Abstract
The neurohypophysial hormone arginine vasopressin (AVP) plays important roles in fluid regulation and vascular resistance. Differences in AVP receptor expression, particularly mediated through variation in the noncoding promoter region of the primary receptor for AVP (AVPR1a), may play a role in social phenotypes, particularly social monogamy, in rodents and humans. Among primates, social monogamy is rare, but is common among New World monkeys (NWM). AVP is a nonapeptide and generally conserved among eutherian mammals, although a recent paper demonstrated that some NWM species possess a novel form of the related neuropeptide hormone, oxytocin. We therefore characterized variation in the AVP and AVPR1a genes in 22 species representing every genus in the three major platyrrhine families (Cebidae, Atelidae and Pitheciidae). For AVP, a total of 16 synonymous substitutions were detected in 15 NWM species. No non-synonymous substitutions were noted, hence, AVP is conserved in NWM. By contrast, relative to the human AVPR1a, 66 predicted amino acids (AA) substitutions were identified in NWM. The AVPR1a N-terminus (ligand binding domain), third intracellular (G-protein binding domain), and C-terminus were variable among species. Complex evolution of AVPR1a is also apparent in NWM. A molecular phylogenetic tree inferred from AVPR1a coding sequences revealed some consensus taxonomic separation by families, but also a mixed group composed of genera from all three families. The overall dN/dS ratio of AVPR1a was 0.11, but signals of positive selection in distinct AVPR1a regions were observed, including the N-terminus, in which we identified six potential positive selection sites. AA substitutions at positions 241, 319, 399 and 409 occurred uniquely in marmosets and tamarins. Our results enhance the appreciation of genetic diversity in the mammalian AVP/AVPR1a system, and set the stage for molecular modeling of the neurohypophyseal hormones and social behavior in primates.
Introduction
The neurohypophysial hormone arginine vasopressin (AVP) is synthesized primarily in the paraventricular nucleus (PVN) and supraoptic nucleus (SON) of the hypothalamus and then transported to the posterior pituitary for release into systemic circulation. Its two primary peripheral functions are related to osmoregulation and vasoconstriction [1], although AVP also exerts important regulatory effects in the central nervous system via neural projections that remain within the brain [2]. AVP is a nonapeptide, an ancient family of neuropeptides found in both vertebrates and invertebrates. All vertebrate nonapeptides, including AVP, are derived from arginine vasotocin (AVT) [3], [4]. The AVT gene was duplicated and subsequently modified to synthesize multiple oxytocin-like peptides in vertebrates. The evolution of vasopressin-like neuropeptides among vertebrates has been more conservative, with AVP replacing AVT in most mammals [3], [5], although pig, tenrec, opossum, and wallaby display variation in amino acid (AA) structure [6], [7]. Nevertheless, the ratio of nonsynonymous-to-synonymous divergence (dN/dS) of AVP is low across mammals (0.005), suggesting very strong conservation of this gene [6].
AVP exerts effects on multiple behavioral systems, including learning and memory, aggression, anxiety, affective states, and social behavior, primarily via interactions with the centrally expressed receptor subtype AVPR1a [2]. A causal role for AVP receptor polymorphisms and social behavior has focused on differences in repetitive microsatellite regions (short tandem repeats; STR) in the 5′ regulatory region of AVPR1a. In prairie voles, variation in the composition and length of these STRs are associated with species-level differences in gene expression in vitro and with differences in receptor expression and distribution in the brain [8]. These 5′ regulatory STRs also predict species differences in social monogamy, with some monogamous voles expressing STRs, and other non-monogamous vole species lacking these STR regions [9], [10]. In humans, there is a complex microsatellite (RS3) in the 5′ regulatory region of AVPR1a [11]. RS3 polymorphisms are associated with a variety of social phenotypes [12]–[18], including pair bonding associated behaviors [19].
However, broad comparative studies of AVPR1a STRs in rodents revealed that the causal links between promoter variation and sociality are not necessarily straightforward. Specifically, STRs are present in multiple species of non-monogamous voles, and are absent in only two non-monogamous species [12]. Further, the presence or absence of STR microsatellites does not account for social variation in other taxa, including Peromyscus [13] and primates [14]–[16].
Recently, attention has turned to variation in the coding regions for AVPR1a, previously considered to be unimportant for functional differences in either AVP signaling or social variation [9], [17], [18]. In 24 species representing the genus Microtus, exceptionally high variation in exon 1 of the AVPR1a gene has been demonstrated, particularly in the N-terminus ligand-binding extracellular and G-protein intracellular regions of the GPCR domain [19]. Similarly, a comparative study of AVPR1a coding regions in Peromyscus documented high rates of nonsynonymous nucleotide substitutions concentrated in the N-terminal extracellular domain within this taxon, with evidence of positive selection at these sites. However, no systematic AA substitutions were reliably associated with social monogamy among Microtus or Peromyscus [13], [19], and the impact of coding variation in AVPR1a signaling activity appears to be minimal [13]. In New World monkeys (NWM), AVPR1a coding variation among four genera (Aotus, Pithecia, Callicebus, and Saimiri) confirms selective evolution of the N-terminus and G-protein binding regions in this taxon sufficient to clearly differentiate this clade from those containing other primates and mammals [16]. No unique signatures in either AVPR1a promoter or coding regions were identified that differentiated monogamous vs. non-monogamous NWM, although only one non-monogamous genus (Saimiri) was included in this study, and the sample was limited to only four of 17 genera that comprise the NWM.
In the present paper, we explore variation in the coding regions of AVP/AVPR1a across primates, with a particular focus on the entire clade of NWM. This taxon represents a particularly important test case for the link between AVP-signaling and sociality for two reasons. First, while social monogamy is rare among mammals and especially among primates [20], [21], this mating system is common in NWM, being present in more than 50% of genera (nine of 17) and 63.2% of species (74 of 117) in this group [21]. Secondly, a novel structural variant of the related neuropeptide oxytocin (OT) has been identified in four genera of NWM (Callithrix, Cebus, Saimiri, Aotus), in which a single in-frame mutation (Thy to Cyt) in the coding region of the OT gene results in an AA substitution from Leu to Pro at position 8 [22]. This suggests there may be unique selection for variability in neuropeptide signaling in this clade, given that both nonapeptides are derived from a duplication of the arginine vasotocin gene [23].
Because there are no comprehensive data regarding AVP/AVPR1a genetic variation throughout the NWM clade, we amplified, sequenced, and analyzed the AVP and AVPR1a coding region in species representing each of 17 genera in the three NWM families (Cebidae, Atelidae, and Pitheciidae). We examined variation in AVPR1a sequences across genera, in multiple species within some genera, and across multiple individuals within selected species. These NWM sequences were then contrasted with those from other primate taxa. Finally, we evaluated whether ligand and/or receptor nucleotide and subsequent AA substitutions were associated with social system variation in NWM.
Materials and Methods
Animals
A total of 22 NWM species were sampled, which covered all three families and eight subfamilies, and at least one species per genus. With the exception of Cacajao, we analyzed genomic DNA from at least two individuals, one male and one female, from each genus (sample range per species: 2–6). The species, DNA source, sex, and institutional source of each sample are presented in Table S1 in File S1. Sequences for AVP and AVPR1a for all other primates (hominoid, Old World, and prosimian primates) were accessed from UCSC Gene Browser/NCBI/Ensembl.
Ethics Statement
All samples were accessed from archival blood or tissue banks, or from extracted DNA samples provided by the institutions listed in Table S1 in File S1. All institutions are licensed and/or accredited by appropriate agencies (e.g., USDA, AZA). IACUC information is also provided in Table S1 in File S1 where relevant.
Amplification and Sequencing Gene Segments
Genomic DNA was extracted from whole blood or tissue samples using the DNeasy Blood and Tissue Kit (Qiagen) following the manufacturer’s protocol. DNA yield from the extractions was quantified using Nanodrop (Thermo Scientific) and DNA quality was measured using 0.8% agarose gel electrophoresis. The same sets of PCR primers were used to amplify AVP and AVPR1a coding regions in all NWM species (Table S2 in File S1). All primers were designed based on the AVP and AVPR1a conserved genomic regions in several taxa including human, chimpanzee and rhesus macaque (UCSC Genome Browser, http://genome.ucsc.edu/). All genomic regions were amplified under the following conditions: each 55 µL PCR reaction contained 100 ng genomic DNA, 4 mM MgCl2, 500 µM of each dNTP, 400 µM of each forward and reverse primers, 1.5 units of Taq DNA polymerase with 5.5 µL 10×PCR buffer. PCR was performed on a Techne 3 Prime Personal thermal cycler (Cole-Parmer) using a touchdown program with the following parameters: initial denaturation for 4 min at 95°C; followed by 12 cycles of 95°C for 35 s, 67°C (−0.6°C per cycle) for 35 s, and 72°C for 50 s; and followed by 23 cycles of 95°C for 35 s, 58°C for 35 s, and 72°C for 50 s; and a final extension at 72°C for 8 min. PCR products were run on 1% agarose gel, and only those with single bands were sequenced. PCR products were purified using QIAquick PCR Purification Kit (Qiagen), and sequenced directly in two directions with forward and reverse primers using an Applied Biosystems (ABI) 3730 48-capillary electrophoresis DNA analyzer, in the High-Throughput DNA Sequencing and Genotyping Core Facility at the University of Nebraska Medical Center.
Sequence Alignment and Molecular Analysis
All coding sequences of AVP and AVPR1a generated in this study were deposited into GenBank (accession numbers: KJ641423 to KJ641466). Genomic sequences of AVP and AVPR1a in humans, nonhuman hominoid primates, Old World monkeys (OWM), and prosimians were acquired from UCSC Genome Bioinformatics. Sequence alignments of codon sequences were performed using MUSCLE in MEGA 6.0 [24]. We determined predicted AA structure for AVP and AVPR1a, and classified AA substitutions as conservative or radical according changes in polarity, charge, and volume: substitutions with a change in one or more categories were classified as radical, while substitutions with no changes in the three categories were classified as conservative [25].
Selection of models of nucleotide substitution was performed with MEGA 6.0 using a maximum likelihood statistical method [24], and the model with the lowest BIC score (Bayesian Information Criterion) was selected. A molecular phylogenetic tree of NWM was then generated using AVPR1a coding sequences with the Maximum Likelihood algorithm (1000 bootstrap, Tamura 3-parameter+G model) in MEGA 6.0 [24], with the prosimian Microcebus AVPR1a sequence identified as an outgroup relative to anthropoid primates. Positive selection on the entire AVPR1a gene was assessed using site models (M8/M8a), branch models (0/2), and branch-site models (A/B) with PAML 4.7 [26]. Evidence of positively selected extracellular, transmembrane, and intracellular regions in AVPR1a were estimated by using a sliding window analysis with a window length of 50 and a step size of 10 with the software DnaSP 5.10 to compare NWM species against those of humans and Microcebus [27].
Classification of Social Monogamy
Social monogamy in mammals refers to a long-term or sequential living arrangement between an adult male and an adult female. This arrangement is frequently defined as: sharing the same territory, high rates of sociosexual behavior between pairmates, and often, but not always, biparental care. The presence or absence of genus-wide social monogamy in primates was assigned based on recent surveys [21], [28], and we note that our definition of social monogamy includes species that are not genetically monogamous [29].
Results
AVP diversity in primates
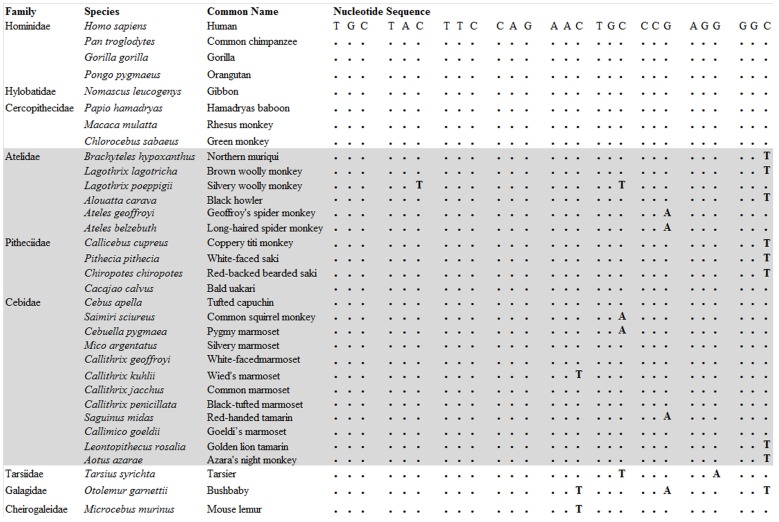
All primate species sampled had coding regions that yielded identical AA sequences for AVP (Cys-Tyr-Phe-Gln-Asn-Cys-Pro-Arg-Gly). In the families Hominidae, Hylobatidae, and Cercopithecidae, AVP coding sequences (27 nucleotides) were conserved, and no substitutions were identified (Figure 1). In NWM, AVP coding sequences were aligned among 22 species and contrasted with the human AVP nucleotide sequences. No non-synonymous substitutions were identified in this parvorder, and the nucleotides associated with AA at positions 1, 3, 4 and 8 were completely conserved across NWM. However, we identified a total of 16 synonymous substitutions in 15 species. The last nucleotide was more variable relative to other nucleotides, with a terminal T in 8 species. Nucleotides coding for the 6th AA exhibited two variants (TGT and TGA) in three genera from two families (Figure 1). Additionally, we noted species-specific nucleotide substitutions in cases in which more than one species per genus was sampled (Callithrix and Lagothrix). In tarsiers and two species of prosimian primates, six synonymous nucleotide substitutions were identified. Otolemur had synonymous substitutions at AA positions 5, 7 and 9, Microcebus had a single substitution at AA position 5, and Tarsius had two substitutions at positions 6 and 8 (Figure 1).
Figure 1. Arginine vasopressin (AVP) coding sequences for primates.
New World monkeys (NWM) are indicated by shaded area. ‘.’ represents identity with the human AVPR1a sequence.
AVPR1a diversity in primates
There were high levels of AA homology with human AVPR1a in hominoid primates and OWM. Nonhuman hominoid primates showed >98.4% sequence homology, and OWM exhibited >97.1% sequence homology. The prosimian Microcebus showed 89.5% homology with human AVPR1a, including a 9-amino acid deletion in the N-terminus. All nonhuman primates differed from human AVPR1a at positions 245 and 319. The amino acid at position 245 resides in the G-protein binding region, and constitutes a radical AA change compared with human. The amino acid at position 319 is located in the fourth extracellular region, and constitutes a radical AA change. Two substitution sites (positions 63 and 414) were unique to OWM.
Nucleotide substitutions in AVPR1a were widespread in NWM, with many substitutions resulting in AA changes throughout the coding region of this gene. Sequence homology with human AVPR1a among species in this taxon ranged from 92.3% (Callithrix and Saguinus) to 94.2% (Chiropotes). Relative to human AVPR1a sequences, we identified a total of 206 nucleotide substitutions in 22 species, which resulted in 66 predicted AA substitutions in the 418 AA that comprise AVPR1a (Figure S1 and S2 in File S1). Of the 66 AA substitutions, 10 were conserved across all NWM species (highlighted in green in Figure S1 in File S1). Identical AA substitutions at eight positions occurred with high frequency (defined as >94%) across NWM genera (positions 3, 5, 25, 26, 43, 264, 321 and 337), and 27 of the substitutions were genus-specific (Figure S1 in File S1).
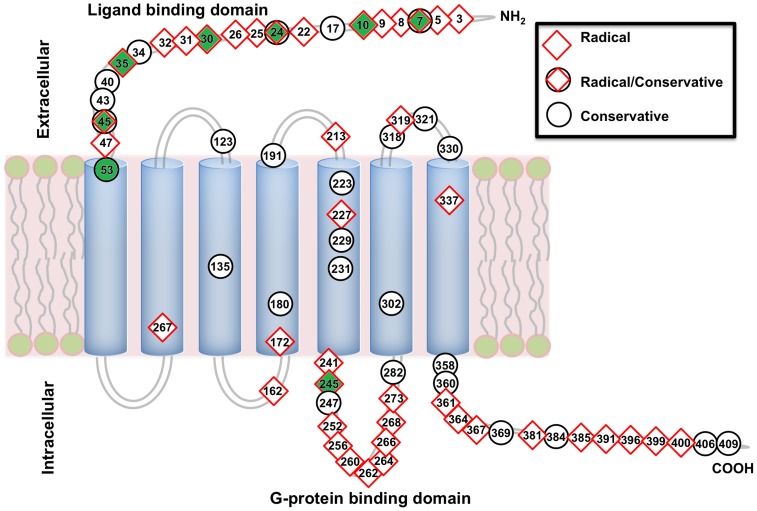
Across each region of AVPR1a for NWM, a higher percentage of AA substitutions were present in the extracellular N-terminus (20/52; 38.5%), the third intracellular region (13/54; 24.1%), and the intracellular C-terminus (15/67; 22.4%) than in other regions of AVPR1a (<22.2%) (Figure S2 in File S1). Transmembrane (TM) regions showed fewer substitutions compared with other regions (Figure S2 in File S1). A total of 47 AA substitutions in NWM AVPR1a involved radical changes, and 27 conservative changes (Figure 2; Table S3 in File S1). In three regions of the AVPR1a, multiple radical AA substitutions were noted across NWM. A total of 18, 13, and 9 radical physico-chemical substitutions were found in the N-terminus, the third intracellular region, and the C-terminus, respectively (Figure 2; Table S3 in File S1). Some AA substitutions had more than one change in physico-chemical category (Table S3 in File S1).
Figure 2. Structural model of AVPR1a.
Numbers reflect amino acid substitutions in NWM relative to the human AVPR1a, and radical physicochemical substitutions are indicated by red diamonds and conservative changes by black circles. Potential positively selected sites are highlighted in green.
In three genera, we sampled and sequenced AVPR1a coding regions for more than one species. In Callithrix (C. jacchus, C. kuhlii, C. geoffroyi, and C. penicillata), the predicted AA for AVPR1a were identical across all four species. In two species sampled from the genus Ateles, we identified only one conservative AA substitution in the N-terminus (position 17). There was greater AVPR1a variation in the two species sampled from Lagothrix, with four AA changes being radical substitutions located in the N-terminus (positions 7, 10, 30 and 47), and the fifth was a conservative substitution located in the first TM region (position 53).
For four species in the genus Callithrix, we sampled 6 individuals per species to quantify intraspecific variation in nucleotide and AA sequences for AVPR1a. We identified only three synonymous nucleotide substitutions in the AVPR1a coding region among individuals in C. kuhlii and C. penicillata (AA positions 137, 151 and 302). No nucleotide substitutions were identified among individuals in C. jacchus or C. geoffroyi. All intraspecific nucleotide substitutions were located in exon 1. Thus, intraspecific individual variation in AVPR1a is low within the genus Callithrix.
AVPR1a phylogeny and evolution in primates
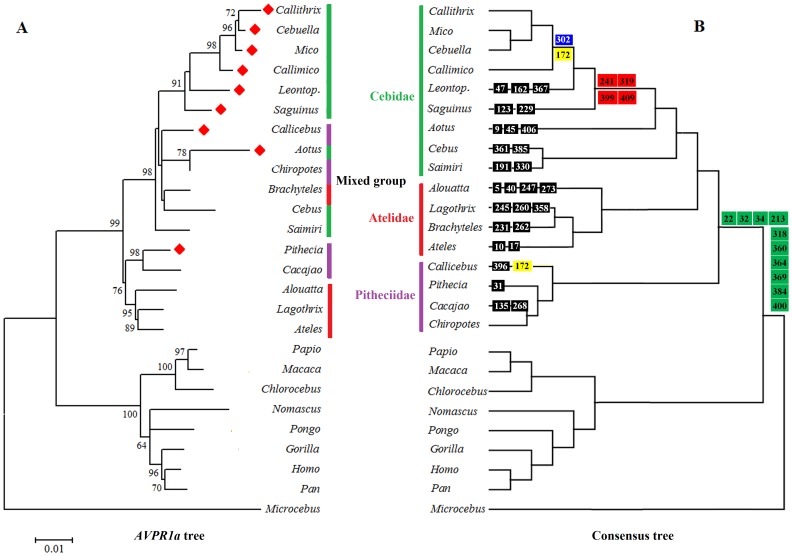
A molecular phylogenetic tree was generated based on AVPR1a coding sequences (Figure 3A) and contrasted with a consensus molecular tree based on 54 nuclear genes ([30]; Figure 3B). Hominoid and OWM clearly clustered together, and were distinctly separate from the one species of prosimian for which the AVPR1a sequence was available. In NWM, all genera in the subfamily Calllitrichinae, and several genera in families Pitheciidae and Atelidae clustered together with high bootstrap support based on AVPR1a sequences. However, in the middle region of the tree, some genera from each family were intermixed, including Cebus, Saimiri and Aotus (Cebidae), Brachyteles (Atelidae), and Callicebus and Chiropotes (Pitheciidae), albeit with low bootstrap support (Figure 3A; bootstrap support with values <60 are not shown). Of the nine socially-monogamous NWM, six of nine genera clustered in one clade, with two additional genera in a sister clade, and Pithecia more distantly related.
Figure 3. Molecular phylogenetic trees in primates.
A. Tree inferred from AVPR1a nucleotide coding sequences in primates. If bootstrap support is <60, no value is shown at nodes. Scale bar indicates the branch length in nucleotide substitutions per site. A red diamond indicates a genus characterized by social monogamy. B. A consensus tree of primates based on 54 nuclear genes (34,927 bp; [30]). Families Cebidae, Atelidae and Pitheciidae are highlighted in green, red and purple lines, respectively. Amino acid substitutions of the AVPR1a gene are plotted on the consensus tree: NWM-specific (green square), Callitrichinae-specific (red square), marmoset-specific (blue square), marmoset and Callicebus-specific (yellow square), and genera-specific (black square).
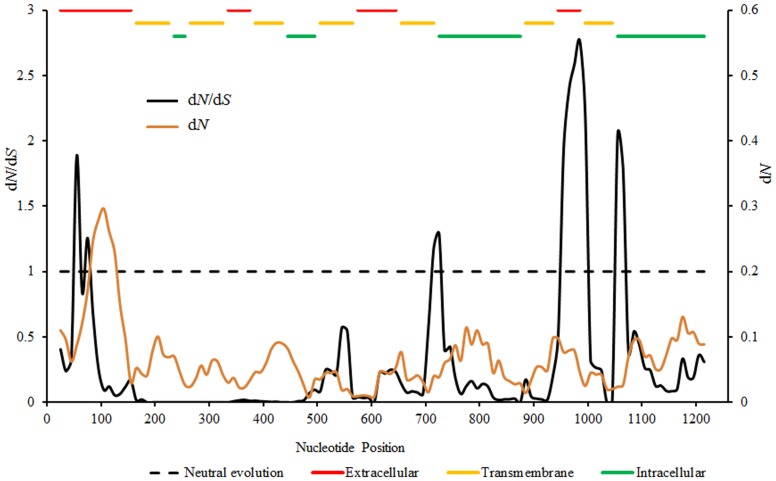
The overall dN/dS ratio for AVPR1a was 0.11, suggesting that purifying selection has acted upon AVPR1a in NWM. However, along the AVPR1a coding region against human AVPR1a, a sliding window analysis showed signals of positive selection (dN/dS >1.0) in multiple elements of the receptor, including the N-terminus, the third intracellular region, the fourth extracellular region, and the C-terminus (Figure 4). Other regions showed comparatively few non-synonymous mutations (Figure S2 in File S1), which led to low dN/dS ratios. PAML identified eight potential positive selection sites (P>0.05), including positions 7, 10, 24, 30, 35, 45, 53 and 245. In addition, sliding window analyses of NWM AVPR1a against Microcebus showed lower regional dN/dS ratios than those comparisons with humans (Figure S3 in File S1).
Figure 4. Sliding window analysis of dN/dS ratios and dN values along the NWM AVPR1a.
The ratio/value are drawn over the midpoint window position (window length 50, step size 10) from whole coding region. A dN/dS ratio above 1 indicates possible positive selection in the region. The elements of AVPR1a are highlighted in red (extracellular), yellow (transmembrane), and green (intracellular) lines.
AVPR1a diversity and social system
All genera within the subfamily Callitrichinae (marmosets and tamarins) are socially monogamous, and the species we analyzed exhibited four unique AA substitutions that differentiate this group from other NWM (Figure S1 in File S1). These substitutions include a radical change in an AA in the G-protein binding domain (Cys241), a radical change in the 4th extracellular region (Asn319), and a radical and conservative change in the C-terminus (positions Val399 and Arg409, respectively). A conservative AA change in position Ile302 in the 6th TM region was noted in marmosets only (Callithrix, Cebuella, Mico, and Callimico). These genera also shared an AA substitution with the monogamous Callicebus (a radical AA change at position 172 in the 4th TM region; Gly172). Other genera-specific substitutions for the remaining monogamous genera (Aotus, Pithecia) can be found in Figure 3B. Thus, while there were no universal substitutions in AVPR1a AA sequences associated with social monogamy, we did identify unique AA substitutions in the Callitrichines, in which social monogamy characterizes the entire subfamily.
Discussion
AVP is conserved in primates
AVP is a neurohypophysial nonapeptide that is highly conserved in mammals [6]. Only two variants in eutherian AVP have been identified: Arg8-AVP is replaced by Lys8- AVP in pigs, opossum, wallabies [6], [7], and Glu4-AVP is replaced by Thr4-AVP in tenrecs [6]. Although a novel form of the related neuropeptide OT was recently identified in NWM [22], our broad survey of this taxon revealed no non-synonymous substitutions in the predicted AA sequence for AVP. Across primates, all species showed consensus amino acids for AVP. Thus, AVP ligand structure at the amino acid level in primates is consistent with that of most other eutherian mammals. We did note that NWM and prosimians possessed multiple synonymous mutations in the coding region for AVP, whereas nonhuman hominoid primates and OWM shared identical nucleotide sequences with humans. To the extent that AVP signaling systems constitute an important determinant of variation of sociality among primates [2], these effects are certainly not dependent on AVP ligand differences among species.
Genetic variation in the AVPR1a gene
The nonhuman hominoid primates and OWM showed high homology with human in the 418 AA that comprise AVPR1a, which is consistent with their close phylogenetic relationship with humans. Two sites were unique to all nonhuman primates sampled (positions 245 and 319) and two were unique to OWM (positions 63 and 414), compared with human. Position 245 is located in the third intracellular region, suggesting possible differences in G-protein binding function in human vs. nonhuman primates. The single genus of prosimian primates characterized in our study (Microcebus) showed lower homology with human AVPR1a, which coincided with the greater phylogenetic genetic distance from humans.
NWM showed high rates of variability in the AVPR1a coding region (Figure S1 and S2 in File S1). Despite the AVP ligand being conserved in NWM, the major ligand recognition and binding region in the receptor (N-terminus) showed multiple radical physico-chemical substitutions. This observation is consistent with, and extends data from previous report showing that variable N-termini exist in the genus Aotus [16]. A short region (from Glu37 to Asn47) in the N-terminus of rat AVPR1a is necessary for AVP recognition and binding [31], [32]. In NWM, we documented substitutions in this specific region, including AA positions 43 and 45. It is not clear whether Lys43 or Glu45 play a role in AVP recognition and binding. The single residue Asn47 was conserved in primates with the exception of Asp47 in Leontopithecus and Lagothrix (Fig S2 in File S1). Both Glu45 and Asp47 constitute radical physico-chemical substitutions (Figure 2). Variable N-termini also were observed in the genus Microtus [19] and Peromyscus [13]. Whether this variation in N-terminus structure has functional significance is not clear, at least for Peromyscus, since signaling assays showed no detectable effect of the substitutions in this region on AVPR1a function [13]. Apart from signaling effects, tests of other functional consequences of N-terminus AA substitutions (e.g., recognition and binding activities between receptor and ligands) are needed to determine whether variability in NWM AVPR1a variants translate to functional differences.
The third intracellular region of AVPR1a plays important roles in signal transduction by binding G-protein [33]. This G-protein binding domain of AVPR1a was characterized by multiple radical substitutions in NWM (Figure 2). At position 256, Callithrix and Alouatta shared a unique substitution (Asn256). In this region, we also found an amino acid that appears to be unique to humans among all primates for which we had samples (Cys245). NWM and OWM showed different substitutions at position 264, both of which being radical changes. The radical changes found in this region might influence signal transduction, since the third intracellular region can lead to differences in receptor activation [34].
The C-terminus also showed variability in NWM, and multiple radical changes were identified. These findings indicated that these substitutions could alter AVPR1a function in NWM. Only five substitutions of 71 C-terminal residuals were observed across the genus Peromyscus [13], and hence C-terminus variation in Peromyscus is lower than in NWM (5/71 vs. 15/67 AA substitutions). No available data regarding C-terminus variation was reported in Microtus [19]. Since the C-terminus plays an important role in signal transduction [35], multiple substitutions in this region could influence the function of AVPR1a.
We assessed AVPR1a sequences in multiple species within three genera of NWM. Low levels of genetic variation were observed within genera, especially in the genus Callithrix. The highest rate of within-genus variation occurred in the contrast of two species of Lagothrix and two species of Ateles. More species per genera should be assessed to provide a more confident estimate of intrageneric variation in AVPR1a. Previous studies on species differences in AVPR1a coding region within single genera in rodents identified substantially higher levels of variation than in NWM. In 24 species in the genus Microtus, AVPR1a AA sequences contained over 100 substitutions, in addition to several deletions and insertions [19]. Within the genus Peromyscus, 27 substitutions were reported among eight species, including two insertions and one deletion [13]. The lower rate of AA substitutions in AVPR1a within single genera of NWM relative to rodents is consistent with the notion that evolution is more rapid in the rodent lineage, relative to the primate lineage [36].
We were able to assess individual polymorphisms in AVPR1a AA sequences in four species of Callithrix. There were no intraspecific differences in AA sequence in any of the four species. We identified only three synonymous nucleotide substitutions in exon 1 of AVPR1a in C. kuhlii and C. penicillata, indicating that AVPR1a is an evolutionarily conserved gene in genus Callithrix. Likewise, two studies on the coding region for human AVPR1a revealed only a synonymous substitution at position 136 in samples of 48 and 125 human participants, respectively [37], [38]. In Peromyscus, there was greater intraspecific variation, with individually-unique AA substitutions at one position identified in three species for which >4 individuals were sequenced [13]. Taken as a whole, therefore, within-species levels of polymorphism in AVPR1a coding regions are low, relative to levels of polymorphisms documented for promoter regions for this gene [14].
AVPR1a evolution
In the phylogenetic tree derived from AVPR1a gene sequences, Hominoid primates, OWM, NWM and prosimians were separated according to conventional phylogenies with high bootstrap values. Three families in NWM generally clustered in a manner similar to the phylogenetic trees generated from 54 nuclear gene regions (Figure 3B) [30], mtDNA phylogenetic tree [39], and morphological characters tree [40]. Within the NWM, however, a mixed clade was observed. This mixed clade exhibited similar variants of AVPR1a in these six species, even though they belong to different families. Diverse traits are demonstrated in these six genera, including social monogamy in Callicebus and Aotus, and small body size in Cebus, Saimiri and Aotus, and different gestation lengths for each species [41]. Likewise, these genera live in diverse habitats ranging from wet topical forests to dry woodland habitats [42]. Thus, there is no consistent predictive variable (social system, body size, fluid regulation [43]) that can account for this cluster. This mixed group of genera in the AVPR1a phylogeny suggests a complexity in the evolution of this gene in NWM that has yet to be explained. The bootstrap values suggest two independent clades with social monogamy (Callicebus, Aotus, and the callitrichines; Pithecia), but further sampling of multiple species per genera, and additional details on the nature of the social phenotype among these taxa, is required to address this possibility.
Evidence for AVPR1a evolution was also indicated by quantification of dN/dS ratios, a sliding window analysis, and identification of potential positive selection sites. These results indicated that despite the low dN/dS for the entire AVPR1a gene, the ligand and G-protein binding regions may be under positive selection. The C-terminus is a critical domain for internalization and signal transduction [35], and dN/dS ratio of this domain indicated that this important element is underlying positive selection. In addition, in the fourth extracellular and the 7th TM regions, though the regional dN/dS ratio also showed a higher value than 1.0, few substitutions were observed in the two regions (Figure S2 in File S1). In fact, selection could act on the synonymous sites [44]–[46]. Similar results were noted in the analysis of N-terminus AA substitutions in Microtus and Peromyscus [13], [19]. The dN/dS ratios again lead to the conclusion of complex of evolution of AVPR1a in NWM.
AVPR1a variation and social monogamy
Social monogamy is generally rare among primates, but it is prevalent in NWM, with more than 50% of genera routinely displaying this social system [21], [28], [42], including all species in the subfamily Callitrichinae. We documented AA substitutions that are associated with some, but not all, monogamous NWM. Substitutions at six positions were noted in Callitrichine primates (marmoset and tamarin; or marmosets only), a taxon in which all species are socially monogamous. Four of the substitutions were radical physico-chemical substitutions, suggesting a possible functional change in AVPR1a. Whether these substitutions in AVPR1a contribute to monogamy in Callitrichine primates, or are simply a consequence of phylogeny, needs to be investigated further. Additionally, among the six substitutions listed above, only one was noted in another monogamous NWM genus (Gly172 in Callicebus; Figure S1 in File S1), but not in Aotus or Pithecia. While there are substitutions in AVPR1a associated with some monogamous NWM, the absence of a consistent pattern of substitutions uniquely associated with social monogamy suggests that if AVPR1a variation contributes to this social phenotype, it does so in complex ways. We note that the majority (six of nine genera) of socially-monogamous NWM clustered together with high bootstrap support in the phylogeny based on AVPR1a nucleotide sequences, suggesting that exploring the link between AVP signaling and social monogamy is worthy of additional attention.
Many of the substitutions in the social monogamous NWM genera were located in the C-terminus, and a region also characterized by a high dN/dS ratio. The C-terminus plays a critical role in internalization and signal transduction [35]. Human AVPR1a contains one proximal protein kinase C motif from position 382 to 384 (SRR). Deletion of this motif caused reduced receptor phosphorylation [47], and inhibited AVP stimulation of DNA synthesis and progression through the cell cycle [35]. We found that all NWM species display one specific substitution in this motif, Lys384. The substitution Arg409 occurs in the protein kinase C motif of marmosets and tamarins characterized by social monogamy. It would be fruitful to explore further whether variation in the C-terminus of AVPR1a contributes to altered receptor function and ultimately variation in social behavior.
Social monogamy is a complex social behavior. Recent hypotheses regarding the selective pressures leading to this trait include the difficulty of male defense of multiple females [21], protection from male infanticide [28], and certainty of paternity/genetic monogamy [29]. While the systematic evaluation of these hypotheses is beyond the scope of the present paper, it is worthwhile noting with regard to the third hypothesis that only one genera of NWM (Aotus) has been demonstrated to exhibit genetic monogamy, yet social monogamy and paternal care is common in NWM [29]. In any event, molecular sequence variation could reflect adaptation to these or other selective forces [16], [21], [28]. Research on Peromyscus suggests that mating system variation in rodents is not a consequence of simple genetic variation, and that social variation is likely to be mediated by multiple genetic mechanisms [13]. Candidate genes that may contribute to social monogamy include genes coding for OT, dopamine, and their respective cellular receptors [48], [49]. Evolutionary analyses in 25 species of Microtus showed that monogamy is not predicted by a single polymorphism in the promoter region of the AVPR1a gene [12]. In the present study, we documented six AA substitutions that were associated with social monogamy in some NWM. However, it is important to point out that these changes were specific to the subfamily Callitrichinae, and hence may represent simple phylogenetic differences and not functional variants vis-à-vis social system.
Supporting Information
Contains Table S1, Sample information for the New World monkeys in this study. Table S2, PCR primers used to amplify genomic coding regions of AVP and AVPR1a (Underlined primer are the nested primers). Table S3, Radical or conservative change for each substitution in AVPR1a of NWM. N-term, N-terminus; TM, transmembrane region; IC, intracellular region; C-term, C-terminus. Reference sequence is human AVPR1a. Figure S1, Alignment of AVPR1a amino acid substitutions in NWM (shaded) and non-NWM relative to human AVPR1a. Green indicates NWM-specific substitutions, red represents unique substitutions in marmoset and tamarin, yellow indicates the substitution in marmoset and titi monkeys, blue indicates the marmoset-specific substitution, and ‘.’ represents identity with human. The vertical numbers in this figure indicate the amino acid position in AVRP 1a protein. AVPR1a sequences of Tarsius syrichta and Otolemur garnettii were not available from public data. The numbers of potential positive selection sites are framed. * indicates social monogamy. Figure S2, Alignment of the AVPR1a predicted amino acids in primates. The dot implicates the identity with human; pink indicates the extracellular regions; yellow represents the transmembrane regions; and blue indicates the intracellular regions. NWM are framed in the figure. Figure S3, Sliding window analysis of the dN/dS ratio and dN value along AVPR1a gene. A. Microcebus and 17 NWM genera. B. Microcebus and Hominoid. C. Old World monkeys against Microcebus. The ratio/value are drawn over the midpoint window position (window length 50, step size 10) from whole coding region. The elements of AVPR1a are highlighted in red (extracellular), yellow (transmembrane), and green (intracellular).
(DOCX)
Acknowledgments
We thank Drs. G. Lu, R. Kellar, and members of the Callitrichid Research Center for comments on previous versions of this manuscript. We gratefully acknowledge the staff at the institutions listed in Table S1 in File S1 for their willingness to share tissues and/or genomic DNA samples with us. This project used biological materials (Saguinus) funded by the National Center for Research Resources (P51 RR013986) and is currently supported by the Office of Research Infrastructure Programs/OD P51 OD011133. For titi monkey tissues (Callicebus), we acknowledge HD053555 and HD071998 to KLB and P51OD01107 to the California National Primate Research Center.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All summary data are contained in the paper and Supporting Information. Original genome sequences developed for this study can be publicly accessed: http://www.ncbi.nlm.nih.gov/nuccore/ using accession numbers KJ641423 to KJ641466.
Funding Statement
This study was supported in part by funds from the NIH (HD042882) and the Nebraska Research Initiative. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Koshimizu TA, Nakamura K, Egashira N, Hiroyama M, Nonoguchi H, et al. (2012) Vasopressin V1a and V1b receptors: from molecules to physiological systems. Physiol Rev 92: 1813–1864. [DOI] [PubMed] [Google Scholar]
- 2. Caldwell HK, Lee HJ, Macbeth AH, Young WS 3rd (2008) Vasopressin: behavioral roles of an “original” neuropeptide. Prog Neurobiol 84: 1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Acher R, Chauvet J (1995) The neurohypophysial endocrine regulatory cascade: precursors, mediators, receptors, and effectors. Front Neuroendocrinol 16: 237–289. [DOI] [PubMed] [Google Scholar]
- 4. Acher R, Chauvet J, Chauvet MT (1995) Man and the chimaera. Selective versus neutral oxytocin evolution. Adv Exp Med Biol 395: 615–627. [PubMed] [Google Scholar]
- 5. Goodson JL (2008) Nonapeptides and the evolutionary patterning of sociality. Prog Brain Res 170: 3–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wallis M (2012) Molecular evolution of the neurohypophysial hormone precursors in mammals: Comparative genomics reveals novel mammalian oxytocin and vasopressin analogues. Gen Comp Endocrinol 179: 313–318. [DOI] [PubMed] [Google Scholar]
- 7. Ferguson DR (1969) The genetic distribution of vasopressins in the peccary (Tayassu angulatus) and Warthog (Phacochoerus aethiopicus). Gen Comp Endocrinol 12: 609–613. [DOI] [PubMed] [Google Scholar]
- 8. Hammock EA, Young LJ (2005) Microsatellite instability generates diversity in brain and sociobehavioral traits. Science 308: 1630–1634. [DOI] [PubMed] [Google Scholar]
- 9. Young LJ, Nilsen R, Waymire KG, MacGregor GR, Insel TR (1999) Increased affiliative response to vasopressin in mice expressing the V1a receptor from a monogamous vole. Nature 400: 766–768. [DOI] [PubMed] [Google Scholar]
- 10. Lim MM, Wang Z, Olazabal DE, Ren X, Terwilliger EF, et al. (2004) Enhanced partner preference in a promiscuous species by manipulating the expression of a single gene. Nature 429: 754–757. [DOI] [PubMed] [Google Scholar]
- 11. Thibonnier M, Graves MK, Wagner MS, Chatelain N, Soubrier F, et al. (2000) Study of V(1)-vascular vasopressin receptor gene microsatellite polymorphisms in human essential hypertension. J Mol Cell Cardiol 32: 557–564. [DOI] [PubMed] [Google Scholar]
- 12. Fink S, Excoffier L, Heckel G (2006) Mammalian monogamy is not controlled by a single gene. Proc Natl Acad Sci U S A 103: 10956–10960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Turner LM, Young AR, Rompler H, Schoneberg T, Phelps SM, et al. (2010) Monogamy evolves through multiple mechanisms: evidence from V1aR in deer mice. Mol Biol Evol 27: 1269–1278. [DOI] [PubMed] [Google Scholar]
- 14. Donaldson ZR, Kondrashov FA, Putnam A, Bai Y, Stoinski TL, et al. (2008) Evolution of a behavior-linked microsatellite-containing element in the 5′ flanking region of the primate AVPR1A gene. BMC Evol Biol 8: 180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rosso L, Keller L, Kaessmann H, Hammond RL (2008) Mating system and avpr1a promoter variation in primates. Biol Lett 4: 375–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Babb PL, Fernandez-Duque E, Schurr TG (2010) AVPR1A sequence variation in monogamous owl monkeys (Aotus azarai) and its implications for the evolution of platyrrhine social behavior. J Mol Evol 71: 279–297. [DOI] [PubMed] [Google Scholar]
- 17. Insel TR, Young LJ (2001) The neurobiology of attachment. Nat Rev Neurosci 2: 129–136. [DOI] [PubMed] [Google Scholar]
- 18. Nair HP, Young LJ (2006) Vasopressin and pair-bond formation: genes to brain to behavior. Physiology (Bethesda) 21: 146–152. [DOI] [PubMed] [Google Scholar]
- 19. Fink S, Excoffier L, Heckel G (2007) High variability and non-neutral evolution of the mammalian avpr1a gene. BMC Evol Biol 7: 176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kleiman DG (1977) Monogamy in mammals. Q Rev Biol 52: 39–69. [DOI] [PubMed] [Google Scholar]
- 21. Lukas D, Clutton-Brock TH (2013) The evolution of social monogamy in mammals. Science 341: 526–530. [DOI] [PubMed] [Google Scholar]
- 22. Lee AG, Cool DR, Grunwald WC Jr, Neal DE, Buckmaster CL, et al. (2011) A novel form of oxytocin in New World monkeys. Biol Lett 7: 584–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Goodson JL (2013) Deconstructing sociality, social evolution and relevant nonapeptide functions. Psychoneuroendocrinology 38: 465–478. [DOI] [PubMed] [Google Scholar]
- 24. Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhang J (2000) Rates of conservative and radical nonsynonymous nucleotide substitutions in mammalian nuclear genes. J Mol Evol 50: 56–68. [DOI] [PubMed] [Google Scholar]
- 26. Yang Z (2007) PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24: 1586–1591. [DOI] [PubMed] [Google Scholar]
- 27. Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25: 1451–1452. [DOI] [PubMed] [Google Scholar]
- 28. Opie C, Atkinson QD, Dunbar RI, Shultz S (2013) Male infanticide leads to social monogamy in primates. Proc Natl Acad Sci U S A 110: 13328–13332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Huck M, Fernandez-Duque E, Babb P, Schurr T (2014) Correlates of genetic monogamy in socially monogamous mammals: insights from Azara's owl monkeys. P Roy Soc B-Biol Sci 281: 20140195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Perelman P, Johnson WE, Roos C, Seuanez HN, Horvath JE, et al. (2011) A molecular phylogeny of living primates. PLoS Genet 7: e1001342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hawtin SR, Wesley VJ, Parslow RA, Patel S, Wheatley M (2000) Critical role of a subdomain of the N-terminus of the V1a vasopressin receptor for binding agonists but not antagonists; functional rescue by the oxytocin receptor N-terminus. Biochemistry 39: 13524–13533. [DOI] [PubMed] [Google Scholar]
- 32. Hawtin SR, Wesley VJ, Parslow RA, Simms J, Miles A, et al. (2002) A single residue (arg46) located within the N-terminus of the V1a vasopressin receptor is critical for binding vasopressin but not peptide or nonpeptide antagonists. Mol Endocrinol 16: 600–609. [DOI] [PubMed] [Google Scholar]
- 33. Kristiansen K (2004) Molecular mechanisms of ligand binding, signaling, and regulation within the superfamily of G-protein-coupled receptors: molecular modeling and mutagenesis approaches to receptor structure and function. Pharmacol Ther 103: 21–80. [DOI] [PubMed] [Google Scholar]
- 34. Liu J, Wess J (1996) Different single receptor domains determine the distinct G protein coupling profiles of members of the vasopressin receptor family. J Biol Chem 271: 8772–8778. [DOI] [PubMed] [Google Scholar]
- 35. Thibonnier M, Plesnicher CL, Berrada K, Berti-Mattera L (2001) Role of the human V1 vasopressin receptor COOH terminus in internalization and mitogenic signal transduction. Am J Physiol Endocrinol Metab 281: E81–92. [DOI] [PubMed] [Google Scholar]
- 36. Li WH, Ellsworth DL, Krushkal J, Chang BHJ, HewettEmmett D (1996) Rates of nucleotide substitution in primates and rodents and the generation time effect hypothesis. Molecular Phylogenetics and Evolution 5: 182–187. [DOI] [PubMed] [Google Scholar]
- 37. Wassink TH, Piven J, Vieland VJ, Pietila J, Goedken RJ, et al. (2004) Examination of AVPR1a as an autism susceptibility gene. Mol Psychiatry 9: 968–972. [DOI] [PubMed] [Google Scholar]
- 38. Saito S, Iida A, Sekine A, Kawauchi S, Higuchi S, et al. (2003) Catalog of 178 variations in the Japanese population among eight human genes encoding G protein-coupled receptors (GPCRs). J Hum Genet 48: 461–468. [DOI] [PubMed] [Google Scholar]
- 39. Finstermeier K, Zinner D, Brameier M, Meyer M, Kreuz E, et al. (2013) A Mitogenomic Phylogeny of Living Primates. PLoS ONE 8: e69504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shoshani J, Groves CP, Simons EL, Gunnell GF (1996) Primate phylogeny: morphological vs. molecular results. Mol Phylogenet Evol 5: 102–154. [DOI] [PubMed] [Google Scholar]
- 41. Harris RA, Tardif SD, Vinar T, Wildman DE, Rutherford JN, et al. (2014) Evolutionary genetics and implications of small size and twinning in callitrichine primates. Proc Natl Acad Sci U S A 111: 1467–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fleagle JG (1998) Primate Adaptation and Evolution. Academic Press. 596.
- 43. Barberis C, Mouillac B, Durroux T (1998) Structural bases of vasopressin/oxytocin receptor function. J Endocrinol 156: 223–229. [DOI] [PubMed] [Google Scholar]
- 44. Chamary JV, Parmley JL, Hurst LD (2006) Hearing silence: non-neutral evolution at synonymous sites in mammals. Nat Rev Genet 7: 98–108. [DOI] [PubMed] [Google Scholar]
- 45. Pond SK, Muse SV (2005) Site-to-site variation of synonymous substitution rates. Mol Biol Evol 22: 2375–2385. [DOI] [PubMed] [Google Scholar]
- 46. Hurst LD, Pal C (2001) Evidence for purifying selection acting on silent sites in BRCA1. Trends Genet 17: 62–65. [DOI] [PubMed] [Google Scholar]
- 47. Berrada K, Plesnicher CL, Luo X, Thibonnier M (2000) Dynamic interaction of human vasopressin/oxytocin receptor subtypes with G protein-coupled receptor kinases and protein kinase C after agonist stimulation. J Biol Chem 275: 27229–27237. [DOI] [PubMed] [Google Scholar]
- 48. Anacker AM, Beery AK (2013) Life in groups: the roles of oxytocin in mammalian sociality. Front Behav Neurosci 7: 185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Garcia JR, MacKillop J, Aller EL, Merriwether AM, Wilson DS, et al. (2010) Associations between dopamine D4 receptor gene variation with both infidelity and sexual promiscuity. PLoS One 5: e14162. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Contains Table S1, Sample information for the New World monkeys in this study. Table S2, PCR primers used to amplify genomic coding regions of AVP and AVPR1a (Underlined primer are the nested primers). Table S3, Radical or conservative change for each substitution in AVPR1a of NWM. N-term, N-terminus; TM, transmembrane region; IC, intracellular region; C-term, C-terminus. Reference sequence is human AVPR1a. Figure S1, Alignment of AVPR1a amino acid substitutions in NWM (shaded) and non-NWM relative to human AVPR1a. Green indicates NWM-specific substitutions, red represents unique substitutions in marmoset and tamarin, yellow indicates the substitution in marmoset and titi monkeys, blue indicates the marmoset-specific substitution, and ‘.’ represents identity with human. The vertical numbers in this figure indicate the amino acid position in AVRP 1a protein. AVPR1a sequences of Tarsius syrichta and Otolemur garnettii were not available from public data. The numbers of potential positive selection sites are framed. * indicates social monogamy. Figure S2, Alignment of the AVPR1a predicted amino acids in primates. The dot implicates the identity with human; pink indicates the extracellular regions; yellow represents the transmembrane regions; and blue indicates the intracellular regions. NWM are framed in the figure. Figure S3, Sliding window analysis of the dN/dS ratio and dN value along AVPR1a gene. A. Microcebus and 17 NWM genera. B. Microcebus and Hominoid. C. Old World monkeys against Microcebus. The ratio/value are drawn over the midpoint window position (window length 50, step size 10) from whole coding region. The elements of AVPR1a are highlighted in red (extracellular), yellow (transmembrane), and green (intracellular).
(DOCX)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All summary data are contained in the paper and Supporting Information. Original genome sequences developed for this study can be publicly accessed: http://www.ncbi.nlm.nih.gov/nuccore/ using accession numbers KJ641423 to KJ641466.