Summary
PARP-1 regulates EMT induction via activating TGF-β signaling in an in-vivo model of prostate cancer progression, resulting in high-grade tumors. This study is of major translational significance in understanding mechanisms of resistance to PARP inhibitors in advanced prostate cancer.
Abstract
Poly (ADP-ribose) polymerase (PARP) is involved in key cellular processes such as DNA replication and repair, gene transcription, cell proliferation and apoptosis. The role of PARP-1 in prostate cancer development and progression is not fully understood. The present study investigated the function of PARP-1 in prostate growth and tumorigenesis in vivo. Functional inactivation of PARP-1 by gene-targeted deletion led to a significant reduction in the prostate gland size in young PARP-1−/− mice (6 weeks) compared with wild-type (WT) littermates. To determine the effect of PARP-1 functional loss on prostate cancer onset, PARP-1−/− mice were crossed with the transgenic adenocarcinoma of the mouse prostate (TRAMP) mice. Pathological assessment of prostate tumors revealed that TRAMP+/−, PARP-1−/− mice exhibited higher grade prostate tumors compared with TRAMP+/− PARP-1+/+ (16–28 weeks) that was associated with a significantly increased proliferative index and decreased apoptosis among the epithelial cells in TRAMP+/− PARP-1−/− prostate tumors. Furthermore tumors harboring PARP-1 loss, exhibited a downregulation of nuclear androgen receptor. Impairing PARP-1 led to increased levels of transforming growth factor-β (TGF-β) and Smads that correlated with induction of epithelial–mesenchymal transition (EMT), as established by loss of E-cadherin and β-catenin and upregulation of N-cadherin and ZEB-1. Our findings suggest that impaired PARP-1 function promotes prostate tumorigenesis in vivo via TGF-β-induced EMT. Defining the EMT control by PARP-1 during prostate cancer progression is of translational significance for optimizing PARP-1 therapeutic targeting and predicting response in metastatic castration-resistant prostate cancer.
Introduction
PARP-1 is the major isoform of poly-ADP-ribose polymerases (PARP); it is a nuclear protein with an ADP-ribosyl transferase catalytic domain with multiple cellular functions including transcriptional regulation and DNA damage repair (1,2). PARP-1 catalyzes the polymerization of ADP-ribose units from donor nicotinamide adenine dinucleotide (NAD+) molecules on DNA strand breaks (1,3). During DNA damage, cells employ multiple types of DNA repair mechanisms: base excision repair, nucleic acid excision repair, homologous recombination, single strand annealing, mismatch repair and non-homologous end joining to repair these damages (3). Under low levels of DNA damage, PARP-1 acts as a survival factor specifically involved in DNA repair while, in the presence of extensive DNA damage, PARP-1 promotes apoptosis (2).
The current understanding that DNA damage-responsive PARPs are the most likely mediators of the therapeutic effect by PARP inhibitors, defines PARP-1 as the primary responder to DNA damage. Targeting PARP-1 has considerable therapeutic value against human cancer. PARP-1 inhibitors are exciting new agents that have shown promise in their toxicity profile, efficacy and resistance mechanism for BRCA-mutated ovarian and breast cancer (4,5). The clinical activity of different PARP inhibitors against tumors from BRCA mutation carriers is established (5,6); more recent studies indicate selective targeting of PARP allosteric regulation to enhance the antitumor therapeutic response (7). Emerging evidence also supports the use of PARP-1 inhibitors as an effective treatment modality in other human cancers harboring BRCA mutations including prostate tumors (8,9).
Prostate cancer development and early onset disease is driven by an aberrant androgen signaling via the androgen receptor (AR) activity for growth promotion and apoptosis inhibition (10,11). Development and progression to advanced metastatic prostate cancer is a consequence of activation of survival processes in direct link with the tumor microenvironment such as apoptosis suppression, resistance to anoikis and epithelial–mesenchymal transition (EMT) (12,13). In preclinical models of prostate cancer EMT endows epithelial cells with invasive properties, induces stem cell properties and prevents apoptosis, thus facilitating cancer metastasis (14–17). Surgical or medical androgen deprivation therapy represents a first line therapy for the treatment of locally advanced prostatic tumors (11,18). Despite the initial success, the majority of patients will relapse and develop castration-resistant prostate cancer (CRPC) and these prostate tumors continue to be dependent on androgen/AR signaling despite being in the presence of castrate androgen levels (18–20). Persistently active AR signaling in CRPC is documented by the antitumor activity exhibited by the new androgen/AR axis inhibitors, such abiraterone and enzalutamide (21,22). Yet the emerging therapeutic resistance to these promising agents (23), underscores the significant challenge of developing novel effective strategies beyond AR-targeted activity, or optimizing its targeting by combination strategies. An additional role for PARP-1 has recently been documented (besides its DNA damage repair function) to regulate AR transcriptional activity in models of CRPC, thus identifying a new therapeutic targeting value for PARP-1 in advanced disease (24).
Therapeutic targeting of PARP-1 via pharmacologic inhibition has promising applications in the clinic, either alone or in combination with radiation therapy, for the treatment of advanced prostate cancer (8,25,26). Dissecting the mechanisms driving resistance to PARP-1 inhibitors will enable the identification of strategic conditions to resensitize tumor cells to PARP-1 inhibition and development of effective combination approaches. In this study we investigated the consequences of physiological impairing of PARP-1 function in vivo, on normal and tumorigenic prostate growth. We found that functional inactivation of PARP-1 by gene-targeted deletion, against a transgenic adenocarcinoma of the mouse prostate (TRAMP) background, results in high-grade prostate tumors due to enhanced proliferative capacity, loss of apoptosis and EMT induction potentially via transforming growth factor-β (TGF-β) signaling.
Materials and methods
Transgenic mouse models
Transgenic and knockout male mice were maintained under environmentally controlled conditions and subject to a 12 h light/dark cycle with food and water ad libitum. The TRAMP mouse model (C57BL/6-Tg-TRAMP-8247Ng/J; Jackson Laboratories, Bar Harbor, ME). (Stock #: 003135) is a well-characterized model of prostate cancer progression to metastasis. The TRAMP transgene is in the C57BL/6J genetic background. PARP-1−/− mouse model (Jackson Laboratories, # 002779) is from 129Sv background. PARP-1−/− mice harbor an inactive PARP-1 protein via the ADPRT mutation (27) that does not affect protein expression levels (Supplementary Figure S2, available at Carcinogenesis Online). By crossing the original PARP-1−/− female mice (background 129Sv) with wild-type (WT) C57BL/6J male mice for six generations, PARP-1−/− in the C57BL/6J background mice were generated. PARP-1−/− males in the C57BL/6J background were crossed to TRAMP heterozygous females. After genotyping, TRAMP+/− PARP-1+/− females were crossed to TRAMP−/−, PARP-1+/− males. Animals (3–6 mice/group) were divided into the following experimental groups: (i) TRAMP−/−, PARP-1−/−; (ii) TRAMP−/−, PARP-1+/−; (iii) TRAMP−/−, PARP-1+/+; (iv) TRAMP+/−, PARP-1−/−; (v) TRAMP+/−, PARP-1+/−; (vi) TRAMP+/−, PARP-1+/+.
Ethics statement
Studies involving animals: For all the studies with mice the institutional and national guidelines for the care and use of animals were followed according to a protocol approved by the University of Kentucky Institutional Review Committee.
Immunohistochemical analysis
Tissue specimens are fixed in 10% (vol/vol) formalin (Sigma–Aldrich, St Louis, MO), and formalin-fixed paraffin-embedded tissue specimens are sectioned (6 μm) using a Finesse microtome (Thermo Shandon, Pittsburgh, PA). Paraffin sections (6 μm) were deparaffinized, rehydrated and stained with hematoxylin and eosin and were subjected to pathologic evaluation by the pathologist (C.H.). Formalin-fixed paraffin-embedded sections of mouse prostates were deparaffinized and subjected to antigen retrieval in citrate buffer (pH 6; Dako, Glostrup, Denmark); The following antibodies were used for detection of specific proteins: rabbit polyclonal antibody against nuclear antigen Ki-67 (Abcam, Cambridge, MA), as a marker of cell proliferation; rabbit polyclonal antibody against E-cadherin (Cell Signaling Technology, Danvers, MA), rabbit polyclonal antibody against the AR and mouse monoclonal antibody against N-cadherin (Santa Cruz Biotechnology, Santa Cruz, CA); the rabbit monoclonal antibody against Smad3 was from Epitomics (Burlingame, CA) and the ZEB-1 antibody was a generous gift from Dr Darlington (University of Louisville, KY). Negative controls consisted of staining with rabbit and mouse IgG (Santa Cruz Biotechnology). Biotinylated goat anti-rabbit and goat anti-mouse IgG and horseradish peroxidase–streptavidin conjugate were used (Millipore, Billerica, MA). Color development was performed using a FAST 3,3′-diaminobenzidine-based kit (Sigma–Aldrich) and counterstained with hematoxylin. Images were captured using an Olympus BX51 microscope system (Olympus America, Center Valley, PA). The number of positive cells (for each protein) over the total number of prostate epithelial cells/field (200–300) was counted by two independent observers (three fields per each section). For the nuclear immunohistochemical quantification of AR, targeted areas on each slide were quantified via digital imaging with an Aperio ScanScope XT whole slide scanner, followed by analysis with Aperio Spectrum Version 11.2.0.80 software (Aperio, Vista, CA).
Apoptosis detection
The incidence of apoptosis was evaluated in situ using the terminal deoxynucleotidyl transferase-mediated dUTP-biotin nick end labeling (TUNEL) assay (Millipore). Prostate tissue sections were counterstained with methyl green and TUNEL-positive cells were counted per high power field (13). Numerical values represent as the average number of positive cells counted from three different fields per section.
Western blot analysis
Prostate tissue was homogenized in TRIzol Reagent (Life Technologies, Grand Island, NY), NE-PER Nuclear and Cytoplasmic Extraction Reagents (Pierce Biotechnology, Rockford, IL). Whole tissue protein (see Supplementary Data, available at Carcinogenesis Online), cytoplasmic and nuclear protein were extracted following the manufacturer’s instructions. Protein expression was determined by immunoblotting using the following specific antibodies: anti β-catenin, anti-Snail and Slug (Cell Signaling Technology); anti-E-cadherin, anti-N-cadherin, anti-Smad3 (same as immunohistochemical analysis), Smad4 and AR (Santa Cruz Biotechnology). Whole and cytoplasmic protein levels were normalized to glyceraldehyde 3-phosphate dehydrogenase expression, using the GAPDH antibody (Cell Signaling Technology). Nuclear protein expression was normalized to histone H3 expression, using the histone H3 antibody (Cell Signaling Technology). Protein content was quantified using the Pierce BCA Protein Assay Kits (Thermo Scientific, Rockford, IL) and protein samples were subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis and transferred to Hybond-C membranes (Amersham Pharmacia Biotech, Piscataway, NJ). Membranes were blocked in 5% milk (2% bovine serum albumin combined 2% goat serum for ZEB-1antibody) in Tris-buffered saline containing 0.05% Tween 20, and following incubation with the respective primary antibody (overnight at 4°C), membranes were exposed to species-specific horseradish peroxidase-labeled secondary antibody (Jackson ImmunoResearch Laboratories, West Grove, PA). Signal detection was achieved with HyGLO Quick Spray Chemiluminescent HRP Antibody Detection Reagent (Denville scientific, Metuchen, NJ). Fold change was determined based on glyceraldehyde 3-phosphate dehydrogenase and histone H3 expression as loading controls for the cytoplasmic and nuclear protein analyses, respectively.
Real-time PCR analysis
Real-time reverse transcription–PCR was used to determine messenger RNA (mRNA) expression for transmembrane protease, serine 2 (TMPRSS2), TGF-β, Smad4, AR, β-catenin, Twist-1, ZEB-1, E-cadherin and N-cadherin genes. Total RNA was extracted from prostate tissue using the TRIzol Reagent (Invitrogen, Life Technologies), and 1 μg was subjected to reverse transcription into complementary DNA using a reverse transcription kit from Promega Corporation (Madison, WI) under the following conditions: 25°C, 10 min; 42°C, 60 min and 95°C, 5 min. Primer pairs and TaqMan probes (Applied Biosystems, Foster City, CA) were used to determine TGF-β, AR, β-catenin, Snail, E-cadherin and N-cadherin gene expression. Primer pairs to assess Smad4, Twist-1 and ZEB-1 mRNA expression were designed using Primer Express software (Applied Biosystems, Branchburg, NJ). The following primers were used: Smad4: forward, GCTTGGGTCAACTCTCCAATG, reverse, TGTGCAACCTCGCTCTCTCA; TMPRSS2: forward, AGGTTCTGGGACAGCAACTGTT, reverse, AGCT GAT GCA TGTGCCTGAAG; Twist-1: forward, CCGGAGACCTAGATGTCATTGTT, reverse, AGTT ATCCAGCTCCAGAGTCTCTAGAC; ZEB-1, forward, GAGA CAC AAATA TGAGCACACAGGTAA, reverse, ATGATGCTTGTGTTTAAATGCCTTT. Real-time PCR was conducted in an ABI Prism 7300 system (Applied Biosystems), in TaqMan Universal PCR Master Mix and Power SYBR green PCR Master Mix. Numerical data (normalized to 18S mRNA and β-actin levels) indicate mean values ± standard error of the mean, n = 4–6.
Statistical analysis
Group I, group II and group III are identified as first littermate pair, second littermate pair and third littermate pair. Each pair of littermates are two offspring (TRAMP+/−, PARP-1−/− and TRAMP+/−, PARP-1+/+) produced by the same pregnancy. Due to different period extracted protein from different littermate pair, there were large variations from one littermate pair to another littermate pair. Considering this heterogeneity among the transgenic mice, the raw data are included for relevant interoffspring comparison among the littermates. The numerical data are analyzed for statistical significance using the unpaired t-test by Graphpadprism4. Values are expressed as the mean ± standard error of the mean. Statistical differences were considered significant at P value of <0.05.
Results
Effect of PARP-1 loss on prostate glandular growth
To assess the effect of PARP-1 deficiency on prostate development, PARP-1−/− (homozygous), PARP-1+/− (heterozygous) and PARP-1+/+ (WT) mice generated by intercrossing of PARP-1+/− mice born according to a Mendelian ratio. The different prostate lobes from the PARP-1−/− (homozygous) mice, 6–12 weeks of age, and age-matched PARP-1+/+ littermates, were subjected to histopathological analysis; the histology appeared phenotypically normal (Figure 1A). The prostate glands from PARP-1−/− mice were underdeveloped relative to WT littermates as revealed by the difference in the prostate weight normalized to the body weight in 6 weeks old mice (Figure 1B). There was also a significant decrease in testicular weight in PARP-1−/− mice compared with WT (Supplementary Data, Figure S1, available at Carcinogenesis Online). Evaluation of the proliferative capacity based on Ki-67 nuclear antigen immunostaining revealed that prostate glands from PARP-1−/− mice (at 8 and 12 weeks) had a significantly decreased proliferative index compared with PARP-1+/+ mice (Figure 1C and D). Figure 1E reveals the incidence of prostate apoptosis from age-matched mice. There was no significant change in the number of TUNEL-positive prostate epithelial cells in tumors from the PARP-1−/− versus the WT control mice.
Fig. 1.
Reduction in prostate glands in PARP-1−/− mice due to decreased proliferative activity. (A) Reveals comparative hematoxylin and eosin staining of prostate tissue sections from PARP-1+/+ and PARP-1−/− of mice during development in early age (6, 8 and 12 weeks). (B) Shows the weights of the prostate glands (all lobes included, ventral, lateral, dorsal and anterior lobe) normalized to body weight from PARP+/+ and PARP−/− mice (6 weeks). (C) indicates the Ki-67 nuclear antigen immunoreactivity in prostate tissue from PARP-1+/+ and PARP-1−/− mice; magnification ×400. (D) Represents the quantitative analysis of the data from C. The number of proliferating cells based on Ki-67 positivity was used to determine the proliferative index as described in Materials and methods. Values indicate average from three independent groups ± standard error of the mean. (E) Reveals TUNEL staining for apoptosis detection in prostate tissue from PARP-1+/+ and PARP−/− mice (age 6, 8 and 12 weeks). (F) Indicates the quantitative analysis of TUNEL-positive cells per section with no significant differences between the two groups. Statistical significance (*) for all the data analyses was determined at a value of P < 0.05. Error bars represent standard error of the mean.
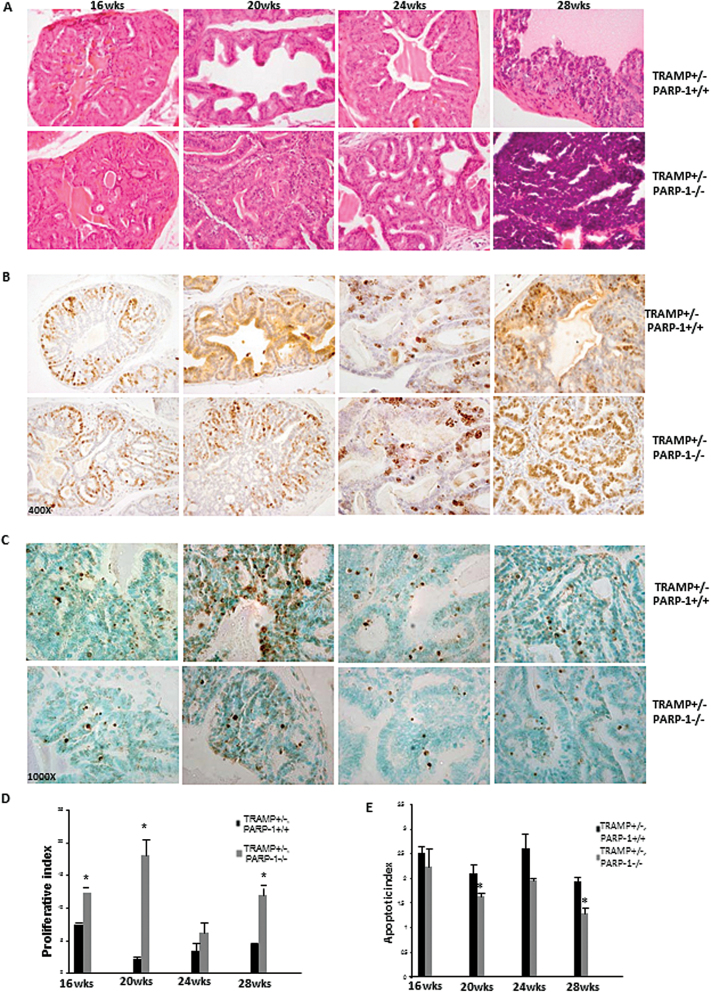
Functional inactivation of PARP-1 accelerates tumor initiation in TRAMP mouse model
Characterizing the time-dependent events between the initiation of prostate premalignant growth and the progression to metastatic disease is critical to optimizing therapeutic targeting of CRPC. To determine the functional involvement of PARP-1 in prostate tumorigenesis, PARP-1−/− mice were crossed with the TRAMP mouse model. Male mice were classified as follows after genotyping: TRAMP+/−, PARP-1−/−; TRAMP+/−, PARP-1+/− and TRAMP+/−, PARP-1+/+.
Pathologic assessment was conducted using a standard grading scale in TRAMP mice (28) (Table I) by a pathologist. As summarized on Table I, prostate tumorigenic lesions from TRAMP+/− PARP-1−/− mice exhibited higher grade compared with lesions in TRAMP+/− PARP-1+/+ for mice 16–28 weeks of age (Figure 2A). Moreover, there was a sharp increase in the size of the tumor compared with same age of TRAMP+/− PARP-1+/+ mice. The differences in the severity and aggressiveness in the tumor grade of the lesions between the TRAMP+/− PARP-1−/− and TRAMP+/− PARP-1+/+ groups were also further validated by increased proliferative capacity among tumor cells as detected by Ki-67 immunoreactivity in serial sections (Figure 2B). A significant increase in the number of Ki-67-positive epithelial cells from prostate tumors from PARP-1−/− mice was detected compared with WT littermates in TRAMP mice (Figure 2B and D). Apoptosis evaluation in serial prostate tumor sections of increasing age revealed a considerable reduction in the number of TUNEL-positive cells in the TRAMP+/− PARP-1−/− derived prostate tumors, compared with WT (Figure 2C). This decrease in apoptosis reached statistical significance for mice at 20 and 28 weeks of age (Figure 2E; P < 0.05).
Table I.
Impact of PARP-1 on prostate tumor grade and aggressiveness
| Pathological evaluation of prostate tumors in TRAMP mice with PARP-1+/+ or PARP-1−/− genotype during age-dependent tumorigenic progression | ||
|---|---|---|
| 16 weeks | TRAMP+/− PARP+/+: | Grade 2–3: Variable filling of lumen with papillary epithelial in growths |
| TRAMP+/− PARP−/−: | Grade 3: Extensive papillary protrusions into the lumen, but not yet cribriforming | |
| 20 weeks | TRAMP+/− PARP+/+: | Grade 3: Papillary projection in all acini, but no cribriform architecture or filling of the lumen |
| TRAMP+/− PARP−/−: | Grade 4–5: Filling and expansion of the lumen, with variably distinct masses beginning to form | |
| 24 weeks | TRAMP+/− PARP+/+: | Grade 3: Robust papillary hyperplasia with some cribriform architecture, but not yet completely filling the lumen and no complex glandular formation. Also, not yet invasive |
| TRAMP+/− PARP−/−: | Grade 5: Large mass of complex, disorganized glandular structures, often in back-to-back arrangement, some microscopic foci of possible early invasion | |
| 28 weeks | TRAMP+/− PARP+/+: | Grade 3: Robust papillary ingrowths, but not yet completely filling the lumen or expanding the overall acinar structure |
| TRAMP+/− PARP−/−: | Grade 6: Scattered foci of unequivocally invasive cancer | |
Fig. 2.
PARP-1 functional loss in TRAMP model increases prostate tumor aggressiveness via enhanced proliferation and reduced apoptosis. (A) Histological evaluation of prostate tissue from TRAMP+/− PARP-1+/+ and TRAMP+/− PARP-1−/− mice of increasing age (16, 20, 24 and 28 weeks) by hematoxylin and eosin staining. (B) Prostate cell proliferative index in tumors from TRAMP+/− PARP-1+/+ and TRAMP+/− PARP-1−/− mice. Ki-67 immunoreactivity was significantly higher in prostate tissue from TRAMP+/− PARP-1−/− compared with TRAMP+/− PARP-1+/+ mice; magnification ×400. (D) Quantitative analysis of Ki-67 staining. (*) indicates statistical significance at P < 0.05. (C) reveals the TUNEL staining of prostate tumor apoptotic cells from TRAMP+/− PARP-1+/+ and TRAMP+/− PARP-1−/− mice (16–28 weeks). Magnification ×1000. (E) Values indicate the average number TUNEL-positive cells per higher field ± standard error of the mean (three fields/point from D).
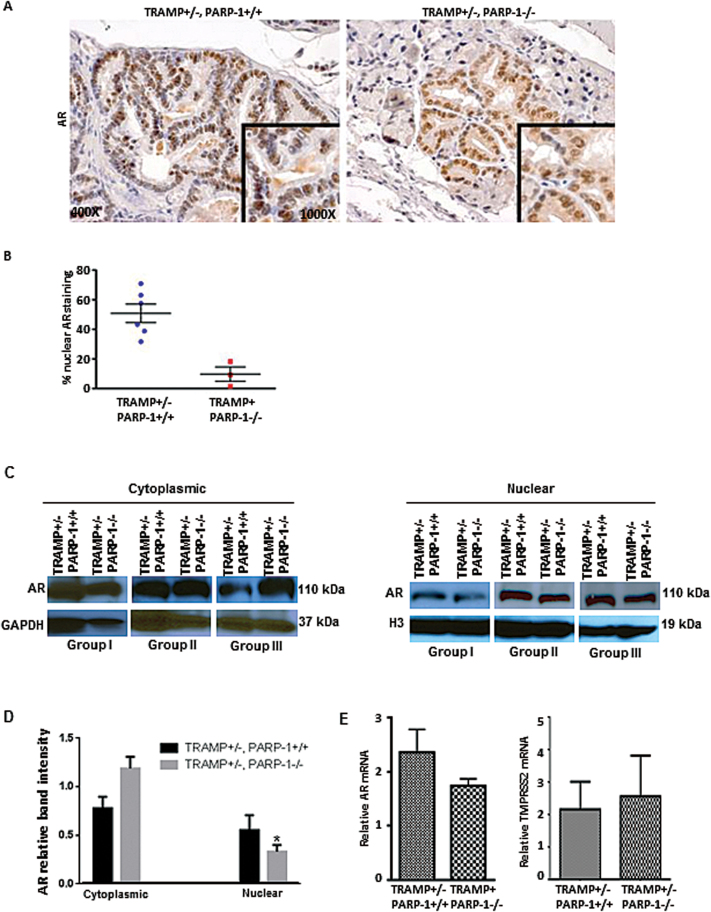
Loss of PARP-1 impairs AR nuclear localization and activity
The AR is a transcription factor and when inactive, it resides in the cell cytoplasm and upon activation by androgens, it translocates to the nucleus where it binds DNA and activates specific gene expression. To determine the functional consequences of impairing PARP-1 on AR activity, we performed analysis of AR expression and cellular localization in prostate tumors from the transgenic mice by immunostaining. The results revealed a significant reduction in nuclear AR in prostate epithelial cells from the TRAMP+/−, PARP-1−/− derived tumors compared with WT (20 weeks) (Figure 3A and B). Figure 3C indicates western blotting of AR distribution in the cytoplasmic and nuclear fractions of prostate tissue lysates. Also shown on Figure 3D is the quantitative densitometric analysis of the protein band intensity from the three independent groups. Data from reverse transcription–PCR analysis indicating mRNA expression of AR and its target gene TMPRSS2 in prostate tumors from TRAMP+/− PARP-1−/− mice compared with TRAMP+/− PARP-1+/+ mice are shown on Figure 3. There was only a modest decrease in AR mRNA in prostate tumors from TRAMP+/− PARP-1−/− compared with WT mice as shown on Figure 3E; however, there was not a significant difference on AR and TMPRSS2 mRNA expression between prostate tumors from TRAMP+/− PARP-1−/− and TRAMP+/− PARP+/+ mice (Figure 3E).
Fig. 3.
Loss of PARP-1 reduces AR nuclear localization and activity. (A) Profiling of AR protein localization and immunoreactivity in prostate tissues from TRAMP+/− PARP+/+ and TRAMP+/− PARP-1−/− mice (20 weeks); magnification ×400. (B) Quantitative analysis of data from A, reveals a significant decrease in nuclear AR in prostate tissue from TRAMP+/−, PARP-1−/− mice compared with controls (P < 0.05). (C) Western blot analysis of cytoplasmic and nuclear fractions from prostate tissue lysates. The blots shown represent the actual results from three independent groups of mice (I, II and III, each group was littermate: TRAMP+/− PARP-1+/+ and TRAMP+/− PARP-1−/−). (D) The barographs on the right indicate the protein band intensity for cytoplasmic and nuclear AR as determined by densitometry and expressed relative to the loading control protein (for each fraction). (E) Shows the numerical data from the reverse transcription–PCR analysis of mRNA expression of AR and its target gene TMPRSS2 in prostate tumors from TRAMP+/− PARP-1−/− mice compared with WT control mice. Values represent average from 4–6 mice per group ± standard error of the mean.
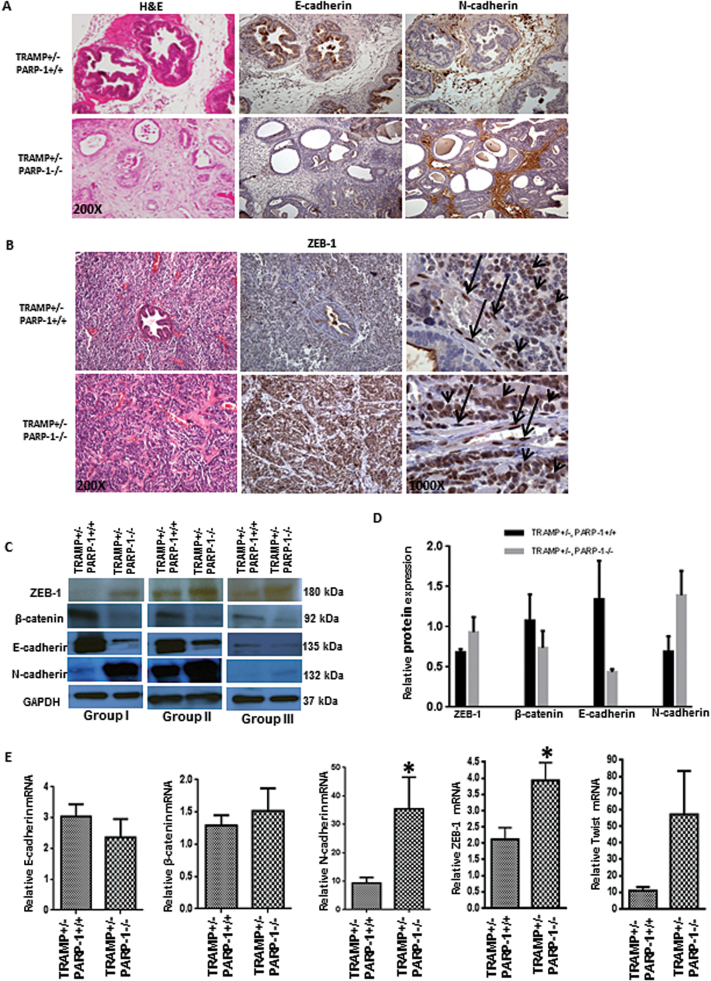
PARP functional loss induces EMT and TGF-β signaling in prostate tumorigenesis model
To identify the potential mechanism driving the more aggressive phenotype observed in the prostate tumors from TRAMP+/−, PARP-1−/− compared with TRAMP+/− PARP-1+/+, we profiled the EMT landscape in the different groups of mice. Expression of epithelial and mesenchymal markers was comparatively analyzed by immunostaining in prostate sections from TRAMP+/− PARP-1−/− and TRAMP+/− PARP-1+/+ mice. Acquisition of mesenchymal characteristics caused by PARP-1 functional loss as indicated by the increased N-cadherin immunoreactivity paralleled by reduced E-cadherin expression in serial sections of prostate cancer tissue from TRAMP+/−, PARP-1 −/− mice (Figure 4A). EMT was confirmed by the high expression and strong nuclear presence of the transcriptional regulator ZEB-1 (Figure 4B, C) (Supplementary Figure S4, available at Carcinogenesis Online), that was associated with the development of high-grade prostate tumors as revealed by hematoxylin and eosin staining (Table I). Although a high expression of ZEB-1 in prostate tumors (Figure 4B, arrow head) and stromal cells (Figure 4B, arrow) derived from PARP-1+/+ mice, the prostate tumors and stromal cells derived from PARP-1−/− mice exhibited a significantly higher intensity of ZEB-1 expression compared with PARP-1+/+ derived tumors. Protein expression profiling (by western blotting), revealed a downregulation of epithelial cell proteins E-cadherin and β-catenin (Figure 4C), that was associated with a marked increase in mesenchymal cell markers, N-cadherin and ZEB-1, in prostate tumors from the transgenic mice TRAMP+/− PARP-1−/− compared with those in control mice (Figure 4C and D). Real time reverse transcription–PCR analysis revealed that upon PARP-1 functional loss there is a significant increase in the mRNA levels for the EMT promoter genes, N-cadherin, ZEB-1 and Twist (Figure 4E), a transcript profile consistent with EMT induction (29). No significant changes were detected in E-cadherin and β-catenin mRNA expression.
Fig. 4.
PARP-1 deficiency yields acquisition of EMT phenotype during prostate tumorigenesis. (A) Expression profile of E-cadherin and N-cadherin in prostate tissue from TRAMP+/− PARP-1+/+ and TRAMP+/− PARP-1−/− mice (28 weeks); magnification ×200. (B) Reveals immunoreactivity pattern for ZEB1 in prostate tumor sections from TRAMP+/−/PARP-1 +/+ and TRAMP+/−/PARP-1−/− mice. Magnification, ×200 and ×1000 (middle and right panels, respectively, ZEB-1 staining). Serial sections were stained with hematoxylin and eosin (left), revealing high grade, poorly differentiated prostate tumor from TRAMP+/− PARP−/− mice. (C) Shows western blot analysis of EMT regulators ZEB-1, E-cadherin, N-cadherin, β-catenin expression in prostate tissue from three different groups of TRAMP+/− PARP-1+/+ and TRAMP+/− PARP-1−/− mice (16 weeks). The three blots are representative of three independent groups of mice (groups I, II and III). (D) The barographs indicate the relative band intensity (from C) as determined by densitometry and expressed relative to glyceraldehyde 3-phosphate dehydrogenase (loading control). (E) Shows mRNA profile by reverse transcription–PCR analysis of E-cadherin, β-catenin, N-cadherin, ZEB-1 and Twist gene expression in TRAMP+/− PARP-1+/+ versus TRAMP+/− PARP-1−/− mice (16 weeks). Statistical significance (*) was determined at a value of P < 0.05. Error bars represent standard error of the mean from the three groups of littermate pairs.
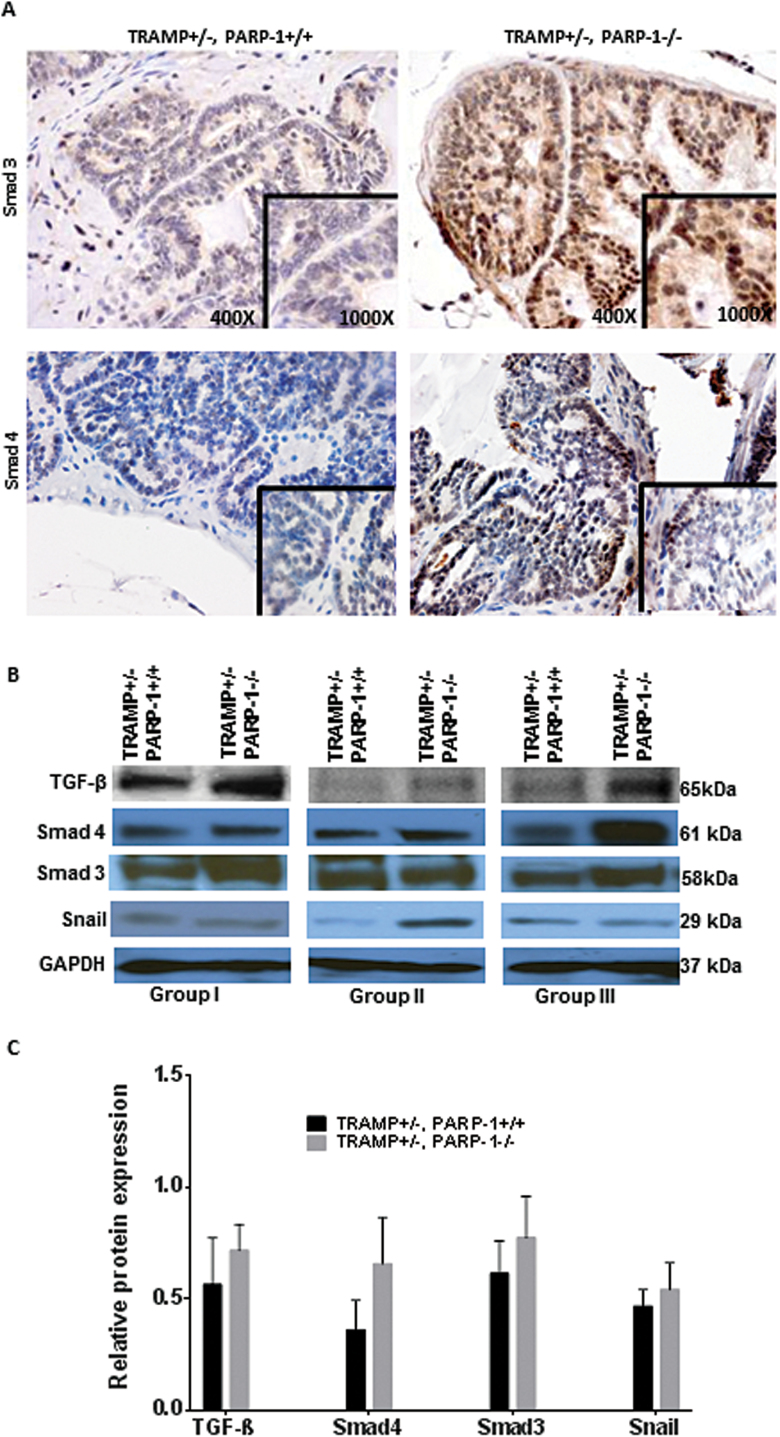
Considering the critical function of TGF-β signaling pathway as a driver of EMT (29,30), we subsequently examined the expression and cellular distribution of TGF-β effectors Smad3 and Smad4. A striking increase in Smad3 immunoreactivity and β-catenin nuclear localization was detected in prostate tumors from the PARP-1−/− compared with WT controls (Figure 5A) (Supplementary Figure S4, available at Carcinogenesis Online). Protein levels for both Smad3 and Smad4 were markedly elevated in prostate tumors from TRAMP+/− PARP-1−/− mice (Figure 5B, C). These changes in the Smad signaling effectors were associated with an increase in TGF-β ligand expression in tumors in TRAMP mice harboring PARP-1 loss (Figure 5B and C). There was also an upregulation in Snail protein levels in prostate tumors consequential to PARP loss (Figure 5B and C).
Fig. 5.
Dysfunctional PARP-1 induces TGF-β-mediated EMT in prostate tumors. Analysis of expression of TGF-β signaling effectors by immunostaining and western blotting in prostate tissue from TRAMP mice harboring PARP-1 functional loss compared with TRAMP+/− PARP-1+/+ mice (A and B, respectively). (A) Reveals intense nuclear immunoreactivity for the TGF-β intracellular effector, Smad3 protein in prostate tumor cells in tissue specimens from TRAMP+/− PARP-1−/− mice, compared with age-matched TRAMP+/− PARP-1+/+ mice; magnification, ×400. (B) Comparative protein profiling by western blot analysis of prostate tumor lysates for TGF-β (ligand) and its main intracellular effectors Smad 3 and Smad4 in TRAMP+/− PARP-1−/− prostate tumors versus WT mice, and Snail, EMT regulator, were also detected by western blotting. Results are shown for each of the three independent groups of mice analyzed (for each genotype) are shown (groups I, II and III). Molecular weights are shown on the right (kDa) for each specific protein. (C) Indicates the results of densitometric analysis from B, with the barographs showing the average value of band intensity relative to the loading control protein.
Discussion
The role of PARP-1 in prostate cancer has not been fully identified despite the use of PARP inhibitors to impair advanced disease. The present study demonstrates that functional inactivation of PARP-1 by gene-targeted deletion, in an in vivo model of prostate cancer progression, leads to EMT induction toward high-grade prostate tumors. The association of the clinical emergence of CRPC with increased PARP-1 activity during disease progression (24,31) and the documented therapeutic efficacy of PARP-1 inhibitors in the treatment of patients with advanced disease (8), implicate a significant involvement of PARP-1 in prostate cancer development and progression. Mounting mechanistic evidence supports a critical role for PARP-1 in EMT control via the ability of PARP-1 to transcriptionally regulate functionally relevant genes, in direct accord with our findings. Specifically PARP-1 directly regulates Smad-mediated transcriptional activation during TGF-β-induced EMT in cancer cells (29). PARP-1 has also been shown to bind to Snail promoter, integrin-linked kinase responsive element (SIRE) to modulate Snail expression (32); in addition, PARP-1 is required for Snail cooperative interaction with LSD1 to functionally repress PTEN (33). Moreover, PARP-1 can transcriptionally activate fibronectin gene expression (34) and regulate Snail1 protein stability (35). A V762A (valine to alanine) polymorphism in the proline rich PPXXP domain-PARP-1 activation domain has been linked to prostate cancer (31).
As a corollary to the consequences of PARP-1 loss on promoting prostate tumorigenesis, we observed the impact of PARP-1 loss on EMT and the potentiation of its key characteristic features, such as E-cadherin loss and ZEB-1 transcription factor nuclear accumulation and enhanced expression of mesenchymal features such as N-cadherin. Recruitment of mesenchymal stem cells into prostate tumors, promotes their conversion into cancer-associated fibroblasts that facilitate metastasis by establishing a supportive tumor microenvironment (13,36,37). The EMT phenotype was identified in the prostates of TRAMP+/−, PARP-1−/− mice as manifested by loss of E-cadherin and β-catenin and upregulation of N-cadherin (mesenchymal marker) and ZEB-1 and Snail transcriptional regulators of EMT. TGF-β induces prostate cancer EMT via signaling through downstream Smad-dependent signaling (38). Smad-mediated TGF-β signaling events induce context-dependent expression of ZEB1, mediated in part by signaling interactions with effectors such as the activated R-Smads and the Snail (SNAI1) transcription factor (39,40). This rapidly growing evidence designates a prospective role for ZEB1 as an attractive target for inhibiting EMT (41), toward impairing metastatic disease and overcoming therapeutic resistance in CRPC. Complete reversion of malignant cells to an epithelial-like phenotype might not be achieved by targeting ZEB transcription factors alone (42), but engaging additional players such as specific microRNAs might be required (43).
Significantly enough, the germline BRCA2 mutation confers the highest genetic risk of prostate cancer at 8.6-fold among younger men (≤65 years) (44). Patients with prostate tumors harboring gBRCAm are more aggressive, with a higher likelihood of distant metastasis and poor survival outcomes (45). The focused pursuit of the PARP family of enzymes has identified only few factors other than damaged DNA to activate PARP-1-mediated protein modification (3). The action of PARP inhibitors in prostate cancer cells with other DNA repair abnormalities besides BRCA mutations, is governed by PTEN defects and ETS gene fusions (45,46).The TMPRSS2-ERG fusion gene, observed in ~50% of human prostate cancers, leads to an androgen-induced ERG expression and promotes tumorigenesis. Preclinical evidence has established that PARP1 directly interacts with ERG to inhibit ETS gene fusion activity and inhibiting PARP1 reduces ETS-positive prostate cancer growth, thus enabling a mechanistic rationale for PARP-1 inhibition in ETS gene fusion-positive prostate cancer similar to that of BRCA1/2 deficiency (46). More recent studies demonstrated preferential targeted radiosensitization with PARP-1 inhibition of TMPRSS2-ERG fusion-gene-positive prostate cancer cells (26), especially those that are PTEN-deficient (47). ERG positivity may therefore have a potential predictive biomarker value for sensitivity to PARP inhibitors in CRPC. However, in our experimental model of prostate cancer progression, there were no significant differences in PTEN protein and TMPRSS2 mRNA expression in the prostate tumors from TRAMP+/− PARP-1+/+ versus TRAMP+/− PARP-1−/− mice (Supplementary Figures S2 and S3, available at Carcinogenesis Online).
Mechanistic evidence supporting that the transcriptional dynamics triggered by TGF-β are predominantly governed by the Smad nuclear localization rather than activity (48), challenges the conventional view of the TGF-β signaling in control of complex key processes in cancer cells. Moreover, molecular studies have identified PARP-1 as a Smad-interacting partner and its ability to regulate Smad transcriptional events and responses to TGF-β, including EMT via ADP ribosylation of Smad 3 and 4 (by causing dissociation of nuclear Smads from Smad binging elements) (29). Such a regulatory effect by PARP-1 on nuclear Smad function, is further supported by our findings indicating a significant increase in nuclear Smad3 upon PARP-1 functional loss that is associated with EMT induction in prostate tumors. We also found a marked decrease in nuclear AR localization and cellular content in prostate tumors from TRAMP+/− PARP-1−/− mice (20 weeks), reflecting a reduced AR activity. This is accordance with two lines of recent preclinical evidence: (i) linking PARP-1 function with the AR transcriptional activity in tumor progression to CRPC (25), and (ii) indicating a requirement of a threshold of AR activity to induce prostate cancer cell EMT and invasive behavior (17). Thus, the emerging EMT landscape in prostate cancer involves the governing by PARP-1 via TGF-β signaling and potential AR contribution toward aggressive disease and therapeutic resistance (Figure 6). Our study gains support from the reported clinical association between prostate tumors with a transcript profile consistent with EMT with early relapse after surgical resection, as well as biochemical recurrence in patients with bone metastasis (49,50).
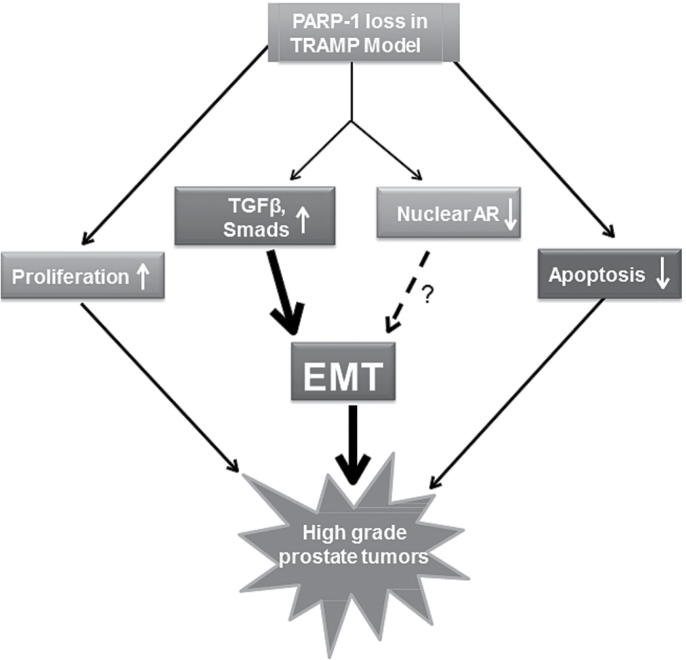
Fig. 6.
Schematic illustration of the impact of PARP-1 loss of function leading to EMT induction and aggressive prostate tumor growth, potentially mediated by Smad-directed TGF-β signaling. A potential cross-effect by nuclear AR depletion may contribute to enhanced EMT.
In summary, our findings demonstrate the ability of PARP-1 to regulate Smad-dependent responses to TGF-β signaling, and potential AR activity, directing both toward EMT in prostate cancer progression (schematically shown on Figure 6). Navigating the EMT–MET cycles via selective inhibition of PARP-1 (under the control of androgen/AR axis and TGF-β signaling) may represent a powerful combination strategy for effectively overcoming resistance and understanding side effects in tissue where PARP-1 is rendered dysfunctional. PARP-1 inhibitors may now have to be optimized for further clinical utility in PARP-1 selective targeting for the treatment of metastatic CRPC, via actions beyond DNA repair deficiencies, by disrupting the EMT cellular process in a TGF-β/AR co-dependent manner. In the pursuit of biomarkers to identify tumors with sensitivity to PARP-1 inhibitors, EMT governing during prostate tumor progression might provide new lead candidates.
Supplementary material
Supplementary Table S1 and Figures S1–S4 can be found at http://carcin.oxfordjournals.org/
Funding
James F. Hardymon Endowment ; NIH grant (R01 DK 083761 to N.K.); NCI K08 grant (CA155764); COBRE (National Institute of General Medical Sciences, 2P20 RR020171 to C.H.); the University of Kentucky College of Medicine Physician Scientist Program (P.J.H., C.H.).
Supplementary Material
Acknowledgements
The authors thank L.Howard (Department of Urology, University of Kentucky) for her expert assistance with the electronic submission of the manuscript. Authors’ contributions: Conception and design: H.P., N.K.; Development and methodology: H.P., C.H.; Acquisition of data: H.P., P.J.H., E.A.M., T.A., C.H.; Writing, review of manuscript: N.K., H.P., P.J.H., C.H.; Administrative support: N.K.; Study supervision: N.K.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations:
- AR
androgen receptor
- CRPC
castration-resistant prostate cancer
- EMT
epithelial–mesenchymal transition
- mRNA
messenger RNA
- PARP-1
poly (ADP-ribose) polymerase-1
- TGF-β
transforming growth factor-β
- TMPRSS2
transmembrane protease, serine 2
- TRAMP
transgenic adenocarcinoma of the mouse prostate
- TUNEL
terminal deoxynucleotidyl transferase-mediated dUTP-biotin nick end labeling
- WT
wild-type.
References
- 1. Hassa P.O., et al. (2008). The diverse biological roles of mammalian PARPS, a small but powerful family of poly-ADP-ribose polymerases. Front. Biosci., 13, 3046–3082 [DOI] [PubMed] [Google Scholar]
- 2. Gibson B.A., et al. (2012). New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat. Rev. Mol. Cell Biol., 13, 411–424 [DOI] [PubMed] [Google Scholar]
- 3. Schreiber V., et al. (2006). Poly(ADP-ribose): novel functions for an old molecule. Nat. Rev. Mol. Cell Biol., 7, 517–528 [DOI] [PubMed] [Google Scholar]
- 4. Ratner E.S., et al. (2012). Poly (ADP-ribose) polymerase inhibitors: on the horizon of tailored and personalized therapies for epithelial ovarian cancer. Curr. Opin. Oncol., 24, 564–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Montoni A., et al. (2013). Resistance to PARP-Inhibitors in Cancer Therapy. Front. Pharmacol., 4, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Steffen J.D., et al. (2014). Targeting PARP-1 allosteric regulation offers therapeutic potential against cancer. Cancer Res., 74, 31–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fong P.C., et al. (2009). Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N. Engl. J. Med., 361, 123–134 [DOI] [PubMed] [Google Scholar]
- 8. Sandhu S.K., et al. (2013). Poly (ADP-ribose) polymerase (PARP) inhibitors for the treatment of advanced germline BRCA2 mutant prostate cancer. Ann. Oncol., 24, 1416–1418 [DOI] [PubMed] [Google Scholar]
- 9. Leongamornlert D., et al. ; UKGPCS Collaborators. (2012). Germline BRCA1 mutations increase prostate cancer risk. Br. J. Cancer, 106, 1697–1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nieto M., et al. (2007). Prostate cancer: re-focusing on androgen receptor signaling. Int. J. Biochem. Cell Biol., 39, 1562–1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Harris W.P., et al. (2009). Androgen deprivation therapy: progress in understanding mechanisms of resistance and optimizing androgen depletion. Nat. Clin. Pract. Urol., 6, 76–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Weischenfeldt J., et al. (2013). Integrative genomic analyses reveal an androgen-driven somatic alteration landscape in early-onset prostate cancer. Cancer Cell, 23, 159–170 [DOI] [PubMed] [Google Scholar]
- 13. Pu H., et al. (2009). Dysfunctional transforming growth factor-beta receptor II accelerates prostate tumorigenesis in the TRAMP mouse model. Cancer Res., 69, 7366–7374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Brennen W.N., et al. (2013). Mesenchymal stem cells as a vector for the inflammatory prostate microenvironment. Endocr. Relat. Cancer, 20, R269–R290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yang J., et al. (2008). Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev. Cell, 14, 818–829 [DOI] [PubMed] [Google Scholar]
- 16. Schmalhofer O., et al. (2009). E-cadherin, beta-catenin, and ZEB1 in malignant progression of cancer. Cancer Metastasis Rev., 28, 151–166 [DOI] [PubMed] [Google Scholar]
- 17. Zhu M.L., et al. (2010). Role of androgens and the androgen receptor in epithelial-mesenchymal transition and invasion of prostate cancer cells. FASEB J., 24, 769–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hu R., et al. (2010). Molecular processes leading to aberrant androgen receptor signaling and castration resistance in prostate cancer. Expert Rev. Endocrinol. Metab., 5, 753–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mohler J.L., et al. (2004). The androgen axis in recurrent prostate cancer. Clin. Cancer Res., 10, 440–448 [DOI] [PubMed] [Google Scholar]
- 20. Attar R.M., et al. (2009). Castration-resistant prostate cancer: locking up the molecular escape routes. Clin. Cancer Res., 15, 3251–3255 [DOI] [PubMed] [Google Scholar]
- 21. Scher H.I., et al. (2005). Biology of progressive, castration-resistant prostate cancer: directed therapies targeting the androgen-receptor signaling axis. J. Clin. Oncol., 23, 8253–8261 [DOI] [PubMed] [Google Scholar]
- 22. Tran C., et al. (2009). Development of a second-generation antiandrogen for treatment of advanced prostate cancer. Science, 324, 787–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ryan C.J., et al. ; COU-AA-302 Investigators. (2013). Abiraterone in metastatic prostate cancer without previous chemotherapy. N. Engl. J. Med., 368, 138–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schiewer M.J., et al. (2012). Dual roles of PARP-1 promote cancer growth and progression. Cancer Discov., 2, 1134–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Han S., et al. (2013). Targeted radiosensitization of ETS fusion-positive prostate cancer through PARP1 inhibition. Neoplasia, 15, 1207–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Barreto-Andrade J.C., et al. (2011). Response of human prostate cancer cells and tumors to combining PARP inhibition with ionizing radiation. Mol. Cancer Ther., 10, 1185–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang Z.Q., et al. (1995). Mice lacking ADPRT and poly(ADP-ribosyl)ation develop normally but are susceptible to skin disease. Genes Dev., 9, 509–520 [DOI] [PubMed] [Google Scholar]
- 28. Suttie A., et al. (2003). A grading scheme for the assessment of proliferative lesions of the mouse prostate in the TRAMP model. Toxicol. Pathol., 31, 31–38 [DOI] [PubMed] [Google Scholar]
- 29. Lönn P., et al. (2010). PARP-1 attenuates Smad-mediated transcription. Mol. Cell, 40, 521–532 [DOI] [PubMed] [Google Scholar]
- 30. Heldin C.H., et al. (2009). Mechanism of TGF-beta signaling to growth arrest, apoptosis, and epithelial-mesenchymal transition. Curr. Opin. Cell Biol., 21, 166–176 [DOI] [PubMed] [Google Scholar]
- 31. Lockett K.L., et al. (2004). The ADPRT V762A genetic variant contributes to prostate cancer susceptibility and deficient enzyme function. Cancer Res., 64, 6344–6348 [DOI] [PubMed] [Google Scholar]
- 32. Lee J.M., et al. (2006). The epithelial-mesenchymal transition: new insights in signaling, development, and disease. J. Cell Biol., 172, 973–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lin Y., et al. (2014). Doxorubicin enhances Snail/LSD1-mediated PTEN suppression in a PARP1 dependent manner. Cell Cycle, 13, 1708–1716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Stanisavljevic J., et al. (2011). The p65 subunit of NF-κB and PARP1 assist Snail1 in activating fibronectin transcription. J. Cell Sci., 124(Pt 24), 4161–4171 [DOI] [PubMed] [Google Scholar]
- 35. Rodríguez M.I., et al. (2011). Poly(ADP-ribose)-dependent regulation of Snail1 protein stability. Oncogene, 30, 4365–4372 [DOI] [PubMed] [Google Scholar]
- 36. Jung Y., et al. (2013). Recruitment of mesenchymal stem cells into prostate tumours promotes metastasis. Nat. Commun., 4, 1795–1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Peinado H., et al. (2007). Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat. Rev. Cancer, 7, 415–428 [DOI] [PubMed] [Google Scholar]
- 38. Deckers M., et al. (2006). The tumor suppressor Smad4 is required for transforming growth factor beta-induced epithelial to mesenchymal transition and bone metastasis of breast cancer cells. Cancer Res., 66, 2202–2209 [DOI] [PubMed] [Google Scholar]
- 39. Fuxe J., et al. (2010). Transcriptional crosstalk between TGF-β and stem cell pathways in tumor cell invasion: role of EMT promoting Smad complexes. Cell Cycle, 9, 2363–2374 [DOI] [PubMed] [Google Scholar]
- 40. Graham T.R., et al. (2008). Insulin-like growth factor-I-dependent up-regulation of ZEB1 drives epithelial-to-mesenchymal transition in human prostate cancer cells. Cancer Res., 68, 2479–2488 [DOI] [PubMed] [Google Scholar]
- 41. Drake J.M., et al. (2009). ZEB1 enhances transendothelial migration and represses the epithelial phenotype of prostate cancer cells. Mol. Biol. Cell, 20, 2207–2217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Das S., et al. (2009). Complete reversal of epithelial to mesenchymal transition requires inhibition of both ZEB expression and the Rho pathway. BMC Cell Biol., 10, 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kim T., et al. (2011). p53 regulates epithelial-mesenchymal transition through microRNAs targeting ZEB1 and ZEB2. J. Exp. Med., 208, 875–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kote-Jarai Z., et al. ; UKGPCS Collaborators. (2011). BRCA2 is a moderate penetrance gene contributing to young-onset prostate cancer: implications for genetic testing in prostate cancer patients. Br. J. Cancer, 105, 1230–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Castro E., et al. (2013). Germline BRCA mutations are associated with higher risk of nodal involvement, distant metastasis, and poor survival outcomes in prostate cancer. J. Clin. Oncol., 31, 1748–1757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Brenner J.C., et al. (2011). Mechanistic rationale for inhibition of poly(ADP-ribose) polymerase in ETS gene fusion-positive prostate cancer. Cancer Cell, 19, 664–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chatterjie P., et al. (2013). PARP inhibition sensitizes to low-dose radiation TMPSS2-ERG fusion gene-expressing and PTEN deficient prostate cancer cells. PLoS One, 8, e60408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Warmflash A., et al. (2012). Dynamics of TGF-β signaling reveal adaptive and pulsatile behaviors reflected in the nuclear localization of transcription factor Smad4. Proc. Natl Acad. Sci. USA, 109, E1947–E1956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sethi S., et al. (2010). Molecular signature of epithelial-mesenchymal transition (EMT) in human prostate cancer bone metastasis. Am. J. Transl. Res., 3, 90–99 [PMC free article] [PubMed] [Google Scholar]
- 50. Behnsawy H.M., et al. (2013). Expression patterns of epithelial-mesenchymal transition markers in localized prostate cancer: significance in clinicopathological outcomes following radical prostatectomy. BJU Int., 111, 30–37 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.