Summary
Targeted loss of E-cadherin and TGFβ receptor II leads to spontaneous squamous cell carcinoma in constitutive and inducible knock-out mouse models. Tumors develop in the oral cavity and forestomach with a long latency and have the potential for regional and distant metastasis.
Abstract
Although the etiology of squamous cell carcinomas of the oral mucosa is well understood, the cellular origin and the exact molecular mechanisms leading to their formation are not. Previously, we observed the coordinated loss of E-cadherin (CDH1) and transforming growth factor beta receptor II (TGFBR2) in esophageal squamous tumors. To investigate if the coordinated loss of Cdh1 and Tgfbr2 is sufficient to induce tumorigenesis in vivo, we developed two mouse models targeting ablation of both genes constitutively or inducibly in the oral–esophageal epithelium. We show that the loss of both Cdh1 and Tgfbr2 in both models is sufficient to induce squamous cell carcinomas with animals succumbing to the invasive disease by 18 months of age. Advanced tumors have the ability to invade regional lymph nodes and to establish distant pulmonary metastasis. The mouse tumors showed molecular characteristics of human tumors such as overexpression of Cyclin D1. We addressed the question whether TGFβ signaling may target known stem cell markers and thereby influence tumorigenesis. From our mouse and human models, we conclude that TGFβ signaling regulates key aspects of stemness and quiescence in vitro and in vivo. This provides a new explanation for the importance of TGFβ in mucosal homeostasis.
Introduction
The treatment of squamous cell cancers (SCC) of the upper aero-digestive tract has not significantly improved despite great strides to better understand the disease. Their resistance to radio-chemotherapy, high recurrence rate, and metastatic behavior are challenges that require novel models and approaches. The aggressive progression of these tumors is associated with increased cell invasion and metastasis due to the loss of cell–cell-adhesion regulated by E-cadherin and transforming growth factor beta (TGFβ) signaling. E-cadherin has emerged as one of the ‘caretakers’ of the epithelial phenotype (1). A direct role for E-cadherin in suppression of tumor invasion has been demonstrated by the reversion of undifferentiated, invasive tumors to a differentiated phenotype after transfection of E-cadherin cDNA in cell culture (2). Furthermore, E-cadherin is a powerful prognostic marker for oral squamous cell carcinomas (OSCCs) showing an inverse relationship with survival (3).
Despite the clear link of reduced levels of E-cadherin with invasive behavior and poor patient survival, the precise mechanisms underlying E-cadherin’s function in the context of metastasis or therapy-resistance are not well established. One possibility is that the adhesive function of E-cadherin prevents cells from dissociating from each other and migrating into adjacent tissues (4). Alternatively, E-cadherin binds and sequesters β-catenin to the membrane, which regulates the cytoplasmic pool of β-catenin and therefore, its role as an essential intracellular mediator of the Wnt signaling pathway (5).
This hypothesis is supported by studies in Drosophila and Xenopus embryos (6,7). Mice lacking E-cadherin in the skin and other squamous epithelia, including the esophagus, either die perinatally due to water barrier defects of the epidermis (8) or show hyperproliferative and degenerative responses, but are viable (9). In the mouse model generated by Tinkle et al. (8), adhesion complexes are unaffected by the loss of E-cadherin through compensation by P-cadherin.
Alternatively, E-cadherin can be viewed as an inhibitor of epithelial–mesenchymal transition (EMT) and by extension of the establishment of highly therapy-resistant cancer stem cells (CSCs).
The role of TGFβ signaling in EMT is well established (10). However, TGFβ exhibits context–dependent effects on keratinocytes ranging from cell cycle arrest to EMT (11–13).
Loss of Tgfbr2 has been modeled in mice through a dominant- negative approach in skin and mammary glands, and by the conditional knock-out of Tgfbr2 (14,15). In these models, tumors develop with a high potential for metastasis, thereby supporting the tumor-suppressive function of Tgfbr2 and intact TGFβ signaling. Hypermethylation of TGFBR1 and TGFBR2 has been reported for high-grade dysplasia and squamous cell carcinoma of the esophagus (16). Moreover restoration of wild-type TGFBR2 expression in colon and breast carcinoma cell lines that lack a functional TGFBR2 allele (17) and overexpression in thyroid carcinomas reinforces this hypothesis (18). Indeed, re-expression of TGFBR2 conferred growth inhibition, suppressed anchorage independence, and abolished tumor formation in nude mice (17).
We previously observed that 70% of human esophageal tumors demonstrated coordinated loss of E-cadherin and TGFBR2 (19). When grown in three-dimensional organotypic cultures, cells with impaired E-cadherin and TGFBR2 function demonstrate fibroblast-dependent invasion into the underlying matrix (20). When analyzing publicly available databases for CDH1 and TGFBR2 expression in head-and-neck squamous cell carcinoma, we observed direct correlations of CDH1 and TGFBR2 status with disease recurrence and overall patient survival. Therefore, we hypothesized that coordinated loss of E-cadherin, Cdh1, and transforming growth factor beta receptor II, Tgfbr2, will lead to tumorigenesis in vivo. We developed a mouse model targeting Cdh1 and Tgfbr2 loss in the oral–esophageal epithelium using the Epstein–Barr virus L2 promoter, ED-L2. For spatio-temporal control, we also generated an inducible mouse model using Cre-ERT under the control of the keratin 14-promoter. We show that the loss of Cdh1 and Tgfbr2 in the absence of carcinogen treatment is sufficient to induce invasive OSCC, although single knock-out mice show no or rare events of tumorigenesis. These double knock-out tumors showed upregulation of stem cell markers, which are known TGFβ targets. Analysis of mouse and human models indicates that TGFβ signaling plays a crucial role in the regulation of proliferation and stemness and eventually tumorigenesis.
Materials and methods
Dataset analysis
To determine the clinical correlation of TGFBR2 and CDH1, a comprehensive analysis of the Human Genome Atlas HNSCC data set was performed via cBio Portal (21,22). A total of 279 HNSCC patients were used in the analysis. Briefly, de-identified patient data, including gene expression and clinical data, were extracted from individual records available on cBio Portal. The collected information from each patient was analyzed in Prism version 6.00 for Mac (GraphPad software, La Jolla, CA, www.graphpad.com). Survival analysis using the Kaplan–Meier method was performed using the Mantel-Cox log-rank test and log rank test for trend. Contingency table analysis of TGFBR2 and CDH1 differential expression and prognosis and survival was performed using Fisher’s exact test. Relative risk (RR) and the corresponding 95% confidence interval (95% CI) are presented. Statistical significance was defined as P < 0.05.
Breeding and genotyping of mice
All animal studies were approved by the Institutional Animal Care and Use Committee at Vanderbilt University. The Epstein–Barr virus promoter ED-L2 was used to target Cre activity to the oral–esophageal squamous epithelium as described previously (23); these mice were a gift from Dr. Jonathan Katz (University of Pennsylvania). ED-L2-Cre transgenic mice on a C57BL/6J background were crossed with loxP-flanked E-cadherin, Cdh1, mice (Jackson Laboratory, Bar Harbor, Maine) and/or with TGFβ receptor II, Tgfbr2 flox/flox mice, both on a C57BL/6J background. We bred ED-L2-Cre (23) and Cdh1 flox/flox mice as well as ED-L2-Cre and Tgfbr2 flox/flox mice to generate single knockout mice (KO), and double knockout (dKO) mice, L2-Cre/Cdh1 flox/flox/Tgfbr2 flox/flox, subsequently referred to as L2/dKO. The Cdh1 flox/flox mice were genotyped according to instruction on http://jaxmice.jax.org/protocolsdb/f?p=116:1:2821520305073265. Tgfbr2 flox/flox mice were genotyped as described previously (24). A second colony was generated using K14-Cre-ERT mice (CD1 background, Jackson Laboratory, Bar Harbor, ME) to generate conditional single and double knock-out mice K14-Cre-ERT/Cdh1 flox/flox/Tgfbr2 flox/flox, hereafter K14/dcKO, for spatio-temporal control of gene knockout using oral gavage or intraperitoneal injection of tamoxifen.
Tamoxifen induction
Tamoxifen (Sigma, St Louis, MO) was diluted in peanut oil (Sigma) and administered three times at a concentration of 1mg/day on alternating days by intraperitoneal injection. Alternatively, (Z)-4-hydroxytamoxifen (Sigma) was administered by oral gavage using a feeding needle. After initial Cre-mediated recombination resulting in the knock-out of Cdh1 and Tgfbr2, mice received a «booster» shot once a month for the first 3 months.
Histology
After sacrificing the mice, tongue, esophagus and forestomach were removed and processed for histology. Additionally, buccal mucosa, lymph nodes, lung, liver and tumors were collected in diseased mice. Briefly, tissues were fixed in 10% formalin and embedded in paraffin. Five micron sections were applied to Probe-on Superfrost Plus slides (Fisher Scientific, Pittsburgh, PA). Slides were stained with hematoxylin and eosin, and images were captured on a Zeiss microscope with a Zeiss Hrc5 camera (Carl Zeiss Microscopy, Thornwood, NY). Matched littermate controls and mutant mice were examined histologically by review of slides in a blinded fashion by our pathologist.
Antibodies
Antibodies against E-cadherin, β-catenin, p120 and integrin β1 (ITGB1) were purchased from BD Biosciences (Franklin Lakes, NJ). Anti-pSMAD2, MECP2, Ki67 (clones 8D5 and D3B5), and YAP/TAZ (clone D24E4) were from Cell Signaling Technology (Danvers, MA). Anticytokeratin K14 was from Thermo Fisher Scientific (Fremont, CA). p63 and α-tubulin antibodies were purchased from Abcam (San Francisco, CA); and Ki67 from Vector Laboratories (Burlingame, CA). Anti-MYC, Cyclin D1, PML and TGFBR2 (clone L21) were from Santa Cruz Biotechnologies (Santa Cruz, CA). Podoplanin (PDPN) was from eBioscience (San Diego, CA), and SOX2 from EMD Millipore (Billerica, MA).
Immunohistochemistry and immunofluorescence
Immunohistochemistry was performed with the Vecta Elite kit (Vector Laboratories, Burlingame, CA) following the manufacturer’s protocol using their reagents. Antigen retrieval was performed by heating paraffin sections in a pressure cooker for 12 min followed by a one-hour incubation in the pressure cooker. Primary antibody was incubated overnight at 4°C and secondary antibody for 30 min at 37°C. Then the signal was developed using the diaminobenzidene substrate kit for peroxidase. For immunofluoresence staining, primary antibody was incubated overnight at 4°C and detected using Texas-Red or fluorescein isothiocyanate-labeled secondary antibody (Vector Laboratories, Burlingame, CA). Stained sections were mounted with 4′,6-diamidino-2-phenylindole (DAPI)-containing mounting medium (Vector Laboratories, Burlingame, CA).
Western blotting
Western blots were performed as described previously (20). Experiments were repeated at least twice.
Results
Coordinated loss of Cdh1 and Tgfbr2 results in squamous cell carcinoma
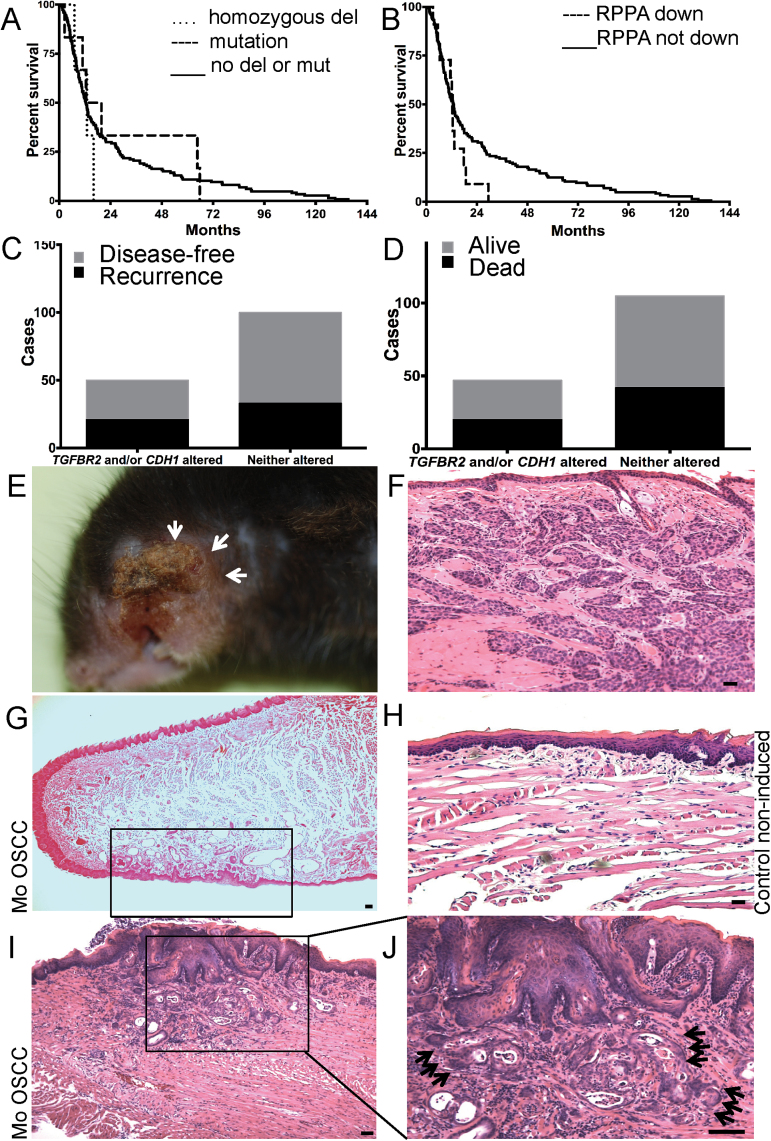
We have demonstrated previously that the coordinated loss of E-cadherin (CDH1) and TGFβ receptor II (TGFBR2) is frequently detected in upper aero-digestive tract cancer (19). We queried the HNSCC dataset of the Cancer Genome Atlas, through the use of cBio Portal (21,22) to correlate CDH1 and TGFBR2 expression with patient disease recurrence and overall survival. HNSCC patients with a homozygous deletion or mutation in TGFBR2 exhibited lower survival compared with patients without homozygous deletion or mutation of TGFBR2 (Mantel-Cox log-rank P = 0.71; log-rank test for trend P = 0.09) (Figure 1A). We next examined E-cadherin protein expression, as examined by reverse phase protein array, and observed no overall survival differences between patients with downregulated E-cadherin, compared with those without downregulated E-cadherin (P = 0.58) (Figure 1B). The lack of statistical significance within these two analyses is probably due to low patient numbers following subgroup stratification. Additionally, we looked at the synergistic effect of TGFBR2 and CDH1 on disease recurrence and overall survival. Though not statistically significant, patients with either or both TGFBR2 or CDH1 altered had higher proportions of disease recurrence or death compared with patients without TGFBR2 or CDH1 altered (Figure 1C and D). The compiled data indicate a negative impact on HNSCC patient prognosis when TGFBR2 and CDH1 are disregulated.
Fig. 1.
Altered TGFBR2 and CDH1 in patient prognosis and mouse model. (A) Overall survival of HNSCC patients when stratifying by TGFBR2 status. Patients with homozygous deletion or mutation of TGFBR2 showed lower survival compared with patients without a homozygous deletion or mutation of TGFBR2. (P = 0.71). (B) Overall survival of HNSCC patients when stratifying by CDH1 reverse phase protein array expression. Patients with downregulation of CDH1 protein expression showed lower survival compared with patients with unaltered CDH1 protein expression. (P = 0.58). (C) Contingency analysis of TGFBR2 and/or CDH1 expression and disease-free survival. Patients with altered TGFBR2 and/or altered CDH1 had a higher proportion of disease recurrence than patients with unaltered TGFBR2 or CDH1 (RR = 1.27, 95% CI = 0.83, 1.96). (D) Contingency analysis of TGFBR2 and/or CDH1 expression and overall survival. Patients with altered TGFBR2 and/or altered CDH1 had no proportional difference in overall survival compared with patients with unaltered TGFBR2 or CDH1 (RR = 1.06, 95% CI = 0.71, 1.60). (E) Cdh1/Tgfbr2 double knockout (dcKO) mice present with lesions that can involve the facial skin, in this case the whisker pad, due to their invasive nature (arrows). (F) Histological analysis of a OSCCs originating in the oral cavity extending to the buccal mucosa and involving the surface of the skin. (G) Low magnification of a representative tongue with focal tumor lesion on the ventral side (rectangle). (H) Normal mucosa of control animal. (I) Invasive SCC of the tongue (J higher magnification) invading into the muscle (black arrows). Scale bar, 50 micron.
To elucidate the functional consequences of the simultaneous E-cadherin and TGFBR2 loss in vivo, we generated mice with targeted deletion of the E-cadherin gene, Cdh1, and Tgfbr2 in the oral cavity, esophagus and forestomach using the Epstein–Barr virus promoter ED-L2. Additionally, we established a tamoxifen-inducible colony using K14-Cre-ERT mice.
The observed ablation of Cdh1 and Tgfbr2 expression was restricted to focal areas and presented in a mosaic knock-out. This pattern resulted in focal SCC formation of the head-and-neck area (Figure 1) as well as forestomach (Supplementary Figure S1, available at Carcinogenesis Online) and death by the age of 18 months or older. Histological analysis of tissues with constitutive and inducible loss of both, Cdh1 and Tgfbr2 in L2/dKO or K14/dcKO, earlier identified a atrophic epithelium in targeted areas of the tongue, oral cavity and esophagus (Figure 2A; Supplementary Figures S2A and S3, available at Carcinogenesis Online). We also observed hyperplasia of selected minor salivary glands of the tongue (data not shown).
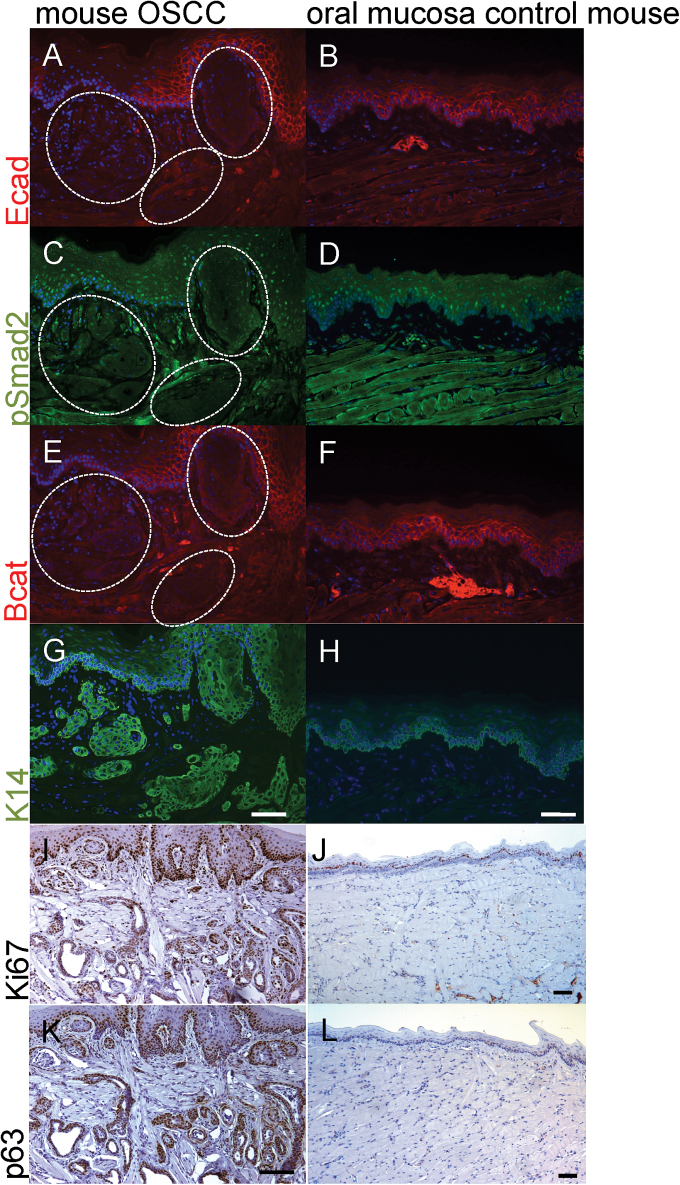
Fig. 2.
Tumors exhibit disruption of adherens junctions and increased proliferation. Focal invasive areas (dashed white circles) of a tongue SCC are negative for E-cadherin (A, red) compared with the expression pattern of the normal tongue epithelium (B). (C) Nuclear pSmad2 (green) signal is lost in invasive areas (dashed white circles). (D) Normal mouse mucosa has positive pSmad2 signal throughout. B-catenin, red, is lost in these invasive cells (dashed white circles (E)), while (G) K14, green, stains positive. The signal is increased compared with the normal tongue epithelium (H) of a non-induced control animal. The tumor cells invading into the center of the tongue are strongly positive for Ki67 (I) and p63 (K) compared with normal tongue, (J) and (L), respectively. Scale bars, 50 micron.
Invasive OSCCs were present in 74% and 77% of the constitutive and induced dKO mice, respectively (Table I). SCCs of the tongue could be detected as early as 10 months of age in dKO animals, but not in animals with deletion of a single gene, or in non-induced K14/dcKO mice. Some tumors progressed to grow into the skin exhibiting their invasive nature (Figure 1E and F) but no multifocal disease was observed. Anatomical subsites of the tumors were the buccal mucosa with involvement of the facial skin (Figure 1E and F), the oropharynx and the apex and dorsal and ventral tongue (Figure 1G, site-specific focal invasion in one area of the ventral tongue, rectangle; H for normal non-induced control animal; 1G, I; representative images of focal advanced disease). Furthermore, in the forestomach, an extension of the squamous esophageal epithelium, tumorigenesis occurred at the squamous-gastric junction and was characterized by well-differentiated areas with keratin-pearl formation (Supplementary Figure S1A–D, available at Carcinogenesis Online).
Table I.
Overview of tumor incidence in mice
| Genotype | Tumor incidence (number of animals) |
|---|---|
| L2/dKO | 74% (17/23) |
| L2/KO | 8% (3/36) |
| K14/dcKO, induced | 77% (27/35) |
| K14/dcKO, non-induced | 6% (2/30) |
| K14/cKO, induced | 10% (3/30) |
Number of double knock-out (dKO) presenting with tumors compared with single knock-out (KO) animals. Knock-out in L2-driven animals is constitutive, K14 is a conditional knock-out (cKO) and tamoxifen-induced.
Mosaic ablation of Cdh1 and Tgfbr2 in the tongue, esophagus and forestomach
To verify loss of Cdh1 and Tgfbr2, we performed immunofluorescence stainings with antibodies against E-cadherin (Figure 2A and B) and pSmad2 (Figure 2C and D), a downstream target of TGFβ signaling. In K14/dcKO mice, tamoxifen-induction led to the mosaic loss of E-cadherin and pSmad2 (Figure 2A and C). Non-induced control K14/dcKO littermates or dKO mice lacking the Cre-recombinase gene displayed membranous E-cadherin staining similar to the unaffected adjacent mucosa of experimental mice or nuclear pSmad2 protein expression (Figure 2B and D). We showed loss of E-cadherin and pSmad2 in the invading tissues (Figure 2A and C, white dashed circles, Supplementary Figure S2, available at Carcinogenesis Online), which was of squamous origin as shown by the K14-positive staining (Figure 2G, serial section of a representative area of focal expression loss).
E-cadherin is a key component of adherens junctions and binds β-catenin and p120 through its cytoplasmic tail. Loss of E-cadherin was accompanied by the loss of β-catenin expression in the tongue, esophagus and forestomach (Figures 2E; SupplementaryFigure S2, available at Carcinogenesis Online). Non-induced control animals did not exhibit the loss of β-catenin (Figure 2F). The areas of E-cadherin and β-catenin loss were still positive for K14 staining, demonstrating the squamous origin of the invasive tissue lesion (Figure 2G). Adjacent oral mucosa (Figure 2A and E) still showed expression similar to normal mucosa in non-induced control mice (Figure 2B and F) suggesting a coordinate loss and disruption of adherens junction function. Similarly, the loss of E-cadherin binding partners β-catenin and p120 was observed in the forestomachs, which exhibit mosaic loss of expression for E-cadherin and pSmad2 (Supplementary Figure S2A, available at Carcinogenesis Online) compared with control animals (Supplementary Figure S2C, available at Carcinogenesis Online) and develop tumors (Supplementary Figure S2C and D, available at Carcinogenesis Online).
E-cadherin can regulate cell growth, and the loss of contact inhibition, which increases cell proliferation. Therefore, we analyzed the mouse tumors for their proliferation potential. Tumors of the oral cavity and the forestomach were strongly positive for both the proliferation marker Ki67 and p63, a squamous lineage marker (Figure 2I and K; Supplementary Figure S1E and G, available at Carcinogenesis Online), when compared with normal mucosa (Figure 2J and L; Supplementary Figure S1F and H, available at Carcinogenesis Online) in control animals.
Interestingly, although we confirmed knockout of E-cadherin and concomitant loss of β-catenin and concomitant loss of pSmad2 expression, the esophagus of L2/dKO and K14/dcKO displayed no evidence of tumor initiation (Supplementary Figure S3A–F, available at Carcinogenesis Online). We observed increased infiltration of immune cells in the submucosa suggesting an inflammatory microenvironment (Supplementary Figure S3A and B, available at Carcinogenesis Online), which appears to lack the ability to promote tumorigenesis.
Occurrence of regional and distant metastasis in dKO mice
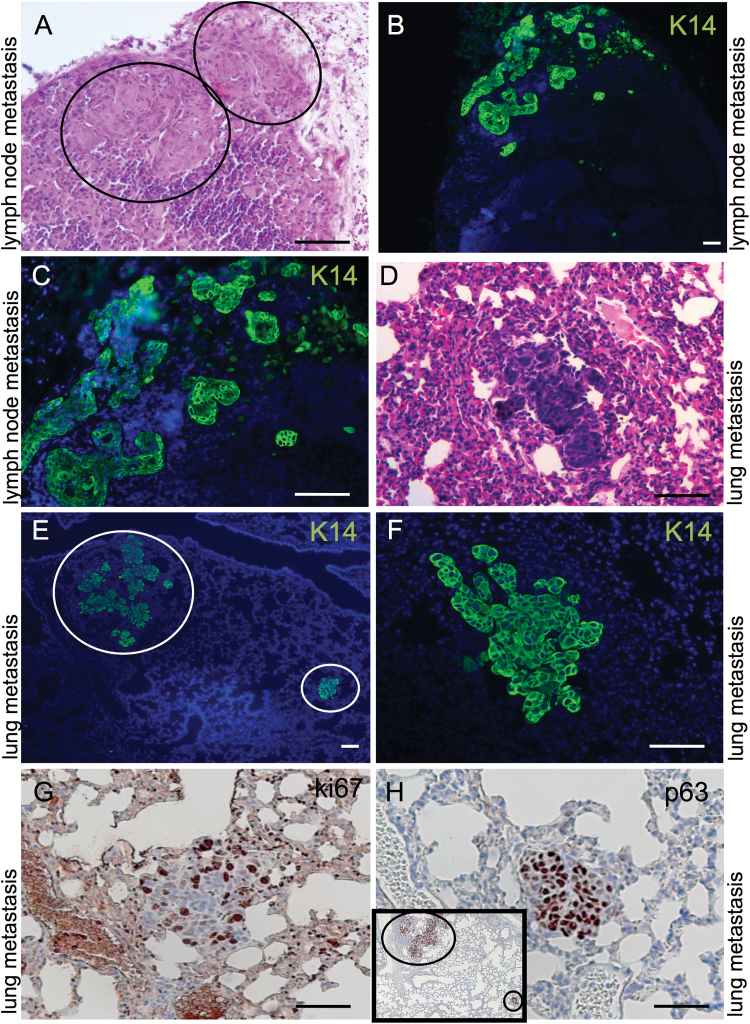
The ability of the tumors in the L2/dKO mice and K14/dcKO mice to metastasize to regional lymph nodes and lungs mimicked the metastatic spread of the human tumors and corroborate their similarities to the human disease (Figure 3). K14-positive cell clusters were identified in regional lymph nodes and the lung (Figure 3, lymph node involvement in A–C and pulmonary metastasis in D–F). Local metastasis to the lymph nodes appears to precede pulmonary metastasis, as not all animals in that we observed K14-postitive clusters in the lymph nodes showed positive lungs. In contrast, we identified positive lymph nodes in all animals with K14-positive cells spread to the lungs. The identified lung metastases were highly proliferative and of epithelial origin as shown by Ki67 immunohistochemistry staining and strong positivity for p63, respectively (Figure 3G and H).
Fig. 3.
OSCCs in Cdh1/Tgfbr2 knockout mice are able to metastasize regionally and distantly. Tumor cell nests could be detected with keratin 14, K14, staining in regional lymph nodes (A, hematoxylin and eosin; B and C, K14 immunofluorescence). Tumors can spread to the lung as seen on lung sections stained with hematoxylin and eosin (D), and anti-K14 antibody in (E, F higher magnification). Lung metastases are Ki67- and p63-positive (G–H). Scale bars, 50 micron.
Upregulation of c-Myc, Cyclin D1 and Yap1 phenocopies human OSCC
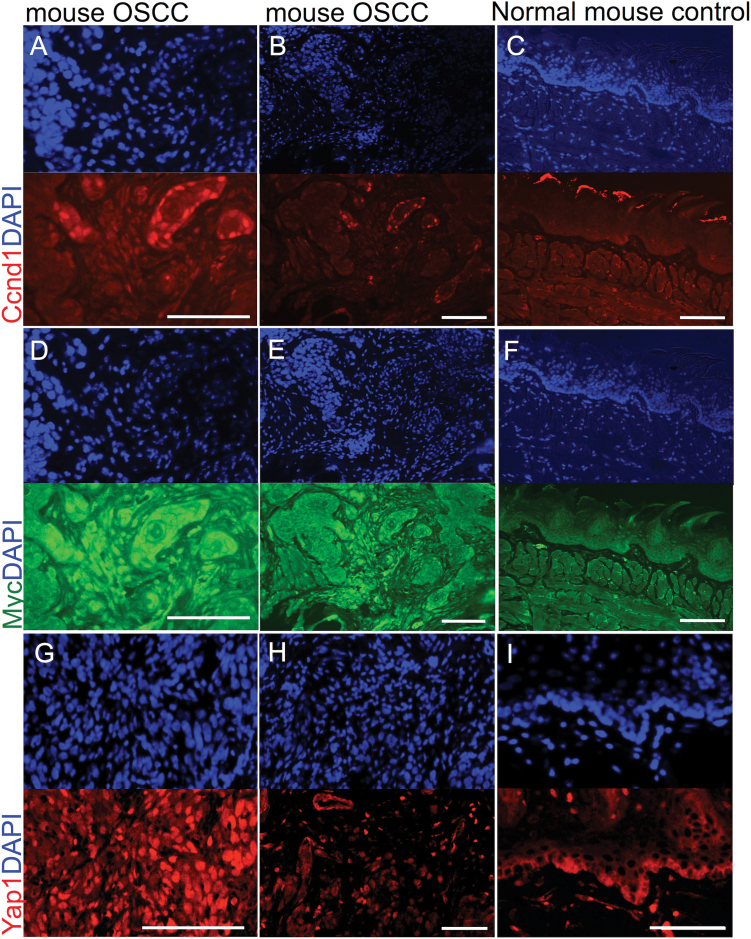
The murine OSCCs demonstrated upregulation of the oncogene Cyclin D1 (Figure 4A–C), which is seen in human SCCs. This observation further highlighted the similarity with human OSCCs. Additionally, the expression of c-Myc and Yap1, another squamous oncogene (25,26), was upregulated in mouse tumors (Figure 4D–I). Interestingly, nuclear localization for c-Myc was similar in all areas of the tumor tissues (Figure 4D and E) but absent in normal control animals (Figure 4F), Cyclin D1 overexpression was restricted to areas of early invasion (Figure 4A and B; C control tongue from normal animal shows unspecific staining in the keratinized layer). Neither c-Myc nor Cyclin D1 was expressed at detectable levels in the mouse tongue epithelium of non-induced control animals. Although Yap1 was highly expressed and localized to the nucleus in the tumor samples (Figure 4G and H), its expression was restricted to the cytoplasm of the basal layer in normal controls (Figure 4I). The upregulation of c-Myc and Cyclin D1 upon loss of E-cadherin and TGFβ signaling was also observed in the squamous tumors of the forestomach (Supplementary Figure S2E–H, available at Carcinogenesis Online).
Fig. 4.
Upregulation of Cyclin D1 (Ccdn1), c-Myc and Yap1 in mouse OSCCs. Mouse tumors are positive for Ccdn1 (A, B; red, DAPI blue), Myc (D, E; green, DAPI blue), and Yap1 (G, H; red, DAPI blue). Ccdn1 and Myc are up-regulation compared with normal mucosa (C and F, respectively). Yap1 (G, H) is localized to the nucleus in tumor tissues compared with cytoplasmic localization in normal oral mucosa (I). Scale bars, 50 micron.
A quiescent basal cell layer in human oral mucosa and its loss during tumorigenesis
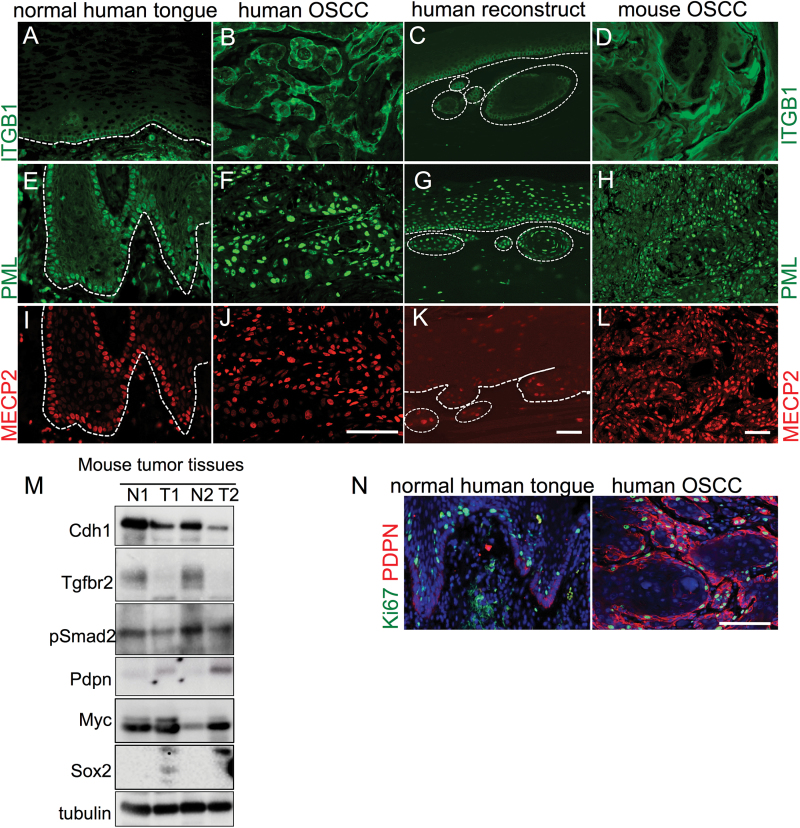
TGFβ signaling and E-cadherin are important regulators of keratinocyte proliferation (12,13,27,28). Furthermore, ITGB1 is a known TGFβ1 target and has been described as a stem cell marker (29,30). In an attempt to determine whether stemness and proliferation are affected by the coordinated loss of E-cadherin and TβRII in the mouse model, we analyzed ITGB1 (Figure 5A–D), PML, a regulator of TGFβ signaling (31,32, Figure 5E–H) and MECP2, which has functions in chromatin organization. All of them were restricted to the basal and quiescent cells in the normal human epithelium (Figure 5I–L). For this analysis we performed immunofluorescence staining of normal human tongue, human OSCC tissues, organotypic three-dimensional reconstructs of human keratinocytes expressing dominant-negative mutants of E-cadherin and TGFBR2 to mimic the coordinated loss established in vivo (20) and compared them with the mouse tumors. We showed that the expression of several stem cell markers is restricted to the basal layer in the human oral mucosa (Figure 5A, E and I, dashed line demarcates the basal membrane). These markers were upregulated in the human (Figure 5B, F and J) and mouse OSCCs (Figure 5D, H and L) and their spatial restriction to the basal layer disrupted in human organotypic reconstruct cultures (Figure 5C, G and K, dashed line demarcates the basal membrane, circles the invasive areas).
Fig. 5.
Changes in putative stem cell marker expression during tumor progression. The expression of ITGB1 (A, green) was restricted to the basal layer in the normal human epithelium compared with human OSCC tissues (B), organotypic reconstructs with functional loss of E-cadherin and TGFBR2 (C) or mouse OSCCs (D). Similarly, PML (green) outlines the basal cell layer in normal human oral mucosa (E), but its signal increases in human (F) and mouse OSCCs (H) as well as loses it restriction to the basal layer in organotypic reconstructs (G). The expression of MECP2 mirrors PML expression and localization (I–L). Scale bars, 50 micron. Western blot analysis (M) of two paired mouse tissues, normal N versus tumor T shows the upregulation of Pdpn, Myc and Sox2. Upregulation of PDPN (red) in human OSCC is shown by immunofluorescence compared with normal human tongue (N, Ki67 is green). Western blots shown in this figure were cropped.
Western blot analysis using paired normal control and tumor samples from the two dKO mouse models (Figure 5M) confirmed lower expression levels of Cdh1, Tgfbr2 and lower level activation of the TGFβ signaling as measured by pSmad2 in the tumor samples. Using antibodies against additional putative stem cell markers such as Pdpn, Myc as well as the transcription factor Sox2, which is involved in the determination of cell fate and stem cell maintenance, we observed elevated protein expression in the mouse tumor samples (Figure 5M). That upregulation of PDPN occurs in human OSCC was shown by immunofluorescence staining compared with normal tongue (Figure 5N, co-staining with the proliferation marker Ki67).
Together, these data indicate a role for TGFβ-mediated regulation of stemness and suggest a mechanism how loss of this signaling pathway could result in tumorigenesis.
Discussion
The nature of the oral mucosal cells that are prone to initiate cancer is still unknown. Important events in this progression are the loss of normal proliferation control and the ability to invade locally and into the lymphatic system to advance to metastatic disease. We believe that TGFβ signaling and the cell adhesion protein E-cadherin play major roles in these processes. However, their contributions are multi-faceted, complex and at times contradictory.
For instance, TGFβ signaling has been described as a tumor suppressor during early stages and as a tumor-promoter in later stages of tumorigenesis (33). However, inactivation of TGFβ signaling through mutations of TGFβ receptors and of their downstream effector molecules, the Smads, is associated with OSCC (34,35). The tissue-specific knockout of components of the TGFβ signaling pathway such as TGFBR1, TGFBR2 and Smad4 has been shown to result in tumor formation providing overwhelming evidence for the tumor suppressive function of this signaling pathway.
TGFβ and inhibition of proliferation in stem cells
TGFβ signaling has several context-dependent effects on keratinocytes with growth inhibition being the primary outcome. Additionally, TGFβ signaling is a potent inducer of EMT. These two major aspects of TGFβ are now frequently discussed in the context of cancer stem cells and normal stemness.
Many normal stem cell populations rely on TGFβ signaling for their quiescence control and their slow-cycling properties (36–39). We speculated that the tumorigenic advantages of elimination of TGFβ signaling are based on relaxing growth control in the stem cell compartment.
Adherens junctions in tumorigenesis beyond cell adhesion
Even in established cancer cell lines, the loss of E-cadherin can be accompanied by the expansion of CSC populations, EMT, and increased colony-forming capacity and tumorigenesis (40). Of interest, CSC populations seem characterized by low levels of E-cadherin (41) and purified squamous stem cells frequently have a low proliferation rate (41,42).
We have shown previously that E-cadherin and TGFBR2 physically interact and that E-cadherin can be an important determinant of TGFBR2 levels (19). Therefore, we speculated that the disruption of both genes may have synergistic effects in regard to malignant transformation in the mucosa. Indeed, in our model we detected an increase in tumor incidence in double compared with single KO mice.
The ED-L2-Cre mouse model has been successfully used to abolish the expression of another cell–cell adhesion molecule, p120, exemplifying that the disruption of adherens junctions is truly a hallmark of epithelial cancers. L2-Cre/p120flox/flox mice presented with a similar phenotype of invasive squamous tumors in the oral cavity and forestomach. However, esophageal SCCs were described (43), which we do not observe in our dKO systems. Interestingly, the p120 model is associated with increased numbers of immature myeloid cells, inflammation and desmoplasia in the tumor microenvironment. Analysis of esophageal tissues from our two models showed infiltrating immune cells in some cases, but no signs of esophageal tumorigenesis. Interestingly, the oral tissues in the mouse seem to be more sensitive to aberrations in TGFβ signaling when compared with the esophagus since treatment with an anti-TGFβ antibody results in oral and to a lesser extent esophageal lesions (44). Similarly, conditional inactivation of Tgfbr2 in mouse fibroblasts induced SCCs in adjacent epithelia including the forestomach, an extension of the squamous esophageal epithelium, but no tumors were observed in the esophagus itself (45).
Synergy between TGFβ signaling and other signaling pathways in oral carcinogenesis
Furthermore, the function of adherens junctions and TGFβ signaling intersect at the process of EMT. However, the overall trend appears to favor a loss of TGFβ signaling in squamous carcinogenesis. For example, TGFBR2 and SMAD4 have been shown to be significantly down-regulated in OSCC tumor tissues compared with normal oral epithelium (46). Smad4−/− mice develop OSCC supporting the importance of the TGFβ pathway in the regulation of epithelial homeostasis (47). Similarly, coordinated Krt5-Cre mediated deletion of Smad4 and Pten showed accelerated tumor formation in the forestomach with a corresponding increase in proliferation in the squamous epithelium of the forestomach and esophagus. Expression of cell cycle regulators such as p21, p16 and p27 was downregulated and Cyclin D1 upregulated (48). Combined loss of Pten and Tgfbr1 also promotes OSCCs with upregulation of putative stem cell markers such as CD44, CD133, Sox2 and Oct4 (49). Interestingly, double knockdown of Pten and Tgfbr1 resulted in activation of the mTOR pathway and increased levels of survivin (50). Therefore, treatment with rapamycin (50) or the mTOR inhibitor PF-04691502 (51) suppressed onset and progression of tumorigenesis. We tested the contributions of the PTEN/Akt pathways in our model without detecting any differential expression or activation in the tumor tissues compared with normal adjacent tissues samples (data not shown).
Tgfbr2 loss alone is not sufficient to induce OSCCs in our model. K14-mediated knock-out of Tgfbr2 alone resulted in spontaneous anal and genital squamous cell carcinoma in a study by Guasch et al. (52). Four major pathways have been identified driving OSCC: mitogenic signaling, NOTCH, cell cycle and TP53 (53). Amongst the prominent driver genes were CCND1 and MYC. TGFβ antimitotic function involves cyclin-dependent kinase inhibitor gene responses and the downregulation of c-Myc for activation of the p15 arrest pathways (54). Given the considerable overlap of the pathways, we analyzed the head-and-neck squamous cell carcinoma TCGA provisional set for alterations in those pathways: p15 and p16 were altered in 39% of cases, most having homozygous deletion of the proteins. Twenty-two percent of the cases show MYC amplification or mRNA upregulation. Crosstalk between TGFβ and Notch-signaling has been described to dependent on ZEB1 and ZEB2 (55). Genetic inhibition of the Notch-mediated transcriptional activity prevented squamous differentiation and enriched for EMT-competent cells. Data from the head-and-neck squamous cell carcinoma TCGA data set show Notch alterations with an overall occurrence in 21% of the cases.
Key targets of the Hippo pathway, YAP and WWTR1, can interact with Smad molecules (56). However, the outcome of this interaction depends largely on the cell context. The YAP/Smad complex can promote EMT (57), and has been shown to induce a pro-invasive and metastatic phenotype in breast cancer (58). The Crumbs polarity complex interacts with YAP, which relays cell density information by promoting cytoplasmic retention of YAP resulting in suppressed TGF-β signaling (59). Interestingly, YAP can mediate cytoplasmic retention of phosphorylated SMAD3 thereby suppressing its function (60). Silencing of both YAP1 and IGF2BP3 can restore TGF-β signaling and also inhibited pluripotent gene expression and tumorigenesis. The adherens junction component α-catenin is also implicated in the regulation of cell density and tumor suppression, and has been shown to be a negative regulator of Yap1 (61). Furthermore, this study showed that activation of Yap1 lead to epidermal stem cell expansion and squamous-cell carcinoma-like tumors. We hypothesize that Cdh1 and Tgfbr2 knockout could initiate cancer initiating cell and CSC generation.
This leads to a novel hypothesis that the combined loss of Cdh1 and Tgfbr2 may in fact initiate tumorigenesis with long latency, since it provides an increased proliferation rate for cells, potentially stem cells, which are prone to malignant transformation over time. Future studies will have to show whether this is a mechanism that underlies early steps of squamous epithelial carcinogenesis and coins the molecular make-up and frequency of CSCs.
Supplementary material
Supplementary Figures S1–S3 can be found at http://carcin.oxfordjournals.org/
Funding
This work was supported in part by grants from the National Institute of Health (DK075379, DK094900) and a pilot grant from the Vanderbilt Digestive Disease Research Center (P30DK058404). Core Services performed through Vanderbilt University Medical Center’s Digestive Disease Research Center are supported by National Institute of Health grant (P30DK058404) Core Scholarships and the Vanderbilt Ingram Cancer Center Support grant (P30CA068485).
Supplementary Material
Acknowledgements
We would also like to thank members of the Beauchamp laboratory, Drs Deane, Means and Beauchamp, as well as Dr Anil K.Rustgi for helpful discussions. We thank Shazia Ansari and Jalal A.Hamaamen for technical assistance with the animal colony. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations:
- CDH1
E-cadherin
- CSC
cancer stem cell
- DAPI
4′,6-diamidino-2-phenylindole
- dKO
double knockout mice
- EMT
epithelial–mesenchymal transition
- KO
single knockout mice
- OSCC
oral squamous cell carcinoma
- PDPN
podoplanin
- SCC
squamous cell cancer
- TGFBR2
transforming growth factor beta receptor II.
References
- 1. Thiery J.P. (2002). Epithelial–mesenchymal transitions in tumour progression. Nat. Rev. Cancer, 2, 442–454 [DOI] [PubMed] [Google Scholar]
- 2. Vleminckx K., et al. (1991). Genetic manipulation of E-cadherin expression by epithelial tumor cells reveals an invasion suppressor role. Cell, 66, 107–119 [DOI] [PubMed] [Google Scholar]
- 3. Bosch F.X., et al. (2005). E-cadherin is a selective and strongly dominant prognostic factor in squamous cell carcinoma: a comparison of E-cadherin with desmosomal components. Int. J. Cancer, 114, 779–790 [DOI] [PubMed] [Google Scholar]
- 4. Behrens J., et al. (1989). Dissecting tumor cell invasion: epithelial cells acquire invasive properties after the loss of uvomorulin-mediated cell–cell adhesion. J. Cell Biol., 108, 2435–2447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Peifer M., et al. (2000). Wnt signaling in oncogenesis and embryogenesis—a look outside the nucleus. Science, 287, 1606–1609 [DOI] [PubMed] [Google Scholar]
- 6. Fagotto F., et al. (1996). Binding to cadherins antagonizes the signaling activity of beta-catenin during axis formation in Xenopus. J. Cell Biol., 132, 1105–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sanson B., et al. (1996). Uncoupling cadherin-based adhesion from wingless signalling in Drosophila. Nature, 383, 627–630 [DOI] [PubMed] [Google Scholar]
- 8. Tunggal J.A., et al. (2005). E-cadherin is essential for in vivo epidermal barrier function by regulating tight junctions. EMBO J., 24, 1146–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tinkle C.L., et al. (2004). Conditional targeting of E-cadherin in skin: insights into hyperproliferative and degenerative responses. Proc. Natl Acad. Sci. U.S.A., 101, 552–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Katsuno Y., et al. (2013). TGF-β signaling and epithelial–mesenchymal transition in cancer progression. Curr. Opin. Oncol., 25, 76–84 [DOI] [PubMed] [Google Scholar]
- 11. Cheng C.F., et al. (2008). Profiling motility signal-specific genes in primary human keratinocytes. J. Invest. Dermatol., 128, 1981–1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Levy L., et al. (2005). Smad4 dependency defines two classes of transforming growth factor beta target genes and distinguishes TGFβ-induced epithelial–mesenchymal transition from its antiproliferative and migratory responses. Mol. Cell Biol., 25, 8108–8125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Vijayachandra K., et al. (2009). Induction of p16ink4a and p19ARF by TGFbeta1 contributes to growth arrest and senescence response in mouse keratinocytes. Mol. Carcinog., 48, 181–186 [DOI] [PubMed] [Google Scholar]
- 14. Forrester E., et al. (2005). Effect of conditional knockout of the type II TGF-beta receptor gene in mammary epithelia on mammary gland development and polyomavirus middle T antigen induced tumor formation and metastasis. Cancer Res., 65, 2296–2302 [DOI] [PubMed] [Google Scholar]
- 15. Gorska A.E., et al. (2003). Transgenic mice expressing a dominant-negative mutant type II transforming growth factor-beta receptor exhibit impaired mammary development and enhanced mammary tumor formation. Am. J. Pathol., 163, 1539–1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dong Z., et al. (2012). Concordant promoter methylation of transforming growth factor-beta receptor types I and II occurs early in esophageal squamous cell carcinoma. Am. J. Med. Sci., 343, 375–381 [DOI] [PubMed] [Google Scholar]
- 17. Wang J., et al. (1995). Demonstration that mutation of the type II transforming growth factor beta receptor inactivates its tumor suppressor activity in replication error-positive colon carcinoma cells. J. Biol. Chem., 270, 22044–22049 [DOI] [PubMed] [Google Scholar]
- 18. Turco A., et al. (1999). Overexpression of transforming growth factor beta-type II receptor reduces tumorigenicity and metastastic potential of K-ras-transformed thyroid cells. Int. J. Cancer, 80, 85–91 [DOI] [PubMed] [Google Scholar]
- 19. Andl C.D., et al. (2006). Coordinated functions of E-cadherin and transforming growth factor beta receptor II in vitro and in vivo . Cancer Res., 66, 9878–9885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Andl C.D., et al. (2010). Cathepsin B is the driving force of esophageal cell invasion in a fibroblast-dependent manner. Neoplasia, 12, 485–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cerami E., et al. (2012). The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov., 2, 401–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gao J., et al. (2013). Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal., 6, pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tetreault M.P., et al. (2010). Esophageal squamous cell dysplasia and delayed differentiation with deletion of krüppel-like factor 4 in murine esophagus. Gastroenterology, 139, 171–181.e9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chytil A., et al. (2002). Conditional inactivation of the TGF-beta type II receptor using Cre:Lox. Genesis, 32, 73–75 [DOI] [PubMed] [Google Scholar]
- 25. India Project Team of the International Cancer Genome Consortium (2013). Mutational landscape of gingivo-buccal oral squamous cell carcinoma reveals new recurrently-mutated genes and molecular subgroups. Nat. Commun., 4, 2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhang L., et al. (2011). Yes-associated protein promotes cell proliferation by activating Fos related activator-1 in oral squamous cell carcinoma. Oral Oncol., 47, 693–697 [DOI] [PubMed] [Google Scholar]
- 27. Kim N.G., et al. (2011). E-cadherin mediates contact inhibition of proliferation through Hippo signaling-pathway components. Proc. Natl Acad. Sci. U.S.A., 108, 11930–11935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Stockinger A., et al. (2001). E-cadherin regulates cell growth by modulating proliferation-dependent beta-catenin transcriptional activity. J. Cell Biol., 154, 1185–1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jones P.H., et al. (1993). Separation of human epidermal stem cells from transit amplifying cells on the basis of differences in integrin function and expression. Cell, 73, 713––24 [DOI] [PubMed] [Google Scholar]
- 30. Seery J.P., et al. (2000). Asymmetric stem-cell divisions define the architecture of human oesophageal epithelium. Curr. Biol., 10, 1447–1450 [DOI] [PubMed] [Google Scholar]
- 31. Lin H.K., et al. (2004). Cytoplasmic PML function in TGF-beta signalling. Nature, 431, 205–211 [DOI] [PubMed] [Google Scholar]
- 32. Tang M.K., et al. (2013). Promyelocytic leukemia (PML) protein plays important roles in regulating cell adhesion, morphology, proliferation and migration. PLoS One, 8, e59477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Piek E., et al. (2001). Suppressor and oncogenic roles of transforming growth factor-beta and its signaling pathways in tumorigenesis. Adv. Cancer Res., 83, 1––54 [DOI] [PubMed] [Google Scholar]
- 34. Chen T., et al. (2001). Novel inactivating mutations of transforming growth factor-beta type I receptor gene in head-and-neck cancer metastases. Int. J. Cancer, 93, 653–661 [DOI] [PubMed] [Google Scholar]
- 35. Sivadas V.P., et al. (2013). Novel mutations and expression alterations in SMAD3/TGFBR2 genes in oral carcinoma correlate with poor prognosis. Genes Chromosomes Cancer, 52, 1042–1052 [DOI] [PubMed] [Google Scholar]
- 36. Chabanon A., et al. (2008). A cross-talk between stromal cell-derived factor-1 and transforming growth factor-beta controls the quiescence/cycling switch of CD34(+) progenitors through FoxO3 and mammalian target of rapamycin. Stem Cells, 26, 3150–3161 [DOI] [PubMed] [Google Scholar]
- 37. Moreno S.G., et al. (2010). TGFbeta signaling in male germ cells regulates gonocyte quiescence and fertility in mice. Dev. Biol., 342, 74–84 [DOI] [PubMed] [Google Scholar]
- 38. Nishimura E.K., et al. (2010). Key roles for transforming growth factor beta in melanocyte stem cell maintenance. Cell Stem Cell, 6, 130–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sakaki-Yumoto M., et al. (2013). TGF-β family signaling in stem cells. Biochim. Biophys. Acta, 1830, 2280–2296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ye J., et al. (2012). Enrichment of colorectal cancer stem cells through epithelial–mesenchymal transition via CDH1 knockdown. Mol. Med. Rep., 6, 507–512 [DOI] [PubMed] [Google Scholar]
- 41. Geng S., et al. (2013). Cancer stem-like cells enriched with CD29 and CD44 markers exhibit molecular characteristics with epithelial–mesenchymal transition in squamous cell carcinoma. Arch. Dermatol. Res., 305, 35–47 [DOI] [PubMed] [Google Scholar]
- 42. La Fleur L., et al. (2012). A CD44high/EGFRlow subpopulation within head and neck cancer cell lines shows an epithelial–mesenchymal transition phenotype and resistance to treatment. PLoS One, 7, e44071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Stairs D.B., et al. (2011). Deletion of p120-catenin results in a tumor microenvironment with inflammation and cancer that establishes it as a tumor suppressor gene. Cancer Cell, 19, 470–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Vitsky A., et al. (2009). Homeostatic role of transforming growth factor-beta in the oral cavity and esophagus of mice and its expression by mast cells in these tissues. Am. J. Pathol., 174, 2137–2149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bhowmick N.A., et al. (2004). TGF-beta signaling in fibroblasts modulates the oncogenic potential of adjacent epithelia. Science, 303, 848–851 [DOI] [PubMed] [Google Scholar]
- 46. Wang X., et al. (2009). Growth inhibition induced by transforming growth factor-beta1 in human oral squamous cell carcinoma. Mol. Biol. Rep., 36, 861–869 [DOI] [PubMed] [Google Scholar]
- 47. Bornstein S., et al. (2009). Smad4 loss in mice causes spontaneous head and neck cancer with increased genomic instability and inflammation. J. Clin. Invest., 119, 3408–3419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Teng Y., et al. (2006). Synergistic function of Smad4 and PTEN in suppressing forestomach squamous cell carcinoma in the mouse. Cancer Res., 66, 6972–6981 [DOI] [PubMed] [Google Scholar]
- 49. Bian Y., et al. (2012). Loss of TGF-β signaling and PTEN promotes head and neck squamous cell carcinoma through cellular senescence evasion and cancer-related inflammation. Oncogene, 31, 3322–3332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sun Z.J., et al. (2012). Chemopreventive and chemotherapeutic actions of mTOR inhibitor in genetically defined head and neck squamous cell carcinoma mouse model. Clin. Cancer Res., 18, 5304–5313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Herzog A., et al. (2013). PI3K/mTOR inhibitor PF-04691502 antitumor activity is enhanced with induction of wild-type TP53 in human xenograft and murine knockout models of head and neck cancer. Clin. Cancer Res., 19, 3808–3819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Guasch G., et al. (2007). Loss of TGFbeta signaling destabilizes homeostasis and promotes squamous cell carcinomas in stratified epithelia. Cancer Cell, 12, 313–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Pickering C.R., et al. (2013). Integrative genomic characterization of oral squamous cell carcinoma identifies frequent somatic drivers. Cancer Discov., 3, 770–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Warner B.J., et al. (1999). Myc downregulation by transforming growth factor beta required for activation of the p15(Ink4b) G(1) arrest pathway. Mol. Cell. Biol., 19, 5913–5922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ohashi S., et al. (2011). A NOTCH3-mediated squamous cell differentiation program limits expansion of EMT-competent cells that express the ZEB transcription factors. Cancer Res., 71, 6836–6847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sudol M. (2012). WW domains in the heart of Smad regulation. Structure, 20, 1619–1620 [DOI] [PubMed] [Google Scholar]
- 57. Zhang H., et al. (2014). Yap1 is required for endothelial to mesenchymal transition of the atrioventricular cushion. J. Biol. Chem., 289, 18681–18692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hiemer S.E., et al. (2014). The transcriptional regulators TAZ and YAP direct transforming growth factor β-induced tumorigenic phenotypes in breast cancer cells. J. Biol. Chem., 289, 13461–13474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Varelas X., et al. (2010). The Crumbs complex couples cell density sensing to Hippo-dependent control of the TGF-β-SMAD pathway. Dev. Cell, 19, 831–844 [DOI] [PubMed] [Google Scholar]
- 60. Chen C.L., et al. (2013). Reciprocal regulation by TLR4 and TGF-β in tumor-initiating stem-like cells. J. Clin. Invest., 123, 2832–2849 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 61. Schlegelmilch K., et al. (2011). Yap1 acts downstream of α-catenin to control epidermal proliferation. Cell, 144, 782–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.