Summary
We show interactive effects of genetic, environmental and physiological factors on genome rearrangements driven by homologous recombination (HR). The glycosylase Aag is a major driver of spontaneous HR. DNA methylation damage and cell proliferation act synergistically on HR in vivo.
Abstract
Cancer susceptibility varies between people, affected by genotoxic exposures, genetic makeup and physiological state. Yet, how these factors interact among each other to define cancer risk is largely unknown. Here, we uncover the interactive effects of genetical, environmental and physiological factors on genome rearrangements driven by homologous recombination (HR). Using FYDR mice to quantify HR-driven rearrangements in pancreas tissue, we show that DNA methylation damage (induced by methylnitrosourea) and cell proliferation (induced by thyroid hormone) each induce HR and together act synergistically to induce HR-driven rearrangements in vivo. These results imply that developmental or regenerative proliferation as well as mitogenic exposures may sensitize tissues to DNA damaging exposures. We exploited mice genetically deficient in alkyl-adenine DNA glycosylase (Aag) to analyse the relative contributions of unrepaired DNA base lesions versus intermediates formed during base excision repair (BER). Remarkably, results show that, in the pancreas, Aag is a major driver of spontaneous HR, indicating that BER intermediates (including abasic sites and single strand breaks) are more recombinogenic than the spontaneous base lesions removed by Aag. Given that mammals have about a dozen DNA glycosylases, these results point to BER as a major source of pressure on the HR pathway in vivo. Taken together, methylation damage, cell proliferation and Aag interact to define the risk of HR-driven sequence rearrangements in vivo. These data identify important sources of sequence changes in a cancer-relevant organ, and advance the effort to identify populations at high-risk for cancer.
Introduction
Cancer is driven by the accumulation of changes in DNA. Damage to DNA, by both endogenous factors and by environmental exposures, can lead to sequence changes in oncogenes and tumor suppressor genes that in turn can result in uncontrolled proliferation and neoplastic transformation (1). Distinct DNA repair pathways have evolved to counter the many kinds of DNA damage, and these pathways have been shown to be essential in protecting against DNA damage-induced carcinogenesis (2). Paradoxically, however, errors made during DNA repair can also create sequence changes that promote cancer (3).
Homologous recombination (HR) is a DNA double-strand break (DSB) repair pathway that is active in the S and G2 phases of the cell cycle (4), and it is the major repair pathway for broken replication forks (5). During HR, DSBs are repaired by copying sequence information from a homologous template, usually the sister chromatid (4). To initiate HR, the DSB is resected to produce a 3′ overhang (4). Subsequently, BRCA2 helps to load RAD51 onto the single-stranded overhang, creating a nucleoprotein filament that is capable of homology searching (4). Sequence information is then copied from the homologous template into the break point to repair the DSB and restore the original sequence. While HR is generally error-free, misalignments between homologous sequences can result in sequence rearrangements such as deletions and insertions, especially at repeated sequences that are abundant in the genome (6). Indeed, HR-driven rearrangements between repeated sequences have been shown to contribute to carcinogenesis (7,8). Further, repair synthesis during HR may increase point mutations (3). Finally, HR between homologous chromosomes can lead to loss of heterozygosity (LOH), a frequent cause of tumor suppressor inactivation. Studies with cultured cells have shown that HR is the underlying cause of LOH 30–70% of the time (9), and this finding was corroborated in clinical studies using tumor samples (10). Thus, virtually all cancers harbor HR-driven LOH events at tumor suppressor loci.
Although the importance of HR in carcinogenesis is increasingly recognized, surprisingly little is known about what actually causes HR in vivo. The role of HR in the repair of broken replication forks (4,5) suggests that proliferating cells are at an increased risk for HR-driven sequence changes. Furthermore, DNA lesions that interrupt replication (e.g. single strand breaks and base damages that block DNA polymerases) are particularly recombinogenic due to their ability to cause replication fork breakdown (11,12). One important DNA repair enzyme with the potential to impact HR is the alkyl-adenine DNA glycosylase (Aag, also called Mpg). Aag removes base lesions that inhibit replication (such as 3-methyladenine), and its yeast paralog Mag1 has been shown to suppress HR induced by 3-methyladenine in vitro (13). Aag removes more than one type of replication blocking lesion (14), making it a candidate for protection against both spontaneous and exposure-induced fork breakdown in mammals in vivo. While Aag substrates can block replication, paradoxically, Aag has also been shown to induce HR in vitro (15). After base removal, the resulting abasic site is cleaved by AP endonuclease to create a single strand break, and the ends are then processed and ligated by downstream enzymes in the base excision repair (BER) pathway (16). Importantly, Aag-induced BER intermediates have the potential to cause replication fork breakdown and thus induce HR (17).
Here, we set out to explore the interplay between Aag, cell proliferation and DNA damage in an effort to integrate our understanding of their individual and combined effects on genomic stability. Using the fluorescent yellow direct repeat (FYDR) mice, the first transgenic mouse model for detection of HR in adult animals (18), we show that DNA methylation damage and cell proliferation interact synergistically to induce HR through replication fork breakdown. Importantly, we show that the synergistic increase in HR in our system is not mediated by O 6-methylguanine lesions but by BER substrates and intermediates. Further, we show that Aag modulates both spontaneous and proliferation-induced HR in vivo. Importantly, these factors vary among people, and their combinatorial effects likely modulate the risk of cancer. Taken together, these studies of a cancer-relevant tissue reveal the underlying mechanisms by which cancer risk factors interact to modulate susceptibility to DSBs and sequence rearrangements in vivo.
Materials and methods
Chemicals
Methylnitrosourea (MNU), bromodeoxyuridine (BrdU), triiodothyronine (T3), soybean trypsin inhibitor and collagenase were purchased from Sigma–Aldrich.
Animals
C57Bl/6 pun FYDR mice (18) were used for all experiments. Aag –/– mice (19) were backcrossed into the C57Bl/6 pun FYDR background for 10 generations. Mice were 5–7 weeks old at the start of experiments, experimental cohorts had a 1:1 male:female ratio. Mice were housed in an AAALAC accredited, specific pathogen free facility under a 12h light/dark cycle and were fed a standard rodent diet (LabDiet RMH 3000, Purina LabDiet) and autoclaved water ad libitum. Litters were split between experimental groups. All animal experiments were conducted according to the Guide for the Care and Use of Laboratory Animals and were approved by the MIT Committee on Animal Care.
T3-induced cell proliferation
Mice were treated with T3 as described in ref. (20). Briefly, mice were fed a diet supplemented with 4 ppm T3 ad libitum. T3 was incorporated into standard rodent diet (LabDiet RMH 3000, Purina LabDiet) by Purina TestDiet, control mice were fed the same diet without T3. To determine the time of peak cell proliferation induced by T3, pancreata were harvested daily after the start of feeding and proliferation was measured by BrdU incorporation detected by flow cytometry. To determine the extent of peak cell proliferation induced by T3, pancreata were harvested at the time of peak proliferation and proliferation was measured by BrdU incorporation detected by flow cytometry, as well as by Ki-67 immunohistochemistry.
BrdU incorporation
Mice were dosed with 75mg/kg BrdU in an intraperitoneal injection. Four hours after receiving BrdU, mice were humanely euthanized and their pancreata were harvested for BrdU detection by antibody labeling and flow cytometry. Pancreata were disaggregated by mechanical chopping and collagenase digestion, and single-cell suspensions were stained with the APC Cell Proliferation Detection Kit (BD Pharmingen) according to the manufacturer’s instructions. Samples were analysed on a FACSCalibur flow cytometer (BD Biosciences) using CellQuest Pro software. On average, 20000 cells were analysed per sample.
Ki-67 immunohistochemistry
Mice were humanely euthanized and pancreata were harvested and fixed in 10% neutral buffered formalin, embedded in paraffin, sectioned at 4 μm and deparaffinized. Heat-induced antigen retrieval was performed using a modified citrate buffer (Dako), and Ki-67 was detected with a rat anti-mouse Ki-67 antibody (Dako) used at a dilution of 1:100 at room temperature for 1h. A biotinylated rabbit anti-rat Ig secondary antibody (Dako) was used at a dilution of 1:100 at room temperature for 20min, and was detected using streptavidin-conjugated peroxidase and DAB. Sections were counter-stained with hematoxylin. For each section, the percentage of Ki-67 positive nuclei was determined in 20 randomly selected images (×20) using custom image analysis software (Visiopharm, Hørsholm, Denmark).
50% lethal dose (LD50) determination
LD50 values for MNU were determined according to ref. (21). Briefly, 6- to 8-week-old mice were given increasing doses of MNU, where one mouse was treated per dose and each dose was 50% greater than the preceding dose. At six doses per experiment, the lowest dose at which a mouse is moribund or dies within 30 days is a good approximation of the LD50 (21). The experiment was repeated two to three times per genotype, and average values are reported.
Induction of methylation damage
Mice were treated with MNU (dissolved in PBS, pH 4) in a single intraperitoneal injection. Wild type and Aag –/– mice were dosed with 25mg/kg MNU, Mgmt –/– mice were dosed with 0.4mg/kg MNU (both doses are ≈20% of the respective LD50 values, Supplementary Table 1). Control mice were dosed with PBS, pH 4. For combined methylation damage and cell proliferation, MNU was given at the peak of T3-induced cell proliferation (after 3 days of feeding for female mice, and after 4 days of feeding for male mice). To determine the timing of DSB formation following T3+MNU treatment, pancreata were harvested at various time points after MNU injection and γH2AX phosphorylation was assessed by Western blotting. To compare DSB formation between mice treated with MNU, T3 and combination treatment, pancreata were harvested 6h after MNU injection and DSB formation was assessed by γH2AX immunofluorescence. To quantify HR induced by methylation damage and increased cell proliferation, pancreata were harvested 3–4 weeks after MNU injection and HR was measured with the FYDR assay.
Western blotting
Flash-frozen pancreata were pulverized in liquid nitrogen, lysed with 2× Laemmli sample buffer and protein concentrations were determined with the RC-DC Protein Assay Kit (Bio-Rad). Samples were separated by SDS–PAGE and transferred to nitrocellulose membranes (Bio-Rad). Membranes were incubated with primary antibodies against pSer139-H2AX (Sigma–Aldrich) at 1:500 dilution and beta-actin (Millipore) at 1:30000 dilution at 4°C overnight. Primary antibodies were detected with an HRP-conjugated secondary antibody (Dako) and the Amersham ECL Prime Detection Reagent (GE Healthcare).
γH2AX and Ki-67 immunofluorescence
Sections (4 μm) of formalin-fixed, paraffin-embedded tissue were deparaffinized and antigen-retrieved using modified citrate buffer (Dako). Sections were incubated with a primary Ki-67 antibody (rabbit polyclonal, Abcam) at a dilution of 1:100 at room temperature for 1h, and a secondary antibody (goat anti-rabbit conjugated with Alexa Fluor® 568, Invitrogen) at a dilution of 1:500 at room temperature for 1h. γH2AX primary antibody (mouse monoclonal, Millipore) was used at a dilution of 1:100 at 4°C for 3h, and was detected with a secondary antibody (goat anti-mouse IgG conjugated with Alexa Fluor® 488, Invitrogen) used at a dilution of 1:500 at room temperature for 1h. Sections were counter-stained with DAPI before imaging. For each section, images of 20 randomly selected image fields were acquired at a magnification of ×40 using ImagePro Plus software (Media Cybernetics). DAPI-stained nuclei were counted using ImagePro Plus. Ki-67 positive nuclei and nuclei containing more than 10 γH2AX foci were counted manually in a blinded fashion.
FYDR assay
In situ imaging.
Freshly harvested pancreata were immediately immersed in ice cold soybean trypsin inhibitor solution (0.01% in PBS). After compressing between glass coverslips separated by 0.5mm spacers, pancreata were imaged on a Nikon 80i epifluorescence microscope (Nikon) with a CCD camera (CoolSNAP EZ, Photometrics) using a ×1 objective at a fixed exposure time (2 s). EYFP was detected in the FITC channel. Multipoint images captured using an automated stage (ProScan II, Prior Scientific) and NIS Elements software (Nikon) were stitched automatically or manually in Adobe Photoshop (Adobe Systems). Brightness and contrast were adjusted identically across treatment groups, and foci were manually counted in a blinded fashion. Areas of pancreata were determined using ImageJ software (NIH) by manually tracing the pancreas outline.
Flow cytometry.
Following imaging, pancreata were disaggregated into single-cell suspensions as described in ref. (18), with minor modifications. Briefly, pancreata were minced with scalpel blades, followed by digestion with collagenase V (2mg/ml in Hanks’ Balanced Salt Solution) for 40min at 37°C. The resulting suspension was gently triturated to increase mechanical separation and filtered through a 70 µm cell strainer (BD Falcon) into an equal volume of media (DMEM F12 HAM with 20% FBS). Cells were collected by centrifugation, resuspended in 350 μl OptiMEM (Invitrogen) and filtered through 35 µm filter caps into flow cytometry tubes (Beckton Dickinson). Samples were analysed on a FACScan cytometer (Beckton Dickinson) using CellQuest Pro software (Beckton Dickinson). On average, 1300000 cells were analysed per sample.
Statistics
Proliferation and DSB indices were compared with Student’s t-test. Numbers of recombinant foci and recombinant cell frequencies do not follow a normal distribution and were compared with a Mann–Whitney U-test. All statistical analyses were two-tailed and were performed in GraphPad Prism, Version 5.02 (GraphPad Software). A P value less than 0.05 was considered statistically significant.
Results
The Aag glycosylase is a major driver of spontaneous HR in vivo
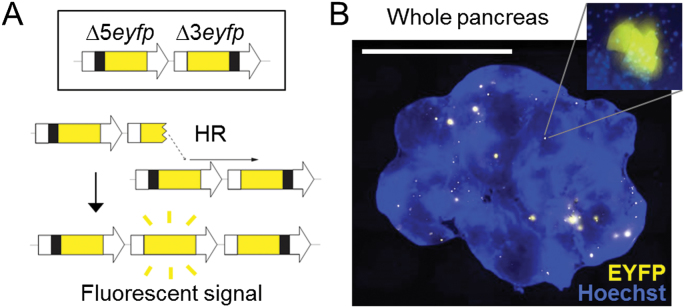
To explore the role of Aag in HR in vivo, we compared HR frequencies between wild type and Aag –/– mice (19) using the FYDR mice, in which two EYFP expression cassettes, each missing essential sequences, have been integrated (18). HR events that reconstitute full-length EYFP coding sequence are detected by fluorescence (Figure 1A). HR frequency in disaggregated tissue can be measured by flow cytometry (18), and furthermore, rare HR events are detectable as distinct fluorescent foci in situ within intact pancreata (Figure 1B).
Fig. 1.
The FYDR mouse detects HR in situ in intact pancreas tissue. (A) Reconstitution of full-length EYFP coding sequence from two truncated copies through replication fork restart by HR. Note that the appearance of fluorescent signal indicates the gain of one repeat unit. Arrows represent expression constructs. EYFP coding sequences are in yellow, promoter and polyadenylation signal sequences are in white, and deleted sequences are in black. Drawing is not to scale. (B) Representative image of a pancreas from a FYDR mouse showing fluorescent foci in situ in intact tissue. Freshly harvested, unfixed whole pancreas was counterstained with Hoechst, compressed to 0.5mm and imaged under a fluorescent microscope. Fluorescence is pseudocolored. Original magnification, ×1. Scale bar = 1cm. Inset: individual focus at ×40 original magnification.
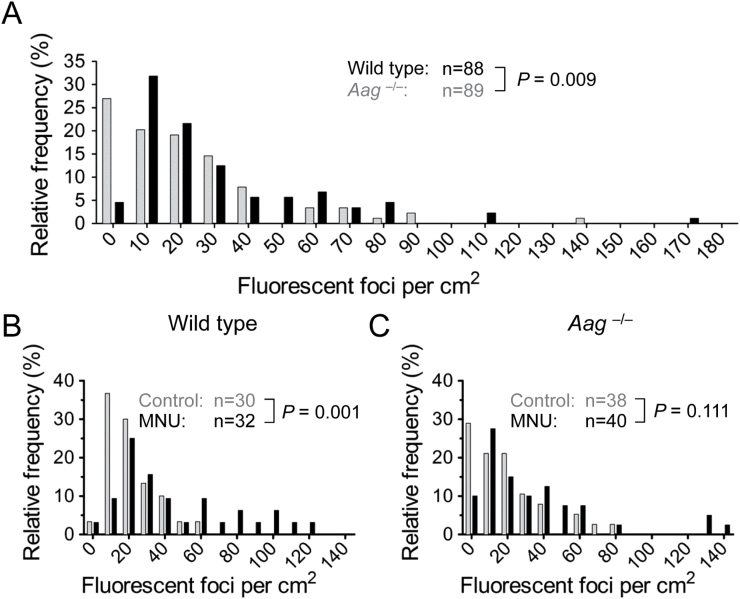
People show a wide range of Aag activities (22–24), and Aag modulates HR in vitro (13). To test the role of Aag on HR in mammals in vivo, we measured HR in wild type and Aag –/– mice by both in situ imaging and flow cytometry. Remarkably, Aag –/– mice had a significantly lower spontaneous frequency of HR (Figure 2A). This result was supported by additional analysis showing that when compared to wild type mice, Aag –/– mice have a reduced frequency of recombinant cells when analysed by flow cytometry (Supplementary Figure 1). As can be seen in Figure 2A, HR frequencies are not normally distributed and display considerable variation. High animal-to-animal variability is well documented in gene-specific mouse mutation assays (25–28), including the FYDR mouse (18). Notably, the inter-animal distribution of DNA base lesion frequencies is surprisingly similar to the distribution of mutation and HR frequencies (29), which points to a potential source of variability. Importantly, statistical analysis of HR data was performed using a two-tailed Mann–Whitney test, a non-parametric test that does not assume a normal distribution and instead compares data in one experimental group to another in a ranks-based fashion (30).
Fig. 2.
Aag glycosylase activity promotes spontaneous and damage-induced HR in vivo. (A) Wild type mice (n = 88, dark bars) have more spontaneous fluorescent foci in their pancreata than Aag –/– mice (n = 89, light bars). The number of fluorescent foci detected in situ per cm2 pancreatic tissue was determined for each animal, and the relative frequencies of mice having the indicated number of foci are shown. (B) Treatment with the methylating agent MNU induces HR in wild type mice. Pancreata were harvested 3–4 weeks after 25mg/kg MNU injection and fluorescent foci were quantified. Frequency distributions of fluorescent foci per cm2 pancreatic tissue in control (n = 30, light bars) and in MNU-treated (n = 32, dark bars) wild type mice are shown. (C) No significant increase in HR in Aag –/– mice treated with MNU (n = 40, dark bars) compared to control (n = 38, light bars). Experimental groups were compared with a two-tailed Mann–Whitney U-test.
The observation that a deficiency in Aag reduces the frequency of HR indicates that Aag-initiated BER is a major driver of spontaneous HR in the pancreas. These results demonstrate that a single DNA glycosylase can have significant impact on HR in vivo. Importantly, mammals have about a dozen different glycosylases (31), calling attention to the BER pathway as a major driver of HR in mammals in vivo.
DNA methylation damage induces HR and is potentiated by Aag
It is well established that exposure to DNA damage can promote cancer (32), however effects are different among people, suggesting the presence of modulating factors (33). Alkylating agents are known to impact cancer risk, and exposure is unavoidable, as they are present in our environment, such as in food, cosmetics and cigarette smoke, as well as being present as cellular metabolites (34,35). The Aag glycosylase removes a wide range of alkylated bases (14). To test the effect of Aag on damage-induced HR in vivo, we exposed wild type and Aag –/– mice to the model alkylating agent MNU. MNU creates the Aag substrate lesions 3-methyladenine and 7-methylguanine, as well as O 6-methylguanine and other minor damage products (35). Most of the MNU-induced lesions are 7-methylguanine, which does not directly inhibit DNA polymerases, but can be converted into recombinogenic BER intermediates (35). In contrast, 3-methyladenine is directly replication blocking, and O 6-methylguanine can block replication when mismatch repair forms gaps during futile cycling (35).
Following MNU exposure, the frequency of fluorescent foci (wherein each focus indicates a single recombination event) was determined by in situ imaging and normalized for tissue area. The proportion of mice with given numbers of foci per cm2 was then plotted, and the resulting frequency distributions were compared between wild type and Aag –/– mice. In wild type mice, the frequency distribution is significantly shifted to the right following MNU exposure (Figure 2B), indicating that MNU induces HR, possibly as a result of either replication-blocking lesions, or their downstream BER intermediates. Interestingly, MNU-induced HR is not apparent in the Aag –/– cohort (Figure 2C). Consistent with these results, we did not detect a significant difference between fluorescent cell frequencies in control and MNU-treated wild type and Aag –/– mice (Supplementary Figure 2).
DNA methylation damage and hormonally induced cell pr oliferation synergize on DSBs in vivo
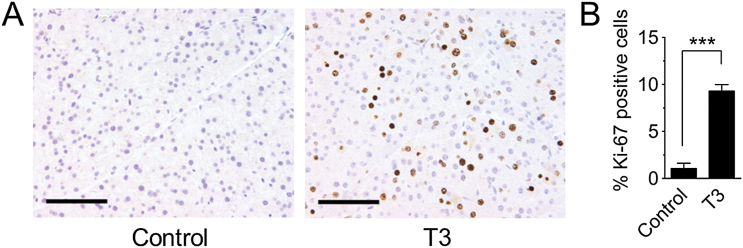
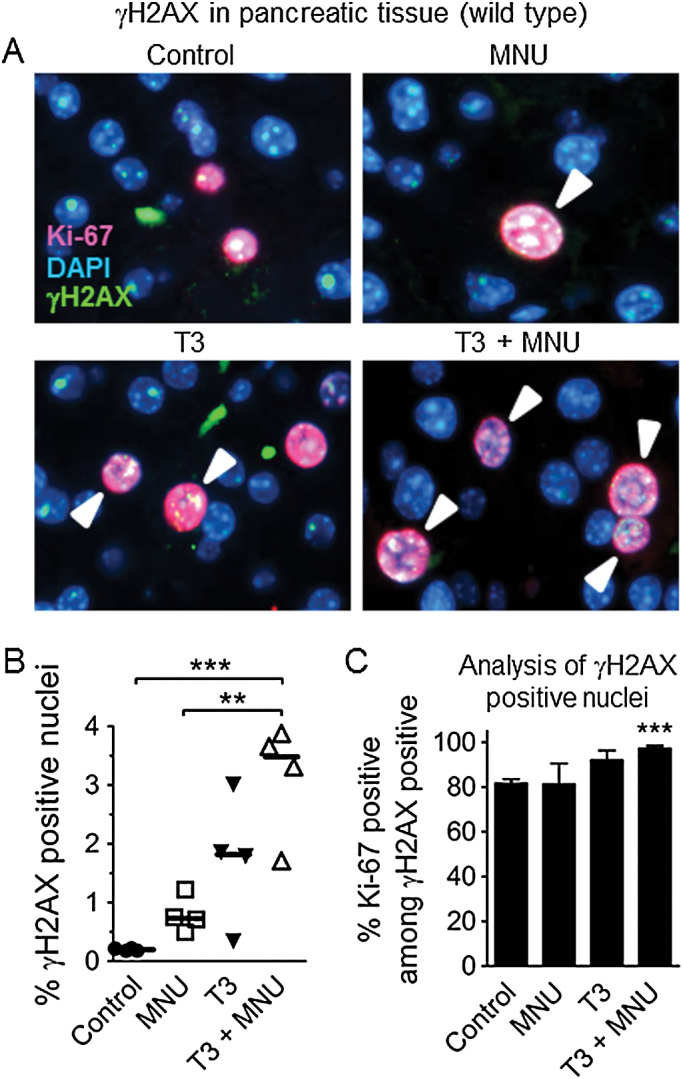
Base damage and single strand breaks are predicted to be particularly problematic during DNA replication, when they can be converted into recombinogenic DSBs. To specifically learn about the impact of cell proliferation on DSB susceptibility in vivo, animals were exposed to thyroid hormone (T3), a potent pancreatic mitogen (20). Consistent with previous studies (20), a diet supplemented with T3 induces cell proliferation in the pancreas, as indicated by Ki-67 (a marker for proliferating cells; Figure 3) and also led to increased BrdU incorporation into the genome (Supplementary Figure 3). To measure DSBs, we evaluated Ser-139 H2AX phosphorylation (γH2AX) in tissue sections. Histone H2AX is phosphorylated along several kilobases adjacent to DSBs and is therefore frequently used as a DSB marker (36). Although in some cases γH2AX foci do not perfectly reflect DSBs, analysis of γH2AX foci nevertheless provides a very powerful approach for evaluating factors that modulate DSBs (36). We first determined the time of relatively high levels of H2AX phosphorylation by western blotting (Supplementary Figure 4) and found it to be 6h after treatment. We then performed γH2AX immunohistochemistry at this time, and quantified the number of nuclei that were positive for γH2AX among 20 images per mouse (a total ~12000 nuclei per condition) (positive cells were defined as having >10 foci; Supplementary Figure 5). Images were collected from dispersed tissue regions to control for possible inconsistencies across the organ, and analysed in a blinded fashion. In pancreata from control mice, γH2AX positive nuclei were very rare. However, the frequency of cells with increased DSBs increased significantly after MNU treatment (Figure 4A,B). Exposure to T3 also led to an increase in the frequency of γH2AX positive nuclei (Figure 4A,B), indicating that proliferation alone is sufficient to induce DSBs in the pancreas in vivo. We predicted that increased cell proliferation would sensitize the pancreas to exposure-induced HR. Indeed, when MNU was administered to mice at the peak of T3-induced cell proliferation, DNA damage and cell proliferation appear to act synergistically to induce DSBs in vivo (Figure 4A,B), which is consistent with DNA damage-induced replication fork breakdown.
Fig. 3.
Hormone-induced cell proliferation in the pancreas. Mice were fed a diet containing 4 ppm thyroid hormone (T3). Pancreata were harvested at peak T3-induced proliferation, after treatment for 3 days (female mice) or 4 days (male mice). (A) Representative images of histological sections stained for the proliferation marker Ki-67. Original magnification, ×20. Scale bar = 100 μm. (B) Quantification of Ki-67 labeling in control mice (n = 24) and in T3-treated mice (n = 28) shows significantly higher labeling index after T3 treatment. Data are mean ± SEM. ***P < 0.001 (Student’s t-test).
Fig. 4.
DNA damage and cell proliferation interact on the formation of replication-associated DSBs. (A) Staining for the DSB marker γH2AX (green) and the proliferation marker Ki-67 (red) in pancreas sections. Mice were treated with thyroid hormone (T3) to induce cell proliferation in the pancreas. At the peak of T3-induced cell proliferation, mice were treated with MNU (25mg/kg) or mock treatment. Six hours after MNU injection, pancreata were harvested for analysis. A representative image from each treatment group is shown. Original magnification, ×40. Fluorescence is pseudocolored, gH2AX positive cells are indicated by arrowheads. (B) Quantification of γH2AX positive nuclei (>10 foci) shows synergy between MNU and T3 on DSBs. Symbols indicate data from individual mice, horizontal bars show median values for each group (n = 4 in all groups). (C) DSBs arise predominantly in proliferating cells. The percentage of γH2AX positive nuclei that are also positive for Ki-67 is plotted for each treatment group. Data are mean ± SEM; n = 4 mice in each group. **P < 0.01, ***P < 0.001 (Student’s t-test).
DNA methylation damage and cell proliferation induce DSBs in vivo in a replication-dependent manner
To further study the relationship between DSBs and DNA replication, tissue sections were co-stained for proliferation (Ki-67; Figure 4A, red) and DSB (γH2AX; Figure 4A, green) markers. Nuclei positive for both Ki-67 and γH2AX were then quantified in 20 randomly selected image fields per tissue section. In all four treatment groups, >80% of the damaged γH2AX positive cells were also positive for Ki-67 (Figure 4C), consistent with replication-driven DSBs. Additionally, under conditions of increased cell proliferation and DNA damage, nearly all of the γH2AX positive nuclei were also Ki-67 positive (T3 + MNU >90%; Figure 4C), which was statistically significantly greater than tissue from the control untreated animals. These results provide direct evidence for DNA damage and cell proliferation acting synergistically to induce DSBs in vivo as a result of replication fork breakdown. As DSBs formed at broken replication forks are repaired by HR, we next set out to measure HR under similar conditions.
Aag promotes damage- and proliferation-induced HR
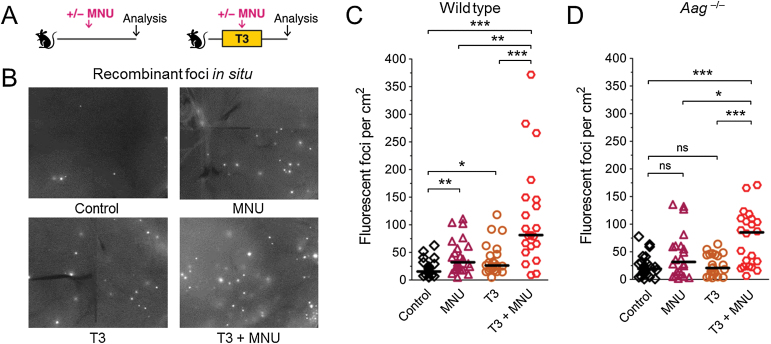
To measure the independent effects of DNA damage and cell proliferation on HR, wild type and Aag –/– mice were exposed to either MNU or T3 (Figure 5A, see below for analysis of their combined effect). In wild type mice, an increase in fluorescent foci after either MNU or T3 treatment was apparent upon microscopic examination (Figure 5B). Indeed, quantification of HR shows that the frequency of fluorescent foci per cm2 tissue area is significantly higher in MNU-treated mice compared to control mice (purple triangles, Figure 5C). When cell proliferation was stimulated with T3 in wild type mice, again we observed a significant increase in HR compared to control mice (yellow circles, Figure 5C). These results show that exogenous DNA damage induces HR in mature animals in vivo, and furthermore that increased cell proliferation alone is sufficient to lead to an accumulation of cells that have undergone HR in adult animals. In contrast to the wild type mice, however, we did not detect a significant increase in the frequency of HR in Aag –/– mice exposed either to MNU or to T3 (Figure 5D). Taken together, both MNU-induced methylation damage and T3-induced cell proliferation induced HR in wild type mice, but these effects were not as strong in Aag –/– mice. Together with the lower levels of spontaneous HR observed in the Aag –/– mice (Figure 2A), these results support a model wherein Aag-generated BER intermediates are more recombinogenic than Aag’s substrates (Figure 6, see Discussion).
Fig. 5.
DNA damage, cell proliferation and Aag interact on HR in vivo. (A) Experimental setup. Mice were fed a diet with thyroid hormone (T3) or control diet. At the peak of T3-induced cell proliferation, mice received MNU (25mg/kg i.p.) or control treatment. Sequence rearrangements were measured 3–4 weeks after MNU injection. (B) Induction of HR detected in situ in pancreata of wild type mice. Freshly harvested pancreata were compressed to 0.5mm and imaged under a fluorescent microscope. Representative details of composite images are shown, fluorescent foci are apparent in situ. Original magnification, ×1. Brightness and contrast have been enhanced identically across images. (C) In wild type mice, MNU-induced damage and T3-induced proliferation both induce HR, and interact in a synergistic fashion. Frequencies of fluorescent foci per cm2 pancreatic tissue are shown in control (n = 21), MNU-treated (n = 23), T3-treated (n = 22) and T3+MNU-treated (n = 21) mice. T3+MNU treatment induces the formation of more fluorescent foci than the sum of the effects of MNU and T3. (D) In Aag –/– mice, the increase in HR in MNU-treated mice (n = 22) compared to control mice (n = 23) does not reach statistical significance, and T3-treated mice (n = 22) show no increase. T3+MNU treatment (n = 22) induces the formation of more fluorescent foci than the sum of the effects of MNU and T3. In B and C, symbols indicate data from individual mice, horizontal bars show median values for each group. ns, not statistically significant, *P < 0.05, **P < 0.01, ***P < 0.001 (Mann–Whitney U-test).
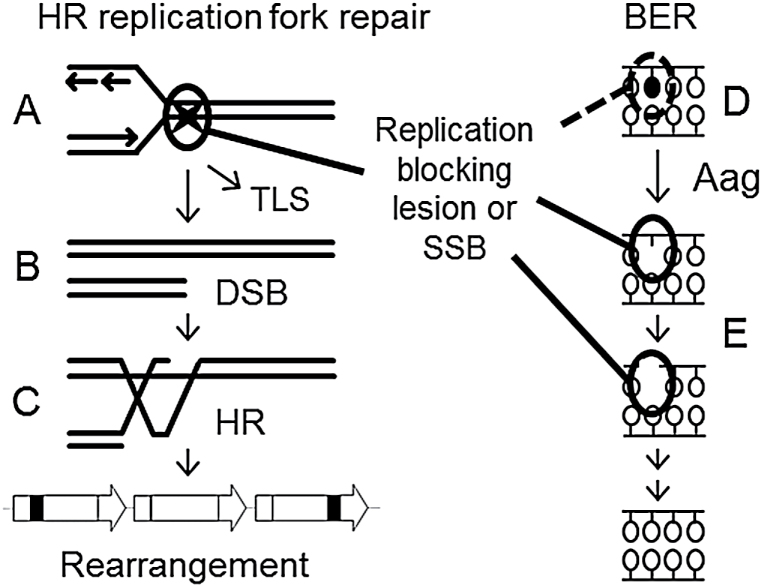
Fig. 6.
Model for the interaction of DNA damage, cell proliferation and Aag-initiated BER on HR-driven sequence rearrangements. (A–C) A replication blocking DNA lesion or SSB (denoted by a star) can induce a sequence rearrangement through replication fork breakdown. (A) When encountered by the replication fork, the lesion may be bypassed by translesion synthesis (TLS) or (B) may lead to replication fork breakdown and the formation of a one-ended DSB. (C) The DSB is then repaired by HR, but a misalignment between repeated sequences can produce a sequence rearrangement. (D) Replication blocking DNA damage, such as the Aag substrate 3-methyladenine, synergizes with cell proliferation to induce replication fork breakdown and the formation of HR-driven rearrangements. (E) BER intermediates are formed after the Aag-mediated excision of both replication blocking and innocuous base lesions. BER intermediates can also block replication and induce rearrangement formation. Thus, an imbalance of Aag activity relative to the activities of downstream BER enzymes can modulate the formation of HR-driven rearrangements induced by DNA methylation damage. SSB, single strand break.
DNA methylation damage and cell proliferation act synergistically to induce HR in vivo
Having observed that methylation damage and cell proliferation each individually induce HR (Figure 5B,C), we next set out to explore their combined effect. In wild type animals, the combination of MNU and T3 resulted in a striking increase in fluorescent foci (Figure 5B,C). Importantly, the combined exposure is more than additive (there is an increase of ~20, ~10 and ~70 foci/cm2 in the MNU, T3, and double exposed cohorts, respectively). Similar to the wild type mice, for the Aag –/– mice, exposure to T3+MNU induces HR in a synergistic fashion when analysed by foci counting (Figure 5D). We observed similar trends when samples were analysed by flow cytometry, however statistical significance was not consistently observed with this sized cohort due to insufficient sensitivity of this approach (data not shown). These data are consistent with the ability of cell proliferation to enhance the recombinogenic effect of both damaged bases and BER intermediates.
In addition to HR induction by BER-dependent mechanisms, increased cell proliferation may also potentiate HR as a consequence of single strand gaps that are formed across from O 6-methylguanine by mismatch repair (35). It is therefore possible that the large increase in HR seen after T3+MNU treatment is partly or wholly due to O 6-methylguanine-induced HR. To test this possibility, we performed the T3+MNU experiment with Mgmt –/– mice that do not repair O 6-methylguanine (37). Importantly, the MNU dose used in these mice was the same percentage of the LD50 as in wild type and Aag –/– mice (Supplementary Table 1). Results showed that Mgmt –/– mice do not show any increase in HR after T3+MNU treatment under conditions that induce HR in wild type mice (Supplementary Figure 6). The lack of HR induction by O 6-methylguanine may be due to the much lower abundance of these lesions compared to the Aag substrates 3-methyladenine and 7-methylguanine following MNU exposure (35). Taken together, the large increase in HR after T3+MNU treatment is likely due to BER intermediates (in wild type mice) and BER substrate lesions (in Aag –/– mice) rather than O 6-methylguanine.
Together with the observation of the replication-dependent induction of γH2AX by MNU (Figure 4), these results collectively support a model wherein replication blocking Aag substrates and downstream BER intermediates act synergistically with cell proliferation to induce DSBs and sequence rearrangements in vivo (Figure 6).
Discussion
HR has emerged as an important source of sequence changes that promote cancer (6,38,39), and yet relatively little has been done to elucidate the major drivers of HR in vivo. Here, we investigated how HR is affected by three key factors in mutagenesis: DNA damage, DNA repair and cell proliferation. While these factors have been studied individually or in pairs in vitro (40,41) and during embryonic development (42), systematic studies to address interactions among these three variables in adult animals were lacking. We found that exposure to an exogenous methylating agent as well as conditions that stimulate cell proliferation lead to increased HR, and that DNA damage and cell proliferation together act synergistically to induce HR in vivo. To learn more about the underlying mechanisms by which DNA damage leads to HR in vivo, we modulated the levels of the Aag glycosylase. Under spontaneous conditions, Aag promotes HR (presumably by creating BER intermediates; Figure 6). Interestingly, exposure to DNA damage (methylation) along with enhanced cell proliferation causes a synergistic increase in HR whether or not Aag is present, which is consistent with the ability of both methylated bases and their downstream BER intermediates to induce replication fork breakdown. Importantly, Aag activities, DNA damage levels and proliferation levels vary among people and also among different tissues. Interactions among these variables may thus contribute to the variation in cancer risk among individuals.
BER is a multistep pathway initiated by the removal of the damaged base by a glycosylase, and proceeding through a series of intermediates until repair is completed (16,31). Importantly, all BER intermediates, as well as some substrate lesions, are replication blocking (see below for a more detailed discussion). Indeed, intricate handing-off mechanisms have been described between successive BER enzymes, presumably to minimize the half-life of BER intermediates and thus to minimize replication breakdown (16). Thus, balance between the activities of successive BER enzymes is critical in avoiding the accumulation of BER intermediates. Consistent with the importance of BER balance, both Aag deficiency and Aag overexpression induce HR in yeast (13), and overexpression of Aag in mammalian cells leads to an increase in DNA breaks and SCEs (15). In surprising contrast with these in vitro results, we find that in the mouse pancreas, endogenous Aag activity increases HR both spontaneously, and also under conditions of increased cell proliferation. These data suggest that endogenous Aag activity leads to the formation of recombinogenic downstream BER intermediates, which is consistent with the observation that the rate-limiting step of BER has been found to be the lyase activity of polymerase beta, which is downstream of Aag (43). Thus, these data suggest that endogenous Aag activity in the mouse pancreas may not be balanced with the activities of downstream BER enzymes. Taken together, the data reported here call attention to the possibility that undisturbed BER is not necessarily balanced in vivo, and that BER imbalance may promote spontaneous genomic instability (see below).
One of the more significant findings from this work is the observation that under normal physiological conditions in vivo, the Aag DNA glycosylase on its own is sufficient to induce HR. While overexpression of Aag and its paralogs have been shown to pressure HR in vitro (13,15,44), this result is the first to point to normal levels of Aag-initiated BER intermediates as drivers of spontaneous HR in mammals in vivo (Figure 6). Further, Aag is just one glycosylase among many. Given that there are about a dozen different DNA glycosylases in mammals (31), the concerted pressure from glycosylases on the HR pathway is likely quite significant. Indeed, HR may have evolved in part to enable replication in the face of BER intermediates, thus calling attention to the importance of inter-pathway interactions in maintaining genomic stability (45).
Alkylating agents are risk factors for cancer (34), and the Aag glycosylase initiates the BER pathway at methylated bases, including 3-methyladenine (≈9% of MNU-induced lesions) which is replication blocking, and the more abundant 7-methylguanine (≈70% of lesions) which does not directly inhibit DNA polymerases (17). Both lesions are converted by Aag into replication blocking BER intermediates that are normally processed by downstream BER enzymes, including AP endonuclease and polymerase beta (16). In addition to protection by excision repair, translesion synthesis (TLS) also prevents fork breakdown by enabling bypass of 3-methyladenine (46). Therefore, when considering the pros and cons of lesion removal, unrepaired lesions under some conditions can be tolerated more effectively than downstream BER intermediates such as single strand breaks, which cannot be bypassed. Endogenous Aag activity may therefore promote spontaneous HR by converting both the replication blocking 3-methyladenine and the abundant but innocuous 7-methylguanine into replication blocking BER intermediates. Thus, differences in Aag activity among people (22–24) may influence the formation of spontaneous rearrangements, and also those formed after exposure to alkylating agents present in food, cosmetics and cigarette smoke (34). Indeed, higher Aag activities have been associated with an increased cancer risk (24,47). Taken together, variable Aag activities in people may modulate the formation of carcinogenic sequence changes driven by HR.
It is well established that HR is active primarily during S and G2, when a sister chromatid is available for repair, and when broken replication forks require HR for accurate restoration (4,48). Therefore, we developed conditions that enabled direct evaluation of the impact of cell proliferation on HR in vivo. We found that a hormone-induced increase in proliferation on its own was sufficient to increase HR. The moderate yet significant induction of HR during increased proliferation is likely due to the short duration and variable extent of proliferation. Specifically, T3-induced cell proliferation lasts less than one week, peaking several days after initiation of exposure. Thus, recombinant foci in T3-fed animals accumulated under variably increased proliferation for <1 week, while foci in control animals accumulated under baseline proliferation for 8–10 weeks. This difference in the duration of accumulation likely underlies the fact that the increase in recombinant foci in T3 treated animals over control is not proportionate to the increase in proliferation at the peak of T3 treatment. Although hormonally induced proliferation had a modest impact on HR, the increase in proliferation led to a dramatic increase in susceptibility to alkylation-induced HR. This result suggests that in people, physiologically relevant increases in the level of proliferation can boost the formation of carcinogenic sequence changes induced by DNA damaging exposures. This has important implications in public health, suggesting that exposures in utero and in childhood can be particularly detrimental to genomic stability. Indeed, cell proliferation during development increases HR-driven deletions induced by DNA damage in vivo (42). Further, conditions that involve regenerative proliferation, such as chronic injury or inflammation, can also increase susceptibility to HR-driven sequence changes (unpublished results). Finally, exposure to hormone-mimicking chemicals that increase proliferation can potentiate methylation-induced carcinogenesis (49). Interestingly, under conditions where there is increased cell proliferation, exposure to exogenous DNA damage synergistically induced HR, regardless of the presence or absence of Aag. Since downstream BER intermediates such as AP sites and single strand breaks are also able to induce HR during replication, forcing cells to replicate their DNA when subjected to increased levels of either Aag substrates or downstream intermediates is anticipated to induce HR. Importantly, given that there are many sources of DNA lesions that inhibit replication, mitogenic changes in physiology are expected to significantly sensitize tissues to HR-driven rearrangements. Furthermore, it is likely that increased cell proliferation also increases other mechanisms of mutagenesis, such as point mutations induced by repair synthesis during HR (3), DNA polymerase errors, mispairing, and mutations introduced during translesion synthesis (50). Taken together, increased cell proliferation may contribute to DNA sequence changes in itself and in combination with DNA damaging exposures, thus increasing the risk of cancer.
There are several mechanisms by which full length Eyfp sequence can be restored during HR at the direct repeat substrate. If there is a fork breakdown during DNA replication, mis-insertion can restore full length Eyfp, at the same time resulting in the gain of one repeat unit (a triplication, i.e. a rearrangement at the FYDR repeat substrate; Figure 1A). Importantly, the length of the FYDR repeat (≈500bp) is similar to the length of Alu repeats in the human genome (≈300bp). Alu repeats make up almost 10% of the human genome and are frequent sites of HR-driven rearrangement formation (51). Indeed, Alu-mediated rearrangements can activate oncogenes in cancer (52) and inactivate tumor suppressor genes such as p53 (53). Further, HR-driven rearrangements between Alu repeats have been shown to drive carcinogenesis both in solid tumors (7) and in hematological malignancies (8). Thus, HR events detected in FYDR mice after replication fork repair indicate the formation of genetic changes that are relevant for carcinogenesis in humans.
The role of HR in carcinogenesis has been highlighted by the discovery that mutations in BRCA1 and BRCA2, which lead to an HR deficiency, also lead to a significant increase in the risk of cancer (54). While reduced HR capacity predisposes to cancer, an increase in HR also predisposes to cancer (e.g. BLM). Furthermore, it is well established that HR itself is a source of sequence rearrangements and LOH (6–10). Thus, both an increase in HR and a deficiency in HR can promote carcinogenic sequence changes. Here, we have delineated some of the key variables that put pressure on the HR pathway—that is, damage, division and BER. For a BRCA carrier who does not yet have cancer, conditions that pressure HR will drive LOH and thus increase the frequency of cells that lose function of the remaining wild type BRCA allele—thus promoting cancer. Importantly, after initiation of cancer, these same pressures on HR will lead to significant sequence rearrangements in the BRCA deficient cancer cells, since they will not have HR to repair broken forks, and thus will be prone to chromosomal aberrations. Thus, while pressures on HR are deleterious for a normal person, they are much worse for a BRCA1/2 carrier both in terms of initiation and promotion.
HR is critical for DSB repair, and it also causes sequence rearrangements known to promote cancer, including insertions, deletions and LOH. The FYDR mouse model provides a valuable tool for revealing conditions that pressure HR. Using this model, we have shown that the Aag glycosylase promotes both spontaneous HR and proliferation-induced HR, revealing BER as a major source of pressure on the HR pathway in vivo. Importantly, by revealing that out of a dozen or so different DNA glycosylases, a single glycosylase on its own is sufficient to increase HR in vivo, these results provide some of the first in vivo evidence that BER is a critical underlying cause of HR in mammals. In addition to studies of spontaneous recombination, we have also explored the impact of DNA damage and cell proliferation. On its own, an increased level of cell proliferation is sufficient to increase the risk of spontaneous HR. Furthermore, cell proliferation and DNA damage act in a synergistic fashion to induce HR, regardless of BER status. Taken together, these studies of inter-pathway interactions between BER and HR have revealed new insights into the combined effects of genes, environment and physiology on genomic instability in mammals in vivo.
Supplementary material
Supplementary Table 1 and Figures 1–6 can be found at http://carcin.oxfordjournals.org/
Funding
National Institute of Health (grants R01-CA079827, R33-CA112151 and P01-CA026731); the MIT Center for Environmental Health Sciences (National Institute of Environmental Health Sciences P30-ES002109); and the National Research Foundation Singapore through the Singapore−MIT Alliance for Research and Technology.
Supplementary Material
Acknowledgements
The authors thank Yvette Miller for histology and Patrick Verdier for imaging support.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations:
- Aag
alkyl-adenine DNA glycosylase
- BER
base excision repair
- BrdU
bromodeoxyuridine
- DSB
DNA double-strand break
- FYDR
fluorescent yellow direct repeat
- HR
homologous recombination
- LD50
lethal dose
- LOH
loss of heterozygosity
- MNU
methylnitrosourea.
References
- 1. Hanawalt P.C. (1998). Genomic instability: environmental invasion and the enemies within. Mutat. Res., 400, 117–125 [DOI] [PubMed] [Google Scholar]
- 2. Hoeijmakers J.H. (2001). Genome maintenance mechanisms for preventing cancer. Nature, 411, 366–374 [DOI] [PubMed] [Google Scholar]
- 3. Hicks W.M., et al. (2010). Increased mutagenesis and unique mutation signature associated with mitotic gene conversion. Science, 329, 82–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Helleday T., et al. (2007). DNA double-strand break repair: from mechanistic understanding to cancer treatment. DNA Repair (Amst)., 6, 923–935 [DOI] [PubMed] [Google Scholar]
- 5. Hanawalt P.C. (2007). Paradigms for the three rs: DNA replication, recombination, and repair. Mol. Cell, 28, 702–707 [DOI] [PubMed] [Google Scholar]
- 6. Bishop A.J., et al. (2000). Homologous recombination as a mechanism for genome rearrangements: environmental and genetic effects. Hum. Mol. Genet., 9, 2427–2334 [DOI] [PubMed] [Google Scholar]
- 7. Pal J., et al. (2011). Genomic evolution in Barrett’s adenocarcinoma cells: critical roles of elevated hsRAD51, homologous recombination and Alu sequences in the genome. Oncogene, 30, 3585–3598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Strout M.P., et al. (1998). The partial tandem duplication of ALL1 (MLL) is consistently generated by Alu-mediated homologous recombination in acute myeloid leukemia. Proc. Natl. Acad. Sci. U. S. A., 95, 2390–2395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gupta P.K., et al. (1997). High frequency in vivo loss of heterozygosity is primarily a consequence of mitotic recombination. Cancer Res., 57, 1188–1193 [PubMed] [Google Scholar]
- 10. James C.D., et al. (1989). Mitotic recombination of chromosome 17 in astrocytomas. Proc. Natl. Acad. Sci. U. S. A., 86, 2858–2862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zeman M.K., et al. (2014). Causes and consequences of replication stress. Nat. Cell Biol., 16, 2–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Saleh-Gohari N., et al. (2005). Spontaneous homologous recombination is induced by collapsed replication forks that are caused by endogenous DNA single-strand breaks. Mol. Cell. Biol., 25, 7158–7169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hendricks C.A., et al. (2002). The S. cerevisiae Mag1 3-methyladenine DNA glycosylase modulates susceptibility to homologous recombination. DNA Repair (Amst)., 1, 645–659 [DOI] [PubMed] [Google Scholar]
- 14. Wyatt M.D., et al. (1999). 3-methyladenine DNA glycosylases: structure, function, and biological importance. Bioessays, 21, 668–676 [DOI] [PubMed] [Google Scholar]
- 15. Coquerelle T., et al. (1995). Overexpression of N-methylpurine-DNA glycosylase in Chinese hamster ovary cells renders them more sensitive to the production of chromosomal aberrations by methylating agents—a case of imbalanced DNA repair. Mutat. Res., 336, 9–17 [DOI] [PubMed] [Google Scholar]
- 16. Prasad R., et al. (2010). Substrate channeling in mammalian base excision repair pathways: passing the baton. J. Biol. Chem., 285, 40479–40488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wyatt M.D., et al. (2006). Methylating agents and DNA repair responses: methylated bases and sources of strand breaks. Chem. Res. Toxicol., 19, 1580–1594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wiktor-Brown D.M., et al. (2006). Age-dependent accumulation of recombinant cells in the mouse pancreas revealed by in situ fluorescence imaging. Proc. Natl. Acad. Sci. U. S. A., 103, 11862–11867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Engelward B.P., et al. (1997). Base excision repair deficient mice lacking the Aag alkyladenine DNA glycosylase. Proc. Natl. Acad. Sci. U. S. A., 94, 13087–13092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ledda-Columbano G.M., et al. (2005). Induction of pancreatic acinar cell proliferation by thyroid hormone. J. Endocrinol., 185, 393–399 [DOI] [PubMed] [Google Scholar]
- 21. Deichmann W.B, et al. (1943). Determination of approximate lethal dose with about six animals. J. Ind. Hyg. Toxicol., 25, 415–417 [Google Scholar]
- 22. Hall J., et al. (1993). Alkylation and oxidative-DNA damage repair activity in blood leukocytes of smokers and non-smokers. Int. J. Cancer, 54, 728–733 [DOI] [PubMed] [Google Scholar]
- 23. Calvo J.A., et al. (2013). Aag DNA glycosylase promotes alkylation-induced tissue damage mediated by Parp1. PLoS Genet., 9, e1003413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Crosbie P.A., et al. (2012). Elevated N3-methylpurine-DNA glycosylase DNA repair activity is associated with lung cancer. Mutat. Res., 732, 43–46 [DOI] [PubMed] [Google Scholar]
- 25. Mientjes E.J., et al. (1998). DNA adducts, mutant frequencies, and mutation spectra in various organs of lambda lacZ mice exposed to ethylating agents. Environ. Mol. Mutagen., 31, 18–31 [PubMed] [Google Scholar]
- 26. Carr G.J., et al. (1995). Statistical design and analysis of mutation studies in transgenic mice. Environ. Mol. Mutagen., 25, 246–255 [DOI] [PubMed] [Google Scholar]
- 27. Piegorsch W.W., et al. (1997). Sources of variability in data from a positive selection lacZ transgenic mouse mutation assay: an interlaboratory study. Mutat. Res., 388, 249–289 [DOI] [PubMed] [Google Scholar]
- 28. Fung K.Y., et al. (1998). Statistical analysis of lacZ mutant frequency data from MutaMouse mutagenicity assays. Mutagenesis, 13, 249–255 [DOI] [PubMed] [Google Scholar]
- 29. Mangerich A., et al. (2012). Infection-induced colitis in mice causes dynamic and tissue-specific changes in stress response and DNA damage leading to colon cancer. Proc. Natl. Acad. Sci. U. S. A., 109, E1820–E1829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Motulsky H. (2013). Intuitive Biostatistics. Oxford University Press, New York, NY [Google Scholar]
- 31. Krokan H.E., et al. (1997). DNA glycosylases in the base excision repair of DNA. Biochem. J., 325 (Pt 1), 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wogan G.N., et al. (2004). Environmental and chemical carcinogenesis. Semin. Cancer Biol., 14, 473–486 [DOI] [PubMed] [Google Scholar]
- 33. Fry R.C., et al. (2008). Genomic predictors of interindividual differences in response to DNA damaging agents. Genes Dev., 22, 2621–2626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hecht S.S. (1997). Approaches to cancer prevention based on an understanding of N-nitrosamine carcinogenesis. Proc. Soc. Exp. Biol. Med., 216, 181–191 [DOI] [PubMed] [Google Scholar]
- 35. Fu D., et al. (2012). Balancing repair and tolerance of DNA damage caused by alkylating agents. Nat. Rev. Cancer, 12, 104–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pilch D.R., et al. (2003). Characteristics of gamma-H2AX foci at DNA double-strand breaks sites. Biochem. Cell Biol., 81, 123–129 [DOI] [PubMed] [Google Scholar]
- 37. Glassner B.J., et al. (1999). DNA repair methyltransferase (Mgmt) knockout mice are sensitive to the lethal effects of chemotherapeutic alkylating agents. Mutagenesis, 14, 339–347 [DOI] [PubMed] [Google Scholar]
- 38. Costantino L., et al. (2014). Break-induced replication repair of damaged forks induces genomic duplications in human cells. Science, 343, 88–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Carr A.M., et al. (2013). Replication stress-induced genome instability: the dark side of replication maintenance by homologous recombination. J. Mol. Biol., 425, 4733–4744 [DOI] [PubMed] [Google Scholar]
- 40. Galli A., et al. (1999). Cell division transforms mutagenic lesions into deletion-recombinagenic lesions in yeast cells. Mutat. Res., 429, 13–26 [DOI] [PubMed] [Google Scholar]
- 41. Nikolova T., et al. (2010). Homologous recombination protects mammalian cells from replication-associated DNA double-strand breaks arising in response to methyl methanesulfonate. DNA Repair (Amst)., 9, 1050–1063 [DOI] [PubMed] [Google Scholar]
- 42. Bishop A.J., et al. (2001). Susceptibility of proliferating cells to benzo[a]pyrene-induced homologous recombination in mice. Carcinogenesis, 22, 641–649 [DOI] [PubMed] [Google Scholar]
- 43. Sobol R.W., et al. (2000). The lyase activity of the DNA repair protein beta-polymerase protects from DNA-damage-induced cytotoxicity. Nature, 405, 807–810 [DOI] [PubMed] [Google Scholar]
- 44. Memisoglu A., et al. (2000). Contribution of base excision repair, nucleotide excision repair, and DNA recombination to alkylation resistance of the fission yeast Schizosaccharomyces pombe . J. Bacteriol., 182, 2104–2112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cavalier-Smith T. (2002). Origins of the machinery of recombination and sex. Heredity (Edinb)., 88, 125–141 [DOI] [PubMed] [Google Scholar]
- 46. Johnson R.E., et al. (2007). A role for yeast and human translesion synthesis DNA polymerases in promoting replication through 3-methyl adenine. Mol. Cell. Biol., 27, 7198–7205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Leitner-Dagan Y., et al. (2012). N-methylpurine DNA glycosylase and OGG1 DNA repair activities: opposite associations with lung cancer risk. J. Natl. Cancer Inst., 104, 1765–1769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Shrivastav M., et al. (2008). Regulation of DNA double-strand break repair pathway choice. Cell Res., 18, 134–147 [DOI] [PubMed] [Google Scholar]
- 49. Desaulniers D., et al. (2001). Modulatory effects of neonatal exposure to TCDD, or a mixture of PCBs, p,p′-DDT, and p-p′-DDE, on methylnitrosourea-induced mammary tumor development in the rat. Environ. Health Perspect., 109, 739–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tombolan F., et al. (1999). Effect of mitogenic or regenerative cell proliferation on lacz mutant frequency in the liver of MutaTMMice treated with 5, 9-dimethyldibenzo[c,g]carbazole. Carcinogenesis, 20, 1357–1362 [DOI] [PubMed] [Google Scholar]
- 51. Kolomietz E., et al. (2002). The role of Alu repeat clusters as mediators of recurrent chromosomal aberrations in tumors. Genes. Chromosomes Cancer, 35, 97–112 [DOI] [PubMed] [Google Scholar]
- 52. Frederick L., et al. (2000). Analysis of genomic rearrangements associated with EGRFvIII expression suggests involvement of Alu repeat elements. Neuro. Oncol., 2, 159–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Slebos R.J., et al. (1998). Inactivation of the p53 tumor suppressor gene via a novel Alu rearrangement. Cancer Res., 58, 5333–5336 [PubMed] [Google Scholar]
- 54. Iqbal J., et al.; Hereditary Breast Cancer Study Group. (2012). The incidence of pancreatic cancer in BRCA1 and BRCA2 mutation carriers. Br. J. Cancer, 107, 2005–2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.