Summary
This study describes the generation of a novel Cyp2abfgs-null mouse model and evidence obtained with this mouse model that supports the critical role of P450 enzymes of the Cyp2abfgs gene subfamilies in NNK-induced lung tumorigenesis.
Abstract
Cytochrome P450 (P450) enzymes encoded by the mouse Cyp2abfgs gene cluster are preferentially expressed in the respiratory tract. Previous studies have demonstrated that pulmonary P450-mediated bioactivation is necessary for lung tumorigenesis induced by the tobacco-specific lung procarcinogen 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK), and that CYP2A5 mediates a noteworthy fraction, but not all, of NNK bioactivation in the lung. The aim of this study was to determine whether other P450s encoded by the Cyp2abfgs gene cluster also play significant roles in NNK lung tumorigenesis. A novel Cyp2abfgs-null mouse was generated, in which all Cyp2a, 2b, 2g, 2f and 2s genes are deleted. The Cyp2abfgs-null mouse was viable, fertile and without discernible physiological abnormalities or compensatory increases in the expression of other P450s. NNK bioactivation in vitro and NNK-induced DNA adduction and lung tumorigenesis in vivo were determined for wild-type (WT) and Cyp2abfgs-null mice; the results were compared with previous findings from Cyp2a5-null mice. The Cyp2abfgs-null mice exhibited significantly lower rates of NNK bioactivation in lung and liver microsomes, compared with either WT or Cyp2a5-null mice. The levels of lung O6-methyl guanine DNA adduct were also substantially reduced in Cyp2abfgs-null mice, compared with either WT or Cyp2a5-null mice. Moreover, the Cyp2abfgs-null mice were largely resistant to NNK-induced lung tumorigenesis at both low (50mg/kg) and high (200mg/kg) NNK doses, in contrast to the WT or Cyp2a5-null mice. These results indicate for the first time that, collectively, the CYP2A, 2B, 2F, 2G, and 2S enzymes are indispensable for NNK-induced lung tumorigenesis.
Introduction
Tobacco smoke contains over 70 carcinogens that affect the respiratory system (1). Cytochrome P450 (P450)-mediated in situ bioactivation of at least some of these carcinogens may be a key event in tobacco smoke-induced lung tumorigenesis (2). Among these carcinogens, the tobacco-specific N-nitrosamine 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) is one of the most potent, inducing lung tumors (mainly adenoma and adenocarcinoma) in rodents in a dose-dependent fashion (3). NNK is also believed to be a major risk factor for lung tumorigenesis in humans (4).
To exert carcinogenicity, NNK requires bioactivation by P450 enzymes, which hydroxylates NNK at the α-methylene or α-methyl position to form electrophilic intermediates that can adduct DNA and eventually cause mutations (3). The methylene hydroxylation pathway generates pro-mutagenic O6-methylguanine (O6-mG) adduct, which is persistent and is a critical determinant in the initiation of lung tumorigenesis in mice (5). The methyl hydroxylation pathway produces pyridyloxobutyl DNA adduct, which also contributes to NNK-induced tumorigenesis by inhibiting O6-alkylguanine-DNA-alkyltransferase, the enzyme that repairs O6-mG (6). Two stable products generated from α-hydroxylation of NNK, 4-oxo-4-(3-pyridyl)-butanone (OPB) and 4-hydroxy-1-(3-pyridyl)-1-butanone (HPB), represent the bioactivating activities that eventually lead to DNA adduction. The DNA adducts can lead to the prevalent mutations of critical oncogenes or tumor suppressor genes, such as k-ras and p53, that are found in lung adenocarcinoma in both mice and humans (7–9).
Previous studies have demonstrated that pulmonary P450s are the major catalyst of NNK α-hydroxylation and that they play an essential role in NNK-induced lung tumorigenesis in rodents (3,10). However, it remains to be determined which P450 enzymes are important for NNK bioactivation and NNK-induced lung tumorigenesis. Such information is valuable for understanding genetic and environmental factors that may predispose an individual to tobacco smoke-induced lung tumorigenesis. In mice, CYP2A5, which is highly expressed in the lung, was found to have high metabolic activity toward NNK metabolism, with K m values of <10 μM (11). Besides CYP2A5, other P450s expressed in mouse lung, including CYP1A, 2A4, 2B, 2E1, 2F2, and 3A, are also active in NNK metabolism, as indicated by in vitro studies using recombinant P450 enzymes and/or chemical inhibitors (11). Enzyme kinetic studies, which have only been conducted for CYP2A4 and CYP2A5, showed that CYP2A4 has much lower catalytic efficiency toward NNK than CYP2A5 (12,13). The enzyme kinetic data for NNK metabolism by other mouse P450s have yet to be obtained.
Several studies have been conducted to delineate the contribution of mouse CYP2A5 in NNK bioactivation and lung tumorigenesis in vivo. NNK-induced lung tumorigenesis in mice was strongly inhibited by 8-methoxypsoralen (14,15), which reportedly acted through inhibition of CYP2A5, but not other CYP2A isoforms (16). However, more recent studies using Cyp2a5-null mice indicated that, while CYP2A5 is the low-K m enzyme for NNK bioactivation in mouse lung, its absence only caused a partial decrease in NNK bioactivation and lung tumorigenesis (17,18). Furthermore, the residual activity toward NNK in Cyp2a5-null mice was inhibited by 8-methoxypsoralen. Together, these findings suggest that P450 enzymes other than CYP2A5, possibly CYP2B and CYP2F2, which share many substrates with CYP2A enzymes, also contribute to NNK-induced lung tumorigenesis in vivo.
The occurrence of many enzymes potentially contributing to NNK metabolisms and the scarcity of biochemical studies of NNK metabolism by individual mouse P450 enzymes make it challenging to identify the additional enzyme(s) responsible for mediating NNK bioactivation and lung tumorigenesis in vivo. The mouse Cyp2a and Cyp2b gene subfamilies each have multiple members; but, only some of the CYP2A or CYP2B isoforms have been characterized. The mouse Cyp2a and Cyp2b genes are interspersed and located within a Cyp2 gene cluster that also contains Cyp2f2, Cyp2g1 and Cyp2s1. This situation provides an opportunity to test the combined roles of all P450 enzymes encoded by this gene cluster in NNK bioactivation and lung tumorigenesis, relative to the already demonstrated specific contribution by CYP2A5 alone (17,18), and to contributions by Cyp genes located elsewhere in the mouse genome, such as Cyp2e1 and Cyp3a.
In this study, a novel Cyp2abfgs-null mouse was generated, in which a 1.4-megabase pair genomic fragment containing 12 Cyp genes in mouse chromosome 7 (including all Cyp2a, Cyp2b, Cyp2f, Cyp2g and Cyp2s subfamilies) was deleted, through Cre-mediated recombination in vivo. Homozygous Cyp2abfgs-null mice were characterized for viability and fertility; growth rates; general histology and potential compensatory expression of other P450 enzymes that may influence NNK bioactivation. The loss of expression of the targeted genes was confirmed in tissues known to express the Cyp genes in wild-type (WT) mice. The kinetic parameters of microsomal NNK metabolism, tissue levels of O6-mG and NNK-induced lung tumor formation were then determined for Cyp2abfgs-null and WT mice. Our data provide the first in vivo evidence for the dominant role of the CYP2A/B/F/G/S enzymes in combination, and the important role of the non-CYP2A5 enzymes encoded by this gene cluster, in NNK bioactivation and lung tumorigenesis. Our findings demonstrate the need to study the role of the corresponding human enzymes, such as CYP2B6, in the susceptibility of human individuals to lung cancer induced by tobacco carcinogens.
Materials and methods
Chemicals and reagents
NNK, 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol (NNAL), 4-(methylnitrosamino)-1-(3-[2,4,5,6-D4]-pyridyl)-1-butanone (D4-NNK), 4-(methylnitrosamino)-1-(3-[2,4,5,6-D4]-pyridyl)-1-butanol (D4-NNAL), HPB, (3,3,4,4-D4)-4-hydroxy-1-(3-pyridyl)-1-butanone (D4-HPB), NNK-N-oxide, OPB, O6-mG, O6-methyl-deoxyguanosine (O6-Me-dG) and O6-trideuteriomethyl-deoxyguanosine (O6-CD3-dG) were from the same sources as described previously (10,18). Sodium bisulfite and reduced β-nicotinamide adenine dinucleotide phosphate were purchased from Sigma–Aldrich (St Louis, MO). All solvents (acetonitrile, methanol and water) were of liquid chromatography-mass spectrometry grade (ThermoFisher Scientific, Waltham, MA).
Generation of the Cyp2abfgs-null (Cyp2s1-Cyp2f2Δ/Δ) mouse
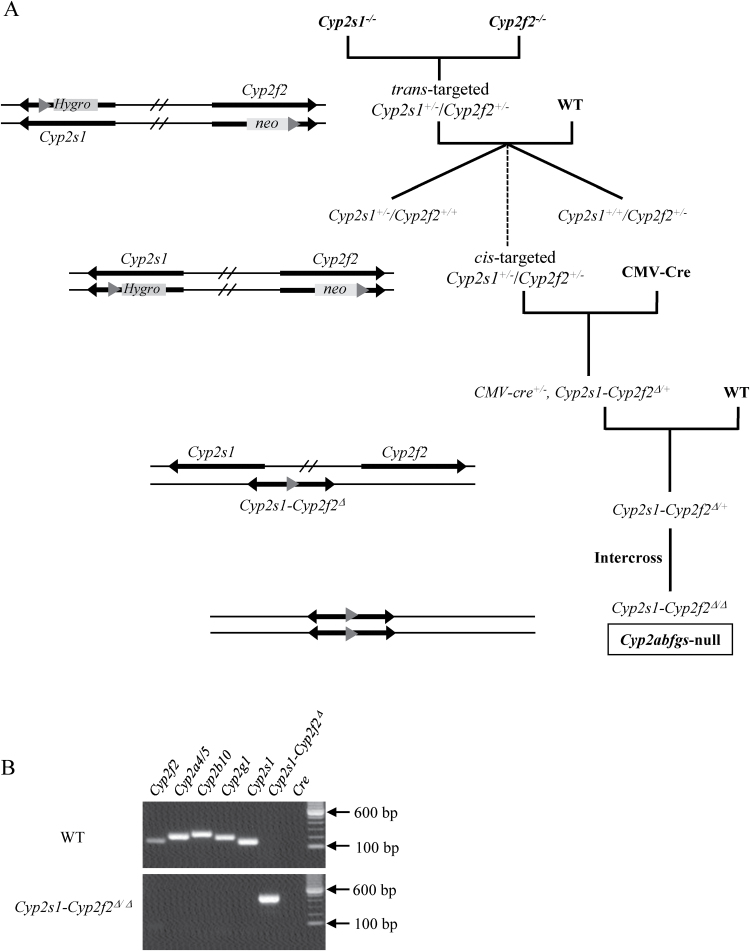
The Cyp2abfgs-null mouse was generated using the in vivo Cre-mediated gene deletion approach (19). The Cyp2f2-null mouse harboring a floxed neo cassette and Cyp2s1-null mouse harboring a floxed hygro cassette were obtained from breeding stocks maintained at the Wadsworth Center and have been described elsewhere (19,20). The LoxP sites in the Cyp2f2-null allele and that in the Cyp2s1-null allele were oriented in the same direction. CMV-Cre transgenic mice (B6.C-Tg(CMV-Cre)1Cgn/J, Stock # 006054), used to cause the deletion of loxP-flanked genes in all tissues, including germ cells (21), were purchased from the Jackson Laboratory. All three mouse strains were on the B6 background.
Animals were bred in five steps, as outlined in Figure 1A. Cyp2s1 - and Cyp2f2 - alleles were identified using PCR, as described (19,20). Male cis-targeted Cyp2s1 +/−/Cyp2f2 +/− mice were crossed with CMV-Cre females, and their pups were screened for the presence of the deleted allele (Cyp2s1-Cyp2f2 Δ) by PCR, with the forward primer F1 (5′-caagccatgtttttgttgga-3′) located in the intron 3 of Cyp2s1, and reverse primer R1 (5′-ctccctgggatgggaactat-3′) at ~300bp downstream of Cyp2f2 exon 4. The PCR product from the Cyp2s1-Cyp2f2 Δ allele was validated by sequence analysis. Cyp2s1-Cyp2f2 Δ/+ mice were crossed with B6 WT mice, and Cyp2s1-Cyp2f2 Δ/+ pups that were negative for Cre were obtained for further intercrossing and production of Cyp2s1-Cyp2f2 Δ/Δ mice.
Fig. 1.
Generation of Cyp2abfgs-null mice. (A) Breeding steps. (B) PCR analysis of genomic DNA from WT and Cyp2s1-Cyp2f2 Δ/Δ mice. PCR was performed using primers for Cyp2f2, Cyp2a4/5, Cyp2b10, Cyp2g1, Cyp2s1, Cyp2s1-Cyp2f2 Δ and Cre. Selected bands of a 100bp DNA size marker are shown. The full length gels are shown in Supplementary Figure S2, available at Carcinogenesis Online.
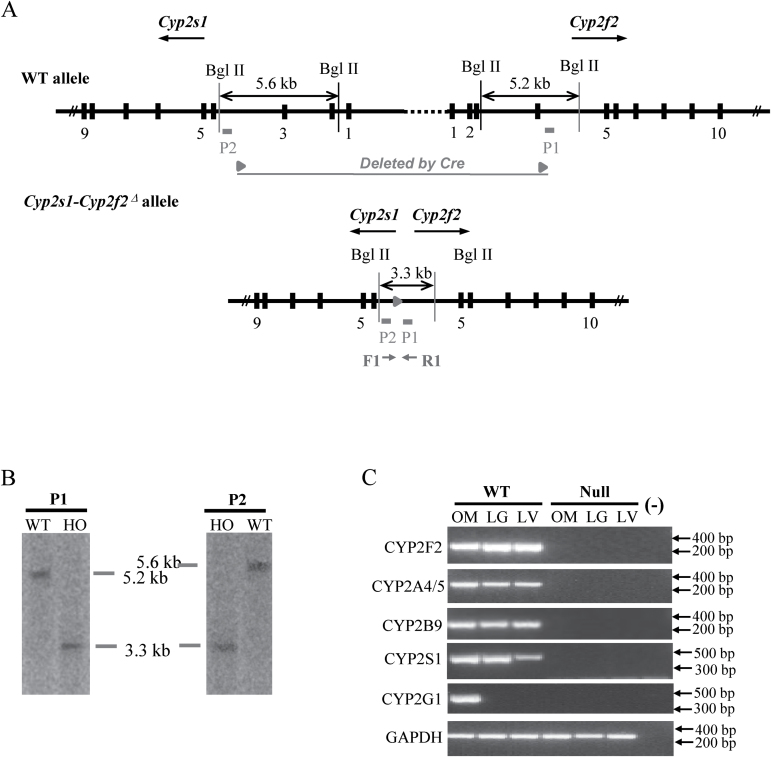
The absence of Cyp2f2, Cyp2a4/5, Cyp2b10, Cyp2g1 and Cyp2s1 genes in the Cyp2s1-Cyp2f2 Δ/Δ mice was confirmed by PCR, using primers described previously (19,20). Southern blot analysis was performed for confirmation of the Cyp2s1-Cyp2f2 Δ structure using two probes: P1, a 727 bp fragment located in Cyp2f2 intron 4, at ~50bp downstream of the residual loxP site, and P2, a 573 bp fragment located in Cyp2s1 intron 3, at ~80bp upstream of the loxP site.
Characterization of the Cyp2s1-Cyp2f2Δ/Δ mouse
Liver, lung and nasal olfactory mucosa (OM) from male and female mice (2-month-old) were pooled for RNA preparation, with use of the RNeasy Mini kit (QIAGEN, Valencia, CA). RNA-PCR analysis was performed using a previously described protocol (22), with use of gene-specific PCR primers for Cyp2f2, Cyp2a4/5, Cyp2b9/10/19, Cyp2g1, Cyp2s1 and glyceraldehyde 3-phosphate dehydrogenase (as an internal control) (19,20,23). Microsomal preparation and immunoblot analysis were carried out essentially as described previously (24,25), using the following antibodies: goat anti-rat CYP1A1/2, rabbit anti-rat CYP3A2 (BD Gentest, Woburn, MA); rabbit anti-rat CYP2C, rabbit anti-rat CPR (Enzo Life Science, Plymouth, PA); rabbit anti-human CYP2E1 (StressGen, Victoria, British Columbia, Canada). Calnexin, a marker protein for the endoplasmic reticulum, was detected using a rabbit anti-human calnexin antibody (GenScript). The intensity of the detected bands was quantified with use of ChemiDoc™ MP Imaging System (Bio-Rad, Hercules, CA). Histological analysis of various tissues from 2-month-old, male and female mice was conducted as described previously (19).
In vitro assay of NNK metabolism
Lung and liver microsomes were prepared from three different batches (each prepared from pooled tissue of three mice) of 2-month old, female Cyp2abfgs-null and WT mice, as described (26). The apparent K m and V max values were determined using a broad range of NNK concentrations (0.5, 1, 2.5, 10, 25, 50, 100, 200, 400 μM), with 0.25mg/ml lung or liver microsomal protein. The reactions were carried out for 10min for liver, at 37°C, conditions under which the rates of product formation were linear with time, in the presence of 5mM sodium bisulfite and 1mM reduced β-nicotinamide adenine dinucleotide phosphate. The methods for sample preparation and liquid chromatography-mass spectrometry analysis were essentially as described (18). The formation of OPB-bisulfite, HPB and NNK-N-oxide was analyzed.
Animal studies
All procedures involving animals were approved by the Wadsworth Center Institutional Animal Care and Use Committee. For pharmacokinetics study and determination of tissue levels of O6-mG, 2-month-old WT and Cyp2abfgs-null female mice (on B6 background) were treated with a single injection of NNK (at 200mg/kg, intraperitoneally) in saline. Blood, lung and liver were collected, as described (18). Plasma levels of NNK and NNAL, and tissue levels of O6-mG, were determined using liquid chromatography-mass spectrometry, as described (10).
For lung tumor bioassay, the Cyp2abfgs-null mice (on B6 background) were backcrossed to the susceptible A/J background for three generations; the A/J-N3 heterozygotes were then intercrossed to obtain A/J-N3 Cyp2abfgs-null and A/J-N3 WT littermates. The tumor bioassay was performed as described (10), starting with 2-month-old female mice and with NNK doses at 0 (saline alone), 50 or 200mg/kg body weight (intraperitoneally). Female mice were studied as they are preferred for NNK tumor bioassays (27). Mice were killed 16 weeks after NNK treatment. Body weights were recorded weekly at 8–10 weeks of age and then biweekly. Lung tumor multiplicity (average number of tumors per mouse) and frequency (percentage of mice with tumors) were determined independently by two researchers, and the averaged results of the two analyses are reported. For histopathological characterization of lung tumors, tissues from saline- and NNK-treated mice (four/strain) were fixed in 10% neutral buffered formalin. Sections were stained with hematoxylin and eosin for tumor type identification.
Other methods
Statistical significance of differences between two groups was examined using Student’s t-test. For body weight and tumor multiplicity differences, two-way analysis of variance was performed, with use of Bonferroni post hoc test for comparisons between strain or treatment groups. For tumor frequency differences, Fisher’s exact test was used to assess statistical significance.
Results
Generation and characterization of the Cyp2abfgs-null (Cyp2s1- Cyp2f2Δ/Δ) mouse
Twelve breeding pairs between trans-targeted Cyp2s1 +/−/Cyp2f2 +/− mice and WT mice were set up and gave birth to a total of 60 pups, of which one (#53) was found to have both Cyp2s1 − and Cyp2f2 − alleles (cis-targeted Cyp2s1 +/−/Cyp2f2 +/− mouse, data not shown) (recombination ratio = ~1.7%). The #53 pup was mated with CMV-Cre mice for deletion of the 1.4 Mb floxed Cyp2abfgs region, and eventual production of the Cyp2s1-Cyp2f2 Δ/Δ mice. Successful target gene deletion was confirmed by PCR analysis of representative Cyp2 genes within the targeted region, including Cyp2f2, Cyp2a4/5, Cyp2b10, Cyp2g1 and Cyp2s1, for which characteristic PCR products were detected in the genomic DNA of WT mice, but not the Cyp2abfgs-null mice (Figure 1B). The deletion of the Cyp2abfgs gene cluster was also confirmed by Southern blot analysis (Figure 2A, B), which showed characteristic bands of 5.2kb (P1) or 5.6kb (P2) for the WT allele, and of 3.3kb (both P1 and P2) for the Cyp2s1-Cyp2f2 Δ allele, and by RNA-PCR, which showed absence of selected CYP2 messenger RNAs (CYP2F2, CYP2A4/5, CYP2B9/10/19, CYP2G1 and CYP2S1) in relevant tissues (liver, lung or OM) of the Cyp2s1-Cyp2f2 Δ/Δ mice (Figure 2C).
Fig. 2.
Validation of the Cyp2abfgs-null mice. (A) Strategy of Southern blot analysis. DNA probes P1 and P2 are shown as solid boxes. The WT Cyp2s1 and Cyp2f2 alleles and the Cyp2s1-Cyp2f2 Δ allele are shown. Following BglII digestion of genomic DNA, P1 would detect a 5.2kb fragment in Cyp2f2 WT allele, whereas P2 would detect a 5.6kb fragment in Cyp2s1 WT allele; both probes would detect a 3.3kb fragment from the Cyp2s1-Cyp2f2 Δ allele. The positions of PCR primers (F and R) used for detection of the Cyp2s1-Cyp2f2 Δ allele and the residual loxP site are also shown. (B) Southern blot analysis. DNA from a WT and a Cyp2s1-Cyp2f2 Δ/Δ (HO) mice (10 μg each) was analyzed. The full length blots are shown in Supplementary Figure S2, available at Carcinogenesis Online. (C) Absence of CYP2F2, CYP2A4/5, CYP2B9/10/19, CYP2G1 and CYP2S1 messenger RNA expression in tissues of the Cyp2abfgs-null mice. RNA-PCR was performed using total RNA prepared from the liver, lung and OM of adult male and female (two each, 2-month-old, pooled) WT or Cyp2abfgs-null mice. PCR products were analyzed on a 1.5% agarose gel and visualized by staining with ethidium bromide. (-), no template control. The positions of selected fragments of a 100bp DNA marker are indicated. The full length gels are shown in Supplementary Figure S2, available at Carcinogenesis Online.
Homozygous Cyp2abfgs-null mice were found to be viable, fertile and normal in general appearance. Genotype frequency among pups derived from intercrosses between Cyp2abfgs-null heterozygotes was 23% (11/48) homozygotes, 52% (25/48) heterozygotes and 25% (12/48) WT, with no significant difference among the three genotypes (P > 0.05, Chi-squared test), suggesting absence of embryonic lethality. Adult (2-month-old) body and organ (liver, kidney, lung, brain, testis and heart for males, and liver, kidney, lung, brain and heart for females) weights were similar between Cyp2abfgs-null and WT mice (data not shown), indicating normal growth of the Cyp2abfgs-null mice. Histological analyses of various tissues, including brain, heart, thymus, lung, nose, stomach, spleen, liver, pancreas, intestine, kidney, skin and the reproductive organs (testis and ovary), revealed no obvious abnormalities (data not shown). Additionally, there were no compensatory changes of liver, lung and OM microsomal levels of CPR or several P450 proteins, including CYP2C, CYP3A, CYP2E1 and CYP1A1/2, in either males or females (data not shown).
NNK metabolism in vitro
Rates of NNK metabolism in lung and liver microsomal reactions were determined for Cyp2abfgs-null (B6) and WT (B6) mice. The apparent K m and V max values and the calculated catalytic efficiency (V max /K m) values are shown in Table I. Compared with WT, the Cyp2abfgs-null mice showed decreases in V max /K m values for the formation of OPB, HPB and NNK-N-oxide of 65, 40 and 60%, respectively, in the lung, and of 70, 90 and 35%, respectively, in the liver. Furthermore, when compared with the previously reported V max /K m values for Cyp2a5-null (B6) mice (18), Cyp2abfgs-null mice exhibited further decreases in V max /K m values for OPB, HPB and NNK-N-oxide formation, by 60, 35 and 50%, respectively, in the lung, and by 50, 35 and 15%, respectively, in the liver. These results indicated that, besides CYP2A5, other members of the Cyp2abfgs gene cluster also play an important role in NNK metabolism.
Table I.
Enzyme kinetic parameters for the formation of NNK metabolites by lung and liver microsomes from WT and Cyp2abfgs-null micea
| Metabolite | Strain | Lung | Liver | ||||
|---|---|---|---|---|---|---|---|
| K m (µM) | V max (pmol/min/mg) | V max/K m | K m (µM) | V max (pmol/min/mg) | V max/K m | ||
| OPB | WT (B6) | 37.0±2.9 | 64.6±1.3 | 1.7 | 40±10 | 247±21 | 6.2 |
| Cyp2abfgs-null (B6) | 65.8±6.1b,c | 42.1±4.4b,c | 0.6 | 114±24b,d | 232±10 | 2.0 | |
| Cyp2a5-null (B6)e | 40.2±2.1 | 61.5±2.0 | 1.5 | 55±5 | 210±10 | 4.0 | |
| HPB | WT (B6) | 2.5±0.4 | 24.6±1.8 | 9.6 | 21±2 | 114±3 | 5.4 |
| Cyp2abfgs-null (B6) | 2.4±0.2 | 13.0±1.4b,c | 5.6 | 82±17b | 61±4b | 0.7 | |
| Cyp2a5-null (B6)e | 2.9±0.3 | 24.4±1.0 | 8.4 | 65±15 | 70±8 | 1.1 | |
| NNK-N-oxide | WT (B6) | 1.4±0.1 | 34.6±3.3 | 23.9 | 90±16 | 29±2 | 0.32 |
| Cyp2abfgs-null (B6) | 1.9±0.4 | 19.9±5.8b,c | 10.2 | 150±27b,d | 32±3c | 0.21 | |
| Cyp2a5-null (B6)e | 1.9±0.3 | 38.7±2.2 | 20.4 | 83±16 | 20±2 | 0.24 | |
aApparent K m and V max values for the microsomal formation of OPB, HPB and NNK-N-oxide were determined as described in Materials and methods. The results shown represent mean ± SD of values determined for three separate microsomal samples, each prepared from tissues pooled from three 2-month-old female mice.
b P < 0.01, compared with WT (B6) mice.
c P < 0.01, compared with Cyp2a5-null (B6) mice.
d P < 0.05, compared with Cyp2a5-null (B6) mice.
eData for Cyp2a5-null (B6) mice (18) are included for comparison.
Pharmacokinetics of plasma NNK and NNAL
Plasma levels of NNK and its major circulating metabolite NNAL were similar between Cyp2abfgs-null (B6) and WT (B6) mice, at various time points following NNK administration at 200mg/kg (Supplementary Figure S1, available at Carcinogenesis Online), except that at the first time point (10min), the NNK and NNAL levels in the Cyp2abfgs-null mice were slightly (~15%) higher than those in the WT mice. But, there was no significant difference in clearance/bioavailability and area under the concentration-time curve values between the two groups, for either NNK or NNAL (data not shown). Overall, this result indicated that enzymes encoded by the Cyp2abfgs gene cluster did not contribute significantly to the systemic clearance of NNK or NNAL.
NNK bioactivation in vivo
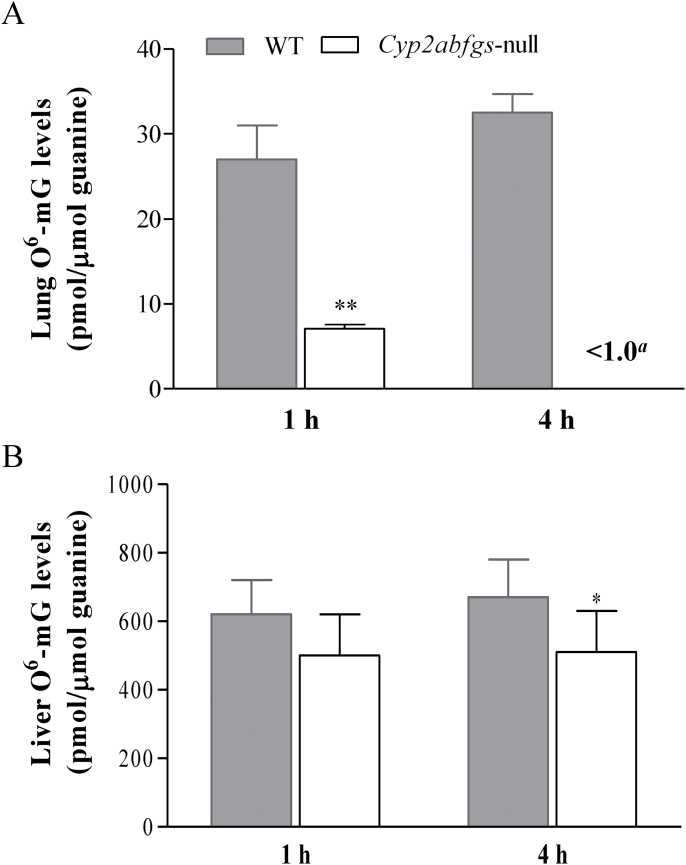
Levels of O6-mG, commonly used as the genotoxic endpoint representing the extent of NNK’s in vivo bioactivation and its lung tumorigenic potential (5), were measured in Cyp2abfgs-null (B6) and WT (B6) mice, at 1 or 4h after NNK injection at 200mg/kg (Figure 3A). In the lung, O6-mG levels were substantially decreased in the Cyp2abfgs-null mice at both the 1 h (by 75%) and 4 h (>99%) time points, compared with WT mice. For comparison, the extent of decrease observed previously in the Cyp2a5-null mice, relatively to WT mice, (at 4h after NNK treatment) were all <50%, at various NNK doses tested (20–200mg/kg); at 200mg/kg, only a 24% decrease was observed. These results indicated that, besides CYP2A5, other members of the Cyp2abfgs gene cluster also play an important role in NNK bioactivation in vivo.
Fig. 3.
NNK-induced O6-mG adduct formation in the lung and liver. Two-month-old female WT (B6) and Cyp2abfgs-null (B6) mice were injected with NNK at a dose of 200mg/kg (intraperitoneally), and lung (A) and liver (B) levels of O6-mG and total guanine were determined at 1 or 4h after the injection, as described in Materials and methods. Data represent mean ± SD (n = 5–8). aDetection limit for lung O6-mG was ~1.0 pmol/μmol guanine. **P < 0.01, *P < 0.05, compared with WT mice.
In the liver, O6-mG levels in NNK-treated Cyp2abfgs-null mice were only marginally lower (by 20–25%), compared with WT mice (Figure 3B); the difference was significant at the 4 h time point. In contrast, no difference in hepatic O6-mG levels was observed in previous studies, between NNK-treated Cyp2a5-null mice and NNK-treated WT mice, at NNK doses ranging from 20 to 200mg/kg (18). This result indicated that members of the Cyp2abfgs gene cluster (other than CYP2A5) also contribute, albeit to a minor extent, to hepatic NNK bioactivation in vivo.
NNK-induced lung tumorigenesis
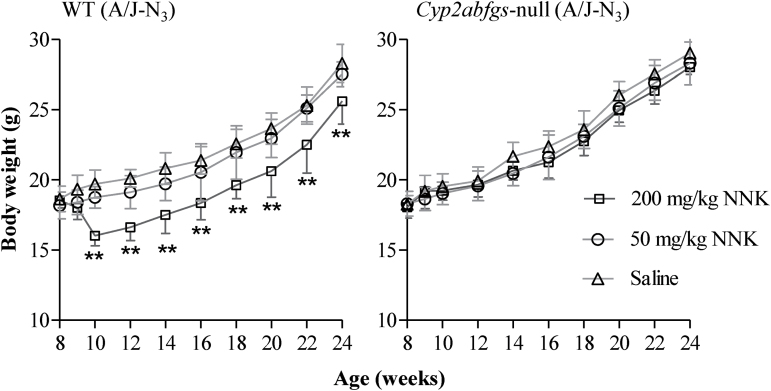
NNK lung tumor bioassays were performed in Cyp2abfgs-null (A/J-N3) and WT (A/J-N3) littermates, at NNK doses of 0 (saline alone), 50 or 200mg/kg. During the 16 weeks after the saline or NNK treatment, all mice exhibited continued weight gain, except for the WT mice in the 200mg/kg dose group, which showed a significant weight loss by 2 weeks after dosing and trailed the other two WT groups (0 and 50mg/kg) in body weights throughout weeks 2–16 (Figure 4). In contrast, the NNK treatment did not induce a weight loss in the Cyp2abfgs-null mice, at either low (50mg/kg) or high (200mg/kg) NNK dose, and no difference in body weight gain was observed among the three dose groups.
Fig. 4.
Body weights of mice after saline or NNK treatment. WT (A/J-N3) and Cyp2abfgs-null (A/J-N3) female mice (8-week old) were treated with a single injection (intraperitoneally) of NNK at 50 or 200mg/kg, or with saline. Body weights were recorded weekly at 8–10 weeks of age, and then biweekly. The results shown are mean ± SD (n = 10–12). Data was analyzed by two-way analysis of variance with Bonferroni post hoc test. **P < 0.01.
Saline-treated mice had a low incidence of spontaneous lung tumors, but there was no significant difference in tumor frequency between the two mouse strains (Table II). In NNK-treated mice, lung tumors were readily detected in all WT mice, but they were rare in Cyp2abfgs-null mice, at either NNK dose. The Cyp2abfgs-null mice showed a substantially decreased tumor multiplicity (by >97%) and tumor frequency (by >80%), at both NNK doses, compared with WT mice; the tumor multiplicity and tumor frequency in the Cyp2abfgs-null mice were similar to those in saline-treated mice. As a comparison, the Cyp2a5-null mice had only a moderate (40–60%) decrease in tumor multiplicity, and no significant change in tumor frequency, at both NNK doses, compared with WT mice (17).
Table II.
NNK-induced lung tumorigenesisa
| Treatment | Strain | Tumor multiplicity (tumors/mouse) | Tumor frequency (%) |
|---|---|---|---|
| Saline | WT (A/J-N3) (n = 10) | 0.1±0.1 | 10 |
| Cyp2abfgs-null (A/J-N3) (n = 10) | 0.1±0.1 | 10 | |
| NNK (50mg/kg) | WT (A/J-N3) (n = 21) | 4.1±0.5b | 100c |
| Cyp2abfgs-null (A/J-N3) (n = 18) | 0.11±0.08d | 11e | |
| NNK (200mg/kg) | WT (A/J-N3) (n = 12) | 29.6±3.4b | 100c |
| Cyp2abfgs-null (A/J-N3) (n = 10) | 0.2±0.4d | 17e |
aWT (A/J-N3) and Cyp2abfgs-null (A/J-N3) (2-month old) female mice were treated with a single injection of saline or NNK at the indicated dose (intraperitoneally). The mice were kept on AIN93G diet after weaning, until they were killed for tumor detection at 4 months after the NNK or saline injection. Tumor multiplicity is shown as mean ± standard error. Tumor frequency was calculated as percentage of mice with tumor in each group.
b P < 0.001, compared with WT/saline group (two-way analysis of variance followed by Bonferroni post hoc test).
c P < 0.001, compared with WT/saline group (Fisher’s exact test).
d P < 0.001, compared with corresponding WT/NNK group (two-way analysis of variance followed by Bonferroni post hoc test).
e P < 0.001, compared with corresponding WT/NNK group (Fisher’s exact test).
Histopathological analysis of NNK-induced lung tumors indicated that they are typical adenomas (data not shown), with the boundary of the lesion well demarcated and alveolar architecture distorted, a result consistent with previous reports (10,28). No tumor or gross pathological changes were observed in the liver, heart, kidney, stomach or intestine of any of the mice studied (data not shown).
Discussion
In the present study, we obtained direct evidence for a dominant role of CYP2A/B/F/G/S enzymes in NNK bioactivation and lung tumorigenesis, by comparing the kinetic parameters of microsomal NNK metabolism, tissue levels of O6-mG and lung tumor incidences between the novel Cyp2abfgs-null mouse and WT control mice. There were large differences in lung tumor incidence between WT and Cyp2abfgs-null mice, which were consistent with similar differences between the two strains in levels of O6-mG in the lung. The validity of this finding was supported by the confirmation that there was no compensatory change in the expression of other P450 enzymes examined in the Cyp2abfgs-null mouse and that the loss of Cyp2abfgs expression did not alter the bioavailability of NNK in its target tissue, lung.
A further comparison of the results from this study with those of two recent studies on NNK bioactivation and tumorigenesis in the Cyp2a5-null mice (17,18) revealed the important role of the non-CYP2A5 enzymes encoded by the Cyp2abfgs gene cluster in NNK bioactivation and NNK-induced lung tumorigenesis. The specific contributions by individual enzymes in the Cyp2abfgs subfamilies, other than CYP2A5, can be further determined in future studies, by characterization of the activities of individual P450 enzymes toward NNK in vitro, and/or by studying NNK lung tumorigenesis in mouse models with deletion of individual (e.g. Cyp2f2-null, Cyp2g1-null, Cyp2s1-null) or several P450 genes (e.g. Cyp2a(4/5)bgs-null) (19,20). In that regard, some predictions can be made based on existing knowledge. It has been established that NNK-induced lung tumorigenesis depends on target tissue bioactivation by pulmonary P450 enzymes (10). Given that CYP2A4 and CYP2A12 are mainly expressed in the liver, CYP2G1 is exclusively expressed in the OM, and CYP2A22 and CYP2S1 are ‘orphan’ enzymes (29,30), we can perhaps restrict the list of important NNK-bioactivating enzymes in the lung to the CYP2B enzymes and CYP2F2, in addition to CYP2A5.
The Cyp2abfgs-null mice showed substantial decreases in catalytic efficiency toward NNK in vitro and ability to form O6-mG DNA adduct in vivo, compared with WT mice (Table I and Figure 3). When comparing lung microsomal activities toward NNK between Cyp2a5-null mice (18) and Cyp2abfgs-null mice, it is apparent that the additional loss of CYP2B and CYP2F2 expression in the Cyp2abfgs-null mice caused further decreases in V max values and in catalytic efficiency, for OPB, HPB and NNK-N-oxide formation in the lung. Mouse CYP2B has been reported to catalyze α-hydroxylation, but not N-oxide formation, in the lung, as indicated by 25–45% inhibitions of OPB and HPB, but not N-oxide, formation, by an antibody to CYP2B (3). Mouse CYP2F2 also seems to contribute to HPB formation (18), but it is not yet clear whether CYP2F2 is also active in the N-oxide formation. Although the catalytic efficiencies of the mouse CYP2B and 2F2 enzymes toward NNK have not been determined directly, previous studies on Cyp2a5-null mice had suggested that the enzymes responsible for the residual activities in the Cyp2a5-null mouse lung were not as efficient as CYP2A5 toward NNK, at least in the formation of OPB and HPB. However, given the relatively high abundance of CYP2B and CYP2F2 in the lung (20,31), it is reasonable to expect that their loss would lead to a significant decrease in the apparent V max. Additionally, the greater decrease in formation of lung O6-mG in Cyp2abfgs-null mice (>99% decrease compared with WT; this study) than that in Cyp2a5-null mice (~25% decrease compared with WT; ref. 17), all treated with NNK at 200mg/kg (intraperitoneally), suggested that the residual mouse CYP2B and 2F2 enzymes played a more important role in bioactivation (via α-hydroxylation) than in detoxification (via N-oxide formation) of NNK in vivo.
There were still residual microsomal activities toward NNK in the lungs of Cyp2abfgs-null mice, which might represent contributions by additional lung P450 enzymes, possibly CYP1A and CYP2E1, which have been shown to be active toward NNK in previous microsomal inhibition studies (3). Notably, these residual activities were apparently insufficient to mediate NNK-induced lung DNA damage and tumorigenesis, as indicated by the very low levels of O6-mG detected in vivo, and the resistance of the Cyp2abfgs-null mice to NNK-induced lung tumorigenesis, even at the high NNK dose used. It was also interesting that the levels of O6-mG in the lungs of the Cyp2abfgs-null mice decreased with time, from 1 to 4h after NNK treatment, in contrast to the time-dependent increase in O6-mG levels seen in the WT mice (Figure 3A). The apparently decreased persistence of O6-mG in the lungs of NNK-treated Cyp2abfgs-null mice may be explained by a presumed reduction in the formation of pyridyloxobutyl DNA adduct, which can inhibit repair of O6-mG by the enzyme O6-alkylguanine-DNA-alkyltransferase (6), concurrent with the observed reduction in the formation of HPB. A large decrease in the amounts of DNA adducts formed, combined with a reduction in the persistence of these adducts, seems to explain the resistance of the Cyp2abfgs-null mice to NNK-induced lung tumorigenesis, despite the occurrence of residual NNK bioactivation activities in the lung.
Lung tumor incidence can be affected by genetic background of the mouse strains utilized for bioassays, with A/J being one of the most sensitive strain to NNK-induced lung tumorigenesis. In this study, the WT and Cyp2abfgs-null mice were both A/J-N3 (i.e. backcrossed to the A/J background three times); A/J-N3 WT mice (87.5% A/J) were previously found to have sufficient susceptibility to NNK-induced lung tumor formation (10). Notably, at the moderate NNK dose of 50mg/kg, the tumor multiplicity in A/J-N3 WT mice (4.1 tumors per mouse; this study) was substantially lower than that in pure A/J WT mice (14.5 tumors/mouse; ref. 17); but, at the high NNK dose of 200mg/kg, the tumor multiplicity was more comparable between A/J-N3 WT (29.6 tumors per mouse) and pure A/J WT (36.1 tumors per mouse). Thus, the results from the high-dose group, and the fact that the Cyp2abfgs-null mice were resistant to lung tumorigenesis at both NNK doses, help to eliminate any doubt that the difference in tumor incidence between A/J-N3 WT and A/J-N3 Cyp2abfgs-null mice was due to variations in genetic background between the two groups.
In conclusion, our results strongly support the hypothesis that, besides CYP2A5, other P450s encoded by the Cyp2abfgs gene cluster also play a significant role in NNK-induced lung tumorigenesis in mice. This finding should stimulate further studies on the role of the corresponding human CYPs in NNK-induced lung tumorigenesis. Multiple human P450 enzymes are capable of metabolizing NNK, including CYP1A1, 1A2, 2A6, 2A13, 2B6, 2E1 and 3A4 (3). Among them, CYP2A13 (an ortholog of mouse CYP2A5), which is selectively expressed in the respiratory tract (32), is the most efficient human P450 in the metabolic activation of NNK (11), with K m values in the low µM range (29). CYP2A13 has been found to be associated with NNK-induced lung adenocarcinoma in both human and animal studies (17,33). Human CYP2B6 may also have a significant impact on NNK-induced lung tumorigenesis, given its expression in the lung (34), its relatively high catalytic efficiency toward NNK (second only to CYP2A13) (11), and its large interindividual variations in expression (35). On the other hand, a role of human CYP2F1 (an ortholog of mouse CYP2F2) in NNK-induced lung tumorigenesis seems less likely, as a heterologously expressed form of CYP2F1 has been reported to have very limited activity toward NNK (36). Nonetheless, further in vivo studies are needed to determine the ability of human CYP2B6 and CYP2F1 in NNK lung tumorigenesis. In that connection, the novel Cyp2abfgs-null mouse, with the eradication of all interference from mouse CYP2A/B/F/G/S enzymes, will be a valuable platform for the generation of the CYP2A, 2B, 2F-humanized mouse models for studies on the role of the specific human CYPs in the bioactivation and toxicity of NNK and other tobacco carcinogens.
Supplementary material
Supplementary Figures S1 and S2 can be found at http://carcin.oxfordjournals.org/
Funding
National Cancer Institute, National Institutes of Health (CA092596).
Supplementary Material
Acknowledgements
We thank Dr X.Zhou for assistance with analytical chemistry and Ms W.Yang for technical support. We gratefully acknowledge the use of the Histopathology Core and the Advanced Light Microscopy and Image Analysis Core Facilities of the Wadsworth Center.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations:
- HPB
4-hydroxy-1-(3-pyridyl)-1-butanone
- NNAL
4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol
- NNK
4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone
- OM
olfactory mucosa
- OPB
4-oxo-4-(3-pyridyl)-butanone
- O6-mG
O6-methylguanine
- P450
cytochrome P450
- WT
wild-type.
References
- 1.IARC. (2004). Tobacco smoke and involuntary smoking. IARC Monogr. Eval. Carcinog. Risks Hum., 83, 1––1438 [PMC free article] [PubMed] [Google Scholar]
- 2. Ding X., et al. (2003). Human extrahepatic cytochromes P450: function in xenobiotic metabolism and tissue-selective chemical toxicity in the respiratory and gastrointestinal tracts. Annu. Rev. Pharmacol. Toxicol., 43, 149–173 [DOI] [PubMed] [Google Scholar]
- 3. Hecht S.S. (1998). Biochemistry, biology, and carcinogenicity of tobacco-specific N-nitrosamines. Chem. Res. Toxicol., 11, 559–603 [DOI] [PubMed] [Google Scholar]
- 4. Wynder E.L., et al. (1995). The changing epidemiology of smoking and lung cancer histology. Environ. Health Perspect., 103 (suppl. 8), 143–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Peterson L.A., et al. (1991). O6-methylguanine is a critical determinant of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone tumorigenesis in A/J mouse lung. Cancer Res., 51, 5557–5564 [PubMed] [Google Scholar]
- 6. Peterson L.A., et al. (1993). Pyridyloxobutyl DNA adducts inhibit the repair of O6-methylguanine. Cancer Res., 53, 2780–2785 [PubMed] [Google Scholar]
- 7. Ahrendt S.A., et al. (2001). Cigarette smoking is strongly associated with mutation of the K-ras gene in patients with primary adenocarcinoma of the lung. Cancer, 92, 1525–1530 [DOI] [PubMed] [Google Scholar]
- 8. Greenblatt M.S., et al. (1994). Mutations in the p53 tumor suppressor gene: clues to cancer etiology and molecular pathogenesis. Cancer Res., 54, 4855–4878 [PubMed] [Google Scholar]
- 9. Ronai Z.A., et al. (1993). G to A transitions and G to T transversions in codon 12 of the Ki-ras oncogene isolated from mouse lung tumors induced by 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) and related DNA methylating and pyridyloxobutylating agents. Carcinogenesis, 14, 2419–2422 [DOI] [PubMed] [Google Scholar]
- 10. Weng Y., et al. (2007). Determination of the role of target tissue metabolism in lung carcinogenesis using conditional cytochrome P450 reductase-null mice. Cancer Res., 67, 7825–7832 [DOI] [PubMed] [Google Scholar]
- 11. Jalas J.R., et al. (2005). Cytochrome P450 enzymes as catalysts of metabolism of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone, a tobacco specific carcinogen. Chem. Res. Toxicol., 18, 95–110 [DOI] [PubMed] [Google Scholar]
- 12. Felicia N.D., et al. (2000). Characterization of cytochrome P450 2A4 and 2A5-catalyzed 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) metabolism. Arch. Biochem. Biophys., 384, 418–424 [DOI] [PubMed] [Google Scholar]
- 13. Jalas J.R., et al. (2003). Stereospecific deuterium substitution attenuates the tumorigenicity and metabolism of the tobacco-specific nitrosamine 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK). Chem. Res. Toxicol., 16, 794–806 [DOI] [PubMed] [Google Scholar]
- 14. Takeuchi H., et al. (2003). Pretreatment with 8-methoxypsoralen, a potent human CYP2A6 inhibitor, strongly inhibits lung tumorigenesis induced by 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone in female A/J mice. Cancer Res., 63, 7581–7583 [PubMed] [Google Scholar]
- 15. von Weymarn L.B., et al. (2005). Effects of 8-methoxypsoralen on cytochrome P450 2A13. Carcinogenesis, 26, 621–629 [DOI] [PubMed] [Google Scholar]
- 16. Miyazaki M., et al. (2005). Mechanisms of chemopreventive effects of 8-methoxypsoralen against 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone-induced mouse lung adenomas. Carcinogenesis, 26, 1947–1955 [DOI] [PubMed] [Google Scholar]
- 17. Megaraj V., et al. (2014). Role of CYP2A13 in the bioactivation and lung tumorigenicity of the tobacco-specific lung procarcinogen 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone: in vivo studies using a CYP2A13-humanized mouse model. Carcinogenesis, 35, 131–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhou X., et al. (2012). Role of CYP2A5 in the bioactivation of the lung carcinogen 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone in mice. J. Pharmacol. Exp. Ther., 341, 233–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wei Y., et al. (2013). Generation and characterization of a novel Cyp2a(4/5)bgs-null mouse model. Drug Metab. Dispos., 41, 132–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li L., et al. (2011). Generation and characterization of a Cyp2f2-null mouse and studies on the role of CYP2F2 in naphthalene-induced toxicity in the lung and nasal olfactory mucosa. J. Pharmacol. Exp. Ther., 339, 62–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schwenk F., et al. (1995). A cre-transgenic mouse strain for the ubiquitous deletion of loxP-flanked gene segments including deletion in germ cells. Nucleic Acids Res., 23, 5080–5081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhou X., et al. (2010). Role of CYP2A5 in the clearance of nicotine and cotinine: insights from studies on a Cyp2a5-null mouse model. J. Pharmacol. Exp. Ther., 332, 578–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang Q.Y., et al. (2003). Characterization of mouse small intestinal cytochrome P450 expression. Drug Metab. Dispos., 31, 1346–1351 [DOI] [PubMed] [Google Scholar]
- 24. Ding X., et al. (1994). Steroid metabolism by rabbit olfactory-specific P450 2G1. Arch. Biochem. Biophys., 315, 454–459 [DOI] [PubMed] [Google Scholar]
- 25. Zhang Q.Y., et al. (2009). An intestinal epithelium-specific cytochrome P450 (P450) reductase-knockout mouse model: direct evidence for a role of intestinal p450s in first-pass clearance of oral nifedipine. Drug Metab. Dispos., 37, 651–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gu J., et al. (1998). Purification and characterization of heterologously expressed mouse CYP2A5 and CYP2G1: role in metabolic activation of acetaminophen and 2,6-dichlorobenzonitrile in mouse olfactory mucosal microsomes. J. Pharmacol. Exp. Ther., 285, 1287–1295 [PubMed] [Google Scholar]
- 27. Anderson L.M., et al. (1991). Tumorigenicity of the tobacco-specific carcinogen 4-(methyl-nitrosamino)-1-(3-pyridyl)-1-butanone in infant mice. Cancer Lett., 58, 177–181 [DOI] [PubMed] [Google Scholar]
- 28. Belinsky S.A., et al. (1992). Role of the alveolar type II cell in the development and progression of pulmonary tumors induced by 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone in the A/J mouse. Cancer Res., 52, 3164–3173 [PubMed] [Google Scholar]
- 29. Su T., et al. (2004). Regulation of the cytochrome P450 2A genes. Toxicol. Appl. Pharmacol., 199, 285–294 [DOI] [PubMed] [Google Scholar]
- 30. Zhuo X., et al. (2004). Targeted disruption of the olfactory mucosa-specific Cyp2g1 gene: impact on acetaminophen toxicity in the lateral nasal gland, and tissue-selective effects on Cyp2a5 expression. J. Pharmacol. Exp. Ther., 308, 719–728 [DOI] [PubMed] [Google Scholar]
- 31. Smith T.J., et al. (1993). Mechanisms of inhibition of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone bioactivation in mouse by dietary phenethyl isothiocyanate. Cancer Res., 53, 3276–3282 [PubMed] [Google Scholar]
- 32. Su T., et al. (2000). Human cytochrome P450 CYP2A13: predominant expression in the respiratory tract and its high efficiency metabolic activation of a tobacco-specific carcinogen, 4-(methylnitrosamino)-1- (3-pyridyl)-1-butanone. Cancer Res., 60, 5074–5079 [PubMed] [Google Scholar]
- 33. Wang H., et al. (2003). Substantial reduction in risk of lung adenocarcinoma associated with genetic polymorphism in CYP2A13, the most active cytochrome P450 for the metabolic activation of tobacco-specific carcinogen NNK. Cancer Res., 63, 8057–8061 [PubMed] [Google Scholar]
- 34. Hukkanen J., et al. (2002). Expression and regulation of xenobiotic-metabolizing cytochrome P450 (CYP) enzymes in human lung. Crit. Rev. Toxicol., 32, 391–411 [DOI] [PubMed] [Google Scholar]
- 35. Mo S.L., et al. (2009). Substrate specificity, regulation, and polymorphism of human cytochrome P450 2B6. Curr. Drug Metab., 10, 730–753 [DOI] [PubMed] [Google Scholar]
- 36. Smith T.J., et al. (1992). Metabolism of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone in human lung and liver microsomes and cytochromes P-450 expressed in hepatoma cells. Cancer Res., 52, 1757–1763 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.