Abstract
Genome sequencing of Chlamydia trachomatis serovar D has identified polymorphic membrane proteins (Pmp) that are a newly recognized protein family unique to the Chlamydiaceae family. Cumulative data suggest that these diverse proteins are expressed on the cell surface and might be immunologically important. We performed phylogenetic analyses and statistical modeling with 18 reference serovars and 1 genovariant of C. trachomatis to examine the evolutionary characteristics and comparative genetics of PmpC and pmpC, the gene that encodes this protein. We also examined 12 recently isolated ocular and urogenital clinical samples, since reference serovars are laboratory adapted and may not represent strains that are presently responsible for human disease. Phylogenetic reconstructions revealed a clear distinction for disease groups, corresponding to levels of tissue specificity and virulence of the organism. Further, the most prevalent serovars, E, F, and Da, formed a distinct clade. According to the results of comparative genetic analyses, these three genital serovars contained two putative insertion sequence (IS)-like elements with 10- and 15-bp direct repeats, respectively, while all other genital serovars contained one IS-like element. Ocular trachoma serovars also contained both insertions. Previously, no IS-like elements have been identified for Chlamydiaceae. Surprisingly, 7 (58%) of 12 clinical isolates revealed pmpC sequences that were identical to the sequences of other serovars, providing clear evidence for a high rate of whole-gene recombination. Recombination and the differential presence of IS-like elements among distinct disease and prevalence groups may contribute to genome plasticity, which may lead to adaptive changes in tissue tropism and pathogenesis over the course of the organism's evolution.
Chlamydia trachomatis is an obligate intracellular bacterium and a major cause of ocular and urogenital human infections worldwide (13). C. trachomatis has been divided into two human biological variants (biovars) on the basis of the nature of the disease that each group causes. To date, 18 serological variants (serovars) of these biovars have been identified by monoclonal antibody (MAb) typing of the major outer membrane protein (MOMP) of the organism. The oculogenital or trachoma biovar consists of serovars A to C and Ba that are responsible for trachoma, a chronic ocular inflammatory disease found in developing countries, and serovars Ba, D, Da, E, F, G, H, I, Ia, J, and K and genovariant Ja that are responsible for urogenital infections. The second biovar consists of four serovars (L1, L2, L2a, and L3) that are the causative agents of lymphogranuloma venereum (LGV). The LGV serovars are responsible for more-invasive diseases, including suppurative lymphadenitis, hemorrhagic proctitis, and ulcer formation (11).
While considerable knowledge has been gained about the pathogenesis of these diseases over the last few decades, there is still a lack of understanding about the association of genetic or protein differences among the various serovars of C. trachomatis and their tissue tropism and pathogenic properties. The major constituent of the outer membrane and the best-characterized protein of the organism is MOMP, which is encoded by the ompA gene. MOMP accounts for approximately 60% of the dry weight of the outer membrane of the infectious particle (termed the elementary body [EB]) and is the most antigenic protein of C. trachomatis. The available phylogenetic data for ompA as well as for omcB, the outer membrane complex B protein gene, do not entirely define or group the serovars according to their tissue tropism or virulence properties (32). Furthermore, the analyses to date of genes within the recently identified plasticity zone of the C. trachomatis D-UW-3 genome (GenBank no. AE001273), including those of the partial tryptophan operon (which differentiated ocular serovars from all others) and the partial cytotoxin gene, have also not been able to fully explain these properties (1, 5).
The genome sequence of D has also provided information about a novel group of genes that are predicted to encode surface proteins. These proteins are members of a large superfamily among the Chlamydia species and have no apparent homologs with other bacteria. C. trachomatis has nine members, which have been assigned the names polymorphic membrane proteins A to I (PmpA to PmpI); Chlamydophila pneumoniae (C. pneumoniae) has 21 members (23, 45); and Chlamydophila psittaci (C. psittaci) (strain GPIC) has 17 members (39). C. trachomatis pmp genes are located in two clusters in the chromosome (pmpA to pmpC and pmpE to pmpI), while pmpD is separate from the other pmp genes. Pmp proteins are large (90 to 187 kDa) and have considerable amino acid sequence heterogeneity. The presence of predicted cleavable signal peptide leader sequences for some of them (PmpC, PmpD, PmpE, and PmpI) suggests their location in the outer membrane. Further, a striking similarity between all Pmp proteins is the conserved motif GGAI present mostly in the amino-terminal half of the protein and a predicted C-terminal region forming a beta-barrel (20, 50). These characteristics suggest that Pmp proteins are autotransporters in which the C-terminal 30-kDa region is incorporated into the outer membrane, forming a pore and allowing the translocation of the N-terminal passenger domain to the bacterial surface (20, 50). In agreement with this is the recognition of the GGAI-containing proteins as major immunogens (7).
Although it is not known whether all pmp genes are translated into functional proteins, there is some evidence that the nine pmp genes are transcribed for serovars D and L2 (27). However, the majority of serovars have not yet been evaluated. According to other in vitro results, PmpD, PmpE, PmpG, and PmpH were detected as major constituents of the outer membrane complex of serovar L2 (33, 48; A. O. Kiselev, M. L. Johnson, L. B. Ballweber, W. E. Stamm, and M. F. Lampe, Abstr. Proc. Tenth Int. Symp. Human Chlamydial Infect., p. 567-570, 2002). Several authors have also reported the immunoreactivity of human sera with recombinant Pmp proteins expressed in vitro or Pmp-specific synthetic peptides (15, 27; R. C. Hsia, I. Ahmed, B. Batteiger, O. Sekkides, G. Ridgway, and P. M. Bavoil, Abstr. Fourth Meet. Eur. Soc. Chlamydia Res., p. 219, 2000).
The cumulative sequence, proteomics, and serologic evidence described above suggests that many of the Pmp proteins are surface exposed. While some data are accumulating on Pmp immunogenicity and evolution, both of which are important in understanding the biologic role of Pmp proteins in chlamydiae, PmpC has not been studied despite evidence for its membrane localization. Further, no Pmp studies to date have included comparative analyses of clinical isolates with reference strains, the latter of which represent laboratory-adapted strains that may not reflect the genetic makeup of chlamydial strains that are presently responsible for human disease. Consequently, we performed phylogenetic analyses and statistical modeling to examine the evolution of this gene and address potential differences in tissue tropism and virulence for 12 ocular and urogenital clinical strains and the 18 C. trachomatis reference serovars in addition to genovariant Ja. We also compared these data with the partial sequences of pmpE, pmpH, and pmpI available through GenBank for 15 reference serovars (47).
MATERIALS AND METHODS
Cell culture.
The following C. trachomatis prototype strains representing 18 serovars and one genovariant, Ja, were used in this study: A(HAR-13), B(TW-5), Ba(Apache-2), C(TW-3), D(UW-3), Da(TW-448), E(Bour), F(IC-Cal3), G(UW-57), H(UW-4), I(UW-12), Ia(UW-202), J(UW-36), Ja(UW-92), K(UW-31), L1(440), L2(434), L2a(TW-396), and L3(404). In addition, 12 trachoma and urogenital strains, representing ompA genotypes C, E, F, G, H, I, J, and Ja (see ompA genotyping data below) that had been recently isolated from the conjunctiva of trachoma patients or the genital tract of patients attending sexually transmitted disease clinics were also examined. Each strain was initially propagated in HeLa 229 cells as previously described (10, 11). Briefly, HeLa cells were plated in T-150 cm2 flasks and allowed to reach ∼60% confluence prior to inoculation with the respective serovar or clinical strain in SPG (0.25 M sucrose, 10 mM Na2HPO4, 5 mM l-glutamic acid, 10 μg of gentamicin/ml, 100 μg of vancomycin/ml, 25 U of nystatin/ml, pH 7.4) at room temperature for 2 h on an orbital shaker. Cells were washed with Hanks' balanced salt solution (Gibco-Invitrogen Corporation, Carlsbad, Calif.) and incubated in culture medium (minimal essential medium [Gibco-Invitrogen Corporation] containing 0.5 μg of cycloheximide/ml and 10% fetal bovine serum [HyClone, Logan, Utah]) at 37°C in 5% CO2. EBs for each serovar were harvested at 48 to 72 h postinoculation and purified by discontinuous density centrifugation in Renografin (4) and resuspended in Tris-EDTA (10 mM Tris-Cl, 1 mM EDTA, pH 8.0). Only the initial isolates (without further propagation) for each clinical strain were used in the experiments described as follows.
Serovar and genotype confirmation by ompA sequencing.
The identities of the 18 C. trachomatis reference strains, genovariant Ja, and the 12 clinical strains were confirmed by ompA sequencing before subsequent assays were performed. DNA from each strain was extracted using a High Pure PCR template preparation kit (Roche Diagnostics, Indianapolis, Calif.) according to the manufacturer's instructions. Each DNA sample was subjected to PCR and nested PCR using primers Nlo and Nro and primers Pctm3 and Sero2A (1,014-bp product) (Table 1), respectively, as previously described (3). Amplified products were visualized in ethidium bromide-stained 1% agarose gels, and those of the correct size were purified using a QIAquick PCR purification kit (QIAGEN, Valencia, Calif.). Primer ompA-3 (Table 1) was used for sequencing variable segments 1, 2, and 3 of the ompA gene, and variable segment 4 was sequenced using primer ompA-4 (Table 1), BigDye terminator chemistry, and capillary sequencing (3700 sequencer; Applied Biosystems, Foster City, Calif.). Sequences were aligned using LaserGene99 software (DNASTAR, Madison, Wis.) and presently available chlamydial sequences from GenBank to confirm the identity of each serovar and clinical strain.
TABLE 1.
Oligonucleotide primers used for PCR and sequencing
| Gene | Primer | Primer sequence (5′ to 3′) | Gene location | Amplicon Size (bp) |
|---|---|---|---|---|
| ompA | Nlo | ATGAAAAAACTCTTGAAATCG | 1 to 21a | 1,129 |
| Nro | CTCAACTGTAACTGCGTATTT | 1128 to 1108a | ||
| ompA | Pctm3 | TCCTTGCAAGCTCTGCCTGTGGGGAATCCT | 55 to 84a | 1,014 |
| Sero2A | TTTCTAGATTTCATTTTGTT | 1068 to 1049a | ||
| ompA | OmpA-3b | CCTGCTGAACCAAGCCTTATG | 82 to 102a | |
| ompA | OmpA-4b | CCAATATGCTCAATCTAAACC | 639 to 659a | |
| pmpC | C-9b | GCTGGTCAAGTTATCTGCGGAGTG | −344 to −321d | 3,015 |
| C-10b | TATTCCCGGAGAAGGTGACAGTTC | 2671 to 2648c | ||
| pmpC | C-11b | TGGAGATAGCGCTGGAGACTCTGA | 2394 to 2417c | 3,115 |
| C-12b | GTTAACGCGTACCGAGGGTTCG | |||
| pmpC | C-7b | AGACAACACAGAGTATCGAG | 153 to 172c | |
| pmpC | C-5b | ATAACTACTCCCCCTCTCATAGGA | 1156 to 1179c | |
| pmpC | C-4b | GGGGGTAGAAGATTCTGGGGTATC | 1362 to 1339c | |
| pmpC | C-8b | CTTATCCCACTATAACTCTG | 1713 to 1732c | |
| pmpC | C-15b | ACTCTCCTACTGTAACCATTG | 2720 to 2740c | |
| pmpC | C-17b | AACAGGTACACAGGCAACTG | 3450 to 3469c | |
| pmpC | C-16b | TGACGGATACTGGAGTATTC | 4940 to 4921c | |
| pmpH | H-1b | TCGGGCATTCAGGAGTGGACAT | −367 to −346e | 776 |
| H-2 | CTTCGCCTGCTCCGGAAATACTC | 408 to 386e | ||
| pmpI | I-1 | GAGGCGATTCTTTGCTGCTACTT | 1095 to 1117f | 1,369 |
| I-2b | AACAAACCAACCCAAAACTAAAAT | −273 to −250f |
Primers designed based on the sequence of strain L2 ompA.
Primers used for automated sequencing.
Primers designed based on the sequence of strain D pmpC.
−, a region upstream from the start codon.
Primers designed based on the sequence of strain D pmpH.
Primers designed based on the sequence of strain D pmpI.
pmpC sequencing for C. trachomatis reference and clinical isolates.
Two overlapping amplicons containing the entire pmpC gene (5,313 bp) were generated by PCR using primer pair C9 and C10 and primer pair C11 and C12 (Table 1). The primers were designed using Primer Select software (DNASTAR) on the basis of the pmpC sequence of serovar D/UW-3 (GenBank accession no. AE001315). The PCR reagents and volume were as follows: 1× buffer (Bioline, Randolph, Mass.), 200 μM deoxynucleoside triphosphates (Promega, Madison, Wis.), 2.8 mM MgCl2, 12.5 pmol of each primer, and 1 U of proof-reading Bio-X-Act DNA polymerase (Bioline) in a 25-μl reaction volume. The thermocycling profile consisted of 2 min at 94°C followed by 10 cycles of 10 s at 94°C, 30 s at 65°C, and 3 min 30 s at 68°C and 15 cycles of 10 s at 94°C, 30 s at 65°C, and 3 min 30 s at 68°C (including a step elongation of 10 s for each extension step during the last 15 cycles). Amplified products were visualized in an ethidium bromide-stained 0.7% agarose gel. Sequencing was performed using pmpC PCR and sequencing primers as described above. All pmpC sequences were confirmed by resequencing newly extracted DNA from the original EB stock for the respective serovar and clinical strain. When a discrepancy was found, the same gene segment was sequenced a third time.
N-terminus sequencing of pmpH and pmpI for C. trachomatis reference serovars.
Approximately 75 bp at the N terminus of pmpH and pmpI for 15 C. trachomatis reference serovars were not included in the published sequences available in GenBank. To evaluate the number of conserved cysteine residues for each gene compared with the results seen with pmpC, we sequenced these regions for the 15 reference serovars. PCR and sequencing primer data for pmpH and pmpI are provided in the Table 1. The primers were designed using Primer Select software (DNASTAR) on the basis of serovar D/UW-3 pmpH and pmpI sequences (GenBank accession no. AE001360 and AE001361, respectively). PCR reagents, volumes, and sequencing were as described for pmpC. The thermocycling profile for pmpH consisted of 5 min at 95°C and 1 min at 60°C, 30 cycles of 1 min at 70°C, 30 s at 95°C, and 30 s at 60°C, and a final elongation of 10 min at 70°C. For pmpI the thermocycling profile was the same except for an extension step of 1 min 30 s.
Molecular, phylogenetic, and statistical analyses of pmp sequences.
pmpC sequences for the 18 reference serovars, genovariant Ja, and the 12 patient strains were aligned using LaserGene99 software (DNASTAR). The signal peptide and cleavage sites in the amino acid sequences were identified using the World Wide Web Prediction Server, Center for Biological Sequence Analysis (SignalIP, version 1.1 [http://www.cbs.dtu.dk]), and the Neural Networks method (36).
A search for a conserved GGAI motif in the amino acid sequences of the four Pmp proteins was performed using EditSeq (DNASTAR). This motif is a common feature of Pmp proteins in chlamydiae (16) and may contribute to Pmp immunogenicity (7).
Reconstruction of the evolutionary history of pmpC was accomplished using neighbor-joining tree topologies generated by molecular evolutionary genetics analysis (MEGA, version 2.1; Institute of Molecular Evolutionary Genetics, Pennsylvania State University [http://www.megasoftware.net]) on the basis of distances estimated using a Kimura two-parameter model (25) for substitution events as previously described (32). Data regarding the pmpC phylogenetic trees obtained by this method were compared with the available data in the literature for other C. trachomatis genes (14, 32, 47).
The evolutionary distances between all serovar sequences were also evaluated using the Nei-Gojobori method (34) to evaluate the overall means of synonymous (dS) and nonsynonymous (dN) substitutions among the strains. A pairwise (p)-distance model was used to normalize these values to the number of potential dS and dN sites, because the number of potential synonymous sites is much smaller than the number of potential nonsynonymous sites. This model for estimating genetic distances can only be computed for protein-coding sequences and is performed on the basis of a codon-by-codon comparison. Significant differences in mean dS and dN substitutions were determined by comparing 95% confidence intervals (CI).
Computation of p distances was performed on the basis of the number of nucleotide and amino acid differences (35) between and within disease groups and within the 18 reference serovars and genovariant Ja; similar analyses were performed for the 12 clinical strains.
For all statistical analyses, the complete-deletion option was used to normalize the number of differences on the basis of the number of valid sites compared (when the sequences contained alignment gaps). Furthermore, bootstrap confidence levels were determined by randomly resampling the sequencing data 1,000 times.
Mean genetic distances between the sequences and the standard error (SE) were determined; the distance was computed between the sequences of the two serovars under comparison for all dissimilar serovar-serovar sequence pairs. The 95% CI was calculated from the SE of each mean value (mean ± 1.96 × SE).
Because the percentage of G+C content can provide information on the likelihood of genetic transfer from other species within the same genus or from other organisms (22, 38), we calculated the percentage of G+C content for the reference serovars for the four pmp genes and compared it with the complete D genome results. The percentage of G+C content for each pmp for all sequences was determined using EditSeq software (DNASTAR).
The same molecular, phylogenetic, and statistical analyses were also performed for the partial sequences of pmpE, pmpH, and pmpI (47) for 15 of the reference serovars. The results were compared with those obtained for pmpC with the same serovars.
Database submission.
The sequences of pmpC determined in this study have been submitted to GenBank under accession numbers AF519747 through AF519765. The sequences of pmpH and pmpI corresponding to the N-terminal region of the proteins have been submitted to GenBank under accession numbers AY357238 through AY357252 and AY357223 through AY357237, respectively.
RESULTS
Sequence analysis of pmp genes.
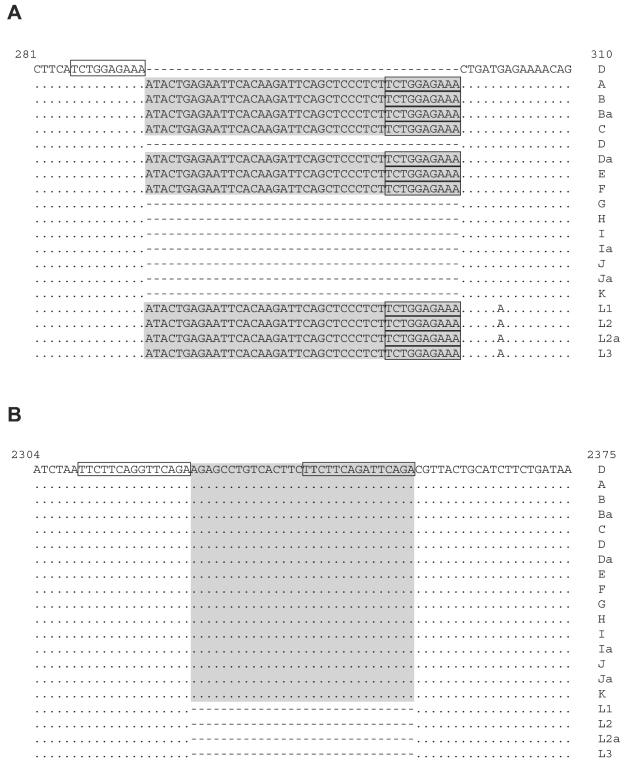
pmpC sequences from prototype serovars of C. trachomatis were aligned with the serovar D/UW-3 pmpC sequences from GenBank (no. AE001315). This gene revealed various sizes of 5,313 to 5,355 bp (Fig. 1). Trachoma and LGV serovars as well as serovars Da, E, and F contained an insertion (while the other serovars had a deletion) of 42 bp at position nucleotide (nt) 295 corresponding to a 14-amino-acid sequence. The insertion included a short, direct-repeat sequence of 10 bp (286TCTGGAGAAA295) in its C terminus also present in the region preceding the insertion (Fig. 1). Furthermore, another insertion of 30 bp corresponding to a 10-amino-acid sequence was located at nt position 2324 (Fig. 1) for all non-LGV serovars. Similarly, this insertion included a direct repeat (DR) of the DNA of 15 bp in its C terminus (2310TTCTTCAG[G/A]TTCAGA2324) which is imperfect compared with the same sequence in the DNA region preceding the insertion. We think that these insertions might reflect insertion sequence (IS)-like elements. If the DRs were created by an IS-like mechanism, then the DRs are caused by duplication of the target sequence and we therefore refer to the sequences as direct target repeats (DTRs).
FIG. 1.
Nucleotide sequences of pmpC domains containing the IS-like elements for all C. trachomatis serovars. (A) First IS element (42-nt insertion); (B) second IS element (30-nt insertion). The sequences were aligned with the GenBank sequence for strain D/UW-3 (accession number AE001315). Numbers at the top of the alignments represent the nucleotide positions. The shaded area represents IS elements. Boxes represent DTR sequences. Dots represent sequence homology; dashes represent the absence of nucleotides. These sequence data are available from GenBank under accession numbers AF519747 through AF519765.
Because of the findings described above, we calculated the probability of observing the two pairs of DRs in the same pmp gene as follows. Nucleotide frequencies were calculated on the basis of a G+C content of 41%. The larger interval between repeats was 32 bps, so the “search space” is an area of 32 bps times the length of pmpC (5.3 kb). The probability of observing an exact match to the first 10-bp sequence that accounts for its G+C content is 1.16 × 10−6. The probability of the 14 bases of the second sequence matching is 6.13 × 10−9; we then multiply this by a factor of 15 to account for the fact that the mismatch could have been anywhere in the sequence (but we conservatively do not give credit for the fact that the mismatched bases are both purines). Thus, the expected number of pairs is the product of the two probabilities named above times the size of the search space squared times the number of pmp genes (nine). Consequently, the probability of finding two DRs (of identical size, G+C content, and distance characteristics) for the pmps was calculated at P = 0.028.
DTRs or possible IS elements were not identified in any other portions of the chromosome for any of the reference serovars and clinical strains. A BLAST search of these sequences against all known bacterial genera identified no significant homology with any other sequences.
The PmpC sequences revealed the presence of a predicted peptidase II cleavage site and also a C-terminal phenylalanine for the reference serovars and clinical strains (data not shown). The search for the tetrapeptide GGAI motif revealed six GGAI motifs for serovars L2, L2a, and L3 and seven for serovars A to L1. Five of them were located in the N-terminal half of the protein (amino acid positions 172 to 175, 447 to 450, 541 to 544, 596 to 599, and 839 to 842), and the other two were very close to this region (917 to 920 and 955 to 958 [data not shown]). We also identified eight motifs for PmpE, two for PmpH (except for LGV group which presents 3 motifs), and six for PmpI. As for PmpC, most of these motifs were located in the N-terminal half of the protein. This is consistent with a previous report in which 2 to 10 motifs were identified among all C. trachomatis pmp genes (for serovar D) (J. Grimwood, M. Wayne, and R. S. Stephens, Abstr. Proc. Ninth Int. Symp. Human Chlamydial Infect., p. 263-266, 1998). Consequently, the presence of the predicted peptidase II cleavage site, the C-terminal phenylalanine, and the GGAI autotransporter motif is consistent with surface localization of PmpC.
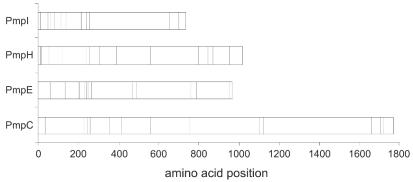
Since cysteine residues may play an important role in the three-dimensional structure of the Pmp proteins, the locations of conserved cysteine residues for the 15 reference serovars were identified and aligned for PmpC, PmpE, PmpH, and PmpI. Figure 2 shows the distribution of these cysteine residues for the four Pmp proteins. The residues were mainly clustered in the N-terminal half of the protein for each Pmp protein. For PmpC, there were a total of 15 cysteines, 13 of which were conserved. One nonconserved cysteine was located at amino acid position 432 and was present only in reference serovars Da, E, and F immediately upstream of one of the GGAI motifs; the other nonconserved cysteine was located at position 1764 in all serovars except for B. For PmpE, there were a total of 16 cysteines, 15 of which were conserved. The nonconserved cysteine was found in reference serovars A, B, Ba, and C at position 309 and was also immediately upstream of one of the GGAI motifs. For PmpH, there were a total of 15 cysteines, and 14 were conserved. The nonconserved cysteine was restricted to the LGV group and located at position 754. Finally, for PmpI, there are a total of 19 cysteines, with one nonconserved cysteine present in the LGV group at position 859.
FIG. 2.
The distribution of conserved cysteine residues for the 15 C. trachomatis reference serovars within each Pmp is denoted by relative amino acid positions indicated as vertical bars. The numbers of conserved cysteines for PmpC, PmpE, PmpH, and PmpI were 13, 15, 14, and 18, respectively.
In general, the distribution of the GGAI motifs along the Pmp proteins seems to be nonrandom, as they occur predominantly in cysteine-rich regions. This is particularly evident for PmpH and PmpI. Two of the three GGAI motifs in PmpH (amino acids 301 to 304 and 384 to 387) were adjacent to cysteine residues (amino acids 305 and 388). For PmpI, four of the six GGAI motifs (amino acids 147 to 150, 203 to 206, 230 to 233, and 266 to 269) were also next to or very close to cysteine residues (amino acids 139, 207, 208, 213, 234, 235, and 251). Considering that PmpI has six GGAI motifs and 16 of 18 cysteines clustered in the N-terminal 270 amino acids (out of a total of 878 amino acids), the distribution is nonrandom at a significance level of P < 0.001.
Percent G+C content.
The mean percentages of G+C content among the 15 reference serovars for pmpC, pmpE, pmpH, and pmpI were 41.2, 41.2, 43.2, and 43.6%, respectively. These values are close to the percentage of G+C content observed for the total genome of serovar D (41.3%) (46). Differences in percentages of G+C content observed for a single serovar compared across pmp genes were higher than those observed across serovars for a single pmp gene. Serovars L1 and L2 had the highest percentages of G+C content among pmp genes, with a 2.7% difference between pmpE and pmpI (P = 0.02). For each pmp gene, minor differences in percentages of G+C content were observed between serovars (<0.5%). The DTR and ISs did not significantly differ in percentages of G+C content compared with the rest of the respective gene or, in the case of serovar D, with the rest of the genome.
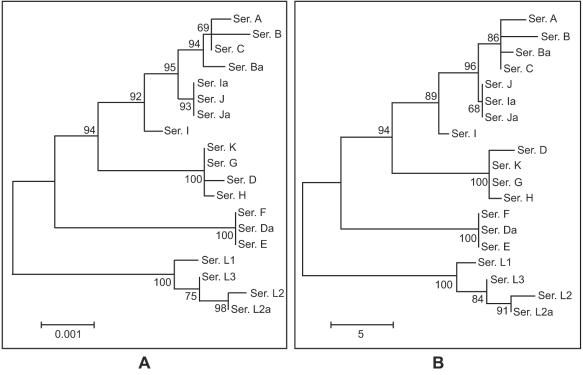
Phylogenetic analysis of pmp genes.
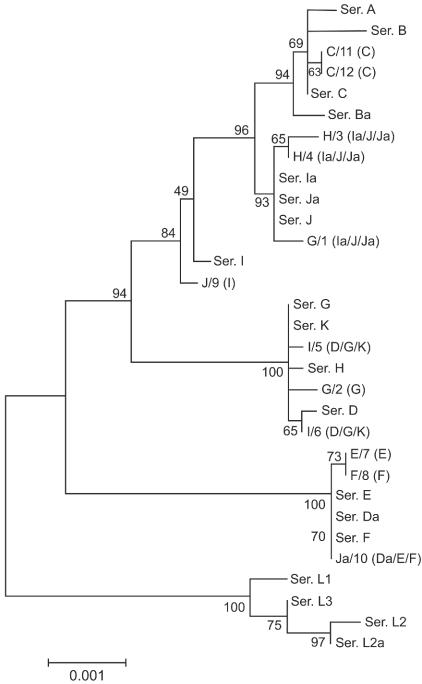
Phylogenetic reconstruction of the nucleotide sequences for pmpC is shown in Fig. 3A (branch lengths are proportional to distances between serovars in the figure). Evolutionary analysis of the inferred amino acid sequences yielded a similar tree for PmpC (Fig. 3B). These reconstructions were similar to that observed for Pmp H (47) but not to that observed for PmpI, with which there was an overall lesser degree of divergence. The pmpC tree revealed a separate clade that clearly distinguishes LGV serovars from all others, as was the case for pmpH. Also, ocular serovars were clustered together for pmpC, although this is more evident for pmpE and pmpH (47).
FIG. 3.
Phylogenetic reconstruction of nucleotide (A) and amino acid (B) sequences, showing the evolutionary history of pmpC by neighbor-joining tree topologies as determined on the basis of distance estimates made using a Kimura two-parameter model for substitution events. These reconstructions were made on the basis of pmpC sequences of the 18 serovars and the genovariant Ja of C. trachomatis. Branch lengths are proportional to distances between serovars. The values at the nodes are the bootstrap confidence levels, representing the percentages of 1,000 bootstrap resamplings for which the strains to the right were separated from the others.
We also found that serovars Da, E, and F evolved separately from the other urogenital serovars, as evidenced by a distant phylogenetic branch from the urogenital group. A similar branch was observed for pmpI (47). This is interesting, because serovars E, F, and D/Da are the most prevalent serovars among sexually transmitted disease populations worldwide. A detailed analysis of the p distances within (Table 2) and between (Table 3) disease groups performed on the basis of the number of nucleotide and amino acid differences (35) supports these findings. An example that illustrates this for pmpC was seen when serovars were divided into ocular, urogenital, and LGV groups. The average nucleotide difference within the urogenital group was 20.7 (SE, 3.2) and the average amino acid difference was 15 (SE, 2.7). When we separate serovars Da, E, and F from the urogenital group and reevaluate the p distances, the mean nucleotide and amino acid differences within the urogenital group were 12.2 (SE, 2.7) and 9.2 (SE, 2.2). Considering the differences between the group averages after the separation of Da, E, and F, these serovars were almost equally distant from the ocular serovars (34.8 [SE, 5.9] nucleotide and 24.5 [SE, 4.7] amino acid differences) as from the other urogenital serovars (33.3 [SE, 5.6] and 23.6 [SE, 4.5], respectively).
TABLE 2.
p distances within disease groups determined based on the number of nucleotide and amino acid differences
| Disease group and compound type | No. (SE; % change) of nucleotide or amino acid changes
|
|||
|---|---|---|---|---|
| pmpC | pmpE | pmpH | pmpI | |
| Ocular | ||||
| Nucleotide | 4.5 (1.5; <0.1) | 4.3 (1.5; 0.2) | 2.0 (0.9; <0.1) | 5.7 (1.9; 0.2) |
| Amino acid | 3.0 (1.2; 0.2) | 2.7 (1.1; 0.3) | 0.5 (0.5; <0.1) | 1.0 (0.7; 0.1) |
| Urogenital | ||||
| Nucleotide | 20.7 (3.2; 0.4) | 33.2 (4.1; 1.2) | 6.7 (1.6; 0.2) | 8.3 (1.9; 0.3) |
| Amino acid | 15 (2.7; 0.8) | 9.9 (2.0; 1.1) | 2.1 (0.9; 0.2) | 4.1 (1.2; 0.5) |
| LGV | ||||
| Nucleotide | 5.3 (1.6; 0.1) | 6.0 (1.9; 0.2) | 2.7 (1.3; <0.1) | 0.7 (0.7; <0.1) |
| Amino acid | 4.2 (1.5; 0.2) | 2.0 (1.1; 0.2) | 1.3 (0.9; 0.1) | 0 (0; 0) |
TABLE 3.
Genetic distances among ocular, urogenital, and LGV disease groups for each pmp gene
| Disease group and compound type | No. (SE; % change) of nucleotide or amino acid changes
|
|||||
|---|---|---|---|---|---|---|
| Ocular | Urogenital | Ocular | Urogenital | |||
|
pmpC
|
pmpE
|
|||||
| Urogenital | ||||||
| Nucleotide | 21.5 (3.6; 0.4) | 119 (9.1; 4.3) | ||||
| Amino acid | 15.3 (2.7; 0.9) | 43.3 (5.7; 4.7) | ||||
| LGV | ||||||
| Nucleotide | 41.8 (6.5; 0.8) | 40.3 (5.2; 0.8) | 115.9 (9.4; 4.2) | 52.5 (5.0; 1.5) | ||
| Amino acid | 32.8 (5.7; 1.9) | 30.7 (4.8; 1.8) | 45.8 (6.2; 5) | 15.8 (2.9; 1.7) | ||
|
pmpH
|
pmpI
|
|||||
| Urogenital | ||||||
| Nucleotide | 145 (11.0; 4.9) | 15.3 (3.2; 0.6) | ||||
| Amino acid | 37.3 (5.8; 3.8) | 8.6 (2.5; 1.0) | ||||
| LGV | ||||||
| Nucleotide | 187.3 (13.1; 6.4) | 168.2 (12.8; 5.7) | 21.8 (4.5; 0.8) | 22.5 (4.2; 0.9) | ||
| Amino acid | 59.6 (7.1; 6.1) | 42.3 (5.6; 4.3) | 7.5 (2.5; 0.9) | 7.5 (2.3; 0.9) | ||
The ratios of mean nonsynonymous to mean synonymous substitutions (dN/dS) within the 15 reference serovars calculated by the Nei-Gojobori method (34) for pmpC, pmpE, pmpH, and pmpI were 0.94 (SE, 0.18), 0.24 (SE, 0.04), 0.15 (SE, 0.02), and 0.25 (SE, 0.08), respectively. The differences between the pmpC gene and the pmpE, pmpH, and pmpI genes were significant for a 95% CI (Fig. 4).
FIG. 4.
Upper and middle graphs show mean synonymous mutation rates and mean nonsynonymous mutation rates, respectively, for each pmp determined by the method of Nei and Gojobori (p-distance model). Minimum and maximum values represent lower and upper limits of the 95% CI of the estimate, while values plotted at the horizontal bar level represent the mean estimates. The lower graph shows the nonsynonymous- to synonymous-mutation ratio for each pmp determined on the basis of the mean estimates shown in the prior two graphs.
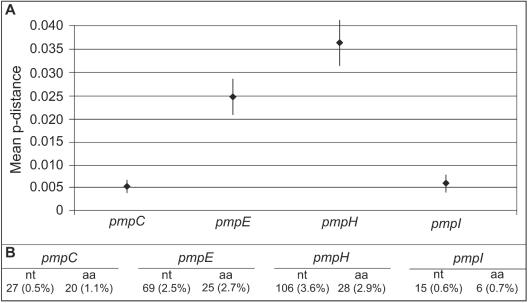
The mean genetic distance among the 15 reference serovars for the respective pmp is shown in Fig. 5. pmpH was the most variable among the serovars (106 [3.6%] nt changes and 28 [2.9%] amino acid changes). Considering the genetic distances among the ocular, urogenital, and LGV disease groups (Table 3), LGV serovars were in general more distant from the other serovars except for pmpE, from which the trachoma serovars exhibited the greatest degree of distance. The evaluation of the nucleotide and amino acid dissimilarities for each pmp gene within disease groups revealed that the LGV serovars were the most closely related (Table 2). For pmpI, in fact, serovars L1 to L3 showed 100% homology in their protein sequences. As expected, the urogenital group was the most heterogeneous, as it encompasses 11 serovars. The nucleotide differences within this group ranged from 6.7 (0.2%) nt (SE, 1.6; 95% CI, 0.13 to 0.33) for pmpH to 33.2 (1.2%) nt (SE, 4.1; 95% CI, 0.90 to 1.48) for pmpE, and the amino acid differences ranged from 2.1 (0.2%) amino acids (SE, 0.9; 95% CI, 0.014 to 0.406) for pmpH to 9.9 (1.1%) amino acids (SE, 2.0; 95% CI, 0.64 to 1.50) for pmpE.
FIG. 5.
Mean p distance within each pmp determined on the basis of the average p distance for all possible pairs of sequences (different serovars) for the same pmp. (A) Minimum and maximum values represent lower and upper limits of the 95% CI of the estimate, while values plotted at horizontal bar level represent the mean estimates. (B) Absolute and percent values for the nucleotide and amino acid mean genetic distance for each pmp. aa, amino acid.
Analysis of pmpC sequence for clinical isolates.
To determine whether there were significant sequence changes among clinical isolates compared with the laboratory-adapted reference serovars for the same serovar, we sequenced pmpC for 12 clinical isolates representing trachoma and urogenital ompA genotypes C, E, F, G, H, I, J, and Ja. A detailed analysis of nucleotide and amino acid differences among the pmpC sequences for these genotypes is shown in Table 4. Surprisingly, seven (58.3%) of these 12 clinical strains revealed a pmpC sequence that did not correspond with the pmpC sequence that was expected given the ompA genotype. The sequence of Ja/10 was one striking example; there were 32 nucleotide differences (corresponding to 23 amino acids) compared with the reference Ja genovariant pmpC sequence. Yet there were no nucleotide or amino acid differences seen when the pmpC sequence of Ja/10 was compared with the pmpC sequences of serovars Da, E, and F; serovars Da, E, and F have identical pmpC sequences. Hence, the Ja/10 strain was labeled Ja/10(Da/E/F). Similarly, H/3(Ia/J/Ja) and H/4(Ia/J/Ja) (both H ompA genotypes) and G/1(Ia/J/Ja) (G ompA genotype) had pmpC sequences almost identical to those of serovars Ia, J, and genovariant Ja whereas patient strains I/5(D/G/K) and I/6(D/G/K) (both I ompA genotypes) had pmpC sequences identical to those of reference serovars D, G, or K. Finally, the clinical isolate J/9(I) (J ompA genotype) had a pmpC sequence identical to that of serovar I. The sequences of the remaining five patient strains revealed few or no nucleotide or amino acid changes compared to the corresponding pmpC reference serovar sequence.
TABLE 4.
Evaluation of pmpC sequences for 12 trachoma and urogenital clinical isolates
| ompA genotype of clinical isolatesa | No. of pmpC sequence differences compared to the reference serovar pmpC sequence
|
Serovar or genovariant to which the pmpC sequence of the clinical sample is most similarb | |
|---|---|---|---|
| No. of nucleotide changes | No. of nonsynonymous amino acid changes | ||
| G/1 (Ia/J/Ja) | 22 | 16 | Ia (2/1); J (2/1); Ja (2/1) |
| G/2 (G) | 2 | 2 | G (2/2) |
| H/3 (Ia/J/Ja) | 24 | 18 | Ia (3/2); J (3/2); Ja (3/2) |
| H/4 (Ia/J/Ja) | 22 | 17 | Ia (1/1); J (1/1); Ja (1/1) |
| I/5 (D/G/K) | 16 | 12 | D (3/3); G (1/1); K (1/1) |
| I/6 (D/G/K) | 16 | 12 | D (1/1); G (1/1); K (1/1) |
| E/7 (E) | 1 | 0 | E (1/0) |
| F/8 (F) | 1 | 0 | F (1/0) |
| J/9 (I) | 7 | 5 | I (2/1) |
| Ja/10 (Da/E/F) | 32 | 23 | Da (0/0); E (0/0); F (0/0) |
| C/11 (C) | 1 | 0 | C (1/0) |
| C/12 (C) | 1 | 0 | C (1/0) |
The first letter of the genotype designation represents the ompA genotype, and the number after the slash represents the identification number; the letter(s) in parentheses represents the serovar(s) to whose sequence the pmpC sequence is most similar.
The initial letter in the serovar or genovariant designation represents the pmpC sequence to which the ompA clinical genotype was most similar; the numbers in parentheses denote the number of nucleotide differences from that pmpC sequence followed by the number of amino acid differences from that pmpC sequence compared to the reference serovar pmpC sequence.
The phylogenetic reconstruction of the nucleotide sequences showing the evolutionary history of pmpC for these strains as well as for the reference serovars is presented in Fig. 6. As expected according to the data presented above, these seven clinical isolates are shown in distant branches from the corresponding ompA prototype serovars and are clustered together with the serovars with pmpC sequences to which their pmpC sequences are similar.
FIG. 6.
Phylogenetic reconstruction of the nucleotide sequences, showing the evolutionary history of pmpC determined on the basis of pmpC sequences of the 18 serovars and the genovariant Ja of C. trachomatis and 12 clinical isolates representing ompA genotypes C, E, F, G, H, I, J, and Ja. Branch lengths are proportional to distances between serovars; bootstrap values are shown at the nodes. In the names of the clinical isolates, the first character represents the ompA genotype and the characters in parentheses represent the serovar(s) to which the pmpC sequence is similar.
DISCUSSION
pmpC revealed base-pair sizes that differed due to what we consider to be the presence of mobile genetic elements (MGEs) for some serovars. MGEs comprise transposons, which carry selectable traits such as drug resistance, and IS-like elements, which affect genetic structure and alter patterns of gene expression (6). While we cannot completely rule out the possible presence of two deletions that may have occurred in pmpC via homologous recombination, the probability of the two sets of DRs (with each set constituting a different sequence) arising by chance in the same gene is quite low (P = 0.028). Because of this low level of probability, an explanation based on selective advantage is warranted. One possibility is that these repeats are maintained by selection so as to allow nonreproductive mutants to present deleted PmpC to the host. Another is that there is a mechanism, perhaps operating via the IS mechanism, by which insertions are created but with the transposed gene partly excised or acting on DNA other than its own. The fact that pmp genes comprise a multiple-gene family across chlamydial genera suggests that this mechanism(s) may be used by the organism to generate diversity.
The existence of IS elements in chlamydiae has never been reported before and would indicate horizontal gene transfer in this intracellular pathogen. It is likely that novel IS elements have not previously been identified in C. trachomatis, because it requires the alignment of sequences from serovars with and without these elements to detect their presence and only the pmpC sequence for serovar D has been available. Further, comparative genomics using C. trachomatis to search for other IS-like elements has not been performed because only the genome sequence of D is available. Another method of detecting IS-like elements would be to search for short DRs, which could find potential DTRs but would not discriminate random repeats from those which, as in our case, are sufficiently active to lead to polymorphism. Analyses of the genome alignments of two C. pneumoniae strains, CWL029 and J138, detected the presence of 44- to 87-bp DRs (44). DRs reflect replicates of DNA segments that are common in nature, and they have been described as differing extremely in size (24). DRs are not associated with IS elements and have been well described for other pathogens, such as mycobacteria, for which there are numerous DRs throughout the genome (51). In contrast, a DTR is limited to 2 to 15 bp, which is characteristic of a given mobile element; this element will generally generate a duplication of fixed length (26, 29).
Additional comparative analyses of Chlamydophila caviae (C. psittaci strain GPIC), Chlamydophila muridarum (C. trachomatis strain MoPn), and C. pneumoniae strain AR39 genomes have also failed to reveal any IS elements (38, 39). This suggests that IS elements are deleterious to the genome, resulting in their elimination from present-day chlamydiae (which is unlikely given that MGEs are widespread among bacterial genera), that C. pneumoniae bacteria are more restricted than bacteria of other chlamydial species or genera in the ability to accept foreign DNA (38, 44), or that there are insufficient chlamydial genome sequences for these types of analyses.
A detailed analysis of the percentage of G+C content which might be helpful in identifying foreign genetic material (due to the specific differences in percentages of G+C content among other species of chlamydiae and other bacteria) was not conclusive in this study. This is not surprising given the base-pair length of the insertions. Both of the IS-like elements lacked terminal inverted repeat sequences (IR) which are also common features of IS elements (6, 29). Since the DTRs represent chromosomal sequences that become duplicated upon insertion of a sequence (29, 30), they provide evidence for an IS. Further, in some cases insertion of MGEs results in a perfect or imperfect terminal DTR without an IR (29, 30).
It is also possible that in similarity to some of the IS elements described for Escherichia coli (17, 31), these putative IS-like elements of C. trachomatis are regionally specific. Indeed, it has been suggested that partial IS elements reflect site specificity and transposition events, with subsequent partial loss of the IS element (29). We think that these short sequences in pmpC constitute the remaining fragment of complete IS elements (ISs range in size from 0.2 to 5.7 kb) that were excised during the evolutionary history of this gene. Possibly a nearly precise excision phenomenon took place, leaving the DTR and remnants of the IS at the site of the insertion (2, 6). If the IS-like elements occurred in a common ancestor before the divergence of the chlamydial strains into monophyletic groups, excision of the first IS for serovars D, G, H, I, Ia, J, Ja, and K and of the second IS for L1-3 and L2a could explain the present-day findings (Fig. 1). The excision mechanism has been well described and involves interactions between the DTR that are promoted by base pairing between the IRs at either end of the IS. This occurs during replication when the DNA is transiently single stranded and normally is not dependent on homologous recombination (6).
Lundblad et al. (28) have also described precise and nearly precise excision phenomena involving IS10 or Tn10 in E. coli K-12 where the pathway for excision depends on mutations in RecA and RecBC. The chlamydial genome contains homologues to these enzymes (i.e., RecA, RecBCD, and RecF) (21, 46, 56) which may function in a similar type of excision event. Millman et al. (32) and Hayes et al. (19) have previously provided significant evidence for intragenic and interspecies recombination for the ompA gene of Chlamydia that likely involves these enzymes. Further, C. trachomatis contains a histone-like protein known as an integration host factor which, in other bacteria, modulates transposition activity of IS elements by binding to sites within or near the terminal IR or DTRs (29). Thus, C. trachomatis has a number of potential mechanisms for facilitating acquisition and excision of IS elements.
In experiments in which selection of IS elements has been maintained for many bacterial generations, these elements have been found to have a major effect on the genetic structure of the population, resulting in rearrangements which alter patterns of gene expression (6). IS elements have been associated with several biological functions of bacteria. Salvatore et al. (42) described the role of IS in pathogenesis of Neisseria meningitidis strains by insertional inactivation of virulence genes encoding membrane proteins. Modulation of capsule expression by a unique IS-based genetic switch mechanism has also been described for N. meningitidis (18). Radnedge et al. (37) reported the importance of IS elements in relation to genes encoding proteins for flagellar synthesis, ABC transport, insect toxicity and bacteriophage functions in Yersinia pestis. Regarding the IS role in gene evolution, Woodford (54) suggested that van clusters (genetic elements responsible for acquired glycopeptide resistance) have been transferred to enterococci on multiple occasions by horizontal dissemination of different van elements which have provided valuable information on the evolution of enterococci.
It is not possible to determine the precise role of the IS elements in pmpC, since there are no data on the complete sequence of these mobile elements or their association with other genetic elements. These remnant IS elements might reflect negative fitness which has limited their spread among chlamydiae and resulted in their nearly precise excision. However, a possible consequence of the presence of IS elements in PmpC evolution is suggested by the higher dN/dS ratio where new amino acid sequences (such as an IS) are introduced and where variable positions are under diversifying selection, are under positive selection for mutations that increase function, or have few functional constraints. In contrast, it had previously been suggested that pmpI is not membrane exposed and thus is not under selective pressure (47), which could explain the high conservation of this sequence. In our study, the lower dN/dS ratio calculated for this protein as well as for PmpE and PmpH was in agreement with the presence of highly conserved proteins that could play important structural or functional roles for which it is essential to preserve their primary and three-dimensional protein structures. Consequently, the lower ratios for PmpE, PmpH, and PmpI are consistent with purifying selection in which deleterious amino acids are selected against.
The analysis of the ocular and urogenital clinical isolates in this study revealed surprising results in that seven (58%) of the isolates contained an entire pmpC gene that did not fit with the expected sequence. Instead, the sequence was from another serovar. Thus, it appears that novel genomes have arisen that contain different combinations of ompA and pmpC genes, suggesting that either ompA or pmpC is involved in the recombination events. This whole-gene or intergenomic recombination phenomenon appears to occur with a much higher frequency than the previously observed intragenic recombination for ompA. This phenomenon was restricted to the urogenital strains, although only two ocular strains were studied. While we do not know the exact site of recombination or whether it involves a single gene (pmpC or ompA) or a larger portion of the genome, chlamydiae do contain the necessary enzymes to enable such an event as discussed above (21, 46, 56). We also know that recombination among urogenital strains is feasible, because mixed infections do occur in vivo (9) and more than one serovar in vitro has been shown to infect a single cell (40). Further, there is some evidence that IS-like elements in bacteria such as E. coli can stimulate Rec-dependent recombination (6). If this is true for chlamydiae, this might explain the recombination noted for the urogenital strains. While it has been suspected that the laboratory-adapted reference strains do not reflect the biological pressure to which clinical strains have been subjected in vivo over time, this is the first evidence that there are indeed significant differences between the two.
Classification of C. trachomatis into serovars has been based on differential MAb recognition of antigenic determinants on MOMP (52, 53). In accordance with serologic and ompA differences, the serovars have been placed in the following serogroup classes: class B (B, Ba, D, Da, E, L1, L2, and L2a), class C (A, C, H, I, Ia, J, K, and L3), and intermediate class (F and G) (55). These classes do not correlate with their known tissue tropism. In general, the phylogenetic reconstructions for pmpC and pmpH indicate that serovar groupings are made on the basis of biologic properties related to tissue tropism and virulence as opposed to phylogenetic grouping by seroclass as for ompA (32). Other genes have been examined that might explain some of the observed tissue tropism for these serovars. Genetic polymorphisms have been noted for the ocular serovars in the tryptophan operon that likely lead to inactivation of the enzyme (5, 12, 43), and the cytotoxin gene appears to be active for serovar D but not L2 (1), although other serovars have not been tested. However, the Pmp proteins are the only proteins to date that have shown a clear distinction that corresponds with tissue specificity and virulence for disease groups.
The evaluation of the nucleotide and amino acid dissimilarities for each pmp within disease groups (Table 2) suggests potential evolutionary coadaptation of strain and tissue specificity. Serovars A, B, Ba, and C, the serovars responsible for ocular infections, were closely related in the phylogenetic analyses for pmpC; this relatedness was more evident for pmpE and pmpH (47). For PmpE, the ocular serovars had an additional nonconserved cysteine residue (amino acids position 309) which was absent from all the other serovars and which was located upstream of a GGAI motif (amino acids 310 to 313). Similarly for PmpH, a conserved cysteine residue (amino acids 305) was located adjacent to a GGAI motif (amino acids 301 to 304) only for the ocular serovars. When we examine the LGV disease group for all Pmp proteins, these serovars were the most closely related and more distant from the other serovars except for PmpE, for which the ocular group was most distant. Notably, both PmpH and PmpI had a nonconserved cysteine residue (amino acids 754 and 859, respectively) in the LGV group which was not present among the other serovars. The N-terminal region where the majority of GGAI motifs are located is also the location of the conserved cysteine residues for all of the reference serovars for the four Pmp proteins and for all of the clinical strains for PmpC. Furthermore, the GGAI motif is nonrandomly distributed. Considering the essential role of conserved cysteines in the three-dimensional structure of the protein and the importance of GGAI motifs in the immunogenic autotransporter proteins, it is likely that these two features are essential in conserving conformational epitopes. Consequently, given the differential presence and location characteristics of cysteine residues and GGAI motifs for the ocular serovars and positions of the cysteine residue for the LGV serovars it is possible that these features impart structural changes in protein conformation resulting in strain differences in adhesion to the host cell, entry into the host cell, or immune selection. Thus, the pmpC and pmpH phylogenetic trees seem congruent with clustering by tissue specificity, suggesting coevolution of the serovar and the tissue that it infects.
The phylogenetically distant branch of serovars Da, E, and F for pmpC and serovars E and F (serovar Da sequences are not available for PmpE, PmpH, and PmpI) for pmpI is surprising, although serovars E, F, and D/Da are the most prevalent serovars worldwide. Of note, the methods most commonly used for C. trachomatis serovar classification (restriction digestion of amplified products [restriction fragment length polymorphism] and serotyping) do not always distinguish between D and Da; for this reason, the real prevalence of serovar Da is not known. After serovars Da, E, and F and serovars E and F were separated from the urogenital group for pmpC and pmpI, respectively, and after the p distances were reevaluated, the distance of these serovars from the ocular disease group was found to be almost equal to the distance from the other genital serovars. The higher level of ecologic success of these prevalent serovars may be related to the host cell adhesion process. Chlamydiae are obligate intracellular bacterial pathogens for which the adherence of the EB, the infectious particle of the organism, to its target is essential for successful infection. Several authors have reported differences among serovars for adherence mechanisms on the basis of studies that used glycosaminoglycan; in these studies, host heparan sulfate appeared to be an important adhesin molecule for L2 but not for serovar E (8, 49).
The genetic specificities of serovars E and F (and probably Da), the most prevalent serovars within pmpC and pmpI, may also explain the ecologic success of these serovars. Da, E, and F were the only genital serovars with two IS elements in the pmpC sequence. Moreover, these three serovars contained an additional nonconserved cysteine at amino acid 432 (replacing a serine), not present in any other serovar, which was adjacent to a GGAI motif. We speculate that these two mobile elements and the additional cysteine residue in proximity to the GGAI motif have induced PmpC structural or functional constraints that are beneficial and that therefore might explain the ecological success of these strains.
The nine pmp genes of C. trachomatis represent 13.6% of the Chlamydia-specific coding capacity (41), which is particularly intriguing in a pathogen showing reductive convergent evolution by elimination of genes that are apparently unnecessary for obligate intracellular existence (57). While there is still a long way to go in terms of deciphering the exact role of Pmp proteins in the biology of C. trachomatis, the results of the present study suggest that pmp sequence polymorphisms, the differential presence of IS elements among different serovars representing distinct disease or prevalence groups, and evidence for recombination of an entire gene (pmpC or ompA) or a larger portion of the genome for over 50% of the clinical isolates contribute to plasticity of the chlamydial genome. This may lead to adaptive changes in tissue tropism and pathogenesis in the course of evolution. Moreover, the unexpected finding of genetic exchange among the urogenital clinical isolates points to the critical need for research that focuses on recent clinical isolates instead of on laboratory-adapted reference strains. These data also support the need for addition genome sequences of C. trachomatis strains to improve our understanding of the mechanisms, including horizontal gene transfer and recombination, driving the evolution and adaptation of this organism.
Acknowledgments
This work was supported by grants from Fundação Para a Ciência e Tecnologia (62%) and FEDER (38%) (POCTI/39822/MGI/2001) (to M.J.B.) and Comissão de Fomento da Investigação em Cuidados de Saúde (187/01) (to J.P.G.) and Public Health Service grants AI39499 and EY/AI12219 from the National Institutes of Health (to D.D.).
REFERENCES
- 1.Belland, R. J., M. A. Scidmore, D. D. Crane, D. M. Hogan, W. Whitmire, G. McClarty, and H. D. Caldwell. 2001. Chlamydia trachomatis cytotoxicity associated with complete and partial cytotoxin genes. Proc. Natl. Acad. Sci. USA 98:13984-13989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blot, M. 1994. Transposable elements and adaptation of host bacteria. Genetica 93:5-12. [DOI] [PubMed] [Google Scholar]
- 3.Borrego, M. J., J. P. Gomes, J. F. Lefebvre, F. Eb, J. Orfila, and M. A. Catry. 1997. Genotyping of Portuguese Chlamydia trachomatis urogenital isolates. Genitour. Med. 73:561-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caldwell, H. D., J. Kromhout, and J. Schachter. 1981. Purification and partial characterization of the major outer membrane protein of Chlamydia trachomatis. Infect. Immun. 31:1161-1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caldwell, H. D., H. Wood, D. Crane, R. Bailey, R. B. Jones, D. Mabey, I. Maclean, Z. Mohammed, R. Peeling, C. Roshick, J. Schachter, A. W. Solomon, W. E. Stamm, R. J. Suchland, L. Taylor, S. K. West, T. C. Quinn, R. J. Belland, and G. McClarty. 2003. Polymorphisms in Chlamydia trachomatis tryptophan synthase genes differentiate between genital and ocular isolates. J. Clin. Investig. 111:1757-1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chalmers, R., and M. Blot. 1999. Insertion sequences and transposons, p. 151-168. In R. L. Charlebois (ed.), Organization of the prokaryotic genome. American Society for Microbiology, Washington, D.C.
- 7.Christiansen, G., A. S. Pedersen, K. Hjerno, B. Vandahl, and S. Birkelund. 2000. Potential relevance of Chlamydia pneumoniae surface proteins to an effective vaccine. J. Infect. Dis. 181(Suppl. 3):S528-S537. [DOI] [PubMed] [Google Scholar]
- 8.Davis, C. H., and P. B. Wyrick. 1997. Differences in the association of Chlamydia trachomatis serovar E and serovar L2 with epithelial cells in vitro may reflect biological differences in vivo. Infect. Immun. 65:2914-2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dean, D., E. Oudens, G. Bolan, N. Padian, and J. Schachter. 1995. Major outer membrane protein variants of Chlamydia trachomatis are associated with severe upper genital tract infections and histopathology in San Francisco. J. Infect. Dis. 172:1013-1022. [DOI] [PubMed] [Google Scholar]
- 10.Dean, D., and V. C. Powers. 2001. Persistent Chlamydia trachomatis infections resist apoptotic stimuli. Infect. Immun. 69:2442-2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dean, D., R. Suchland, and W. Stamm. 2000. Evidence for long-term cervical persistence of Chlamydia trachomatis by omp1 genotyping. J. Infect. Dis. 182:909-916. [DOI] [PubMed] [Google Scholar]
- 12.Fehlner-Gardiner, C., C. Roshick, J. H. Carlson, S. Hughes, R. J. Belland, H. D. Caldwell, and G. McClarty. 2002. Molecular basis defining human Chlamydia trachomatis tissue tropism. A possible role for tryptophan synthase. J. Biol. Chem. 277:26893-26903. [DOI] [PubMed] [Google Scholar]
- 13.Fields, P. I., and R. C. Barnes. 1992. The genus Chlamydia, p. 3691-3709. In A. Balows, H. G. Truper, M. Dworkin, W. Harder, and K. H. Schleifer (ed.), The prokaryotes. Springer-Verlag, New York, N.Y.
- 14.Fitch, W. M., E. M. Peterson, and L. M. de la Maza. 1993. Phylogenetic analysis of the outer-membrane-protein genes of Chlamydiae, and its implication for vaccine development. Mol. Biol. Evol. 10:892-913. [DOI] [PubMed] [Google Scholar]
- 15.Goodall, J. C., G. Yeo, M. Huang, R. Raggiaschi, and J. S. Gaston. 2001. Identification of Chlamydia trachomatis antigens recognized by human CD4+ T lymphocytes by screening an expression library. Eur. J. Immunol. 31:1513-1522. [DOI] [PubMed] [Google Scholar]
- 16.Grimwood, J., L. Olinger, and R. S. Stephens. 2001. Expression of Chlamydia pneumoniae polymorphic membrane protein family genes. Infect. Immun. 69:2383-2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hallet, B., R. Rezsohazy, J. Mahillon, and J. Delcour. 1994. IS231A insertion specificity: consensus sequence and DNA bending at the target site. Mol. Microbiol. 14:131-139. [DOI] [PubMed] [Google Scholar]
- 18.Hammerschmidt, S., R. Hilse, J. P. van Putten, R. Gerardy-Schahn, A. Unkmeir, and M. Frosch. 1996. Modulation of cell surface sialic acid expression in Neisseria meningitidis via a transposable genetic element. EMBO J. 15:192-198. [PMC free article] [PubMed] [Google Scholar]
- 19.Hayes, L. J., P. Yearsley, J. D. Treharne, R. A. Ballard, G. H. Fehler, and M. E. Ward. 1994. Evidence for naturally occurring recombination in the gene encoding the major outer membrane protein of lymphogranuloma venereum isolates of Chlamydia trachomatis. Infect. Immun. 62:5659-5663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henderson, I. R., and A. C. Lam. 2001. Polymorphic proteins of Chlamydia spp.—autotransporters beyond the Proteobacteria. Trends Microbiol. 9:573-578. [DOI] [PubMed] [Google Scholar]
- 21.Hsia, R. C., and P. M. Bavoil. 1996. Homologs of Escherichia coli recJ, gltX and of a putative “early” gene of avian Chlamydia psittaci are located upstream of the “late” omp2 locus of Chlamydia psittaci strain guinea pig inclusion conjunctivitis. Gene 176:163-169. [DOI] [PubMed] [Google Scholar]
- 22.Hueck, C. J. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62:379-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kalman, S., W. Mitchell, R. Marathe, C. Lammel, J. Fan, R. W. Hyman, L. Olinger, J. Grimwood, R. W. Davis, and R. S. Stephens. 1999. Comparative genomes of Chlamydia pneumoniae and C. trachomatis. Nat. Genet. 21:385-389. [DOI] [PubMed] [Google Scholar]
- 24.Kawano, M., T. Oshima, H. Kasai, and H. Mori. 2002. Molecular characterization of long DR (LDR) sequences expressing a stable mRNA encoding for a 35-amino-acid cell-killing peptide and a cis-encoded small antisense RNA in Escherichia coli. Mol. Microbiol. 45:333-349. [DOI] [PubMed] [Google Scholar]
- 25.Kimura, M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111-120. [DOI] [PubMed] [Google Scholar]
- 26.Kleckner, N. 1981. Transposable elements in prokaryotes. Annu. Rev. Genet. 15:341-404. [DOI] [PubMed] [Google Scholar]
- 27.Lindquist, E. A., and R. S. Stephens. 1998. Transcriptional activity of a sequence variable protein family in Chlamydia trachomatis, p. 259-262. In R. S. Stephens, G. I. Byrne, G. Christiansen, I. N. Clarke, J. T. Grayston, R. G. Rank, G. L. Ridgway, P. Saikku, J. Schachter, and W. E. Stamm (ed.), Chlamydial Infections. Proceedings of the Ninth International Symposium on Human Chlamydial Infection. International Chlamydia Symposium, Napa, Calif.
- 28.Lundblad, V., A. F. Taylor, G. R. Smith, and N. Kleckner. 1984. Unusual alleles of recB and recC stimulate excision of inverted repeat transposons Tn10 and Tn5. Proc. Natl. Acad. Sci. USA 81:824-828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mahillon, J., and M. Chandler. 1998. Insertion sequences. Microbiol. Mol. Biol. Rev. 62:725-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Melcher, U. 1999. Molecular genetics: overview, p. 232-236. In R. McClenaghan (ed.), Encyclopedia of genetics. Salem Press, Pasadena, Calif.
- 31.Mendiola, M. V., and F. de la Cruz. 1989. Specificity of insertion of IS91, an insertion sequence present in alpha-haemolysin plasmids of Escherichia coli. Mol. Microbiol. 3:979-984. [DOI] [PubMed] [Google Scholar]
- 32.Millman, K. L., S. Tavaré, and D. Dean. 2001. Recombination in the ompA gene but not the omcB gene of Chlamydia contributes to serovar-specific differences in tissue tropism, immune surveillance, and persistence of the organism. J. Bacteriol. 183:5997-6008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mygind, P. H., G. Christiansen, P. Roepstorff, and S. Birkelund. 2000. Membrane proteins PmpG and PmpH are major constituents of Chlamydia trachomatis L2 outer membrane complex. FEMS Microbiol. Lett. 186:163-169. [DOI] [PubMed] [Google Scholar]
- 34.Nei, M., and T. Gojobori. 1986. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3:418-426. [DOI] [PubMed] [Google Scholar]
- 35.Nei, M., and S. Kumar. 2000. Molecular evolution and phylogenetics. Oxford University Press, New York, N.Y.
- 36.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 37.Radnedge, L., P. G. Agron, P. L. Worsham, and G. L. Andersen. 2002. Genome plasticity in Yersinia pestis. Microbiology 148:1687-1698. [DOI] [PubMed] [Google Scholar]
- 38.Read, T. D., R. C. Brunham, C. Shen, S. R. Gill, J. F. Heidelberg, O. White, E. K. Hickey, J. Peterson, T. Utterback, K. Berry, S. Bass, K. Linher, J. Weidman, H. Khouri, B. Craven, C. Bowman, R. Dodson, M. Gwinn, W. Nelson, R. DeBoy, J. Kolonay, G. McClarty, S. L. Salzberg, J. Eisen, and C. M. Fraser. 2000. Genome sequences of Chlamydia trachomatis MoPn and Chlamydia pneumoniae AR39. Nucleic Acids Res. 28:1397-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Read, T. D., G. S. Myers, R. C. Brunham, W. C. Nelson, I. T. Paulsen, J. Heidelberg, E. Holtzapple, H. Khouri, N. B. Federova, H. A. Carty, L. A. Umayam, D. H. Haft, J. Peterson, M. J. Beanan, O. White, S. L. Salzberg, R. C. Hsia, G. McClarty, R. G. Rank, P. M. Bavoil, and C. M. Fraser. 2003. Genome sequence of Chlamydophila caviae (Chlamydia psittaci GPIC): examining the role of niche-specific genes in the evolution of the Chlamydiaceae. Nucleic Acids Res. 31:2134-2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ridderhof, J. C., and R. C. Barnes. 1989. Fusion of inclusions following superinfection of HeLa cells by two serovars of Chlamydia trachomatis. Infect. Immun. 57:3189-3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rockey, D. D., J. Lenart, and R. S. Stephens. 2000. Genome sequencing and our understanding of chlamydiae. Infect. Immun. 68:5473-5479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Salvatore, P., C. Pagliarulo, R. Colicchio, P. Zecca, G. Cantalupo, M. Tredici, A. Lavitola, C. Bucci, C. B. Bruni, and P. Alifano. 2001. Identification, characterization, and variable expression of a naturally occurring inhibitor protein of IS1106 transposase in clinical isolates of Neisseria meningitidis. Infect. Immun. 69:7425-7436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shaw, A. C., G. Christiansen, P. Roepstorff, and S. Birkelund. 2000. Genetic differences in the Chlamydia trachomatis tryptophan synthase alpha-subunit can explain variations in serovar pathogenesis. Microbes Infect. 2:581-592. [DOI] [PubMed] [Google Scholar]
- 44.Shirai, M., H. Hirakawa, M. Kimoto, M. Tabuchi, F. Kishi, K. Ouchi, T. Shiba, K. Ishii, M. Hattori, S. Kuhara, and T. Nakazawa. 2000. Comparison of whole genome sequences of Chlamydia pneumoniae J138 from Japan and CWL029 from USA. Nucleic Acids Res. 28:2311-2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shirai, M., H. Hirakawa, K. Ouchi, M. Tabuchi, F. Kishi, M. Kimoto, H. Takeuchi, J. Nishida, K. Shibata, R. Fujinaga, H. Yoneda, H. Matsushima, C. Tanaka, S. Furukawa, K. Miura, A. Nakazawa, K. Ishii, T. Shiba, M. Hattori, S. Kuhara, and T. Nakazawa. 2000. Comparison of outer membrane protein genes omp and pmp in the whole genome sequences of Chlamydia pneumoniae isolates from Japan and the United States. J Infect. Dis. 181(Suppl. 3):S524-S527. [DOI] [PubMed] [Google Scholar]
- 46.Stephens, R. S., S. Kalman, C. Lammel, J. Fan, R. Marathe, L. Aravind, W. Mitchell, and L. Olinger. 1998. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science 282:754-759. [DOI] [PubMed] [Google Scholar]
- 47.Stothard, D. R., G. A. Toth, and B. E. Batteiger. 2003. Polymorphic membrane protein H has evolved in parallel with the three disease-causing groups of Chlamydia trachomatis. Infect. Immun. 71:1200-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tanzer, R. J., and T. P. Hatch. 2001. Characterization of outer membrane proteins in Chlamydia trachomatis LGV serovar L2. J. Bacteriol. 183:2686-2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Taraktchoglou, M., A. A. Pacey, J. E. Turnbull, and A. Eley. 2001. Infectivity of Chlamydia trachomatis serovar LGV but not E is dependent on host cell heparan sulfate. Infect. Immun. 69:968-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vandahl, B. B., A. S. Pedersen, K. Gevaert, A. Holm, J. Vandekerckhove, G. Christiansen, and S. Birkelund. 26 November 2002, posting date. The expression, processing and localization of polymorphic membrane proteins in Chlamydia pneumoniae strain CWL029. BMC Microbiol. 2:36. [Online.] http://www.biomedcentral.com/1471-2180/2/36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van Embden, J. D., T. van Gorkom, K. Kremer, R. Jansen, B. A. van Der Zeijst, and L. M. Schouls. 2000. Genetic variation and evolutionary origin of the direct repeat locus of Mycobacterium tuberculosis complex bacteria. J. Bacteriol. 182:2393-2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang, S. P., and J. T. Grayston. 1991. Three new serovars of Chlamydia trachomatis: Da, Ia, and L2a. J. Infect. Dis. 163:403-405. [DOI] [PubMed] [Google Scholar]
- 53.Wang, S. P., C. C. Kuo, R. C. Barnes, R. S. Stephens, and J. T. Grayston. 1985. Immunotyping of Chlamydia trachomatis with monoclonal antibodies. J. Infect. Dis. 152:791-800. [DOI] [PubMed] [Google Scholar]
- 54.Woodford, N. 2001. Epidemiology of the genetic elements responsible for acquired glycopeptide resistance in enterococci. Microb. Drug Resist. 7:229-236. [DOI] [PubMed] [Google Scholar]
- 55.Yuan, Y., Y. X. Zhang, N. G. Watkins, and H. D. Caldwell. 1989. Nucleotide and deduced amino acid sequences for the four variable domains of the major outer membrane proteins of the 15 Chlamydia trachomatis serovars. Infect. Immun. 57:1040-1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang, D. J., H. Fan, G. McClarty, and R. C. Brunham. 1995. Identification of the Chlamydia trachomatis RecA-encoding gene. Infect. Immun. 63:676-680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zomorodipour, A., and S. G. Andersson. 1999. Obligate intracellular parasites: Rickettsia prowazekii and Chlamydia trachomatis. FEBS Lett. 452:11-15. [DOI] [PubMed] [Google Scholar]