Abstract
Haemophilus somnus strain 649 was found to acquire iron from ovine, bovine, and goat transferrins (Tfs). Expression of Tf receptors, as evaluated by solid-phase binding assays, required the organisms to be grown under iron-restricted conditions in the presence of Tf. Competition binding assays revealed the presence of two distinct Tf-binding receptor systems, one specific for bovine Tf and the other capable of binding all three ruminant Tfs. Affinity isolation procedures using total membranes yielded three putative bovine Tf-binding polypeptides and one putative ovine and goat Tf-binding polypeptide. PCR amplification followed by DNA sequence analyses revealed that H. somnus strain 649 possesses genes that encode a bipartite TbpA-TbpB receptor along with a homolog of the Histophilus ovis single-component TbpA receptor. Expression of TbpB and the single-component TbpA would appear to be subject to a form of phase variation involving homopolymeric nucleotide tracts within the structural genes.
Iron-requiring pathogens are able to survive in the extremely iron-limiting extracellular environments of their hosts (9) by virtue of their abilities to acquire iron from host components, such as transferrin (Tf). Many members of the Pasteurellaceae acquire Tf-bound iron by using a receptor-mediated mechanism that involves a bipartite receptor complex composed of two outer membrane proteins, TbpA and TbpB (8). In most organisms, the genes encoding the Tbps are arranged in an operon, with tbpB preceding tbpA (e.g., see references 6, 7, 11, and 18). In Pasteurella multocida and Histophilus ovis, the acquisition of Tf-bound iron involves a single-component outer membrane receptor, also referred to as TbpA (4, 17). While these single-component receptors are related to the TbpA proteins of bipartite systems, they represent a new subfamily of TonB-dependent receptors (17), and for the sake of clarity, we propose that a single-component TbpA receptor and its encoding gene be referred to as TbpA2 and tbpA2, respectively. This terminology is used below.
Unlike the expression of most Tbps, which are induced by conditions of iron restriction (e.g., see references 14, 15, 17, 20, and 21), the TbpA2 proteins of some strains of H. ovis appear to be expressed only when the organisms are grown under iron-restricted conditions in the presence of a suitable Tf (3). Transcription of the genes encoding such proteins is regulated by the amount of iron in the growth medium (4), suggesting the involvement of ferric uptake regulator (Fur) proteins, which, in the presence of iron, would repress the transcription of the tbpA2 genes, and for one strain of H. ovis (strain 3384Y), the apparent requirement for Tf in the growth medium for the expression of TbpA2 has been linked to the number of guanine (G) residues in a specific poly(G) tract within tbpA2 (5). When this strain is grown under iron-replete conditions, the poly(G) tract contains nine G's, introducing a premature stop codon into the reading frame, but when grown under iron-restricted conditions in the presence of bovine Tf, the poly(G) tract contains 8 G's, allowing the translation of a full-length protein (5). Presumably, the latter growth conditions select for a subpopulation of cells that possess the appropriate number of G's.
H. ovis and the bovine pathogen Haemophilus somnus are very closely related (19, 23, 24), and it has now been proposed that they be assigned to a single species and renamed Histophilus somni (1). While interstrain differences are not unexpected, it is notable that while H. ovis is capable of acquiring iron from ovine, bovine, and goat Tfs (3), H. somnus is reported to be capable of acquiring iron only from bovine Tf (26). These differences, and the finding that Tf receptors can be subject to phase variation (5), prompted us to initiate studies relating to the acquisition of Tf-bound iron by the H. somnus strains that we have in our possession, the objective being to determine if these strains resemble H. ovis or the H. somnus strains described by Yu et al. (26). The present communication deals with H. somnus strain 649. This strain was isolated from an aborted bovine fetus and has been shown to cause abortion experimentally (25).
Iron acquisition from transferrins.
Plate assays, as described previously (3), were used to investigate the ability of H. somnus strain 649 to acquire iron from a variety of iron-saturated Tfs (40-μl volumes; 4 mg/ml). Obvious growth of H. somnus strain 649 was noted around disks containing bovine, ovine, and goat Tf but not around disks containing porcine or human Tf (results not shown). These results were in contrast to those described previously for other strains of H. somnus (26) and demonstrated that strain 649 exhibits a transferrin specificity that mimics that of H. ovis (3), P. multocida (17), and also Mannheimia (Pasteurella) haemolytica (26).
Transferrin-binding assays.
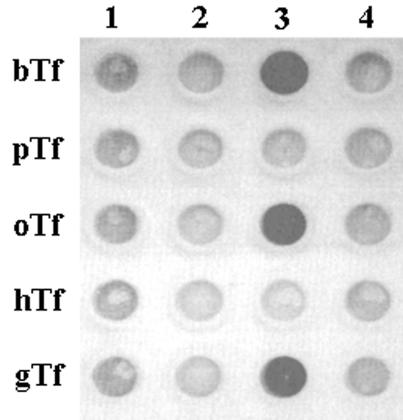
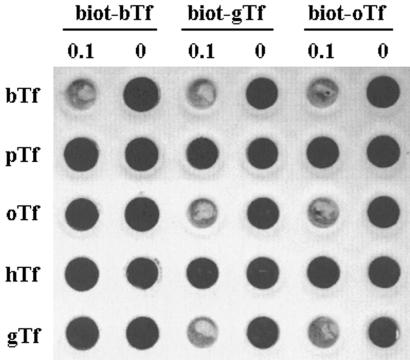
H. somnus strain 649 was grown under iron-replete and iron-restricted conditions and in the presence and absence of bovine Tf (to 80 mg/liter), essentially as described previously for H. ovis (3). For growth under iron-restricted conditions, the basic growth medium (supplemented, HEPES-buffered tryptone-yeast extract medium [3]) was supplemented with ∼15 μM ethylenediamine di-o-hydroxyphenylacetic acid, and for growth under iron-restricted conditions in the presence of Tf, sTYE-H was supplemented with 25 μM ethylenediamine di-o-hydroxyphenylacetic acid. Total membranes were prepared from these cells, essentially as described by Niven et al. (13), and solid-phase binding assays, using biotinylated Tfs as ligands and streptavidin-horseradish peroxidase as a detection agent, were performed as described by Ekins and Niven (3). Interestingly, Tf binding, with a specificity that reflected that observed in the growth assays, was evident only with membranes derived from organisms grown under iron-restricted conditions in the presence of bovine Tf (Fig. 1). These results suggest that in H. somnus strain 649, as in H. ovis strain 3384Y (3, 4, 5), the expression of Tf receptors is iron regulated and that the presence of a suitable Tf in the growth medium selects for a subpopulation of receptor-producing cells. To determine if the ruminant Tfs were bound by the same receptor(s), competition binding assays were initiated using membranes derived from organisms grown under iron-restricted conditions in the presence of bovine Tf. Excess native bovine Tf, but not ovine or goat Tf, was able to block the binding of biotinylated bovine Tf, and when biotinylated ovine or goat Tf was used as the labeled ligand, all three native ruminant Tfs were able to block the binding of the biotinylated Tf (Fig. 2). In effect, it would appear that H. somnus strain 649 possesses two distinct Tf receptors, one specific for bovine Tf and the other capable of binding any of the tested ruminant Tfs.
FIG. 1.
Dot blot demonstrating binding of the indicated Tfs by total membranes from H. somnus strain 649 grown under iron-replete (lane 1) and iron-restricted (lane 2) conditions and under iron-replete (lane 4) and iron-restricted (lane 3) conditions in the presence of bovine Tf. The Tfs are abbreviated as follows: b, bovine; p, porcine; o, ovine; h, human; g, goat.
FIG. 2.
Dot blot demonstrating competition between native Tfs and biotinylated bovine, goat, and ovine Tfs for binding sites on total membranes from H. somnus strain 649 grown under iron-restricted conditions in the presence of bovine Tf. The competing Tfs, abbreviated as in Fig. 1, are indicated on the left side of the figure, and the presence (0.1 [mg]) and absence (0) of the competing Tfs are indicated along the top.
Isolation and identification of transferrin-binding polypeptides.
Tf-binding polypeptides were isolated from total membranes of H. somnus strain 649 by using the affinity procedure developed by Schryvers and Morris (22), as modified by Ricard et al. (20). The membranes used in these experiments were from organisms grown under iron-restricted conditions in the presence of Tf, since such growth conditions were the only ones that yielded organisms exhibiting significant Tf-binding activity. Notably, while the use of biotinylated bovine Tf as the binding ligand in the affinity procedure allowed the isolation of five polypeptides (∼112, 82, 76, 70, and 64 kDa), the use of biotinylated ovine or goat Tf allowed the isolation of only two (∼82 and 64 kDa) (Fig. 3). Based on the apparent molecular masses of the Tbps of H. somnus (14, 27), H. ovis (3), and P. multocida (17), it would not seem unreasonable to suggest that the 112- and 70-kDa polypeptides that were isolated only with bovine Tf represent TbpA and TbpB homologs, respectively, and that the 82-kDa polypeptide, isolated with all three ruminant Tfs, represents a TbpA2 homolog. Regarding the 76-kDa polypeptide, we have demonstrated, in comparable experiments, that the biotinylated binding ligand can be present in the affinity-isolated materials (2), and we suspect that this is also the case here. The 64-kDa polypeptide that was isolated using all three ruminant Tfs is reminiscent of the 66-kDa polypeptide that was isolated from H. ovis under comparable conditions (3), but as with the H. ovis polypeptide, the significance of the 64-kDa H. somnus polypeptide remains obscure. In brief, we believe that a classical bipartite TbpA-TbpB receptor complex is affinity isolated with bovine, but not ovine or goat, Tf and that a single-component TbpA2 receptor is isolated when any of the three ruminant Tfs is used as the binding ligand.
FIG. 3.
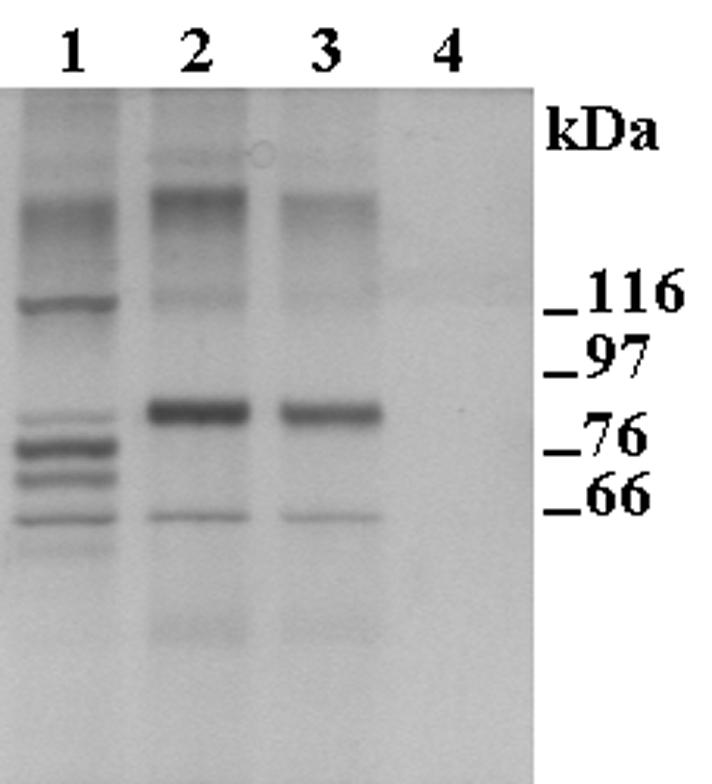
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis of polypeptides affinity isolated from total membranes of H. somnus strain 649 grown under iron-restricted conditions in the presence of bovine Tf. The polypeptides were isolated using biotinylated bovine (lane 1), goat (lane 2), and ovine (lane 3) Tfs as binding ligands. Lane 4 represents a control sample obtained when the affinity isolation procedure was performed in the absence of biotinylated Tf. The numbers refer to the sizes and positions of protein standards.
Identification and sequencing of a tbpA2 homolog.
To investigate the suggested existence of a tbpA2 homolog in H. somnus strain 649, primers (forward, 5′-GATTAAAAAACTTGACAGTTAGGGC; reverse, 5′-GATATTGCCATTGTGACGTTCG) based on the nucleotide sequence of H. ovis tbpA2 (4) (accession no. AY040784) were used in PCR with H. somnus DNA. Amplification yielded a product with a size that was consistent with the presence of a tbpA2 homolog, and additional primers, also based on the nucleotide sequence of H. ovis tbpA2, were then used in PCR to amplify additional portions of the H. somnus strain 649 tbpA2. Direct genomic sequencing (10) was used to obtain single-stranded sequence upstream and downstream of tbpA2, and primers based on the resulting sequences were used to amplify these upstream and downstream regions. The resulting PCR products, purified using the QIAquick PCR purification kit (QIAGEN, Mississauga, Ontario, Canada), were used directly as templates in double-stranded sequencing (BigDye sequencing kit; PE Biosystems, Foster City, Calif.). While assembling the sequence of H. somnus strain 649 tbpA2, we encountered a stretch of eight G's in the coding sequence that would introduce a premature stop codon into the reading frame of the gene. Based on our experiences with H. ovis strain 3384Y tbpA2 (5), it was hypothesized that the expression of TbpA2 by H. somnus strain 649, as in H. ovis strain 3384Y, may be subject to a form of phase variation involving the poly(G) tract within the coding region of the gene. The DNA used to amplify the tbpA2 gene was isolated from organisms grown under iron-replete conditions, and under these conditions there would be no selective pressure for the organism to produce a full-length TbpA2 precursor and functional TbpA2. To investigate this possibility, buffered cell suspensions (1-μl aliquots) of organisms grown under iron-restricted conditions in the presence of goat Tf were used directly in PCR with primers flanking the poly(G) tract. The resulting amplification product was sequenced on both strands, and sequence analysis revealed that the poly(G) tract now contained nine G's, eliminating the premature stop codon and allowing the production of a full-length TbpA2 precursor. A putative cleavage site, resulting in a mature TbpA2 protein, was identified using SignalP V2.0.b2 (http://www.cbs.dtu.dk/services/SignalP-2.0/) (12), and the deduced amino acid sequence of the predicted mature TbpA2 protein was found to share 84% and 73% identity with the TbpA2 proteins of H. ovis (protein accession no. AAK68653.1 and AAK68655.1) and P. multocida (protein accession no. AAG15587.1), respectively. Interestingly, the putative promoter region of H. somnus strain 649 tbpA2 (accession no. AY184230) was also found to be identical to the promoter regions of the two sequenced H. ovis tbpA2 genes (accession no. AY040784 and AY040785), and the uncommon start codon, TTG, is also predicted to initiate translation. Finally, the molecular mass of the predicted mature TbpA2 of H. somnus strain 649 was calculated (Compute pI/Mw [http://ca.expasy.org/tools/pi_tool.html]) to be 83.8 kDa, suggesting strongly that the 82-kDa polypeptide that was isolated with all three ruminant Tfs does represent the TbpA2 protein.
Identification and sequencing of tbpA and tbpB homologs.
Since the presence of two systems for the acquisition of Tf-bound iron is quite novel, we were concerned that such an effect might be due to a mixed culture of organisms, with one organism expressing a bovine Tf-specific receptor and another expressing a Tf receptor capable of binding all three ruminant Tfs. In order to exclude this possibility, samples from an isolated colony of H. somnus strain 649, grown on solid sTYE-H, were used as templates in PCR with primers specific for either tbpA2 or tbpBA. The tbpA2-specific primers (forward, 5′-TGGGTTATCTTGGTTAGAAACAGC; reverse, 5′-CTTGGCGTGACATCTTTCACGTTC) were based on tbpA2 of H. ovis strain 9L (accession no. AY040784). The tbpBA-specific primers were based on the nucleotide sequence of tbpBA of a bovine isolate of H. somnus (strain HS25) along with the conserved amino acid sequences that were used by Ogunnariwo and Schryvers (16) to design degenerate primers 193 (within tbpB) and 223 (within tbpA). The tbpBA sequence (H. somnus strain HS25) has yet to be published and was generously provided to us by A. A. Potter (Veterinary Infectious Disease Organization, Saskatoon, Saskatchewan, Canada). PCR, using the material from the isolated colony as a template and the tbpA2- and tbpBA-specific primer pairs, resulted in the amplification of appropriately sized fragments of ∼750 and ∼1,300 bp, respectively. To ensure that the single ∼1,300-bp amplification product did indeed represent tbpB and tbpA homologs, this product was used directly in sequencing reactions. The acquired nucleotide sequence was found to be highly homologous to that of H. somnus strain HS25 tbpBA as well as to the corresponding sequences in other organisms possessing these genes (results not shown). While these results indicated that H. somnus strain 649 does indeed possess tbpA and tbpB homologs, in addition to a tbpA2 homolog, sequencing of tbpA, tbpB, and flanking regions was continued to permit a comparison of deduced molecular masses of predicted, mature TbpA and TbpB proteins with the apparent molecular masses of the affinity-isolated polypeptides and perhaps to provide some insight into the Tf-dependent expression of Tf-binding activity. DNA from strain 649 grown under iron-replete conditions was used as a template, and sequencing and sequence analyses were performed essentially as described above for tbpA2. Based on the acquired sequences (accession no. AY260102), tbpA was predicted to encode a TbpA precursor yielding a mature TbpA with a deduced molecular mass of 104.9 kDa, but tbpB was predicted to encode a truncated TbpB precursor consisting of only 60 amino acids. Interestingly, however, while a poly(C) tract upstream of the apparently premature stop codon was noted to contain 10 C's, the comparable poly(C) tract in tbpB of H. somnus strain HS25 contains only 8 C's. Since 8 C's appear to maintain the correct reading frame of the gene, it was suspected that the expression of TbpB by strain 649 is also subject to a form of phase variation and that the apparently Tf-dependent expression of Tf-binding activity involves the poly(C)tract within tbpB. To determine if this is the case, strain 649 was grown under iron-restricted conditions in the presence of bovine Tf, and culture samples were serially diluted and spread on solid sTYE-H. Following incubation, isolated colonies were suspended in 10 mM HEPES, pH 7.4 (100-μl volumes), and 5-μl volumes of these suspensions were used directly in PCR with primers flanking the poly(C) tract. The resulting amplification products were sequenced on both strands, and sequence analyses revealed that depending on the colony, the poly(C) tract now contained 9, 10, or 11 C's. Notably, 11 C's would eliminate the premature stop codon and allow the production of a full-length TbpB precursor. The molecular mass of the corresponding, mature TbpB was calculated to be 66.6 kDa, and this, plus the deduced molecular mass of the predicted, mature TbpA (104.9 kDa), suggest strongly that the 112- and 70-kDa affinity-isolated polypeptides do represent TbpA and TbpB, respectively.
Finally, an examination of the partially complete H. somnus strain 129PT genome sequence (DOE Joint Genome Institute; available at www.ncbi.nlm.nih.gov) suggests that the expression of two systems for the acquisition of Tf-bound iron may not be restricted to H. somnus strain 649. Strain 129PT would appear to possess tbpA and tbpB homologs and also a predicted open reading frame of 445 amino acids that shares 98% identity with the last two-thirds of the predicted TbpA2 protein of strain 649. It is tempting to speculate that this stretch of 445 amino acids represents a truncated form of the putative TbpA2 of strain 129PT, with truncation, as in H. somnus strain 649 and H. ovis strain 3384Y (5), being related to a frame shift within the reading frame of the structural gene. Also, since it would appear now that the phase-variable expression of Tf receptors is not restricted to H. ovis, it is possible that the production of bipartite (TbpA-TbpB) and single-component (TbpA2) Tf receptor systems may be more widespread than we realize. In brief, it seems prudent to suggest that in all future studies relating to the production of Tf receptors, the receptor complement should also be determined following growth of the bacteria in the presence of an appropriate Tf.
Nucleotide sequence accession numbers.
The nucleotide sequences of tbpA2 and tbpBA of H. somnus strain 649 were submitted to GenBank and have been assigned accession numbers AY184230 and AY260102, respectively.
Acknowledgments
We are grateful to L. B. Corbeil for provision of H. somnus strain 649, to A. A. Potter for access to the nucleotide sequence of tbpBA of H. somnus strain HS25, and to A. B. Schryvers for critical reading of the manuscript.
This work was supported by grants from the Natural Sciences and Engineering Research Council of Canada (NSERC). A.S. and D.M. were the grateful recipients of NSERC Undergraduate Student Research Awards, and A.E. was the grateful recipient of Postgraduate Scholarships from NSERC and the Fonds pour la Formation de Chercheurs et l'Aide à la Recherche.
REFERENCES
- 1.Angen, Ø., P. Ahrens, P. Kuhnert, H. Christensen, and R. Mutters. 2003. Proposal of Histophilus somni gen. nov., sp. nov. for the three species incertae sedis “Haemophilus somnus,” “Haemophilus agni” and “Histophilus ovis.” Int. J. Syst. Evol. Microbiol. 53:1449-1456. [DOI] [PubMed] [Google Scholar]
- 2.Bahrami, F., A. Ekins, and D. F. Niven. 2003. Iron acquisition by Actinobacillus suis: identification and characterization of transferrin receptor proteins and encoding genes. Vet. Microbiol. 94:79-92. [DOI] [PubMed] [Google Scholar]
- 3.Ekins, A., and D. F. Niven. 2001. Production of transferrin receptors by Histophilus ovis: three of five strains require two signals. Can. J. Microbiol. 47:417-423. [DOI] [PubMed] [Google Scholar]
- 4.Ekins, A., and D. F. Niven. 2002. Identification of fur and fldA homologs and a Pasteurella multocida tbpA homolog in Histophilus ovis and the effects of iron availability on their transcription. J. Bacteriol. 184:2539-2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ekins, A., and D. F. Niven. 2003. Transferrin-dependent expression of TbpA by Histophilus ovis involves a poly G tract within tbpA. FEMS Microbiol. Lett. 220:95-98. [DOI] [PubMed] [Google Scholar]
- 6.Gonzalez, G. C., R.-H. Yu, P. R. Rostek, Jr., and A. B. Schryvers. 1995. Sequence, genetic analysis, and expression of Actinobacillus pleuropneumoniae transferrin receptor genes. Microbiology 141:2405-2416. [DOI] [PubMed] [Google Scholar]
- 7.Gray-Owen, S. D., S. Loosmore, and A. B. Schryvers. 1995. Identification and characterization of genes encoding the human transferrin-binding proteins from Haemophilus influenzae. Infect. Immun. 63:1201-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gray-Owen, S. D., and A. B. Schryvers. 1996. Bacterial transferrin and lactoferrin receptors. Trends Microbiol. 4:185-191. [DOI] [PubMed] [Google Scholar]
- 9.Griffiths, E. 1987. The iron-uptake systems of pathogenic bacteria, p. 69-137. In J. J. Bullen and E. Griffiths (ed.), Iron and infection. Molecular, physiological and clinical aspects. John Wiley & Sons Ltd., Chichester, United Kingdom.
- 10.Heiner, C. R., K. L. Hunkapiller, S.-M. Chen, J. I. Glass, and E. Y. Chen. 1998. Sequencing multimegabase-template DNA with BigDye terminator chemistry. Genome Res. 8:557-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Legrain, M., V. Mazarin, S. W. Irwin, B. Bouchon, M.-J. Quetin-Millet, E. Jacobs, and A. B. Schryvers. 1993. Cloning and characterization of Neisseria meningitidis genes encoding the transferrin-binding proteins Tbp1 and Tbp2. Gene 130:73-80. [DOI] [PubMed] [Google Scholar]
- 12.Nielson, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 13.Niven, D. F., J. Donga, and F. S. Archibald. 1989. Responses of Haemophilus pleuropneumoniae to iron restriction: changes in the outer membrane protein profile and the removal of iron from porcine transferrin. Mol. Microbiol. 3:1083-1089. [DOI] [PubMed] [Google Scholar]
- 14.Ogunnariwo, J. A., C. Cheng, J. Ford, and A. B Schryvers. 1990. Response of Haemophilus somnus to iron limitation: expression and identification of a bovine-specific transferrin receptor. Microb. Pathog. 9:397-406. [DOI] [PubMed] [Google Scholar]
- 15.Ogunnariwo, J. A., and A. B. Schryvers. 1990. Iron acquisition in Pasteurella haemolytica: expression and identification of a bovine-specific transferrin receptor. Infect. Immun. 58:2091-2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ogunnariwo, J. A., and A. B. Schryvers. 1996. Rapid identification and cloning of bacterial transferrin and lactoferrin receptor genes. J. Bacteriol. 178:7326-7328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ogunnariwo, J. A., and A. B. Schryvers. 2001. Characterization of a novel transferrin receptor in bovine strains of Pasteurella multocida. J. Bacteriol. 183:890-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ogunnariwo, J. A., T. K. W. Woo, R. Y. C. Lo, G. C. Gonzalez, and A. B. Schryvers. 1997. Characterization of the Pasteurella haemolytica transferrin receptor genes and the recombinant receptor proteins. Microb. Pathog. 23:273-284. [DOI] [PubMed] [Google Scholar]
- 19.Piechulla, K., R. Mutters, S. Burbach, R. Klussmeier, S. Pohl, and W. Mannheim. 1986. Deoxyribonucleic acid relationships of “Histophilus ovis/Haemophilus somnus,” Haemophilus haemoglobinophilus, and “Actinobacillus seminis.” Int. J. Syst. Bacteriol. 36:1-7. [Google Scholar]
- 20.Ricard, M. A., F. S. Archibald, and D. F. Niven. 1991. Isolation and identification of a putative porcine transferrin receptor from Actinobacillus pleuropneumoniae biotype 1. J. Gen. Microbiol. 137:2733-2740. [DOI] [PubMed] [Google Scholar]
- 21.Schryvers, A. B., and L. J. Morris. 1988. Identification and characterization of the transferrin receptor from Neisseria meningitidis. Mol. Microbiol. 2:281-288. [DOI] [PubMed] [Google Scholar]
- 22.Schryvers, A. B., and L. J. Morris. 1988. Identification and characterization of the human lactoferrin-binding protein from Neisseria meningitidis. Infect. Immun. 56:1144-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stephens, L. R., J. D. Humphrey, P. B. Little, and D. A. Barnum. 1983. Morphological, biochemical, antigenic, and cytochemical relationships among Haemophilus somnus, Haemophilus agni, Haemophilus haemoglobinophilus, Histophilus ovis, and Actinobacillus seminis. J. Clin. Microbiol. 17:728-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walker, R. L., E. L. Biberstein, R. F. Pritchett, and C. Kirkham. 1985. Deoxyribonucleic acid relatedness among “Haemophilus somnus,” “Haemophilus agni,” “Histophilus ovis,” “Actinobacillus seminis,” and Haemophilus influenzae. Int. J. Syst. Bacteriol. 35:46-49. [Google Scholar]
- 25.Widders, P. R., L. G. Paisley, R. P. Gogolewski, J. F. Evermann, J. W. Smith, and L. B. Corbeil. 1986. Experimental abortion and the systemic immune response to “Haemophilus somnus” in cattle. Infect. Immun. 54:555-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu, R.-H., S. D. Gray-Owen, J. Ogunnariwo, and A. B. Schryvers. 1992. Interaction of ruminant transferrins with transferrin receptors in bovine isolates of Pasteurella haemolytica and Haemophilus somnus. Infect. Immun. 60:2992-2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu, R.-H., and A. B. Schryvers. 1994. Transferrin receptors on ruminant pathogens vary in their interaction with the C-lobe and N-lobe of ruminant transferrins. Can. J. Microbiol. 40:532-540. [DOI] [PubMed] [Google Scholar]