Abstract
Vibrio cholerae, the causative agent of cholera, requires iron for growth. One mechanism by which it acquires iron is the uptake of heme, and several heme utilization genes have been identified in V. cholerae. These include three distinct outer membrane receptors, two TonB systems, and an apparent ABC transporter to transfer heme across the inner membrane. However, little is known about the fate of the heme after it enters the cell. In this report we show that a novel heme utilization protein, HutZ, is required for optimal heme utilization. hutZ (open reading frame [ORF] VCA0907) is encoded with two other genes, hutW (ORF VCA0909) and hutX (ORF VCA0908), in an operon divergently transcribed from the tonB1 operon. A hutZ mutant grew poorly when heme was provided as the sole source of iron, and the poor growth was likely due to the failure to use heme efficiently as a source of iron, rather than to heme toxicity. Heme oxygenase mutants of both Corynebacterium diphtheriae and C. ulcerans fail to use heme as an iron source. When the hutWXZ genes were expressed in the heme oxygenase mutants, growth on heme was restored, and hutZ was required for this effect. Biochemical characterization indicated that HutZ binds heme with high efficiency; however, no heme oxygenase activity was detected for this protein. HutZ may act as a heme storage protein, and it may also function as a shuttle protein that increases the efficiency of heme trafficking from the membrane to heme-containing proteins.
Vibrio cholerae is the causative agent of the severe diarrheal disease cholera. V. cholerae has the ability to survive in a variety of environments, including fresh and marine waters, as well as in the human host. Cholera is transmitted to humans from contaminated water and food. Following ingestion, V. cholerae colonizes the lower small intestine, where it secretes cholera toxin. The toxin is responsible for the severe diarrhea characteristic of cholera.
This pathogen has an absolute requirement for iron and must acquire iron in each of the environments in which it grows. Accordingly, it has evolved several mechanisms for iron acquisition, including the production and secretion of the catechol siderophore vibriobactin (9). This iron-chelating compound binds iron with high affinity and is then transported back into the cell. V. cholerae also uses several siderophores produced by other organisms, including ferrichrome (9, 32), enterobactin (22, 53), and schizokinen (35).
In the human host, heme is an abundant potential source of iron. Although siderophores bind iron with high affinity, they cannot remove the iron from heme or heme compounds. Hence, many pathogenic bacteria, including V. cholerae, have systems for the direct utilization of heme, either as free heme or from various heme-containing proteins (8, 39, 46). Heme transport genes have been cloned and characterized from a number of gram-negative pathogens (for reviews, see references 8, 42, and 46).
Proteins involved in the transport of heme into the cell have been identified in V. cholerae. Three distinct outer membrane receptors, HutA, HutR, and HasR (14, 15, 21), specifically transport heme across the outer membrane. This transport requires the activity of TonB, which, together with its accessory proteins ExbB and ExbD, transduces energy from the inner membrane to heme receptors in the outer membrane. Two complete sets of tonB system genes are present in the V. cholerae genome (24). The tonB1 exbB1 exbD1 operon is located on the small chromosome, and the predicted protein sequences have low similarity to TonB, ExbB, and ExbD from enteric bacteria. In contrast, TonB2 is encoded on the large chromosome and has higher sequence similarity to the TonB proteins from other enteric bacteria (12, 24). The hutBCD genes are cotranscribed with the tonB1 system genes. HutBCD comprise a periplasmic binding protein-dependent ABC transport system that is thought to transport heme across the inner membrane. Strains carrying mutations in hutB or hutC can still use heme, suggesting that there must be at least one other mechanism for heme to cross the cytoplasmic membrane (24).
It appears that the intact heme moiety is transported across the outer membrane. This is supported by the observation that supplying heme transport genes from V. cholerae (15) or other organisms (40) to an Escherichia coli strain that is defective in heme biosynthesis confers the ability to grow normally when heme is provided in the medium (15, 40). In addition, a V. cholerae mutant strain defective in the synthesis of heme grows normally in media supplemented with heme or hemoglobin (13). This indicates that heme can be taken up and incorporated into cytochromes and presumably other heme proteins. Transport studies with Yersinia enterocolitica and 14C-labeled hemin have also indicated that the entire heme molecule was transported into the cell (41). The manner in which the transported heme is transferred from the heme transport proteins to cellular heme proteins is not known.
Most heme-transporting organisms can use heme as an iron source, since heme promotes their growth on iron-restricted media. In a few pathogenic bacterial species, including Corynebacterium diphtheriae (33, 49), Neisseria meningitidis (34, 55, 56), Pseudomonas aeruginosa (30), and Staphylococcus aureus (37), heme oxygenase, an enzyme that removes the iron from the heme moiety, has been identified. In these organisms, heme oxygenase is required for the use of heme as a source of iron. However, no heme oxygenase activity has been identified in V. cholerae or any other enteric bacteria, and no gene with homology to known heme oxygenases is present in the sequenced genomes. It is not known whether these bacteria have an unidentified enzyme to remove iron from heme or whether the bacteria can use heme in the absence of such an activity. For example, the efficient incorporation of the newly transported heme into heme proteins may reduce the amount of free iron the bacteria would need to transport from the medium. If this requirement for free iron was reduced to a sufficient level, it may allow the bacteria to grow on iron-restricted media.
In this work we show that HutZ (open reading frame [ORF] VCA0907) is required for efficient heme utilization in V. cholerae. As observed for other heme utilization proteins, the synthesis of HutZ is negatively regulated by iron. HutZ was purified and shown to bind heme with high efficiency. We propose that HutZ may function as a heme shuttling or storage protein or both.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The strains and plasmids used in this study are listed in Table 1. The iron chelator ethylenediamine di(ortho-hydroxyphenylacetic acid) (EDDA) was deferrated by the method of Rogers (31). When added, the antibiotic concentrations used were 250 μg of carbenicillin per ml, 50 μg of kanamycin per ml, and 50 (E. coli) or 5 (V. cholerae) μg of chloramphenicol per ml. Corynebacterium strains were routinely grown in heart infusion broth containing 0.2% (vol/vol) Tween 80 (HIB-Tween).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Reference |
|---|---|---|
| Bacterial strains | ||
| V. cholerae | ||
| Lou15 | El Tor clinical isolate | 36 |
| EWV104 | Lou15 vibB::cam | 51 |
| EWV105 | Lou15 hutW::kan | This study |
| EWV107 | Lou15 vibB::cam hutW::kan | This study |
| EWV115 | Lou15 vibB::cam hutZ::kan | This study |
| C. diphtheriae HC1-7 | HC1 hmuO mutant | 5 |
| C. ulcerans CU29 | CU712 hmuO mutant | 33 |
| E. coli | ||
| DH5α | Host for subcloning; ent+ | 10 |
| SY327λpir | Host for pGP704 derivatives | 23 |
| DH5αλpir | Host for pCVD442 | 23 |
| BL21(DE3) | Host for HutZ expression | 43 |
| Plasmids | ||
| pBluescript SK− | Cloning vector | Stratagene |
| pGEM-Teasy | Cloning vector | Promega |
| pWSK29 | Cloning vector | 47 |
| pWKS30 | Cloning vector | 47 |
| pET30a | T7 cloning vector | Novagen |
| pCmZ | Shuttle vector for E. coli and corynebacteria | 5 |
| pUC18K | Nonpolar aphA-3 cassette | 20 |
| pUC4K | Kanamycin resistance cassette | Pharmacia |
| pACSac | pACYC184 containing sacB | 24 |
| pCVD442 | pGP704 containing sacB | 4 |
| pHUT10 | 10.3-kb HindIII fragment with tonB1 system, hutBCD and hutWXZ in HindIII site of pACYC184 | 15 |
| pEHutZ | pET-30a containing Vent PCR fragment with hutZ | This study |
| pVHT105 | BclI fragment from pHUT10 with hutWXZ in BamHI site of pWSK29 | This study |
| pVHT106 | ApaI deletion of pVHT105, containing only hutW | This study |
| pVHT108 | BclI fragment from pHUT10 with hutWXZ in BamHI site of pCmZ | This study |
| pVHT109 | BclI fragment from pHUT10 with hutWXZ in BamHI site of pCmZ, opposite orientation of insert as pVHT108 | This study |
| pVHT111 | pVHT105; hutX::aph | This study |
| pVHT116 | pVHT105; hutZ::aph | This study |
| pVHT122 | pVHT105; hutW::aph | This study |
| pVHT124 | MluI/NotI deletion of pVHT105, containing only hutZ | This study |
| pVHT128 | pCmZ containing BclI fragment with hutWXZ; hutW::aph | This study |
| pVHT130 | pCmZ containing BclI fragment with hutWXZ; hutX::aph | This study |
| pVHT132 | pCmZ containing BclI fragment with hutWXZ; hutZ::aph | This study |
| pVHT139 | pCmZ containing Pfu PCR fragment with hutZ | This study |
Growth assays.
Cultures were grown overnight in L broth and then diluted 200-fold into minimal medium. The M9 medium was modified and contained 42 mM Na2PO4, 22 mM KH2PO4, 18 mM NH4Cl, 85 mM NaCl, 2.5 mM MgSO4, and 0.4% (wt/vol) sucrose or another carbon source as indicated. All cultures were grown with aeration at 37°C. To determine the ability of the hutZ mutant to grow in iron-restricted medium following growth with heme as the sole iron source, strains (EWV104, EWV115, and EWV115/pVHT124) were grown with shaking at 37°C to late log phase in M9 medium with 200 μg of EDDA per ml and 2.5 μM hemin. Cultures were centrifuged, washed with saline, and resuspended to an A650 of approximately 0.07 in M9 medium containing 200 μg of EDDA per ml. Cultures were grown with aeration at 37°C, and A650 was monitored.
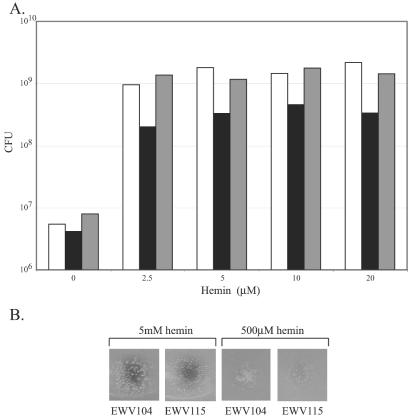
For the heme toxicity studies, overnight cultures were diluted 500-fold into L broth containing 600 μg of EDDA per ml and the indicated quantities of hemin. Cultures were grown with shaking at 37°C. After 3.5 h, the A650 of each culture was determined, and serial dilutions of the culture were plated to determine the number of CFU present. The mean inoculum size for EWV104/pWSK29 was 2.12 × 107; for EWV115/pWKS30 it was 1.95 × 107, and for EWV115/pVHT124 it was 1.89 × 107. For the spot toxicity assays, overnight cultures were diluted 100-fold into L broth and 25 μl was mixed with 20 ml of L agar containing 300 μg of EDDA per ml and poured into a 100-mm-diameter petri plate. After solidification of the agar, 5 μl of either a 5 mM or 500 μM hemin solution was spotted onto the surface of the agar. Growth around the heme spot was photographed following incubation for 24 h at 37°C.
For C. ulcerans heme utilization assays, cultures were grown overnight in HIB-Tween and plated on HIB agar containing 200 μg of EDDA per ml, 5 μM heme, and 2 μg of chloramphenicol per ml.
Western blot assays.
Overnight cultures were diluted 100-fold into L broth containing the indicated concentration of EDDA or hemin. When the A595 reached approximately 0.6, 1 ml of culture was centrifuged for 1 min in a Microfuge, and the pellet was resuspended in 0.2 ml of sodium dodecyl sulfate (SDS) gel sample buffer. The samples were boiled for 10 min and loaded onto an SDS-15% polyacrylamide gel. Following transfer to nitrocellulose, HutZ was visualized with a rabbit antiserum raised against purified HutZ.
DNA sequencing.
DNA was sequenced with an Applied Biosystems Prism 377 DNA sequencer (Perkin-Elmer Corp.). Routine DNA sequence analysis was performed with the program DNA Strider (18). Amino acid sequence alignments were performed with the ClustalW feature of the MacVector package (25). The BLAST program (1) was used to search the National Center for Biotechnology Information protein database.
Mutant strain construction.
To construct EWV107, the kanamycin resistance gene from plasmid pUC4K was inserted as a BamHI fragment into the BglII site in hutW carried on plasmid pHUT10. The SnaBI/HpaI fragment containing hutW::kan was subcloned into the NruI site of pACSac. The resulting plasmid was introduced into V. cholerae strain Lou15 by electroporation, and an allelic exchange mutant named EWV105 was obtained as previously described (52). A vibB::cam mutation was introduced into EWV105 as previously described (51) to give hutW::kan vibB::cam mutant strain EWV107.
To construct EWV115, the region containing hutZ was amplified from Lou15 DNA with Taq DNA polymerase and the primers Liz 211 (5′TTATGGCGAAGCATCATCTGC) and Liz 212 (5′AAGAAGCGGTCAATGGGTGC). The resulting fragment was subcloned into pGEM-Teasy, and the kan gene from pUC4K was inserted as a HincII fragment into the AfeI site in hutZ. The fragment containing hutZ::kan was obtained by digestion with NotI, made blunt with Klenow, and subcloned into the SmaI site of pCVD442. Marker exchange mutations were obtained as previously described (21).
Plasmid construction.
A series of nonpolar mutations in the hutWXZ genes was constructed as follows. The SmaI fragment containing the aphA-3 cassette from plasmid pUC18K was inserted into the StyI site in hutW, the SphI site in hutX, or the Eco47III site in hutZ. The StyI and SphI sites were made blunt with Klenow prior to ligation with the aphA-3 cassette fragment. In each case, the DNA sequence around the 3′ junction of the aphA-3 cassette was determined to confirm the orientation of the insert and that the hut sequence was in frame. The plasmids carrying these inserts are listed in Table 1. Western blotting confirmed that HutZ is produced by pVHT111 (hutX::aph) and pVHT122 (hutW::aph), and the amount produced was similar to that made from the plasmid carrying the wild-type operon (pVHT105). As expected, HutZ was not produced by pVHT116 (hutZ::aph).
pVHT139 was constructed by PCR amplification of hutZ from V. cholerae strain Lou15 with Pfu DNA polymerase (Stratagene) and primers Liz197 (5′-CGGGATCCTATCGCCGAAAAAACAAGC) and Liz198 (5′-CGGGATCCTTCTTTACCGCTCAAGGTGAAAAC). The PCR product was cleaved with BamHI and cloned into the BamHI site of pCmZ. The resulting clone was sequenced to confirm that the cloned fragment did not contain PCR errors. pEHutZ was constructed by PCR amplification of hutZ from pVHT105 with Vent DNA polymerase (New England Biolabs) and a forward primer containing an NdeI site (5′-GGAATTCCATATGGATCAGCAAGTTAAGCA) and a reverse primer containing an XhoI site (5′-CCGCTCGAGTTAGCCATTAGAAATCTTAC). The PCR product was cleaved with NdeI and XhoI and cloned into pET30a cleaved with NdeI and XhoI.
Expression and purification of HutZ and preparation of HutZ antiserum.
An overnight culture of E. coli BL21(DE3) carrying plysS and pEHutZ was diluted into 100 ml of Luria-Bertani (LB) medium contain ampicillin and grown at 37°C to mid-log phase. Ten milliliters of the culture was then diluted into 1 liter of LB medium containing ampicillin, and when the cells reached mid-log phase isopropyl-β-d-thiogalactopyranoside was added to a final concentration of 1 mM. The cells were grown for an additional 4 to 5 h at 30°C and harvested by centrifugation at 10,000 × g for 20 min. Cells were lysed by sonication in buffer containing 50 mM Tris-HCl (pH 7.8), 1 mM EDTA, and 1 mM phenylmethylsulfonyl fluoride, and the suspension was centrifuged at 27,000 × g for 40 min.
The supernatant was applied to a Sepharose-Q Fast Flow column (1.5 by 10 cm) previously equilibrated with 20 mM Tris-HCl (pH 7.5). The column was washed with 3 column volumes of 20 mM Tris-HCl (pH 7.5) containing 50 mM NaCl. Protein was then eluted with the same buffer with a linear gradient of 50 to 500 mM NaCl. HutZ eluted at a concentration of 200 to 250 mM NaCl, and the peak fractions were pooled and dialyzed against 10 mM KPO4 (pH 7.4) at 4°C. HutZ was stored at −80°C or reconstituted with heme as described below. A portion of the purified protein was used for the preparation of anti-HutZ antiserum in rabbits. The antiserum was prepared at Cocalico Biologicals, Inc. (Reamstown, Pa.), in accordance with standard protocols.
Reconstitution of HutZ with heme.
The heme-HutZ complex was prepared as follows. Hemin was added to the purified HutZ protein at a final 2:1 heme-to-protein ratio. The sample was then applied to a Bio-Gel HTP column (1.5 by 6 cm) preequilibrated with 20 mM KPO4 (pH 7.8) buffer. The column was washed with the same buffer (5 volumes). The protein was then eluted with a linear gradient of 10 mM KPO4 (pH 7.8) to 150 mM KPO4 (pH 7.8). The protein fractions were pooled and dialyzed against 20 mM Tris-HCl (pH 7.8) at 4°C. The protein was concentrated with an Amicon filtration unit and stored at −80°C.
Absorption spectroscopy of the HutZ protein.
The UV and visible spectra of wild-type HutZ were recorded in 20 mM Tris (pH 7.5) on a Cary 100Bio spectrophotometer. The millimolar extinction coefficient (ɛ405) for the heme-HutZ complex was determined as previously described (7). The A412 of a purified heme-HutZ sample was measured. An excess of dithionite was added, and the spectrum of the reduced ferrous pyridine hemochrome was recorded. The concentration was calculated from the absorbance maxima at 418.5, 526, and 555 nm with millimolar extinction coefficient values of 170, 17.5, and 34.4, respectively.
RESULTS
Identification of an operon containing heme utilization genes.
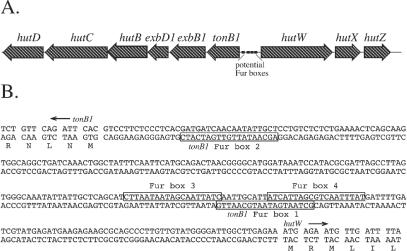
To identify genes that could be involved in the utilization of heme following its entry into the cytoplasm, we examined genes that are closely linked to known heme transport genes and have no known function. Three genes linked to the V. cholerae tonB1 operon have homology with genes in other heme transport loci (Table 2). These genes, named hutW, hutX, and hutZ, form an apparent operon that is divergently transcribed from the tonB1 operon (12, 24) (Fig. 1A). The promoter region for these genes contains several potential binding sites for the iron regulatory protein Fur. The sequences of these potential Fur boxes are indicated in Fig. 1B. Given this information, we decided to determine whether HutWXZ has any role in heme utilization in V. cholerae.
TABLE 2.
Homologies of HutWXZ to selected proteins
| Protein (ORF)a and homolog | Identity (%) | Similarity (%) | Refer- ence(s) |
|---|---|---|---|
| HutW (VCA0909) | |||
| Vibrio parahaemolyticus PhuW (VPA427) | 65 | 80 | 17, 26 |
| Vibrio vulnificus HupW (VV21615) | 64 | 78 | 3 |
| Plesiomonas shigelloides HugW | 51 | 68 | 16 |
| Yersinia pestis YPO0286 | 34 | 52 | 28 |
| E. coli O157:H7 ChuW | 33 | 51 | 11, 29 |
| E. coli K-12 HemN | 20 | 38 | 45 |
| V. cholerae HemN | 20 | 37 | 12 |
| E. coli K-12 ORF yggW (o459) | 20 | 31 | 2 |
| V. cholerae yggW homologue (VC0455) | 17 | 31 | 12 |
| HutX (VCA0908) | |||
| V. parahaemolyticus PhuX (VPA428) | 74 | 85 | 17 |
| V. vulnificus HupX (VV21616) | 72 | 83 | 3 |
| Pasteurella multocida PM0298 | 43 | 67 | 19 |
| P. shigelloides HugX | 43 | 62 | 16 |
| Y. pestis YPO0285 | 34 | 53 | 28 |
| Shigella dysenteriae ShuX | 33 | 55 | 50 |
| HutZ (VCA0907) | |||
| V. vulnificus HupZ (VV21617) | 86 | 93 | 3 |
| V. parahaemolyticus PhuZ (VPA429) | 80 | 91 | 17 |
| P. shigelloides HugZ | 59 | 76 | 16 |
| P. multocida PM0299 | 56 | 74 | 19 |
| P. multocida PM0042 | 45 | 65 | 19 |
| Campylobacter jejuni Cj1613c | 24 | 38 | 27 |
| Haemophilus influenzae Rd HI0854 | 24 | 35 | 6 |
| Helicobacter pylori 26695 HP0318 | 21 | 34 | 44 |
ORF numbering is according to the TIGR database (12).
FIG. 1.
(A) Map of the V. cholerae hutWXZ region. The direction of transcription is indicated by the arrows, and the smaller boxes indicate the potential Fur boxes. (B) Sequence of the tonB1-hutW intergenic region. The boxes indicate the locations of potential Fur boxes. Expression of the tonB1 operon was previously shown to be repressed by iron, and the transcription start was mapped near tonB1 Fur box 1 (25). Potential Fur boxes identified in that work are labeled tonB1 Fur box 1 and tonB1 Fur box 2. Two additional potential Fur boxes are labeled Fur box 3 and Fur box 4.
HutW is a member of the radical SAM superfamily of S-adenosylmethionine-dependent enzymes (38). This large family of proteins uses a radical-based mechanism to catalyze diverse chemical reactions. HutW has the highest homology with proteins encoded by genes linked to heme transport genes of Vibrio parahaemolyticus (26), Plesiomonas shigelloides (16), Yersinia pestis (28), and E. coli O157:H7 (Table 2). HutW has lower homology with HemN, the oxygen-independent form of the heme biosynthetic enzyme coproporphyrinogen oxidase (54), and also with yggW, an ORF of unknown function present in the genome of E. coli K-12 (2) and several other bacterial species. The two downstream genes (Fig. 1A) were also analyzed. The predicted amino acid sequences of HutX and HutZ lack any motifs or signature sequences that would suggest an activity for these proteins. Both proteins, however, have homology with proteins encoded within heme transport regions in other organisms (Table 2). The functions of these proteins in other organisms are also unknown.
The homology of HutW with HemN suggested that it might have coproporphyrinogen oxidase activity. To test this, a Salmonella enterica serovar Typhimurium hemN mutant was transformed with plasmid pVHT105, which contains the hutWXZ operon. This clone did not complement the hemN mutation (data not shown), indicating that hutW is unlikely to encode an oxygen-independent coproporphyrinogen oxidase activity. In a positive control, the hemN gene of E. coli O157:H7 complemented the Salmonella hemN mutation. HutW and several other HutW homologues encoded in heme transport loci are generally annotated as hemN (oxygen-independent coproporphyrinogen oxidases). The annotation of this group of proteins may need to be reviewed.
Genetic analysis of hutWXZ.
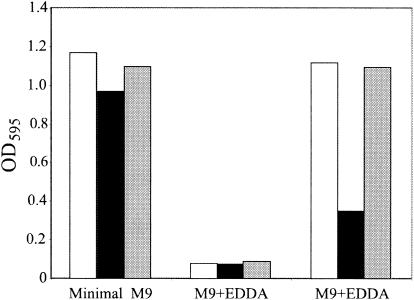
Because the sequence analysis did not suggest a role for the genes in the hutWXZ operon, we used allelic exchange to construct a chromosomal mutant in which a kanamycin resistance cassette was inserted into the hutW gene (see Materials and Methods). We anticipated that this hutW::kan insertion would be polar on hutX and hutZ, allowing us to identify a role of any or all of these genes in heme utilization. A mutation in the vibriobactin biosynthetic gene vibB was also introduced into this strain to reduce growth due to vibriobactin utilization in iron-restricted media. The resulting strain, EWV107, grew similarly to the isogenic vibB mutant strain EWV104 in both LB and minimal media, indicating that the hutWXZ genes do not encode functions required for growth in these media. However, EWV107 was defective in heme utilization (Fig. 2). In this experiment, all strains grew well in minimal medium with no added iron, and their growth was inhibited in the same medium containing the iron chelator EDDA. When the M9-EDDA medium was supplemented with hemin, the hutW::kan mutant grew poorly relative to the isogenic vibB mutant (Fig. 2). Supplying the hutWXZ genes on a plasmid restored the ability of the mutant to use heme efficiently. This result was not dependent on the carbon source used, since the hutW vibB::cam mutant had a defect in heme utilization when glucose, sucrose, lactate, succinate, or glycerol was used as the sole carbon source. In contrast to the result obtained with minimal medium, the hutW::kan vibB::cam mutant was able to use heme as an iron source in L broth containing EDDA and hemin (data not shown). The reason for this difference in heme utilization between L broth and minimal medium is not known.
FIG. 2.
Growth of hutW mutant with heme as the sole iron source. Overnight cultures were diluted 1/100 into M9 medium that was either unsupplemented (minimal M9), supplemented with 200 μg of the iron chelator EDDA per ml (M9+EDDA), or supplemented with 200 μg of EDDA per ml and 2.5 μM hemin (M9+hemin). The optical density at 595 nm (OD595) of each culture was determined after 6 h of growth at 37°C. The strains were the vibB::cam mutant EWV104 (white), the vibB::cam hutW::kan mutant EWV107 (black), and EWV107 carrying hutWXZ on plasmid pVHT105 (gray). Although the relative differences between strains were consistent between experiments, the absolute values for each strain varied owing to subtle changes in the medium and assay conditions, and thus results of only one representative experiment are shown.
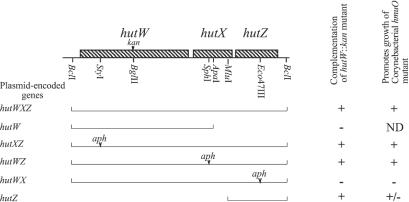
Since the kan insertion in hutW is likely to be polar on hutX and hutZ, additional experiments were performed to determine which of these genes is required for optimal heme utilization (Fig. 3). A plasmid encoding only hutW did not restore that ability of the hutW::kan strain to grow in minimal medium with heme as an iron source. This showed that hutW was not sufficient to complement the hutW::kan mutation and supported the model in which the hutWXZ genes form an operon. The hutW::kan mutant was then transformed with a series of plasmids containing the hutWXZ operon, in which one each of these genes was disrupted with the nonpolar aph cassette (20). The plasmids carrying insertions in hutW and hutX complemented the mutation, but the hutZ insertion plasmid did not. In a further experiment, a plasmid containing only hutZ complemented the hutW::kan mutation, indicating that hutZ is necessary and sufficient for efficient heme utilization by this mutant (Fig. 3).
FIG. 3.
Complementation of a V. cholerae hutW::kan mutant and suppression of corynebacterial hmuO mutants. For complementation of hutW::kan, V. cholerae strain EWV107 was transformed with a plasmid carrying the indicated genes and then tested for growth in M9 minimal medium supplemented with EDDA and hemin as described in the legend to Fig. 2. Where indicated, one of the genes was disrupted with the nonpolar aph gene. The plasmids tested for complementation of hutW::kan were as follows: hutWXZ, pVHT105; hutW, pVHT106; hutXZ, pVHT122; hutWZ, pVHT111; hutWX, pVHT116; hutZ, pVHT124. For suppression of hmuO mutants, either C. diphtheriae strain HC1-7 or C. ulcerans strain CU29 was transformed with plasmids carrying the indicated genes and plated on brain heart infusion medium containing EDDA and hemin. +, colony size of >1 mm, ±, colony size between 0.1 and 1 mm; −, no colonies were observed. The plasmids tested for suppression of the hmuO mutations were as follows: hutWXZ, pVHT108 and pVHT109; hutW, pVHT106; hutXZ, pVHT128; hutWZ, pVHT130; hutWX, pVHT132; hutZ, pVHT139.
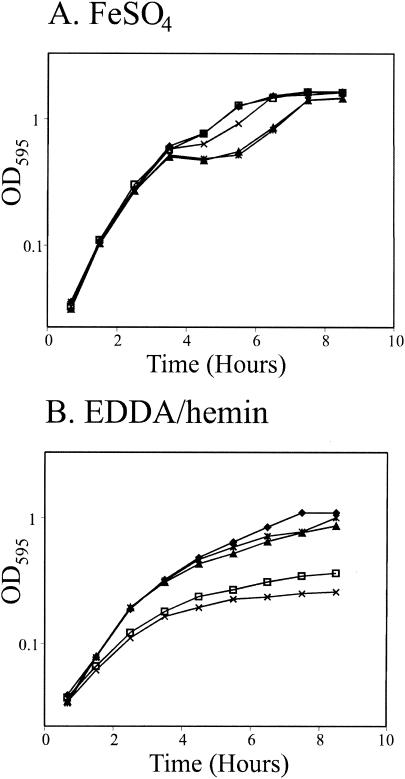
To further characterize hutZ, we constructed a chromosomal hutZ::kan mutation in the vibB genetic background. Like the hutW::kan mutant EWV107, EWV115 had a heme utilization defect. A representative experiment is shown in Fig. 4. All of the strains grew well in M9 medium supplemented with 40 μM iron (Fig. 4A). However, when heme was the sole iron source, the hutZ mutant grew more slowly and reached a lower final cell density than its parent strain (Fig. 4B). The hutZ::kan mutant grew more slowly than the previously characterized hutW::kan mutant in M9-EDDA-hemin medium, and although the difference was small, it was consistently observed in all experiments. Unlike the hutW::kan mutant, the hutZ::kan mutant used heme poorly in L broth (data not shown). Providing hutZ on a plasmid restored the ability to grow with heme as the sole iron source, indicating that the growth defect on heme-containing media was due to loss of hutZ. These data directly demonstrate that hutZ is required for optimal heme utilization by V. cholerae.
FIG. 4.
Time course of growth of the hutZ::kan mutation. Overnight cultures were diluted 1/200 into M9 minimal medium supplemented with 20 μM iron (A) or 200 μg of EDDA per ml and 2.5 μM hemin (B). The optical density at 595 nm (OD595) was measured at the indicated times. Symbols: ⧫, vibB mutant strain EWV104 carrying plasmid vector pWSK29; □, vibB hutW mutant strain EWV107 carrying pWSK29; ▴, strain EWV107 carrying hutZ on plasmid pVHT124; ×, vibB hutZ mutant strain EWV115 carrying pWKS30;  , strain EWV115 carrying hutZ on plasmid pVHT124.
, strain EWV115 carrying hutZ on plasmid pVHT124.
The growth defect on heme is not due to heme toxicity.
Reduced growth on heme as the sole iron source could be due either to failure to efficiently use the heme as a source of iron or to heme toxicity. To test for heme toxicity, overnight cultures of the hutZ mutant, the isogenic HutZ+ strain, and the hutZ mutant complemented with hutZ on a plasmid were diluted into iron-restricted media containing increasing concentrations of hemin and incubated for 3.5 h (Fig. 5A). No growth of the cells was observed in the absence of an added iron source; in fact, some cell death occurred since the number of viable cells present at 3.5 h was lower than that of the original inoculation. When the medium was supplemented with hemin, all three strains grew, although the final titers of the hutZ mutant cultures were consistently lower than those of the HutZ+ strains (Fig. 5A), and the extent of this difference was consistent with the reduced optical density observed in the growth curve (Fig. 4). If heme were toxic to the hutZ mutant, it might be expected that the number of viable hutZ mutant cells would decrease with increasing heme concentrations. The data show that the growth of the hutZ mutant was essentially unchanged across the range of heme concentrations normally used for heme utilization and toxicity assays.
FIG. 5.
Effect of heme on cell viability. (A) Overnight cultures were diluted to approximately 2 × 107/ml into L broth containing 600 μg of EDDA per ml and the indicated concentration of hemin. The cultures were incubated at 37°C with aeration for 3.5 h, and serial dilutions were plated to determine the number of CFU. The strains were the vibB::cam mutant EWV104/pWSK29 (white), the vibB::cam hutZ::kan mutant EWV115/pWKS30 (black), and EWV115 carrying hutWXZ on plasmid pVHT105 (gray). (B) Cultures of the indicated strains were mixed with L agar containing 300 μg of EDDA per ml. Following solidification of the agar, 5 μl of the indicated concentration of hemin was spotted on the surface and plates were incubated at 37°C for 24 h.
Bioassays also were performed in which heme at a concentration of 5 mM or 500 μM was spotted onto iron-restricted agar containing hutZ mutant or HutZ+ cells (Fig. 5B). Cell growth was observed around both heme concentrations, showing that both strains were able to use heme as an iron source. The colony size of the hutZ mutant was smaller than that of the HutZ+ strain, consistent with the reduced growth of the mutant observed in liquid assays. However, even at the highest heme concentration, colonies of the hutZ mutant strain were present at the center of the growth zone, directly under the spot inoculated with heme. Some of the colonies directly under the inoculation were slightly smaller than those at the edge of the inoculated area, but this effect was also present in the HutZ+ strain. Taken together, the results of these experiments do not support the model in which the growth defect of the hutZ mutant observed in our assays is due to increased susceptibility to heme toxicity.
Visualization of HutZ and regulation of hutZ by iron.
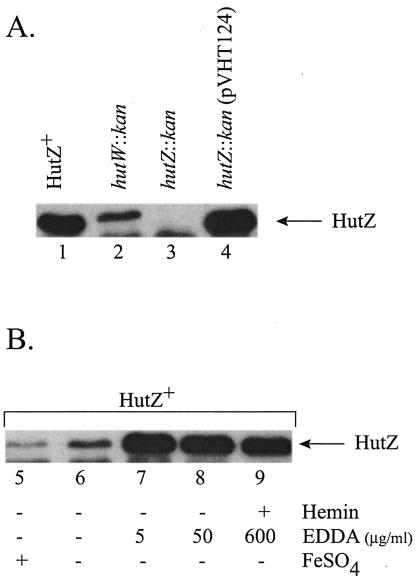
Purified HutZ was used to raise a polyclonal antiserum in rabbits (see Materials and Methods). In a Western blot assay, a 20,000-Mr band was visualized in the sample of cellular protein from the HutZ+ strain (Fig. 6, lane 1). The amount of HutZ present was reduced, but not totally eliminated, in the hutW::kan mutant strain (EWV107), while HutZ was not observed in the hutZ::kan strain (EWV115) (Fig. 6, lane 3). This residual expression of HutZ in the hutW::kan mutant may explain why it had a less severe phenotype than the hutZ::kan mutant. HutZ expression was restored when hutZ was provided on a plasmid (Fig. 6, lane 4). In these immunoblots, a band migrating slightly faster than HutZ was consistently visualized. The identity of this protein is unknown, but since it was also recognized by the preimmune serum, it is unlikely to be related to HutZ.
FIG. 6.
Visualization of HutZ and iron regulation of HutZ production. Extracts of total cellular protein were prepared as described in Methods and Materials, separated on an SDS-15% polyacrylamide gel, and visualized with antiserum to HutZ. (A) The following strains were grown in L broth containing 25 μg of EDDA per ml: EWV104 (vibB::cam) (lane 1), EWV107 (vibB::cam hutW::kan) (lane 2), EWV115 (vibB::cam hutZ::kan) (lane 3), and EWV115/pVHT124 (lane 4). (B) EWV104 (vibB::cam) was grown in LB medium containing the following supplement(s): 40 μM FeSO4 (lane 5), no supplement (lane 6), 5 μg of EDDA per ml (lane 7), 50 μg of EDDA per ml (lane 8), or 600 μg of EDDA per ml and 5 μM hemin (lane 9).
The presence of potential Fur boxes upstream of the hutW promoter suggested that this operon would be regulated by iron. The Western blot shows that only a low level of HutZ is produced when the culture was grown in LB medium supplemented with 40 μM FeSO4 (Fig. 6, lane 5) and that the amount of HutZ present during growth in unsupplemented L broth was only slightly greater than during growth with added iron (lane 6). When the iron chelator EDDA was added to a final concentration of either 5 or 50 μg/ml, the amount of HutZ present was greatly increased (Fig. 6, lanes 7 and 8). These data are consistent with the idea that the synthesis of HutZ is negatively regulated by iron. To determine whether the synthesis of HutZ is also regulated by hemin, we grew a culture in the presence of EDDA and hemin. The amount of HutZ present when the strains were grown in media containing EDDA and hemin was similar to the amount synthesized in the presence of EDDA alone (Fig. 6, lane 9). Thus, HutZ synthesis was neither induced nor repressed by the presence of heme. Addition of heme to L broth in the absence of added iron chelators did not affect HutZ synthesis, indicating that heme also did not influence HutZ synthesis under iron-replete conditions (data not shown).
The hutWXZ genes stimulate growth of corynebacterial hmuO mutants.
The gene for heme oxygenase, hmuO, has been identified in C. diphtheriae and in C. ulcerans, and hmuO mutants fail to grow when hemin is the sole iron source (33). To determine whether the hutWXZ genes might have a function similar to that of hmuO and allow growth of hmuO mutants on heme, we subcloned the hutWXZ genes into the corynebacterial shuttle vector pCmZ (5). The presence of the hutWXZ genes restored the ability of the corynebacterial hmuO mutants to grow with hemin as the sole source of iron (Fig. 3). The ability of the hutWXZ genes to restore growth of the Corynebacterium mutants was similar to that seen when the mutations were complemented with the cloned C. diphtheriae hmuO gene (data not shown). To determine which ORF is required for restoration of growth of the hmuO mutant, the series of nonpolar aph cassette insertions was subcloned into the pCmZ shuttle vector. Plasmids carrying insertions in the hutW and hutX genes restored the growth of the Corynebacterium hmuO mutants with heme as an iron source, but a plasmid with an insertion in hutZ did not (Fig. 3). This indicates that hutZ was necessary for restoration of growth on hemin in this assay. To determine whether hutZ is sufficient for growth, we cloned hutZ into pCmZ. C. diphtheriae or C. ulcerans hmuO mutants carrying the resulting plasmid, pVHT139, grew with heme as the sole source of iron, but the colony size was smaller than that of the hmuO mutants carrying either the complete hutWXZ operon or the cloned C. diphtheriae hmuO gene.
Purification and characterization of HutZ.
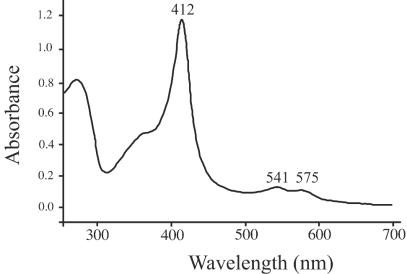
To determine whether HutZ has heme binding or heme oxygenase activity, the HutZ protein was expressed and purified as the apoprotein with no heme bound. Reconstitution of the purified protein with heme resulted in a protein with a Soret at 412 nm and visible bands at 541 and 575 nm (Fig. 7). The spectrum of HutZ closely resembles that of the previously characterized heme binding protein ShuS from Shigella dysenteriae (48). The spectrum suggests that the heme is a ferric six-coordinate low-spin heme in which the proximal ligand to the heme is most likely a histidine. We have recently characterized the heme-ShuS protein by magnetic circular dichroism and shown that the heme in the ShuS complex is coordinated by a histidine with a hydroxide ligand in the distal position at pH 7.8 (A. Wilks, unpublished data). The millimolar extinction coefficient (ɛ412) of the heme-HutZ complex was calculated to be 166 mM−1.
FIG. 7.
Absorbance of the HutZ-heme complex. Purified apo-HutZ was reconstituted with heme, and the absorbance spectrum was recorded.
The heme-HutZ complex was analyzed for HO activity with ascorbate or the NADPH cytochrome P450 reductase system as previously described (30, 49, 56). In the presence of either as a source of reducing equivalents, formation of verdoheme was observed. While verdoheme is an intermediate in the heme oxygenase-catalyzed pathway to biliverdin, it can be formed as a result of nonspecific action of hydrogen peroxide formed during the reaction. This nonspecific reaction can be prevented by addition of catalase to the reaction mixture. In the presence of catalase, the known heme oxygenases catalyze reduction of the iron and binding of oxygen prior to the formation of Fe3+-verdoheme and subsequently biliverdin. In contrast, when catalase was added to the reaction mixture, the heme-HutZ complex did not result in the formation of either verdoheme or biliverdin.
HutZ is needed for growth at low iron concentrations following growth with heme as the sole iron source.
The ability of HutZ to bind heme suggested that it might function as a heme storage protein. If this is the case, we would predict that the hutZ vibB mutant would have lower levels of internal iron stores than the vibB parental strain following growth with heme as the sole iron source. To test this, we diluted overnight cultures of the vibB::cam mutant EWV104, the hutZ::kan vibB::cam mutant EWV115, and EWV115 carrying hutZ on a plasmid into M9 medium containing heme and the iron chelator EDDA and grew the cultures overnight. The cultures were then diluted into M9 medium containing EDDA, and growth was monitored. EWV104 and EWV115 carrying hutZ on a plasmid grew slowly with a doubling time of approximately 3 h, while EWV115 did not grow at all. This is evidence that EWV115 is depleted of its internal iron stores (data not shown).
DISCUSSION
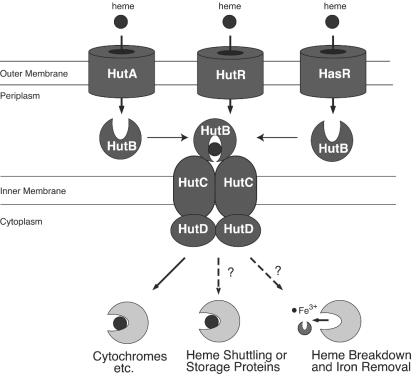
The current model for heme transport in V. cholerae is shown in Fig. 8. In this model, heme is transported across the outer membrane in a TonB-dependent manner by one of the three heme receptors. HutBCD likely functions as a periplasmic binding protein-dependent ABC transporter to transfer heme across the inner membrane (24).
FIG. 8.
Model for heme utilization in V. cholerae. Heme crosses the outer membrane through one of three outer membrane receptors, HutA, HutR, or HasR (14, 21), and this transport requires the activity of one of the two TonB systems (24) (not shown). Heme is then transported across the inner membrane most likely by the HutBCD transport system (24). The fate of the heme moiety following transport is not known, although at least some of the heme is likely incorporated into cytochromes and other heme proteins.
Although many of the proteins that transport heme into the cell have been identified, the fate of heme after it enters the cell is not well understood (42). In principle, transported heme could have one of three possible fates. It could be incorporated directly into heme proteins, it could be degraded to release the iron, or it could be stored for future use. It is known that in V. cholerae the transported heme can be incorporated into cytochromes and possibly other heme proteins, but neither heme storage nor heme degradation has been demonstrated in this or a related organism. An additional question is how heme moves from the heme transport proteins in the membrane to heme-containing proteins located throughout the cell. Since unbound heme is toxic and poorly soluble in water, it is not anticipated that significant quantities of heme would be freely diffusing through the cytoplasm. Either the apo-heme proteins would have to be recruited to the heme transport system or there must be a carrier to take the heme to these proteins.
As a soluble, heme-binding protein, HutZ has the properties expected of a heme carrier or storage protein. Its role in heme storage is supported by the reduced growth of the hutZ mutant in iron-restricted medium following growth with heme as the sole iron source. Furthermore, biochemical experiments have indicated that heme is efficiently transferred from HutZ to the Neisseria heme oxygenase HmuO (Wilks, unpublished). This suggests a possible role of HutZ in heme trafficking within the cell. We propose that HutZ may function to bind the transported heme, storing it in a nontoxic and bioavailable form. Then, in response to unknown cellular signals, the heme could be transferred to cellular heme proteins as needed.
The presence of the hutWXZ operon on a plasmid conferred the ability to grow with heme as the sole iron source on corynebacterial hmuO mutants, and hutZ was required for this effect. This suggested that HutZ could be a heme oxygenase, but purified HutZ did not have detectible heme oxygenase activity. One possibility is that HutZ binds heme and this sequesters the heme and prevents heme toxicity. However, corynebacterial hmuO mutants are not sensitive to high levels of heme, suggesting that lack of growth of these mutants is due to lack of usable iron, rather than toxicity of transported heme. Two additional possibilities are that the corynebacterial strains have an second, extremely weak heme oxygenase that is stimulated in the presence of HutZ and that HutZ allows the cells to bypass the requirement for a heme oxygenase activity in heme utilization. Either of these possibilities may have implications for how organisms like V. cholerae use heme as a source of iron in the absence of an apparent heme oxygenase homologue.
In the hutZ mutant, the ability to use heme was reduced but not eliminated. It is possible that HutZ performs a nonessential function that allows more efficient utilization of heme. Alternatively, HutZ may perform an essential function, but an additional gene encoding a similar function may be present. There is considerable redundancy in the V. cholerae heme utilization genes. There are three heme receptors, two TonB systems, and probably multiple systems for the transport of heme across the inner membrane. At this time, there is no candidate gene with a function redundant with respect to that of hutZ. BLAST searches of the V. cholerae genome sequence (12) have not revealed another ORF with homology to HutZ. The S. dysenteriae ShuS protein binds heme in a manner similar to that of HutZ (48), suggesting that it might have a function similar to that of HutZ. However, no ShuS homologue was identified in a BLAST search of the V. cholerae genome. Little is known about proteins that are involved in the trafficking of endogenously synthesized heme. It is possible that any of the proteins that usually bind endogenously synthesized heme could partially fulfill the role of HutZ.
Many of the characterized heme transport loci contain genes with homology to hutWXZ. An ORF with homology with hemN was first identified within the heme transport loci of S. dysenteriae and E. coli O157:H7 (50), and additional hemN homologues were later found linked to heme transport genes in V. parahaemolyticus (26), V. vulnificus (3), P. shigelloides (16), Y. pestis (28), and V. cholerae (12). Each of the loci listed above also contains a homologue of hutX, but only the P. shigelloides and the other Vibrio species loci contain a hutZ homologue. Several other species contain a hutZ homologue but lack a hutW and hutX homologue, and Pasteurella multocida lacks a hutW homologue but has two hutZ homologues and one hutX homologue (19). The significance of this diversity is not understood, but the sequence conservation among these ORFs is comparable to the conservation observed in heme receptors. Genetic analysis has not indicated that hutW or hutX homologues are required for heme utilization. It is not known whether this indicates that the hutW and hutX homologues do not participate in heme utilization or whether redundant functions are present.
Prior to this work, an effect of hutWXZ homologues was observed in the following functional assay. When the P. shigelloides heme transport genes were used to reconstruct heme transport in E. coli, bacteria carrying clones that contained the heme receptor, the tonB system, and the hutWXZ homologues (called hugWXZ) used heme efficiently as an iron source. When the hugWXZ genes were deleted from the plasmid, the strain only used heme weakly and was sensitive to high levels of heme (16). The ORF(s) required was not identified. Although these data are consistent with the findings reported here for the V. cholerae hutWXZ genes, the hutZ mutant did not appear to be sensitive to high concentrations of heme. This may be due to differences in the genetic backgrounds of the V. cholerae hutZ mutant and E. coli expressing heme utilization proteins from plasmids.
Acknowledgments
We thank Doug Henderson and Alex Mey for helpful discussions and critical reading of the manuscript and Carolyn Fisher and Stacey Smith for technical assistance. We also thank Tom Elliott for providing strains.
The work was supported by National Institutes of Health grants AI50669 to S.M.P. and AI48551 to A.W.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Blattner, F. R., G. I. Plunkett, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 3.Chen, C. Y., K. M. Wu, Y. C. Chang, C. H. Chang, H. C. Tsai, T. L. Liao, Y. M. Liu, H. J. Chen, A. B. Shen, J. C. Li, T. L. Su, C. P. Shao, C. T. Lee, L. I. Hor, and S. F. Tsai. 2003. Comparative genome analysis of Vibrio vulnificus, a marine pathogen. Genome Res. 13:2577-2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Donnenberg, M. S., and J. B. Kaper. 1991. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection vector. Infect. Immun. 59:4310-4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Drazek, E. S., C. A. Hammack, and M. Schmitt. 2000. Corynebacterium diphtheriae genes required for acquisition of iron from haemin and haemoglobin are homologous to ABC haemin transporters. Mol. Microbiol. 36:68-84. [DOI] [PubMed] [Google Scholar]
- 6.Fleischmann, R. D., M. D. Adams, O. White, R. A. Clayton, E. F. Kirkness, A. R. Kerlavage, C. J. Bult, J. F. Tomb, B. A. Dougherty, J. M. Merrick, et al. 1995. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269:496-512. [DOI] [PubMed] [Google Scholar]
- 7.Fuhrop, J. H., and K. M. Smith. 1975. Laboratory methods, p. 804-807. In K. M. Smith (ed.), Porphyrins and metalloporphyrins. Elsevier Scientific Publishing Co., New York, N.Y.
- 8.Genco, C., and D. W. Dixon. 2001. Emerging strategies in microbial haem capture. Mol. Microbiol. 39:1-11. [DOI] [PubMed] [Google Scholar]
- 9.Griffiths, G. L., S. P. Sigel, S. M. Payne, and J. B. Neilands. 1984. Vibriobactin, a siderophore from Vibrio cholerae. J. Biol. Chem. 259:383-385. [PubMed] [Google Scholar]
- 10.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 11.Hayashi, T., K. Makino, M. Ohnishi, K. Kurokawa, K. Ishii, K. Yokoyama, C. G. Han, E. Ohtsubo, K. Nakayama, T. Murata, M. Tanaka, T. Tobe, T. Iida, H. Takami, T. Honda, C. Sasakawa, N. Ogasawara, T. Yasunaga, S. Kuhara, T. Shiba, M. Hattori, and H. Shinagawa. 2001. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 8:11-22. [DOI] [PubMed] [Google Scholar]
- 12.Heidelberg, J. F., J. A. Eisen, W. C. Nelson, R. A. Clayton, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, J. D. Peterson, L. Umayam, S. R. Gill, K. E. Nelson, T. D. Read, H. Tettelin, D. Richardson, M. Ermolaeva, J. Vamathevan, S. Bass, H. Qin, I. Dragoi, P. Sellers, L. McDonald, T. Utterback, R. D. Fleishmann, W. C. Nierman, O. White, S. L. Salzberg, H. O. Smith, R. R. Colwell, J. J. Mekalanos, J. C. Venter, and C. M. Fraser. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406:477-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Henderson, D. P. 1993. Heme iron utilization in Vibrio cholerae: genetics and role in virulence. Ph.D. dissertation. The University of Texas, Austin.
- 14.Henderson, D. P., and S. M. Payne. 1994. Characterization of the Vibrio cholerae outer membrane heme transport protein HutA: sequence of the gene, regulation of expression, and homology to the family of TonB-dependent proteins. J. Bacteriol. 176:3269-3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Henderson, D. P., and S. M. Payne. 1993. Cloning and characterization of the Vibrio cholerae genes encoding the utilization of iron from haemin and haemoglobin. Mol. Microbiol. 7:461-469. [DOI] [PubMed] [Google Scholar]
- 16.Henderson, D. P., E. E. Wyckoff, C. E. Rashidi, H. Verlei, and A. L. Oldham. 2001. Characterization of the Plesiomonas shigelloides genes encoding the utilization of iron from heme. J. Bacteriol. 183:2715-2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Makino, K., K. Oshima, K. Kurokawa, K. Yokoyama, T. Uda, K. Tagomori, Y. Iijima, M. Najima, M. Nakano, A. Yamashita, Y. Kubota, S. Kimura, T. Yasunaga, T. Honda, H. Shinagawa, M. Hattori, and T. Iida. 2003. Genome sequence of Vibrio parahaemolyticus: a pathogenic mechanism distinct from that of V. cholerae. Lancet 361:743-749. [DOI] [PubMed] [Google Scholar]
- 18.Marck, C. 1988. “DNA strider”: a “C” program for the fast analysis of DNA and protein sequences on the Apple Macintosh family of computers. Nucleic Acids Res. 16:1829-1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.May, B. J., Q. Zhang, L. L. Li, M. L. Paustian, T. S. Whittam, and V. Kapur. 2001. Complete genomic sequence of Pasteurella multocida, Pm70. Proc. Natl. Acad. Sci. USA 98:3460-3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Menard, R., P. J. Sansonetti, and C. Parsot. 1993. Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J. Bacteriol. 175:5899-5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mey, A. R., and S. M. Payne. 2001. Haem utilization in Vibrio cholerae involves multiple TonB-dependent haem receptors. Mol. Microbiol. 42:835-849. [DOI] [PubMed] [Google Scholar]
- 22.Mey, A. R., E. E. Wyckoff, A. Oglesby, E. Rab, R. K. Taylor, and S. M. Payne. 2002. Identification of the Vibrio cholerae enterobactin receptors VctA and IrgA: IrgA is not required for virulence. Infect. Immun. 70:3419-3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Occhino, D. A., E. E. Wyckoff, D. P. Henderson, T. J. Wrona, and S. M. Payne. 1998. Vibrio cholerae iron transport: haem transport genes are linked to one of two sets of tonB, exbB, exbD genes. Mol. Microbiol. 29:1493-1507. [DOI] [PubMed] [Google Scholar]
- 25.Olson, S. A. 1994. MacVector: an integrated sequence analysis program for the Macintosh. Methods Mol. Biol. 25:195-201. [DOI] [PubMed] [Google Scholar]
- 26.O'Malley, S. M., S. L. Mouton, D. A. Occhino, M. T. Deanda, J. R. Rashidi, K. L. Fuson, C. E. Rashidi, M. Y. Mora, S. M. Payne, and D. P. Henderson. 1999. Comparison of the heme iron utilization systems of pathogenic vibrios. J. Bacteriol. 181:3594-3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parkhill, J., B. W. Wren, K. Mungall, J. M. Ketley, C. Churcher, D. Basham, T. Chillingworth, R. M. Davies, T. Feltwell, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Moule, M. J. Pollen, C. W. Penn, M. A. Quall, M.-A. Rajandream, K. M. Rutherford, A. H. M. van Vliet, S. Whitehead, and B. G. Barrell. 2000. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403:665-668. [DOI] [PubMed] [Google Scholar]
- 28.Parkhill, J., B. W. Wren, N. R. Thomson, R. W. Titball, M. T. Holden, M. B. Prentice, M. Sebaihia, K. D. James, C. Churcher, K. L. Mungall, S. Baker, D. Basham, S. D. Bentley, K. Brooks, A. M. Cerdeno-Tarraga, T. Chillingworth, A. Cronin, R. M. Davies, P. Davis, G. Dougan, T. Feltwell, N. Hamlin, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Leather, S. Moule, P. C. Oyston, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Genome sequence of Yersinia pestis, the causative agent of plague. Nature 413:523-527. [DOI] [PubMed] [Google Scholar]
- 29.Perna, N. T., G. Plunkett III, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 30.Ratliff, M., W. Zhu, R. Deshmukh, A. Wilks, and I. Stojiljkovic. 2001. Homologues of neisserial heme oxygenase in gram-negative bacteria: degradation of heme by the product of the pigA gene of Pseudomonas aeruginosa. J. Bacteriol. 183:6394-6403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rogers, H. J. 1973. Iron-binding catechols and virulence in Escherichia coli. Infect. Immun. 7:445-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rogers, M. B., J. A. Sexton, G. J. DeCastro, and S. B. Calderwood. 2000. Identification of an operon required for ferrichrome iron utilization in Vibrio cholerae. J. Bacteriol. 182:2350-2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schmitt, M. P. 1997. Utilization of host iron sources of Corynebacterium diphtheriae: identification of a gene whose product is homologous to eukaryotic heme oxygenases and is required for acquisition of iron from heme and hemoglobin. J. Bacteriol. 179:838-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schuller, D. J., W. Zhu, I. Stojiljkovic, A. Wilks, and T. L. Poulos. 2001. Crystal structure of heme oxygenase from the gram-negative pathogen Neisseria meningitidis and a comparison with mammalian heme oxygenase-1. Biochemistry 40:11552-11558. [DOI] [PubMed] [Google Scholar]
- 35.Seliger, S. S., A. R. Mey, A.-M. Valle, and S. M. Payne. 2001. The two TonB systems in Vibrio cholerae: redundant and specific functions. Mol. Microbiol. 39:801-812. [DOI] [PubMed] [Google Scholar]
- 36.Sigel, S. P., and S. M. Payne. 1982. Effect of iron limitation on growth, siderophore production and expression of outer membrane proteins of Vibrio cholerae. J. Bacteriol. 150:148-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Skaar, E. P., A. H. Gaspar, and O. Schneewind. 2004. IsdG and IsdI, heme-degrading enzymes in the cytoplasm of Staphylococcus aureus. J. Biol. Chem. 279:436-443. [DOI] [PubMed] [Google Scholar]
- 38.Sofia, H. J., G. Chen, B. G. Hetzler, J. F. Reyes-Spindola, and N. E. Miller. 2001. Radical SAM: a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms: functional characterization using new analysis and information visualization methods. Nucleic Acids Res. 29:1097-1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stoebner, J. A., and S. M. Payne. 1988. Iron-regulated hemolysin production and utilization of heme and hemoglobin by Vibrio cholerae. Infect. Immun. 56:2891-2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stojiljkovic, I., and K. Hantke. 1992. Hemin uptake system of Yersinia enterocolitica: similarities with other TonB-dependent systems in gram-negative bacteria. EMBO J. 11:4359-4367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stojiljkovic, I., and K. Hantke. 1994. Transport of haemin across the cytoplasmic membrane through a haemin-specific periplasmic binding-protein-dependent transport system in Yersinia enterocolitica. Mol. Microbiol. 13:719-732. [DOI] [PubMed] [Google Scholar]
- 42.Stojiljkovic, I., and D. Perkins-Balding. 2002. Processing of heme and heme-containing proteins by bacteria. DNA Cell Biol. 21:281-295. [DOI] [PubMed] [Google Scholar]
- 43.Studier, F. W., and B. A. Moffatt. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189:113-130. [DOI] [PubMed] [Google Scholar]
- 44.Tomb, J. F., O. White, A. R. Kerlavage, R. A. Clayton, G. G. Sutton, R. D. Fleischmann, K. A. Ketchum, H. P. Klenk, S. Gill, B. A. Dougherty, K. Nelson, J. Quackenbush, L. Zhou, E. F. Kirkness, S. Peterson, B. Loftus, D. Richardson, R. Dodson, H. G. Khalak, A. Glodek, K. McKenney, L. M. Fitzegerald, N. Lee, M. D. Adams, J. C. Venter, et al. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539-547. [DOI] [PubMed] [Google Scholar]
- 45.Troup, B., C. Hungerer, and D. Jahn. 1995. Cloning and characterization of the Escherichia coli hemN gene encoding the oxygen-independent coproporphyrinogen III oxidase. J. Bacteriol. 177:3326-3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wandersman, C., and I. Stojiljkovic. 2000. Bacterial heme sources: the role of heme, hemoprotein receptors and hemophores. Curr. Opin. Microbiol. 3:215-220. [DOI] [PubMed] [Google Scholar]
- 47.Wang, R. F., and S. R. Kushner. 1991. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100:195-199. [PubMed] [Google Scholar]
- 48.Wilks, A. 2001. The ShuS protein of Shigella dysenteriae is a heme-sequestering protein that also binds DNA. Arch. Biochem. Biophys. 387:137-142. [DOI] [PubMed] [Google Scholar]
- 49.Wilks, A., and M. P. Schmitt. 1998. Expression and characterization of a heme oxygenase (HmuO) from Corynebacterium diphtheriae. J. Biol. Chem. 273:837-841. [DOI] [PubMed] [Google Scholar]
- 50.Wyckoff, E. E., D. Duncan, A. G. Torres, M. Mills, K. Maase, and S. M. Payne. 1998. Structure of the Shigella dysenteriae haem transport locus and its phylogenetic distribution in enteric bacteria. Mol. Microbiol. 28:1139-1152. [DOI] [PubMed] [Google Scholar]
- 51.Wyckoff, E. E., S. L. Smith, and S. M. Payne. 2001. VibD and VibH are required for late steps in vibriobactin biosynthesis in Vibrio cholerae. J. Bacteriol. 183:1830-1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wyckoff, E. E., J. A. Stoebner, K. E. Reed, and S. M. Payne. 1997. Cloning of a Vibrio cholerae vibriobactin gene cluster: identification of genes required for early steps in siderophore biosynthesis. J. Bacteriol. 179:7055-7062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wyckoff, E. E., A.-M. Valle, S. L. Smith, and S. M. Payne. 1999. A multifunctional ABC transporter system from Vibrio cholerae transports vibriobactin and enterobactin. J. Bacteriol. 181:7588-7596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu, K., J. Delling, and T. Elliott. 1992. The genes required for heme synthesis in Salmonella typhimurium include those encoding alternative functions for aerobic and anaerobic coproporphyrinogen oxidation. J. Bacteriol. 174:3953-3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhu, W., D. J. Hunt, A. Richardson, and I. Stojiljkovic. 2000. Use of heme compounds as iron sources by pathogenic neisseriae requires the product of the hemO gene. J. Bacteriol. 182:439-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhu, W., A. Wilks, and I. Stojiljkovic. 2000. Degradation of heme in gram-negative bacteria: the product of the hemO gene of neisseriae is a heme oxygenase. J. Bacteriol. 182:6783-6790. [DOI] [PMC free article] [PubMed] [Google Scholar]