincidently, suppression of GPC3 in this study did not change pAKT in the absence of rGEP and FBS in the two GEP-low cell lines, HepG2- and GEP-suppressed Hep3B (data not shown). Therefore, an assistant role of GPC3 is hypothesized in the rGEP-mediated activation of AKT. Moreover, suppression of GPC3 in Hep3B inhibited cell migration (Supplementary Figure S5, available at Carcinogenesis Online), whereas the PI3K pathway has been shown to support Hep3B migration in previous studies (50).
In summary, rGEP was expressed from mammalian cells and purified to homogeneity. This exogenous rGEP was found to trigger MAPK and PI3K signaling transduction pathways in liver cancer cell lines. We identified HS as a novel cell surface-binding partner of GEP while the heparin-binding domain of GEP was mapped to the RRH(555-557). GEP and GPC3 expressions were significantly correlated in clinical samples. Suppression of GPC3 and EXT1 in liver cancer cell lines reduced the cell surface binding of rGEP and rGEP-mediated signaling transduction. Like other HS-binding growth factors, interaction with HS might be essential to the multiple biological roles of GEP in HCC as well as other physiological mechanisms (Supplementary Figure S6, available at Carcinogenesis Online). It would be critical to further investigate the functional roles of HS in GEP-mediated tumorigenesis.
Supplementary material
Supplementary Figures S1–S6 can be found at http://carcin.oxfordjournals.org/
Funding
Sun C.Y. Research Foundation for Hepatobiliary and Pancreatic Surgery, Hong Kong Research Grants Council (GRF 764111, 764112, HKU7/CRG/09); Health and Medical Research Fund (01121776); Seed Funding Program for Basic Research and Small Project Funding Program of the University of Hong Kong.
Conflict of Interest Statement: The University of Hong Kong has filed patent applications for the described works. The authors P.F.Y.C., S.T.F. and S.T.C. are inventors of these patents. S.T.C. has received Pfizer collaborative research grants. The sponsors had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript. The other authors declare no conflict of interest.
Supplementary Material
Glossary
Abbreviations:
- EDTA
ethylenediaminetetraacetic acid
- EGF
epidermal growth factor
- EXT
exostosin
- FBS
fetal bovine serum
- FGF
fibroblast growth factor
- FITC
fluorescein isothiocyanate
- GEP
granulin-epithelin precursor
- GPC
glypican
- HCC
hepatocellular carcinoma
- HGF
hepatocyte growth factor
- HS
heparan sulfate
- HSPG
heparan sulfate proteoglycan
- NC
negative control
- PBS
phosphate-buffered saline
- PDGF
platelet-derived growth factor
- rGEP
recombinant granulin-epithelin precursor
- SDS–PAGE
sodium dodecyl sulfate–polyacrylamide gel electrophoresis
- TNF
tumor necrosis factor
- VEGF
vascular endothelial growth factor.
References
- 1. El-Serag H.B., et al. (2007). Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology, 132, 2557–2576 [DOI] [PubMed] [Google Scholar]
- 2. Cao H., et al. (2012). Improved chemotherapy for hepatocellular carcinoma. Anticancer Res., 32, 1379–1386 [PubMed] [Google Scholar]
- 3. Yeo W., et al. (2005). A randomized phase III study of doxorubicin versus cisplatin/interferon alpha-2b/doxorubicin/fluorouracil (PIAF) combination chemotherapy for unresectable hepatocellular carcinoma. J. Natl. Cancer Inst., 97, 1532–1538 [DOI] [PubMed] [Google Scholar]
- 4. Fan S.T., et al. (2011). Continuous improvement of survival outcomes of resection of hepatocellular carcinoma: a 20-year experience. Ann. Surg., 253, 745–758 [DOI] [PubMed] [Google Scholar]
- 5. American Cancer Society. (2011) Cancer Facts & Figures 2011. American Cancer Society, Atlanta, GA [Google Scholar]
- 6. Zhu J., et al. (2002). Conversion of proepithelin to epithelins: roles of SLPI and elastase in host defense and wound repair. Cell, 111, 867–878 [DOI] [PubMed] [Google Scholar]
- 7. Bateman A., et al. (2009). The granulin gene family: from cancer to dementia. Bioessays, 31, 1245–1254 [DOI] [PubMed] [Google Scholar]
- 8. Daniel R., et al. (2003). Progranulin (acrogranin/PC cell-derived growth factor/granulin-epithelin precursor) is expressed in the placenta, epidermis, microvasculature, and brain during murine development. Dev. Dyn., 227, 593–599 [DOI] [PubMed] [Google Scholar]
- 9. He Z., et al. (2003). Progranulin is a mediator of the wound response. Nat. Med., 9, 225–229 [DOI] [PubMed] [Google Scholar]
- 10. Baker M., et al. (2006). Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature, 442, 916–919 [DOI] [PubMed] [Google Scholar]
- 11. Cruts M., et al. (2006). Null mutations in progranulin cause ubiquitin-positive frontotemporal dementia linked to chromosome 17q21. Nature, 442, 920–924 [DOI] [PubMed] [Google Scholar]
- 12. Cheung S.T., et al. (2004). Granulin-epithelin precursor overexpression promotes growth and invasion of hepatocellular carcinoma. Clin. Cancer Res., 10, 7629–7636 [DOI] [PubMed] [Google Scholar]
- 13. Cheung P.F., et al. (2011). Granulin-epithelin precursor is an oncofetal protein defining hepatic cancer stem cells. PLoS One, 6, e28246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cheung S.T., et al. (2011). Granulin-epithelin precursor and ATP-dependent binding cassette (ABC)B5 regulate liver cancer cell chemoresistance. Gastroenterology, 140, 344–355 [DOI] [PubMed] [Google Scholar]
- 15. Ho J.C., et al. (2008). Granulin-epithelin precursor as a therapeutic target for hepatocellular carcinoma. Hepatology, 47, 1524–1532 [DOI] [PubMed] [Google Scholar]
- 16. He Z., et al. (2002). Progranulin (PC-cell-derived growth factor/acrogranin) regulates invasion and cell survival. Cancer Res., 62, 5590–5596 [PubMed] [Google Scholar]
- 17. Monami G., et al. (2006). Proepithelin promotes migration and invasion of 5637 bladder cancer cells through the activation of ERK1/2 and the formation of a paxillin/FAK/ERK complex. Cancer Res., 66, 7103–7110 [DOI] [PubMed] [Google Scholar]
- 18. Wang W., et al. (2003). PC cell-derived growth factor (granulin precursor) expression and action in human multiple myeloma. Clin. Cancer Res., 9, 2221–2228 [PubMed] [Google Scholar]
- 19. Zanocco-Marani T., et al. (1999). Biological activities and signaling pathways of the granulin/epithelin precursor. Cancer Res., 59, 5331–5340 [PubMed] [Google Scholar]
- 20. Hu F., et al. (2010). Sortilin-mediated endocytosis determines levels of the frontotemporal dementia protein, progranulin. Neuron, 68, 654–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tang W., et al. (2011). The growth factor progranulin binds to TNF receptors and is therapeutic against inflammatory arthritis in mice. Science, 332, 478–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Park B., et al. (2011). Granulin is a soluble cofactor for toll-like receptor 9 signaling. Immunity, 34, 505–513 [DOI] [PubMed] [Google Scholar]
- 23. Gandhi N.S., et al. (2008). The structure of glycosaminoglycans and their interactions with proteins. Chem. Biol. Drug Des., 72, 455–482 [DOI] [PubMed] [Google Scholar]
- 24. Cardin A.D., et al. (1989). Molecular modeling of protein-glycosaminoglycan interactions. Arteriosclerosis, 9, 21–32 [DOI] [PubMed] [Google Scholar]
- 25. Nguyen A.D., et al. (2013). Secreted progranulin is a homodimer and is not a component of high density lipoproteins (HDL). J. Biol. Chem., 288, 8627–8635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sakata H., et al. (1997). Heparin binding and oligomerization of hepatocyte growth factor/scatter factor isoforms. Heparan sulfate glycosaminoglycan requirement for Met binding and signaling. J. Biol. Chem., 272, 9457–9463 [DOI] [PubMed] [Google Scholar]
- 27. Capurro M., et al. (2003). Glypican-3: a novel serum and histochemical marker for hepatocellular carcinoma. Gastroenterology, 125, 89–97 [DOI] [PubMed] [Google Scholar]
- 28. Culouscou J.M., et al. (1993). Biochemical analysis of the epithelin receptor. J. Biol. Chem., 268, 10458–10462 [PubMed] [Google Scholar]
- 29. Feyzi E., et al. (1997). Characterization of heparin and heparan sulfate domains binding to the long splice variant of platelet-derived growth factor A chain. J. Biol. Chem., 272, 5518–5524 [DOI] [PubMed] [Google Scholar]
- 30. Schilling D., et al. (1998). Loop III region of platelet-derived growth factor (PDGF) B-chain mediates binding to PDGF receptors and heparin. Biochem. J., 333 (Pt 3), 637–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Thompson L.D., et al. (1994). Energetic characterization of the basic fibroblast growth factor-heparin interaction: identification of the heparin binding domain. Biochemistry, 33, 3831–3840 [DOI] [PubMed] [Google Scholar]
- 32. Lyon M., et al. (1994). Hepatocyte growth factor/scatter factor: a heparan sulphate-binding pleiotropic growth factor. Biochem. Soc. Trans., 22, 365–370 [DOI] [PubMed] [Google Scholar]
- 33. Fairbrother W.J., et al. (1998). Solution structure of the heparin-binding domain of vascular endothelial growth factor. Structure, 6, 637–648 [DOI] [PubMed] [Google Scholar]
- 34. Kenig M., et al. (2008). Identification of the heparin-binding domain of TNF-alpha and its use for efficient TNF-alpha purification by heparin-Sepharose affinity chromatography. J. Chromatogr. B. Analyt. Technol. Biomed. Life Sci., 867, 119–125 [DOI] [PubMed] [Google Scholar]
- 35. Blaber M., et al. (1996). X-ray crystal structure of human acidic fibroblast growth factor. Biochemistry, 35, 2086–2094 [DOI] [PubMed] [Google Scholar]
- 36. Bernfield M., et al. (1999). Functions of cell surface heparan sulfate proteoglycans. Annu. Rev. Biochem., 68, 729–777 [DOI] [PubMed] [Google Scholar]
- 37. Ashikari-Hada S., et al. (2004). Characterization of growth factor-binding structures in heparin/heparan sulfate using an octasaccharide library. J. Biol. Chem., 279, 12346–12354 [DOI] [PubMed] [Google Scholar]
- 38. Ashikari-Hada S, et al. (1995). Characterization of heparan sulfate oligosaccharides that bind to hepatocyte growth factor. J. Biol. Chem., 270, 29586–29593 [DOI] [PubMed] [Google Scholar]
- 39. Jemth P., et al. (2002). Biosynthetic oligosaccharide libraries for identification of protein-binding heparan sulfate motifs. Exploring the structural diversity by screening for fibroblast growth factor (FGF)1 and FGF2 binding. J. Biol. Chem., 277, 30567–30573 [DOI] [PubMed] [Google Scholar]
- 40. Cochran S., et al. (2009). A surface plasmon resonance-based solution affinity assay for heparan sulfate-binding proteins. Glycoconj. J., 26, 577–587 [DOI] [PubMed] [Google Scholar]
- 41. Lacy H.M., et al. (2002). 6xHis promotes binding of a recombinant protein to heparan sulfate. Biotechniques, 32, 254, 256, 258. [DOI] [PubMed] [Google Scholar]
- 42. Bateman A., et al. (1990). Granulins, a novel class of peptide from leukocytes. Biochem. Biophys. Res. Commun., 173, 1161–1168 [DOI] [PubMed] [Google Scholar]
- 43. Tolkatchev D., et al. (2008). Structure dissection of human progranulin identifies well-folded granulin/epithelin modules with unique functional activities. Protein Sci., 17, 711–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nagamine S., et al. (2012). Organ-specific sulfation patterns of heparan sulfate generated by extracellular sulfatases Sulf1 and Sulf2 in mice. J. Biol. Chem., 287, 9579–9590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lyon M., et al. (1994). Liver heparan sulfate structure. A novel molecular design. J. Biol. Chem., 269, 11208–11215 [PubMed] [Google Scholar]
- 46. Sasisekharan R., et al. (2002). Roles of heparan-sulphate glycosaminoglycans in cancer. Nat. Rev. Cancer, 2, 521–528 [DOI] [PubMed] [Google Scholar]
- 47. Midorikawa Y., et al. (2003). Glypican-3, overexpressed in hepatocellular carcinoma, modulates FGF2 and BMP-7 signaling. Int. J. Cancer, 103, 455–465 [DOI] [PubMed] [Google Scholar]
- 48. Zhang L. (2010). Glycosaminoglycan (GAG) biosynthesis and GAG-binding proteins. Prog. Mol. Biol. Transl. Sci., 93, 1–17 [DOI] [PubMed] [Google Scholar]
- 49. Feng M., et al. (2013). Therapeutically targeting glypican-3 via a conformation-specific single-domain antibody in hepatocellular carcinoma. Proc. Natl Acad. Sci. USA, 110, E1083–E1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Nakanishi K., et al. (1999). Hepatocyte growth factor promotes migration of human hepatocellular carcinoma via phosphatidylinositol 3-kinase. Clin. Exp. Metastasis, 17, 507–514 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.