Abstract
Cellulosomes and noncellulosomal (hemi)cellulolytic enzymes are produced by Clostridium cellulovorans to degrade plant cell walls. To understand their synergistic relationship, changes in mRNA and protein expression in cellulosomes and noncellulosomal (hemi)cellulolytic enzymes (hereafter called noncellulosomal enzymes) of cultures grown on cellobiose, cellulose, pectin, xylan, and corn fiber or mixtures thereof were examined. Cellulase expression, favored particularly by the presence of Avicel, was found with all substrates. Comparison of cellulosome and noncellulosomal enzymes showed that expression profiles were strongly affected by the carbon source. High xylanase or pectate lyase expression was observed when C. cellulovorans was grown on xylan or pectin, respectively. Mixed carbon substrates (cellulose-pectin-xylan mixture or corn fiber) induced a wider variety of enzymes than a single carbon source, such as cellobiose, pectin, or xylan. Cellulosomal proteome profiles were more affected by the carbon source than the noncellulosomal enzymes. Transcription and protein analyses revealed that cellulosomes and noncellulosomal enzymes were expressed simultaneously on mixed carbon sources, but their degree of inducibility varied when the substrate was either cellulose or cellobiose. Cellulosomes and noncellulosomal enzymes had synergistic activity on various carbon substrates. These results indicated that expression of plant cell wall-degrading enzymes is highly influenced by the available carbon source and that synergy between cellulosomes and noncellulosomal enzymes contribute to plant cell wall degradation.
Lignocellulose, the most abundant renewable resource in nature, is composed of the polymers cellulose, hemicellulose, pectin, and lignin (37). The main carbohydrate constituents of lignocellulosic material, i.e., cellulose, xylan, and pectin, consist of main chains of β-1,4-linked pyranosyl units which can be variously substituted. For efficient degradation of cellulose, hemicellulose, and pectin, a number of different enzyme activities are necessary, including activities to cleave the glycosidic bonds, and to remove substituent groups (46).
Clostridium cellulovorans is a mesophilic, anaerobic, spore-forming bacterium which can utilize cellulose, xylan, pectin, and several other carbon sources (7, 35). C. cellulovorans produces an extracellular enzyme complex (called a cellulosome) containing a variety of (hemi)cellulolytic subunits attached to the nonenzymatic scaffolding protein CbpA (2, 7, 31). All cellulosomal enzymatic subunits contain a twice-repeated sequence called the dockerin domain that is generally lacking in noncellulosomal (hemi)cellulolytic enzymes (we will refer to these enzymes as noncellulosomal enzymes). The dockerin domains bind to the hydrophobic domains of CbpA termed cohesins (43, 47). The formation of these complexes is necessary for the efficient degradation of crystalline cellulose (2, 3, 7, 31, 33). However, some noncellulosomal enzymes of C. cellulovorans, such as EngD (14), EngF (32), ArfA (19), and BgaA (19), are not bound to this multienzyme complex. These proteins appear to be part of an ancillary enzyme system for the hydrolysis of polysaccharides. The roles of noncellulosomal cellulases in the degradation of crystalline cellulose are still unknown. So far, we have identified 13 cellulosomal subunits and 4 noncellulosomal enzymes produced by C. cellulovorans (Table 1). The most abundant enzymes analyzed previously are these known enzymes, although there may be a few additional unidentified ones (5, 6, 19-21, 23, 28, 32, 38-41). The genome sequence analysis of Clostridium acetobutylicum ATCC 824 revealed at least 11 proteins that are confidently identified as cellulosome components (30). Despite the extensive research that has been devoted to the components of the cellulosomal enzyme subunits, there is comparatively little information available about the noncellulosomal enzymes of C. cellulovorans. Furthermore, no studies have been performed at the gene and protein levels concerning the regulation and relationship between the cellulosome and the noncellulosomal enzymes.
TABLE 1.
Cellulosomes and noncellulosomal protein subunits of C. cellulovorans
| Gene product | Spot no.a | Modular structureb | No. of amino acids | Mol wt | Exptl pIc (calculated) | Hydrophobic/ hydrophilic ratiod | GenBank accession no. |
|---|---|---|---|---|---|---|---|
| CbpA | I-1 | CBD-HLD-(HBD)2-HLD-(HBD)6-(HLD)2-HBD | 1,848 | 189,149 | 4.8 (4.76) | 3.47 | P38058 |
| EngE | I-2 | (HLD)3-GH5-X-DS | 1,030 | 111,777 | 4.7 (4.77) | 2.14 | AAD39739 |
| EngK | I-3 | CBDIV-Ig-GH9-DS | 892 | 97,024 | 5.7 (5.71) | 1.83 | AAF06107 |
| EngM | ND | CBDIV-Ig-GH9-DS | 876 | 96,373 | NT (5.15) | 1.84 | AAF06111 |
| PelA | I-4 | X-CBDIII-PL4-DS | 914 | 94,458 | NT (4.98) | 2.43 | AAG59609 |
| ExgS | I-5 | GH48-DS | 727 | 80,485 | 5.2 (5.70) | 1.75 | AAC38571 |
| EngY | ND | GH9-DS | 738 | 80,290 | NT (5.00) | 1.91 | AAG59608 |
| EngH | I-6 | GH9-CBDIII-DS | 715 | 79,321 | 5.1 (5.18) | 1.60 | AAC38572 |
| EngL | I-7 | GH9-DS | 522 | 57,629 | 5.0 (5.62) | 1.70 | AAF06109 |
| XynA | I-8 | GH11-DS-NodB | 520 | 57,037 | 6.9 (7.93) | 2.07 | AAN32825 |
| EngB | I-9 | GH5-DS | 440 | 48,823 | 6.6 (7.32) | 1.94 | P28621 |
| ManA | I-10 | GH5-DS | 425 | 47,156 | 5.2 (5.57) | 2.11 | AAF06110 |
| HbpA | ND | HLD-HBD | 240 | 24,940 | NT (4.28) | 2.00 | AAF06108 |
| BgaA | II-1 | GH42 | 659 | 76,462 | NT (5.12) | 1.39 | AAN05452 |
| EngF | II-2 | GH5-CBDXVII | 557 | 60,130 | 5.0 (4.99) | 2.64 | AAB40891 |
| EngD | II-3 | GH5-CBDII | 515 | 55,974 | 6.7 (8.69) | 2.52 | P28623 |
| ArfA | II-4 | GH51 | 492 | 55,730 | 5.3 (4.98) | 1.57 | AAN05450 |
Cellulosomes (I) and noncellulosomal proteins (II) are indicated. Spots are numbered as in Fig. 4. ND, not determined.
Catalytic modules are shown in boldface type. Module abbreviations: CBD, cellulose-binding domain family; GH, glycosyl hydrolase family; DS, dockerin domain; HLD, hydrophilic domain; HBD, hydrophobic domain; Ig, immunoglobulin-like domain; X, unknown domain.
Ratio of the percentage of hydrophobic amino acids to the percentage of hydrophilic amino acids in the protein.
The synergy between cellulosomes and noncellulosomal enzymes is assumed to be necessary for effective plant cell wall degradation (6), but how the expression of these fractions is coordinated under different growth conditions is not known. The cellulosomal enzyme subunit EngB (11) has high homology to a noncellulosomal enzyme, EngD (15), in the NH2-terminal region, which suggested that a comparison of these two enzymes would be fruitful. With the emergence of high-throughput transcription and proteomic technologies, it is now possible to monitor, in parallel, the expression levels of many genes and proteins under different growth conditions (48). A systematic display of the entire system of cellulase enzymes found in Clostridium cultures has not been reported. In this paper, we report the application of a combination of transcriptional and proteomic analyses to cellulosomal and noncellulosomal subunits, such as EngB and EngD. We also show the proteomic profiles, subunit patterns, and synergy of cellulosomes and noncellulosomal proteins.
MATERIALS AND METHODS
Bacterial strain and growth conditions.
C. cellulovorans ATCC 35296 was grown under strictly anaerobic conditions at 37°C in round-bottom flasks containing a previously described medium (33, 35), which included 0.5% (wt/vol) cellobiose, 1.0% (wt/vol) Avicel (microcrystalline cellulose; FMC Corporation), 1.0% xylan (birch wood; Sigma), 1.0% pectin (apples; Sigma), and/or 1% corn fiber (CF). CF and cellulose-arabinoxylan (CAX) complexes provided by David B. Johnston of the U.S. Department of Agriculture were used as plant cell wall substrates (8-10). CAX is produced from CF by removing fat and starch. The determination of cell mass in cultures was based on bacterial protein estimates as described previously (16).
Nucleic acid isolation.
Chromosomal DNA of C. cellulovorans was isolated by using a genomic DNA purification kit (Promega) according to the manufacturer's instructions. Total RNA was extracted from C. cellulovorans broth cultures using an RNeasy kit (Qiagen) with the additional step of treatment with RNAlater RNA stabilization reagent (Ambion) and RNase-free DNase (Promega) according to the manufacturers' instructions.
Northern blot analysis.
RNA samples (10 μg) were denatured in RNA sample buffer at 65°C for 10 min. RNA sample buffer consists of 250 μl of formamide, 83 μl of 37% (wt/vol) formaldehyde, 83 μl of 6× loading dye (Promega), 50 μl of 10× MOPS (morpholinepropanesulfonic acid) buffer (20 mM MOPS, 5 mM sodium acetate, 1 mM EDTA [pH 7.0]), and 34 μl of distilled water. The RNAs were separated through 1% agarose gels in MOPS buffer with 2% (vol/vol) formaldehyde. DNA probes were synthesized by PCR by using specific oligonucleotides derived from the C. cellulovorans sequence as a template (Table 2). The probes were nonradioactively labeled by random priming using digoxigenin (DIG) High Prime (Roche). To add the correct amount of probe for hybridization, serial dilutions of each probe (0.05 to 10 pg) were spotted onto a nylon membrane, and the labeling sensitivity (amount of labeled DNA per spot) was determined. RNA was transferred overnight to a positively charged nylon membrane (Roche) by capillary transfer using 20× standard saline-citrate (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate [pH 7]). Hybridization was performed for 16 to 20 h at 50°C in DIG Eazy Hyb buffer solution (Roche). Washing of the membrane and detection of specific transcripts on the blots were performed by using the DIG luminescence detection kit (Roche) and its recommended protocol.
TABLE 2.
PCR primers used for synthesis of the gene-specific probes
| Gene | Enzyme encoded | 5′ primer | 3′ primer | GenBank accession no. (reference) |
|---|---|---|---|---|
| cbpA | Cellulose-binding protein | ATGCAAAAAAAGAAATCGCTG | GGTTGATGTTGGGCTTGCTGTT | M73817 (34) |
| engE | Endoglucanase E | TACTGATGACTGGGCTTGGATGAG | GTTGCTTTCGCTGCTGC | AF105331 (40) |
| xynA | Xylanase | TGTTAGCCTCTTCTGC | GATTCCAAGTGCCATAGC | AF435978 (21) |
| pelA | Pectate lyase A | GGCGTTGTTGTTCCACCTGT | ACTGTTGAAGGTGCTAAAAG | AF105330 (39) |
| engB | Endoglucanase B | CTACAGCTAAGACAGG | ACTTCAGCCTCATTGG | M75706 (11) |
| engD | Endoglucanase D | GGAGAAACTAACTGGGGA | CTGGTCTTGGCTCATTCA | M37434 (14) |
| engF | Endoglucanase F | TGGTCTACAATGGTTTCCTGGG | GCATCATTCGTTACTCCACC | U37056 (32) |
| arfA | α-l-Arabinofurasnosidase | ATGGAGGATTTTGGGTTGGG | TCGGTGACTCTCCATC | AY128945 (19) |
| bgaA | α-l-Arabinopyranosidase | CAATACCCCTCCACTTTG | GCTGTTACTTGCGACCA | AY128945 (19) |
RNA slot blot analysis.
Total RNAs were diluted into appropriate concentrations with water and followed by adding two times the volume of the RNA sample buffer. After 10-min incubation at 65°C to denature the RNA, the samples were applied to a positively charged nylon membrane (Roche) by using a Hybri-slot apparatus (Gibco-BRL), and the membrane was baked for 30 min at 120°C under vacuum. Filters were hybridized with specific probes as described above for Northern blot analysis.
Preparation of cellulosome and noncellulosomal enzymes.
The cellulosome was purified from culture supernatants of C. cellulovorans cells as described previously (33). The culture supernatants (250 or 1,000 ml) were obtained by centrifugation after 3 to 5 days of growth with different carbon sources. The supernatants were concentrated by 80% ammonium sulfate saturation and dialyzed. The extracellular material was then mixed with Avicel, which resulted in binding of the cellulosome complex and some noncellulosomal enzymes to Avicel. After incubation for 1 h at 4°C, the suspension was poured into a column. The column was washed with 3 volumes of 100 mM phosphate buffer (pH 7.0) to elute the unattached fractions. These unattached fractions were saved as the noncellulosomal fraction after concentration by ultrafiltration using an Ultrafree Biomax centrifugal filter unit (Millipore) with a 10-kDa-cutoff membrane. The bound fraction was eluted from the cellulose column with deionized water and concentrated with an Ultrafree Biomax centrifugal filter unit (10-kDa cutoff; Millipore). In most cases, the noncellulosomal enzymes, which were mainly EngF and EngD, bound to Avicel was less than 5% of the total Avicel-binding protein, which was analyzed by gel filtration on a Sephacryl S-200 column (2.6 by 75 cm; Pharmacia). The concentration of purified protein was measured by the method of Bradford (4) with a protein assay kit from Bio-Rad, using bovine serum albumin as the standard.
SDS-PAGE and Western blot analysis.
Isoelectric focusing (IEF) gels were cast using ReadyPrep rehydration sample buffer (Bio-Rad) (1, 12). Aliquots of sample containing 100 μg of the protein were loaded onto each gel. Each sample was subjected to two-dimensional (2-D) gel electrophoresis in duplicate to control for gel-to-gel variations. After IEF, the gels were equilibrated in two equilibration buffers (6 M urea, 2% sodium dodecyl sulfate [SDS], 0.375 M Tris-HCl [pH 8.8], 20% glycerol). Dithiothreitol (2%, wt/vol) was added to the first equilibration buffer, and iodoacetamide (2.5%, wt/vol) was added to the second equilibration buffer. The second-dimension gels were cast using a linear gradient of 4 to 15% polyacrylamide. The equilibrated tube gels were secured to the 2-D gels using agarose, and SDS-polyacrylamide gel electrophoresis (SDS-PAGE) was performed as described previously (12, 22). Proteins were fixed in the gels by soaking in a solution containing 40% (vol/vol) methanol and 10% (vol/vol) acetic acid for approximately 1 h and subsequently visualized by Coomassie blue staining (Genomic Solutions). For Western blot (immunoblot) analysis, proteins were separated by SDS-PAGE and blotted onto a polyvinylidene difluoride membrane (Immobilon-P; Millipore). The membrane was treated with antibody (diluted 1:5,000) and stained as described previously (40).
Mass spectrometry analysis.
Mass spectrometric analysis was performed to identify the cellulosome and noncellulosomal proteins separated by 2-D PAGE (44). Proteins of interest were excised from the gel and subjected to in-gel digestion with trypsin. Peptide mass spectra were acquired using a matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometer (Biflex III; Bruker), and corresponding proteins were identified by peptide mass finger printing (http://www.matrixscience.com). MS/MS spectra for the digested peptides were also acquired by nanospraying the peptide mixtures into a Q-TOF mass spectrometer (QSTAR; Sciex). Further identification of proteins was made with de novo sequencing of the obtained MS/MS spectra followed by BLAST search. All protein identifications were verified by comparison with theoretical molecular weights and isoelectric points.
Assays of enzymatic activity.
The enzymatic activities on Avicel (for cellulase), carboxymethyl cellulose (CMC) (for endoglucanase), pectin (for pectate lyase), xylan (for xylanase), CAX (for mixed enzymes), and CF (for mixed enzymes) were assayed at pH 6.0 and 37°C by measuring the liberated reducing sugars as d-glucose equivalents by the Somogyi-Nelson method (36, 45). CAX preparations consisted mainly of 21% cellulose and 68% arabinoxylan, and CF preparations consisted mainly of 15% cellulose, 35% arabinoxylan, 8% lignin, and 20% starch. Each reaction mixture consisted of 250 μl of a 1% substrate solution, 100 μl of 250 mM sodium acetate buffer (pH 6.0), and 150 μl of an enzyme solution. The incubation times were 30 min for endoglucanase, pectate lyase, xylanase, and CAXase activities and 18 h for cellulase and CFase enzyme activities. One unit of each enzyme activity was defined as the amount of enzyme which released 1 μmol of reducing sugar per min under the condition indicated, except for Avicelase and CFase assays, where 1 unit of enzyme activity is the amount of enzyme which released 1 μmol of reducing sugar per h.
RESULTS
Induction of cellulosomal and noncellulosomal gene transcription under different growth conditions.
Expression of the five cellulosomal genes cbpA, engE, pelA, xynA, and engB was studied by Northern blot analysis together with four noncellulosomal genes, engD, engF, arfA, and bgaA. The first experiments were performed with medium containing cellobiose, cellulose, pectin, or xylan as the only carbon source, and the results were compared to those obtained with cellulose plus pectin, cellulose plus xylan, cellulose plus pectin and xylan, and cellulose plus pectin and xylan. Because the primary aims were to obtain data on general regulatory patterns of the genes under conditions where the bacterium is growing in the presence of chosen carbon sources and to avoid possible repression, the total RNA was isolated from cells in mid-log growth phase. Accordingly, all probes used were of similar length (approximately 350 bp) (Table 2) and were labeled to a similar sensitivity in order to obtain results which would allow comparison of the relative expression levels of the genes.
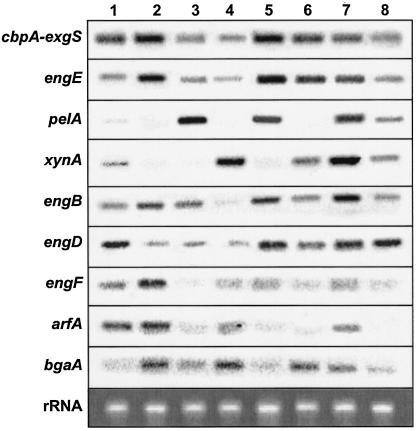
Cellobiose caused expression of all the genes tested, except for pelA and bgaA, which were not detected under the conditions used (Fig. 1, lane 1). Significant expression of most of the genes could be observed with cellulose (Fig. 1, lane 2). With cellulose, the expression of cbpA and engE was particularly high. The pectate lyase gene pelA was expressed strongly with the pectin-based medium; pectin was the only substrate that allowed significant expression of pelA (Fig. 1, lane 3). Pectin also induced cbpA, engE, and engB. Xylan caused especially high expression of the xylanase (xynA), α-l-arabinofuranosidase (arfA), and α-l-arabinopyranosidase (bgaA) genes and was also the most potent carbon source resulting in induction of these particular genes (Fig. 1, lane 4). When cellulose was mixed with pectin and/or xylan, many of the genes were expressed (Fig. 1, lanes 5 to 8). No expression of pelA and xynA occurred with only the cellulose medium, but addition of pectin (Fig. 1, lane 5) and/or xylan (Fig. 1, lane 6) to the cellulose medium efficiently induced pelA and/or xynA gene expression (Fig. 1, lanes 5 to 8). Interestingly, the ratio between cellulose, pectin, and xylan had an effect on cellulase induction. A higher ratio of cellulose to xylan (cellulose/pectin/xylan ratio of 3:1:1 [wt/wt]) (Fig. 1, lane 7) induced most of the genes more strongly than a lower ratio of cellulose to xylan (cellulose/pectin/xylan ratio of 2:1:2 [wt/wt]) (Fig. 1, lane 8).
FIG. 1.
Northern blot analysis of expression of five cellulosomal and four noncellulosomal genes in C. cellulovorans grown on different carbon sources. Total RNA was isolated from C. cellulovorans cultivated on medium containing 0.5% cellobiose (lane 1), 1% cellulose (Avicel) (lane 2), 1% pectin (lane 3), 1% xylan (lane 4), 1% cellulose-pectin mixture (cellulose/pectin ratio of 3:2 [wt/wt]) (lane 5), 1% cellulose-xylan mixture (cellulose/xylan ratio of 3:2 [wt/wt]) (lane 6), 1% cellulose-pectin-xylan mixture (cellulose/pectin/xylan ratio of 3:1:1 [wt/wt]) (lane 7), or 1% cellulose-pectin-xylan mixture (cellulose/pectin/xylan ratio of 2:1:2 [wt/wt]) (lane 8) as the sole carbon sources. RNA (5 μg) was subjected to electrophoresis through 1.5% formaldehyde gels and transferred to nylon membranes, which were subsequently probed with the DIG-labeled specific probes (Table 2). The gene-specific probes used are indicated on the left. Ethidium bromide staining of rRNA is shown as a loading control.
It was confirmed that higher concentrations of xylan to cellulose in the medium repressed cellulase and hemicellulase induction (cellulose/xylan ratio of 1:3 [wt/wt]) (data not shown). Some of the substrates clearly induced stronger expression of only a certain set of genes. Although it is a general inducing compound, cellulose induced the expression of cellulase genes, such as cbpA-exgS and engE, most strongly. The media commonly used to promote high expression of cellulase and hemicellulase are based on mixtures of plant materials. The artificial carbon mixture of cellulose, pectin, and xylan strongly provoked cellulase and hemicellulase expression (Fig. 1, lanes 7 and 8), whereas cellulose, pectin, or xylan alone did not. The xylanase and pectate lyase genes of C. cellulovorans showed a markedly different pattern of expression from that of the cellulases. High xylanase or pectate lyase gene expression was observed only when xylan or pectin was the growth substrate, respectively.
Relative transcriptional levels of cellulosomal engB and noncellulosomal engD under different growth conditions.
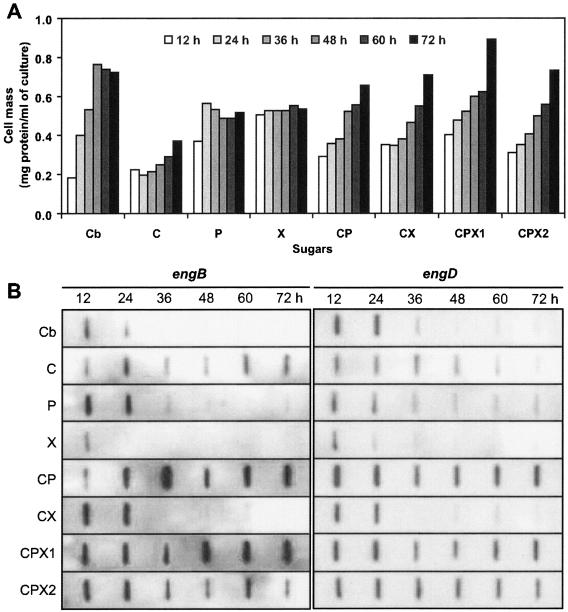
To determine whether EngB and EngD are regulated coordinately when cells are grown on different carbon sources, changes in the expression levels of the genes were monitored during the cultivation of C. cellulovorans with either cellobiose, cellulose, pectin, xylan, cellulose plus pectin, cellulose plus xylan, or cellulose plus pectin and xylan as the carbon source (Fig. 2A). The mRNA levels of engB in bacteria grown on medium containing pectin remained clearly higher than the levels in bacteria grown on medium containing cellobiose, cellulose, or xylan (Fig. 2B). In a separate slot blot experiment, the expression level of engD from cells grown on medium containing 0.5% cellobiose was shown to be higher than that of cells grown on medium containing cellulose (Fig. 2B). Like engB expression, strong induction of engD by cellulose plus pectin was observed, and cellulose plus xylan also induced engD mRNA, although less effectively. Low levels of engD mRNA were detected at different stages of growth with medium containing only pectin or xylan (Fig. 2B).
FIG. 2.
Relative levels of engB and engD transcripts in C. cellulovorans grown on different sugars and at different growth phases. (A) C. cellulovorans growth curve. RNA slot blot analyses were conducted with the same concentrations of RNA (0.5 μg) isolated from C. cellulovorans cultivated on medium containing 0.5% cellobiose (Cb), 1% cellulose (C) (Avicel), 1% pectin (P), 1% xylan (X), 1% cellulose-pectin mixture (CP) (cellulose/pectin ratio of 3:2 [wt/wt]), 1% cellulose-xylan mixture (CX) (cellulose/xylan ratio of 3:2 [wt/wt]), 1% cellulose-pectin-xylan mixture with a cellulose/pectin/xylan ratio of 3:1:1 (wt/wt) (CPX1) or 2:1:2 (wt/wt) (CPX2) as the sole carbon sources. The times (in hours) in the growth curve in panel A are shown in panel B also. The DIG-labeled engB and engD probes were prepared (each from 1 μg of template) by random primed labeling (see Materials and Methods). The carbon source in the culture medium is indicated to the left of the blots.
It is noteworthy that expression of engB and engD was similar at different stages of growth in the presence of certain carbon sources, such as cellulose-pectin mixtures. Also, the presence of cellulose, pectin, and xylan from the beginning of the cultivation induced engB and engD transcription (Fig. 2B), and this level was comparable or slightly higher than that obtained by cultivation with cellulose plus pectin. These results indicate that the engB and engD genes were expressed simultaneously, while their degree of induction varied slightly depending on different carbon sources, such as cellobiose or cellulose.
Relative expression levels of cellulosomal (EngB) and noncellulosomal (EngD) enzyme subunits under different growth conditions.
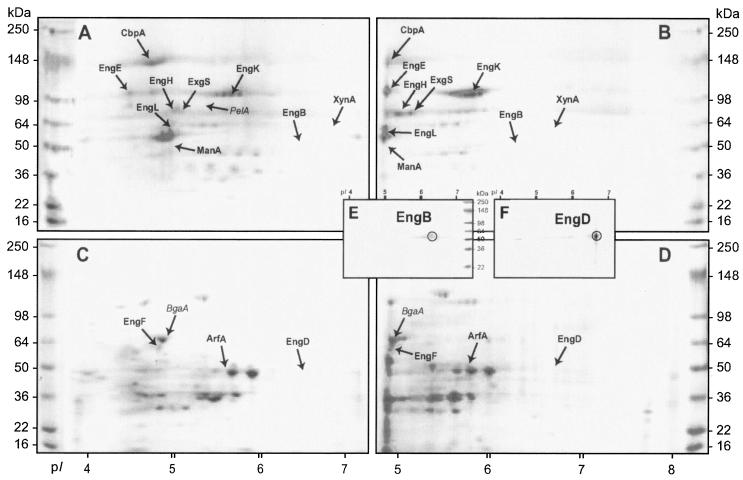
The 2-D gels (SDS-polyacrylamide gels) for the cellulosomal enzymes (Fig. 3A and B) indicated a large population of dense protein spots. Most distinctive spots were identified as known cellulosomal enzymes (Fig. 3A and B) and noncellulosomal enzymes (Fig. 3C and D) by Western blot analysis (Fig. 3E) and/or by the 2-D locations of recombinant proteins (Fig. 3F). On average, about 20 cellulosomal and 30 noncellulosomal spots were detected. The mass spectrometry technique was also used to identify proteins separated on 2-D gels. The peptide mass fingerprinting of CbpA, EngE, ExgS, PelA, EngH, and EngL obtained from the MALDI-TOF analysis unambiguously identified the spots found on the 2-D gels (data not shown). The putative EngK protein migrated to two spots located at molecular mass of 97,024 daltons and pI 5.7 in the 2-D gel in Fig. 3A. The mass spectra of EngK showed the amino acid sequences of tryptic peptides VNQVGYLPGVAK, ATMVSVGELLR, QVPTTLDQTFEFR, and TPLNWYLK, which perfectly matched the entire sequence of EngK (41). The spot presumably identified as EngF (Fig. 3C) was also confirmed by analysis of five tryptic peptides, TNPLSTVDTNR, VTNNFVAQTDGTYK, LSNDWNSNVLR, LNSLTSLDPGSDK, and ATPLVQLLR.
FIG. 3.
2-D PAGE of C. cellulovorans cellulosomal (A and B) and noncellulosomal (C and D) enzyme preparations visualized with Coomassie blue. Panels A to D show the gel zone defined by pH 3 to 7 (A and B) and pH 4 to 8 (C and D) and molecular mass of 16 to 250 kDa. The arrows point to the locations of the enzymes. (E) 2-D Western blot of cellulosomal fractions of crude culture supernatants from cells grown on 1% cellulose incubated with anti-EngB antibody. (F) 2-D gel electrophoresis of the purified recombinant EngD (1 μg) (14). The locations of the EngB (E) and EngD (F) proteins are circled.
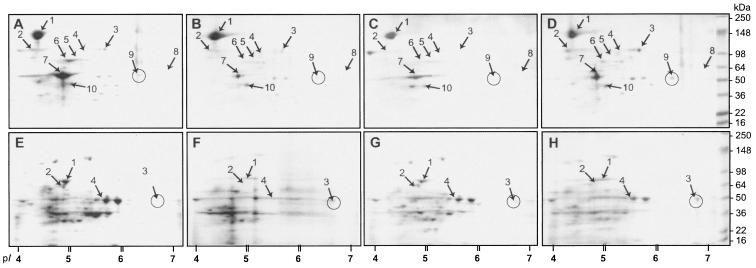
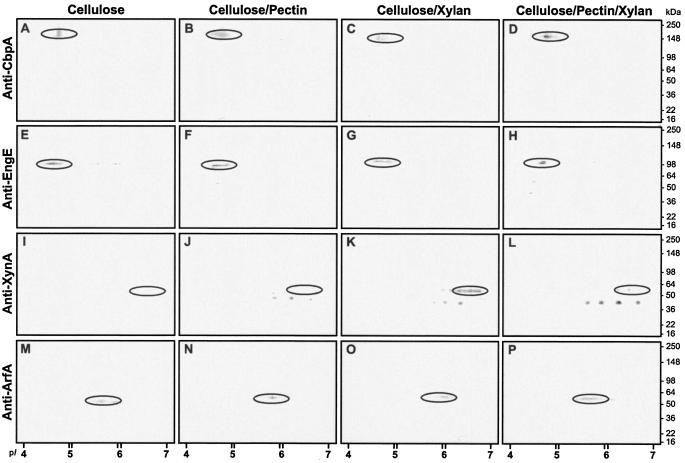
To complement the transcriptional analysis, 2-D PAGE patterns of cellulosomal and noncellulosomal enzymes from cells grown in the presence of cellulose, pectin, and/or xylan were compared. The cellulosomal and noncellulosomal protein spots reproducibly showed differences in abundance under growth with various carbon substrates (Fig. 4). Several cellulosomal proteins appeared to be up-regulated in the cellulose- and cellulose-plus-pectin-induced cultures (Fig. 4A to D). Arrows in Fig. 4 identify all the previously reported proteins, such as CbpA, EngE, EngK, ExgS, and EngL (Table 1). Differences in the amounts of each protein are obvious. The synthesis of CbpA was significantly increased in the presence of cellulose or cellulose plus pectin (Fig. 4A to D, spots 1). Using polyclonal rabbit antibodies directed against CbpA, 2-D Western blots showed a similar high expression pattern of CbpA with cells grown on cellulose or cellulose plus pectin (Fig. 5A to D). The EngE expression pattern was not different in cells grown on cellulose or samples containing cellulose (Fig. 4A to D, spots 2, and 5E to H). The 2-D Western blot analysis showed that XynA was induced only in medium containing xylan (Fig. 5K and L). When subjected to 1-D SDS-PAGE, the relative proportions of certain constituents were dramatically different in cellulosomes from different carbon sources (Fig. 6A). Particularly with the cellulose cultures, the relative proportion of the 60-kDa fraction was the highest, and that of the 65-kDa fraction was the lowest (Fig. 6A, lane 1). The most distinguishable pattern among different carbon source fractions was found with cellulosomes isolated from the CF fraction (Fig. 6A, lane 7).
FIG. 4.
2-D PAGE of cellulosomal (A to D) and noncellulosomal (E to H) fractions of crude culture supernatants from cells grown on various carbon sources. Cells were grown on 1% cellulose (Avicel) (A and E), 1% cellulose-pectin mixture (3:2 [wt/wt]) (B and F), 1% cellulose-xylan mixture (3:2 [wt/wt]) (C and G), and 1% cellulose-pectin-xylan mixture (3:1:1 [wt/wt]) (D and H). The gel images are oriented with the IEF dimension shown on the horizontal axis and the SDS-PAGE dimension on the vertical axis. The approximate pI is indicated along the horizontal axis. The positions of the SDS-PAGE molecular mass standards (in kilodaltons) are presented along the vertical axis. The locations of the enzymes are indicated by the arrows and numbers as follows: in panels A to D, 1, CbpA; 2, EngE; 3, EngK; 4, PelA; 5, ExgS; 6, EngH; 7, EngL; 8, XynA; 9, EngB; and 10, ManA; in panels E to H, 1, BgaA; 2, EngF; 3, EngD; and 4, ArfA. The locations of the EngB (protein 9 in panels A to D) and EngD (protein 3 in panels E to H) proteins are circled.
FIG. 5.
2-D Western blot of C. cellulovorans cellulosomes and noncellulosomal proteins incubated with anti-CbpA (A to D), anti-EngE (E to H), anti-XynA (I to L), and anti-ArfA (M to P). Cellulosomal (A to L) and noncellulosomal (M to P) fractions were isolated from crude culture supernatants from medium containing 1% cellulose (A, E, I, and M), 1% cellulose-pectin mixture (3:2 [wt/wt]) (B, F, J, and N), 1% cellulose-xylan mixture (3:2 [wt/wt]) (C, G, K, and O), and 1% cellulose-pectin-xylan mixture (3:1:1 [wt/wt]) (D, H, L, and P). The locations of the enzyme subunits are circled.
FIG. 6.
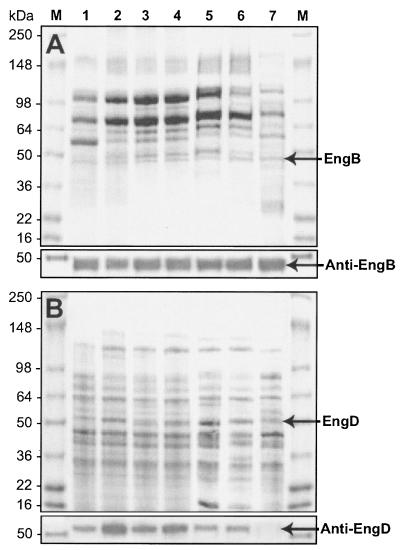
SDS-PAGE analysis and Western blot profile of cellulosomal (A) and noncellulosomal (B) components from cells grown on different carbon sources. Lanes: 1, 1% cellulose (Avicel); 2, 1% cellulose-pectin mixture (3:2 [wt-wt]); 3, 1% cellulose-xylan mixture (3:2 [wt/wt]); 4, 1% cellulose-pectin-xylan mixture (3:1:1 [wt/wt]); 5, 1% xylan; 6, 1% pectin; 7, 1% CF. SDS-PAGE was performed with a 4 to 15% gradient polyacrylamide gel. After electrophoresis, the gels were either stained with Coomassie blue or immunoblotted with anti-EngB or anti-EngD antiserum. The positions of molecular mass markers (M) (in kilodaltons) are indicated to the left of the gels.
Noncellulosomal proteome profiles were also dramatically different in the presence or absence of certain carbon sources such as pectin (Fig. 4E to H). The expression patterns of cells grown on cellulose or cellulose plus xylan (Fig. 4E, G, and H) were very similar. However, the cellulose-pectin mixture-induced culture showed different enzyme profiles (Fig. 4F). Some spots of these cultures were absent (Fig. 4F). In 2-D Western blot analyses, ArfA was detected in most samples tested (Fig. 5M to P). The putative BgaA spot (Fig. 4E to H, spot 1) was significantly different depending on the carbon source. The putative EngF spot was highly induced only by cellulose (Fig. 4E to H, spots 2). The proteins that were absent in certain cultures were probably repressed. The 30-, 45-, 50-, 65-, and 120-kDa polypeptides were more abundant in the noncellulosomal fraction than in the cellulosomal fraction (Fig. 6B). These results showed that the noncellulosomal pattern of proteins was quite different from that of the cellulosomal fraction. The absence or presence of certain protein bands in samples indicated that the occurrence of certain noncellulosomal proteins was dependent on different carbon sources (Fig. 4E to H and 6B).
The appearance of the EngB spot was strongly dependent upon cellulose or cellulose-plus-pectin induction (Fig. 4A, B, and D). The noncellulosomal EngD spot was also regulated by cellulose-plus-pectin induction, but not by cellulose induction (Fig. 4E to H). When subjected to the Western blot analysis, anti-EngB antibody reacted with cellulosomal enzymes from all carbon sources (Fig. 6A). Anti-EngD antibody reacted most with EngD present in the noncellulosomal fractions, except for the noncellulosomal fraction from cells grown on CF (Fig. 6B). The proteins derived from cells grown in cellulose plus pectin reacted most strongly with anti-EngD antibody (Fig. 6B, lanes 2 and 4).
The proteomic profiles of both cellulosomes and noncellulosomal enzymes were dependent on different carbon cultures. The patterns of expression of EngB and EngD were observed to be similar, regardless of the carbon source, even though the EngD band was very prominent when the medium contained the cellulose-pectin mixture.
Synergistic effects between cellulosomes and noncellulosomal proteins during growth on different carbon sources.
To determine whether there is a synergistic relationship between cellulosomes and noncellulosomal enzymes on different carbon sources, we measured degradation activity by using cellulosomal and noncellulosomal enzyme mixtures (1:1, vol/vol). When cellulosome and noncellulosomal enzyme mixtures were incubated with CMC, Avicel, pectin, xylan, CAX, and CF, the amount of reducing sugar released increased significantly compared with that released by the cellulosomal or noncellulosomal fraction alone, and a large synergistic effect was observed on CMC for all cultures grown on different carbon sources (Table 3).
TABLE 3.
Comparison of enzyme activities of cellulosomes and noncellulosomal enzymes and their synergistic effects with different carbon sources
| Carbon source and fractiona | Activity in each fractionb
|
|||||
|---|---|---|---|---|---|---|
| CMCase | Avicelase | Pectate lyase | Xylanase | CAXase | CFase | |
| Cellulose | ||||||
| Cellulosomes | 4.85 | 3.39 | 1.33 | 0.48 | 0.06 | 0.68 |
| Noncellulosomal proteins | 1.29 | 4.22 | 1.60 | 0.62 | 0.15 | 0.84 |
| C-NC mixture | 11.10 | 7.87 | 3.47 | 0.48 | 0.80 | 2.30 |
| Synergy | 1.81 | 1.03 | 1.18 | 0.43 | 3.86 | 1.52 |
| Cellulose-Pectin | ||||||
| Cellulosomes | 3.04 | 1.64 | 0.40 | 0.55 | 0.14 | 0.15 |
| Noncellulosomal proteins | 2.96 | 4.11 | 2.50 | 1.16 | 1.03 | 1.29 |
| C-NC mixture | 14.38 | 8.80 | 3.37 | 3.37 | 1.21 | 1.26 |
| Synergy | 2.40 | 1.53 | 1.16 | 1.96 | 1.04 | 0.87 |
| Cellulose-xylan | ||||||
| Cellulosomes | 4.49 | 2.15 | 0.40 | 1.64 | 0.27 | 0.51 |
| Noncellulosomal proteins | 2.54 | 4.13 | 1.51 | 2.61 | 0.08 | 0.32 |
| C-NC mixture | 17.04 | 9.29 | 3.32 | 4.28 | 0.64 | 1.40 |
| Synergy | 2.42 | 1.48 | 1.73 | 1.01 | 1.83 | 1.68 |
| Cellulose-pectin-xylan | ||||||
| Cellulosomes | 2.44 | 1.14 | 0.45 | 0.49 | 0.05 | 0.19 |
| Noncellulosomal proteins | 2.39 | 3.01 | 2.40 | 1.43 | 0.26 | 1.33 |
| C-NC mixture | 11.19 | 6.94 | 1.49 | 0.60 | 0.76 | 2.59 |
| Synergy | 2.32 | 1.67 | 0.52 | 0.31 | 2.42 | 1.71 |
| Pectin | ||||||
| Cellulosomes | 2.01 | 1.37 | 0.41 | 0.36 | 0.41 | 0.06 |
| Noncellulosomal proteins | 3.83 | 5.69 | 3.67 | 0.52 | 1.26 | 0.40 |
| C-NC mixture | 15.13 | 8.88 | 3.61 | 1.34 | 1.80 | 1.23 |
| Synergy | 2.59 | 1.26 | 0.89 | 1.52 | 1.08 | 2.68 |
| Xylan | ||||||
| Cellulosomes | 2.06 | 0.81 | 0.04 | 0.54 | 0.56 | 0.03 |
| Noncellulosomal proteins | 2.43 | 2.87 | 2.10 | 1.91 | 0.22 | 0.80 |
| C-NC mixture | 10.18 | 5.61 | 1.95 | 3.11 | 2.10 | 0.94 |
| Synergy | 2.26 | 1.53 | 0.91 | 1.27 | 2.67 | 1.13 |
| CF | ||||||
| Cellulosomes | 2.17 | 1.05 | 0.24 | 0.05 | 0.27 | 0.11 |
| Noncellulosomal proteins | 4.87 | 2.20 | 2.04 | 0.93 | 1.73 | 0.47 |
| C-NC mixture | 16.74 | 6.18 | 3.47 | 1.37 | 2.01 | 2.62 |
| Synergy | 2.38 | 1.90 | 1.53 | 1.40 | 1.01 | 4.53 |
The fractions were purified from C. cellulovorans cultivated on medium containing 0.5% cellobiose, 1% cellulose (Avicel), 1% pectin, 1% xylan, 1% cellulose-pectin mixture (cellulose/pectin ratio of 3:2 [wt/wt]), 1% cellulose-xylan mixture (cellulose/xylan ratio of 3:2 [wt/wt]), 1% cellulose-pectin-xylan mixture (cellulose/pectin/xylan ratio of 3:1:1 [wt/wt]), or 1% CF as the sole carbon source. The fractions are cellulosomes, noncellulosomal proteins, and a mixture of cellulosomes and noncellulosomal proteins (C-NC mixture). The synergy in a mixture of cellulosomes and noncellulosomal proteins (cellulosome/noncellulosomal protein ratio of 1:1 [wt/wt]) was calculated by dividing the activity in the mixture by the activity in cellulosomes and noncellulosomal proteins.
The activities of CMCase, pectate lyase, xylanase, and CAXase are shown in milliunits per milliliter. The Avicelase activity and CFase activity are shown in milliunits per hour.
On the other hand, although the pectate lyase or xylanase synergistic effects by cellulosomal and noncellulosomal enzyme mixtures were not detected for some substrate polymers, increases of synergistic coefficient values were observed for cellulose-pectin and CF when the fraction mixtures were used. Of special interest was the increase of degradation activity observed with cellulosomal and noncellulosomal enzyme mixtures from the culture grown on cellulose, pectin, and xylan when CAX and CF were the substrates (Table 3). These results suggest that the cellulosomes and noncellulosomal enzymes may be induced, depending on the carbon source in the growth medium, and that both cellulosomes and noncellulosomal enzymes synergistically contribute to plant cell wall depolymerization by C. cellulovorans.
DISCUSSION
Cellulose, xylan, and pectin exist in nature in close proximity in plant cell walls, and C. cellulovorans has enzymes to ensure efficient utilization of these complex polymers (16, 17, 19, 20, 28). Various natural substrates may induce secreted enzymes that can degrade very precisely particular combinations of polysaccharides and chemical bonds found in a carbon substrate (13, 18, 24, 26, 27, 29). In order to further elucidate the influence of the composition of complex plant cell walls on enzyme production, artificial mixed substrates containing xylan and/or pectin with cellulose were investigated in this study. C. cellulovorans was grown on these different substrates alone or in mixture, and the relationship between carbon source, growth, and enzyme composition was investigated. The data presented in this paper demonstrate specific and common regulatory patterns in the expression of cellulosomes and noncellulosomal enzymes by C. cellulovorans and additionally due to simultaneous analyses, a picture of their relative expression levels under different culture conditions. It was reported previously that carbon sources in the medium have an effect on the subunit composition of the cellulosome (25, 42). Our recent report described the transcriptional analysis of cellulases and their regulation at the mRNA level of C. cellulovorans (16, 17). However, the desired end point for elucidation of the systematic regulation of cellulosomes and noncellulosomal enzymes is not only the analysis of mRNA transcript levels but also the accurate measurement of protein expression profiles and their respective activities under different growth conditions. This study examined the relationship between mRNA and protein expression levels along with their activity for cellulosomes and noncellulosomal enzymes under different culture conditions.
The largest number of cellulosomal enzymes was observed when cells were grown on mixtures containing pectin and xylan, indicating that the inducers in mixed substrates stimulated wider gene expression (Fig. 4). The highest levels of cellulosomal subunits, such as CbpA, EngE, and ExgS, occurred during cultivation on cellulose or when cellulose was one of the components in a substrate mixture (Fig. 1, 4, and 6). It was found that the levels of major enzyme subunits were generally higher in cells grown on cellulose-pectin mixed substrates than those of cells grown on a single substrate. The general trend observed was that a higher ratio of cellulose in the mixture resulted in higher levels of cellulase production (Fig. 1 and 2). The highest Avicelase activity was also found in cellulosomal fractions purified from Avicel cultures (Table 3). On the other hand, the CMCase activity for the cellulosomal fractions did not show a large variation with the different carbon sources used. These results suggested that the major cellulosomal subunits, CbpA, EngE, and ExgS, are constitutively expressed and can be induced by certain carbon sources, such as Avicel. Very low levels of xynA transcripts were shown to be present in uninduced cells, and these transcripts were induced in the presence of xylan (Fig. 1 and 5). Significantly higher levels of pelA were measured only with cultures grown on pectin or a cellulose-pectin mixture (Fig. 1). Thus, there appear to be (hemi)cellulose-degrading mechanisms in C. cellulovorans which mediate partial and strict control of expression of various genes encoding different extracellular hydrolases.
To analyze the synergistic effect between cellulosomes and noncellulosomal enzymes, various enzyme fractions were mixed and tested for activity on different carbon sources. A broad range of cellulosomes and noncellulosomal enzymes from cells grown on different carbon sources retained relatively high activity against CMC, Avicel, pectin, xylan, CAX, and CF, as shown in Table 3. Interestingly, the enzyme mixture of the cellulosomal fraction and noncellulosomal fraction showed the highest specific activity and synergy degrees against natural substrates, such as CF. These results implied that there is an advantage of associating cellulosomes and noncellulosomal enzymes for the efficient degradation of a mixed carbon source, such as plant cell walls. It was found that higher enzyme levels generally resulted when the carbon source used for growth was a mixture of the carbon substrates. On the other hand, the synergism between cellulosomal enzymes (EngE, EngH, EngM, and ExgS) and noncellulosomal enzymes (EngD and EngF) on the degradation of CMC can be considered the most dominant effect on the degradation of CMC by mixed endoglucanases. These results imply that synergy between cellulosomes and noncellulosomal enzymes allows C. cellulovorans to vary the cellulosome and noncellulosomal subunit composition while retaining high activity against certain carbon substrates.
Little is known about the regulatory mechanism of cellulosomes and noncellulosomal enzymes in C. cellulovorans, but this study showed that both enzyme fractions were coordinately expressed with specific carbon sources. Certain carbon sources provoked high levels of expression of one protein or set of proteins, while the effect on expression of other proteins was weaker or insignificant, and this pattern could be reversed with other carbon sources. Understanding the roles and relationships of cellulosomes and noncellulosomal proteins acting on specific substrates is important for the development of an efficient artificial enzyme system for the conversion of cellulosic biomass to valuable sugars. One of our ultimate goals is the preparation of designer cellulosomes, which could degrade cellulose efficiently for industrial purposes. Studying the roles and relationships of enzyme components from the C. cellulovorans cellulase system acting on complex substrates is a key to the development of artificial cellulase and hemicellulase combinations targeted to biomasses with particular carbon compositions.
Acknowledgments
We thank David B. Johnston of the U.S. Department of Agriculture for the gift of CF and CAX. We are also grateful to Young Jin Lee and Young Moo Lee for providing access to the molecular structure facility.
This research was supported in part by the Research Institute of Innovative Technology for the Earth (RITE), Japanese Ministry of Economy, Trade and Industry (METI), and grant DE-DDF03-92ER20069 from the U.S. Department of Energy.
REFERENCES
- 1.Anderson, N. G., and N. L. Anderson. 1978. Analytical techniques for cell fractions. XXI. Two-dimensional analysis of serum and tissue proteins: multiple isoelectric focusing. Anal. Biochem. 85:331-340. [DOI] [PubMed] [Google Scholar]
- 2.Bayer, E. A., Y. Shoham, and R. Lamed. 2000. Cellulose-decomposing bacteria and their enzyme systems, p. 1-74. In M. Dworkin, S. Falkow, E. Rosenberg, K.-H. Schleifer, and E. Stackebrandt (ed.), The prokaryotes: an evolving electronic resource for the microbiological community, 3rd ed. Springer-Verlag, New York, N.Y.
- 3.Boisset, C., H. Chanzy, B. Henrissat, R. Lamed, Y. Shoham, and E. A. Bayer. 1999. Digestion of crystalline cellulose substrates by the Clostridium thermocellum cellulosome: structural and morphological aspects. Biochem. J. 340:829-835. [PMC free article] [PubMed] [Google Scholar]
- 4.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 5.Doi, R. H., A. Kosugi, K. Murashima, Y. Tamaru, and S. O. Han. 2003. Cellulosomes from mesophilic bacteria. J. Bacteriol. 185:5907-5914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doi, R. H., J. S. Park, C. C. Liu, L. M. Malburg, Y. Tamaru, A. Ichiishi, and A. Ibrahim. 1998. Cellulosome and noncellulosomal cellulases of Clostridium cellulovorans. Extremophiles 2:53-60. [DOI] [PubMed] [Google Scholar]
- 7.Doi, R. H., and Y. Tamaru. 2001. The Clostridium cellulovorans cellulosome: an enzyme complex with plant cell wall degrading activity. Chem. Rec. 1:24-32. [DOI] [PubMed] [Google Scholar]
- 8.Doner, L. W., H. K. Chau, M. L. Fishman, and K. B. Hicks. 1998. An improved process for isolation of corn fiber gum. Cereal Chem. 75:408-411. [Google Scholar]
- 9.Doner, L. W., and K. B. Hicks. 1997. Isolation of hemicellulose from corn fiber by alkaline hydrogen peroxide extraction. Cereal Chem. 74:176-181. [Google Scholar]
- 10.Doner, L. W., D. B. Johnston, and V. Singh. 2001. Analysis and properties of arabinoxylans from discrete corn wet-milling fiber fractions. J. Agric. Food. Chem. 49:1266-1269. [DOI] [PubMed] [Google Scholar]
- 11.Foong, F., T. Hamamoto, O. Shoseyov, and R. H. Doi. 1991. Nucleotide sequence and characteristics of endoglucanase gene engB from Clostridium cellulovorans. J. Gen. Microbiol. 137:1729-1736. [DOI] [PubMed] [Google Scholar]
- 12.Gorg, A., C. Obermaier, G. Boguth, A. Harder, B. Scheibe, R. Wildgruber, and W. Weiss. 2000. The current state of two-dimensional electrophoresis with immobilized pH gradients. Electrophoresis 21:1037-1053. [DOI] [PubMed] [Google Scholar]
- 13.Greenberg, N. M., R. A. Warren, D. G. Kilburn, and R. C. Miller, Jr. 1987. Regulation and initiation of cenB transcripts of Cellulomonas fimi. J. Bacteriol. 169:4674-4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hamamoto, T., F. Foong, O. Shoseyov, and R. H. Doi. 1992. Analysis of functional domains of endoglucanases from Clostridium cellulovorans by gene cloning, nucleotide sequencing and chimeric protein construction. Mol. Gen. Genet. 231:472-479. [DOI] [PubMed] [Google Scholar]
- 15.Hamamoto, T., O. Shoseyov, F. Foong, and R. H. Doi. 1990. A Clostridium cellulovorans gene, engD, codes for both endo-β-1,4-glucanase and cellobiosidase activities. FEMS Microbiol. Lett. 72:285-288. [Google Scholar]
- 16.Han, S. O., H. Yukawa, M. Inui, and R. H. Doi. 2003. Regulation of expression of cellulosomal cellulase and hemicellulase genes in Clostridium cellulovorans. J. Bacteriol. 185:6067-6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han, S. O., H. Yukawa, M. Inui, and R. H. Doi. 2003. Transcription of Clostridium cellulovorans cellulosomal cellulase and hemicellulase genes. J. Bacteriol. 185:2520-2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ilmen, M., A. Saloheimo, M. L. Onnela, and M. E. Penttila. 1997. Regulation of cellulase gene expression in the filamentous fungus Trichoderma reesei. Appl. Environ. Microbiol. 63:1298-1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kosugi, A., K. Murashima, and R. H. Doi. 2002. Characterization of two noncellulosomal subunits, ArfA and BgaA, from Clostridium cellulovorans that cooperate with the cellulosome in plant cell wall degradation. J. Bacteriol. 184:6859-6865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kosugi, A., K. Murashima, and R. H. Doi. 2001. Characterization of xylanolytic enzymes in Clostridium cellulovorans: expression of xylanase activity dependent on growth substrates. J. Bacteriol. 183:7037-7043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kosugi, A., K. Murashima, and R. H. Doi. 2002. Xylanase and acetyl xylan esterase activities of XynA, a key subunit of the Clostridium cellulovorans cellulosome for xylan degradation. Appl. Environ. Microbiol. 68:6399-6402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 23.Liu, C. C., and R. H. Doi. 1998. Properties of exgS, a gene for a major subunit of the Clostridium cellulovorans cellulosome. Gene 211:39-47. [DOI] [PubMed] [Google Scholar]
- 24.Lockington, R. A., L. Rodbourn, S. Barnett, C. J. Carter, and J. M. Kelly. 2002. Regulation by carbon and nitrogen sources of a family of cellulases in Aspergillus nidulans. Fungal Genet. Biol. 37:190-196. [DOI] [PubMed] [Google Scholar]
- 25.Matano, Y., J. S. Park, M. A. Goldstein, and R. H. Doi. 1994. Cellulose promotes extracellular assembly of Clostridium cellulovorans cellulosomes. J. Bacteriol. 176:6952-6956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mitchell, W. J. 1998. Physiology of carbohydrate to solvent conversion by clostridia. Adv. Microb. Physiol. 39:31-130. [DOI] [PubMed] [Google Scholar]
- 27.Mittendorf, V., and J. A. Thomson. 1995. Transcriptional induction and expression of the endoglucanase celA gene from a ruminal Clostridium sp. (“ C. longisporum”). J. Bacteriol. 177:4805-4808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murashima, K., A. Kosugi, and R. H. Doi. 2002. Determination of subunit composition of Clostridium cellulovorans cellulosomes that degrade plant cell walls. Appl. Environ. Microbiol. 68:1610-1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nochur, S. V., M. F. Robert, and A. L. Demain. 1993. True cellulase production by Clostridium thermocellum grown on different carbon sources. Biotechnol. Lett. 15:641-646. [Google Scholar]
- 30.Nolling, J., G. Breton, M. V. Omelchenko, K. S. Makarova, Q. Zeng, R. Gibson, H. M. Lee, J. Dubois, D. Qiu, J. Hitti, Y. I. Wolf, R. L. Tatusov, F. Sabathe, L. Doucette-Stamm, P. Soucaille, M. J. Daly, G. N. Bennett, E. V. Koonin, and D. R. Smith. 2001. Genome sequence and comparative analysis of the solvent-producing bacterium Clostridium acetobutylicum. J. Bacteriol. 183:4823-4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schwarz, W. H. 2001. The cellulosome and cellulose degradation by anaerobic bacteria. Appl. Microbiol. Biotechnol. 56:634-649. [DOI] [PubMed] [Google Scholar]
- 32.Sheweita, S. A., A. Ichi-ishi, J. S. Park, C. Liu, L. M. Malburg, Jr., and R. H. Doi. 1996. Characterization of engF, a gene for a noncellulosomal Clostridium cellulovorans endoglucanase. Gene 182:163-167. [DOI] [PubMed] [Google Scholar]
- 33.Shoseyov, O., and R. H. Doi. 1990. Essential 170-kDa subunit for degradation of crystalline cellulose by Clostridium cellulovorans cellulase. Proc. Natl. Acad. Sci. USA 87:2192-2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shoseyov, O., M. Takagi, M. A. Goldstein, and R. H. Doi. 1992. Primary sequence analysis of Clostridium cellulovorans cellulose binding protein A. Proc. Natl. Acad. Sci. USA 89:3483-3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sleat, R., R. A. Mah, and R. Robinson. 1984. Isolation and characterization of an anaerobic, celluloytic bacterium, Clostridium cellulovorans sp. nov. Appl. Environ. Microbiol. 48:88-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Somogyi, M. 1952. Notes on sugar determination. J. Biol. Chem. 195:19-23. [PubMed] [Google Scholar]
- 37.Taiz, L., and E. Zeiger. 1991. Plant and cell architecture, p. 9-25. In Plant physiology. The Benjamin Cummings Publishing Company, Inc., Redwood City, Calif.
- 38.Tamaru, Y., and R. H. Doi. 2000. The engL gene cluster of Clostridium cellulovorans contains a gene for cellulosomal manA. J. Bacteriol. 182:244-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tamaru, Y., and R. H. Doi. 2001. Pectate lyase A, an enzymatic subunit of the Clostridium cellulovorans cellulosome. Proc. Natl. Acad. Sci. USA 98:4125-4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tamaru, Y., and R. H. Doi. 1999. Three surface layer homology domains at the N terminus of the Clostridium cellulovorans major cellulosomal subunit EngE. J. Bacteriol. 181:3270-3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tamaru, Y., S. Karita, A. Ibrahim, H. Chan, and R. H. Doi. 2000. A large gene cluster for the Clostridium cellulovorans cellulosome. J. Bacteriol. 182:5906-5910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tamaru, Y., S. Ui, K. Murashima, A. Kosugi, H. Chan, R. H. Doi, and B. Liu. 2002. Formation of protoplasts from cultured tobacco cells and Arabidopsis thaliana by the action of cellulosomes and pectate lyase from Clostridium cellulovorans. Appl. Environ. Microbiol. 68:2614-2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tokatlidis, K., P. Dhurjati, and P. Beguin. 1993. Properties conferred on Clostridium thermocellum endoglucanase CelC by grafting the duplicated segment of endoglucanase CelD. Protein Eng. 6:947-952. [DOI] [PubMed] [Google Scholar]
- 44.Wilm, M., A. Shevchenko, T. Houthaeve, S. Breit, L. Schweigerer, T. Fotsis, and M. Mann. 1996. Femtomole sequencing of proteins from polyacrylamide gels by nano-electrospray mass spectrometry. Nature 379:466-469. [DOI] [PubMed] [Google Scholar]
- 45.Wood, W. A., and K. M. Bhat. 1988. Methods for measuring cellulose activities. Methods Enzymol. 160:87-112. [Google Scholar]
- 46.Ximenes, E., and F. Filho. 1998. Hemicellulases and biotechnology, p. 165-176. In S. G. Pandalai (ed.), Recent research developments in microbiology. Research Signpost, Trivandrum, Kerala, India.
- 47.Yaron, S., L. J. Shimon, F. Frolow, R. Lamed, E. Morag, Y. Shoham, and E. A. Bayer. 1996. Expression, purification and crystallization of a cohesin domain from the cellulosome of Clostridium thermocellum. J. Biotechnol. 51:243-249. [DOI] [PubMed] [Google Scholar]
- 48.Yoshida, K., K. Kobayashi, Y. Miwa, C. M. Kang, M. Matsunaga, H. Yamaguchi, S. Tojo, M. Yamamoto, R. Nishi, N. Ogasawara, T. Nakayama, and Y. Fujita. 2001. Combined transcriptome and proteome analysis as a powerful approach to study genes under glucose repression in Bacillus subtilis. Nucleic Acids Res. 29:683-692. [DOI] [PMC free article] [PubMed] [Google Scholar]