Abstract
Single-molecule force spectroscopy enables mechanical testing of individual proteins, however low experimental throughput limits the ability to screen constructs in parallel. We describe a microfluidic platform for on-chip protein expression and measurement of single-molecule mechanical properties. We constructed microarrays of proteins covalently attached to a chip surface, and found that a single cohesin-modified cantilever that bound to the terminal dockerin-tag of each protein remained stable over thousands of pulling cycles. The ability to synthesize and mechanically probe protein libraries presents new opportunities for high-throughput mechanical phenotyping.
Mechanical forces play a pivotal role in biological systems by performing tasks such as guiding cell adhesion1, inducing gene expression patterns2, and directing stem cell differentiation3. At the molecular level, mechano-sensitive proteins act as sensors and transducers, communicating the presence and direction of applied forces to downstream signaling cascades. Conformational changes in response to mechanical forces4 and energetic barriers along unfolding pathways can be probed by SMFS4. Optical tweezers, magnetic tweezers, and atomic force microscopy (AFM) have been used to interrogate high-affinity receptor-ligand binding5, measure unfolding and refolding dynamics of individual protein domains6-8, observe base-pair stepping of RNA polymerases9, and identify DNA stretching and twisting moduli10.
Despite these successes, until now SMFS experiments have been significantly limited by low throughput. Experimental data sets typically contain a majority of unusable force-distance traces due to multiple molecular interactions in parallel, or no specific interactions at all. Typical yields of interpretable single-molecule interaction traces in SMFS experiments vary between 1-25 %. The incapacity of SMFS to quickly screen libraries of molecular variants has hindered progress toward understanding sequence-structure-function relationships at the single molecule level. In particular, preparing each protein sample and cantilever separately increases experimental workload and gives rise to calibration uncertainties. Therefore, the ability to interrogate the mechanical behavior of different proteins in a parallel and streamlined format with the same cantilever would be a distinct advantage. Such a screening system could characterize single-molecule properties like unfolding forces, interdomain mechanical signatures, and mechanically-activated catch bond behavior1. Screening of these properties could find applications in biotechnology and human health studies where mechanical dysregulation or misfolding is suspected to play a role in pathology11.
Here we developed a platform for parallel characterization of protein mechanics in a single experiment (Fig. 1). Microspotted gene arrays were utilized to synthesize fusion proteins in situ using cell-free gene expression. Proteins were covalently immobilized inside multilayer microfluidic circuits. A single cantilever was then positioned above the protein array, and used to probe the mechanical response of each protein via a common C-terminal Dockerin (Doc) fusion tag. Genes of interest were chosen such that each gene product exhibited an identifiable unfolding pattern when loaded from the N- to C-terminus. Each target protein was expressed with an N-terminal 11 amino acid ybbR tag, which was used to covalently and site-specifically link it to the surface via Sfp Synthase-catalyzed reaction with coenzyme A (CoA)12. At the C-terminus the proteins contained a 75 amino acid cellulosomal Doc from Clostridium thermocellum (C.t.) as specific handle targeted by the Cohesin (Coh)-modified cantilever.
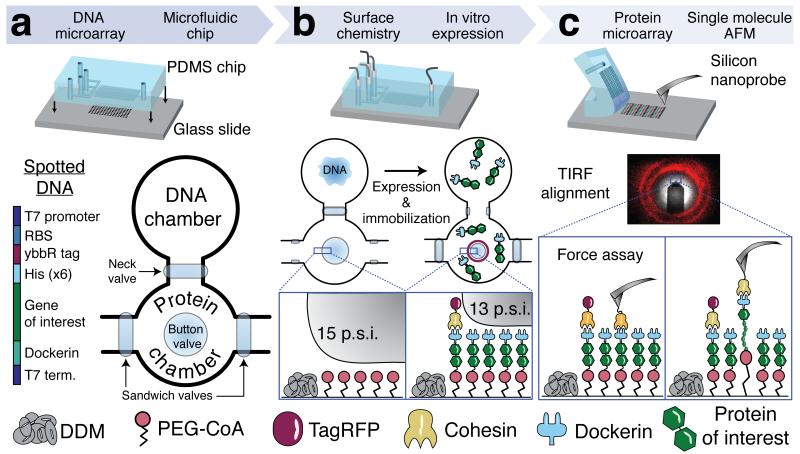
Figure 1.
Method workflow from gene array to single-molecule mechanics.
(a) A gene array was spotted onto a glass slide. Genes were designed with a common set of flanking sequences, including a T7 promoter region, a ybbR tag, a Dockerin tag and a T7 terminator. The multilayer microfluidic chip featuring 640 unit cells was aligned to the DNA microarray and bonded to the glass slide. Each unit cell comprised a DNA chamber, a protein chamber, and superseding elastomeric control valves actuated by pneumatic pressure. (b) Control valves were utilized for spatially selective surface modification of each protein chamber with PEG-CoA, and for fluidic isolation of each chamber prior to in vitro expression of the microspotted DNA. Fluorescent labeling with TagRFP-Cohesin was achieved by partial button valve pressurization, leaving only an outer concentric ring of immobilized gene products exposed to the labeling solution. (c) After removal of the microfluidic device, the resulting well-defined, covalently attached protein microarray was accessed from above with a Cohesin-functionalized AFM cantilever. Single-molecule unfolding traces of each of the investigated protein constructs were thus acquired sequentially at each corresponding array address with a single cantilever in a single experiment.
The gene microarray was aligned and reversibly bonded to a microfluidic chip known as MITOMI (mechanically induced trapping of molecular interactions). The chip has been used in the past for screening transcription factors13,14, and mapping interaction networks15. More recently, our group employed MITOMI chips for molecular force assays16. In this work, MITOMI chips featured 640 dumbbell shaped unit cells in a flow layer and 2,004 micromechanical valves in a control layer. Each unit cell was equipped with pneumatic ‘neck’, ‘sandwich’, and ‘button’ valves (Fig. 1a) according to design principles of soft lithography17. Each neck valve protected the microspotted DNA in the back chamber from exposure to other reagents during surface patterning in the front chamber. The sandwich valves prevented chamber-to-chamber cross contamination, ensuring only a single protein variant being present in each sample spot. For surface chemistry in the front chamber, the button valves were actuated to shield the sample spots, allowing n-Dodecyl β-D-maltoside (DDM) passivation in the surrounding area. Releasing the button valves allowed subsequent functionalization with CoA-PEG in the sample area under the buttons serving as protein immobilization site. The genes were expressed by incubating an in vitro transcription and translation cell extract at 37 °C with the spotted DNA in the back chamber. The synthesized proteins then diffused to the front chamber where they were covalently linked to the surface via Sfp-catalyzed reaction of surface-bound CoA with solution phase N-terminal ybbR peptide tags (Fig. 1b). Partial pressurization of the button valve18 was used for tagging an outer concentric portion of the sample area with a fluorescently labeled Coh that specifically bound to the C-terminal Doc tag of each target protein, thereby confirming successful protein synthesis and surface immobilization (Supplementary Fig. 1). Finally, the microfluidic device was removed from the glass slide, providing access to the protein array from above. This way we generated microarrays of site-specifically and covalently immobilized proteins for subsequent SMFS experiments, starting from a conventional gene array.
An inverted three-channel total internal reflection fluorescence/ atomic force microscope (TIRF-AFM)19 was used to position the cantilever in the center of the fluorescent rings in the protein array and perform SMFS measurements (Fig. 1c). The Coh-modified cantilever was used to probe the surface for expressed target proteins containing the C-terminal Doc tag. Upon surface contact of the cantilever, formation of a Coh-Doc complex allowed measurement of target protein unfolding in a well-controlled pulling geometry (N- to C-terminus). We retracted the probe at constant velocity and recorded force-extension traces that characterized the unfolding fingerprint of the target protein. This approach-retract process could be repeated many times at each array address to characterize each expression construct.
Several unique features of the C-terminal Doc tag made it particularly suitable in this application as a protein handle for SMFS-MITOMI. This tag natively serves as a conserved module of cellulolytic enzymes of the anaerobic bacterium C.t.20. Its small size of 8 kDa did not notably add to the molecular weight of the gene products, which was advantageous for cell-free expression. Additionally, Doc exhibits a specific and high affinity interaction with Coh domains from the C.t. scaffold protein CipA. Coh was used both for fluorescence detection of the expression constructs and for modification of the cantilever. Based on our prior work, the Coh-Doc interaction is characterized to be high affinity, with a Kd in the low nanomolar range and rupture forces >125 pN at a loading rate of 10 nN/s21. Our prior work also indicated that upon forced dissociation, the Doc exhibited a characteristic double sawtooth rupture peak with a contour length increment of 8 nm separating the two peaks. We used this two-pronged double rupture event at the end of each force-extension trace as a positive indicator that the gene of interest was completely expressed through to the C-terminus. Furthermore this double rupture peak indicated that the interaction with the Coh-modified cantilever was specific, and that the pulling geometry was strictly controlled such that force was applied to the molecule of interest from the N to C-terminus.
As a validation and demonstration of our SMFS-MITOMI approach, we expressed genes of interest comprising well-known fingerprint domains in the SMFS literature. We produced multimeric polyproteins including tetrameric human type-III fibronectin (FBN)22 and dimeric chicken brain α-spectrin (SPN)23. We also synthesized monomers of endo-1,4-xylanase T6 from Geobacillus stearothermophilus (XYL)21, superfolder green fluorescent protein (GFP)24, and twitchin kinase25. In all cases, surface immobilization and SMFS assay were enabled by N-terminal ybbR and C-terminal Doc tags on the target proteins. Unfolding data for FBN, SPN, XYL and GFP were obtained with a single cantilever on a single microarray (Figs. 2 and 3). Twitchin kinase was found not to express in sufficient yield to provide reliable unfolding statistics.
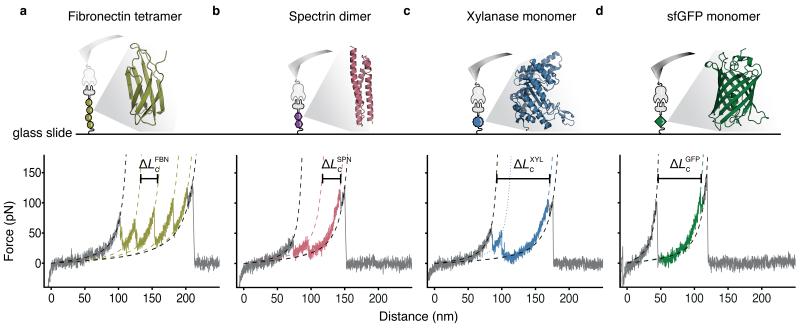
Figure 2.
Single-molecule force traces recorded in different protein spots on a single chip with a single cantilever.
(a-d) Four proteins of interest, anchored between the CoA-functionalized surface and the Cohesin-functionalized cantilever were probed: fibronectin tetramer (a, olive), spectrin dimer (b, red), xylanase monomer (c, blue), and sfGFP monomer (d, green). The crystal structure and pulling configuration (top) are shown for each construct. Each single-molecule force-distance trace (bottom) shows the individual unfolding fingerprint of the respective protein of interest followed by a common, final double sawtooth peak (grey), characteristic of the Cohesin-Dockerin rupture. Experimental data were fitted with the wormlike chain model (dashed lines). Unfolding intermediates were also observed (only fitted for xylanase in c; dotted colored line).
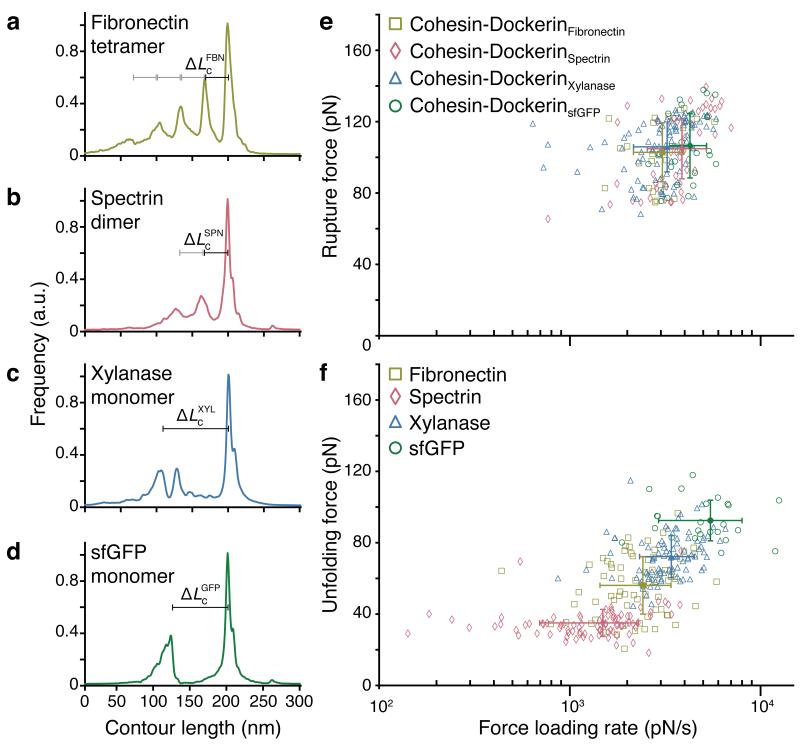
Figure 3.
Unfolding and rupture statistics from multiple force traces.
(a-d) relative frequency of observing given contour lengths determined by transforming and aligning multiple force traces into contour length space via the WLC model. Shown are diagrams for the fibronectin tetramer (a) (n = 27, ΔLcFBN = 33 nm), spectrin dimer (b) (n = 50, ΔLcSPN = 34 nm), xylanase monomer (c) (n = 91, ΔLcXYL = 93 nm) and sfGFP monomer (d) (n = 25, ΔLcGFP = 79 nm). (e) Rupture force vs. loading rate scatter plot of final Cohesin-Dockerin dissociation event. (f) Unfolding force vs. loading rate scatter plot for each protein of interest. The populations in e and f were fitted with 2D Gaussians. Respective means and s.d. are plotted in the corresponding colors as solid symbols and error bars.
We transformed force-extension data (Fig. 2) into contour length space26 using the worm-like chain model, and compared the measured contour length increments with the amino acid sequence lengths of each protein and literature values. The observed contour lengths and rupture forces were consistent with our expectations. FBN showed a four-fold sequence of rupture peaks at contour length increments of 32 nm (Fig. 2a, ΔLcFBN), frequently interrupted by an intermediate peak at 10-12 nm, both characteristic of FBN22. SPN showed two regular sawtooth-like peaks with contour lengths of 33 nm (Fig. 2b, ΔLcSPN)23. XYL exhibited a decreasing multi-peaked unfolding fingerprint with a contour length increment of 92 nm (Fig. 2c, ΔLcXYL), occasionally showing additional increments corresponding to unfolding of remaining XYL sub-domains, a result consistent with the prior study and accounting for N-terminal immobilization of XYL21. GFP unfolding showed a contour length increment of 74 nm (Fig. 2d, ΔLcGFP)24. Thus, at known locations in the array, we could record unfolding traces of individual custom encoded proteins, stretching them from the surface with a Coh-modified cantilever by grabbing the high-affinity and mechanically stable C-terminal Doc tags. Since each protein in the array contained the same C-terminal Doc tag, the final two rupture peaks in all force traces represented rupture of the Coh-Doc complex regardless of the protein of interest.
In our system, surface densities of expressed proteins were comparable to conventional SMFS experiments, resulting in yields of interpretable curves of up to 5 %. By collecting multiple unfolding traces, we assembled contour length diagrams for each protein of interest (Fig. 3a-d)26,27, and confirmed the predicted contour length increments based on the encoded amino acid sequences in each DNA spot. Coh-Doc handle rupture events for all protein constructs in the array clustered to the same population in the force-loading rate plot, independent of the preceding rupture peaks from the protein of interest (Fig. 3e). The Coh-Doc ruptures agree with previously reported values at similar loading rates21. The unfolding events of the proteins of interest produced distinct populations in the force-loading rate plots (Fig. 3f). The unfolding events depended on the internal structure and the unfolding pathway of the fingerprint domain when stretched between its N- and C-termini. SPN, for example, an elongated 3-helix bundle, was previously reported to exhibit a broader energy well (Δx = 1.7 nm23) and showed a flatter distribution of unfolding forces compared with the more compact globular FBN domain with a shorter steeper potential (Δx = 0.4 nm22).
In summary, the described system is highly flexible and efficiently streamlines protein expression, purification, and SMFS into a single integrated platform. The approach is generally viable with other expression systems including extracts derived from insects, rabbit reticulocytes, and human cell lines, and it is capable of introducing post-translational modifications and non-natural amino acids, further widening the pool of target proteins, for example screening of site-directed mutants. Our system allows for synthesis of cytotoxic proteins, or proteins with a tendency to form inclusion bodies during bulk expression. In addition to greatly improved throughput, our system has the advantage of measuring multiple constructs with one cantilever thereby eliminating errors introduced by performing multiple calibrations on different samples with uncertainties of ~10 %28. Detecting subtle differences in mechanical stability with this high-throughput approach could therefore be used to perform mechanical phenotyping experiments on similarly stable families of mutant proteins. This workflow opens the door to large-scale screening studies of protein nanomechanical properties, a possibility that was until now not achievable.
Online Methods
Chip fabrication
Ready-to-use wafers for flow and control layers of the 640-chamber MITOMI design from13 (design name DTPAd, Stanford Microfluidics Foundry). The flow wafer features 15 μm high features, rounded by photoresist reflow, whereas the control wafer features a rectangular cross-section.
Microfluidic chips were cast in polydimethylsiloxane (PDMS) from these wafers. For the control layer, Sylgard 184 (Dow Corning) base and curing agent were mixed at a ratio of 5:1 by weight, poured onto the wafer, degassed, and partially cured for 20 min at 80 °C. For the flow layer wafer, a 20:1 base to curing agent mixture of Sylgard 184 was spin-coated for 75 s at 1600 rpm and partially cured for 30 min at 80 °C. The control layer chips were cut out, inlet holes were punched and the chips were aligned onto the spin-coated PDMS on the flow layer wafer. After baking the two-layer chips for 90 min at 80 °C, they were cut, removed from the wafer and inlet/outlet holes were punched. Microfluidic chips were stored for up to 6 weeks.
Cloning
For the construction of the fusion proteins, Gibson Assembly29 was used. A ratio of 0.07 pmol vector to 0.3 pmol of insert was used for the fusion reaction. The primer sequences are provided in Supplementary Table 1. A pET28a plasmid was linearized with primers 1 and 2. The Dockerin Type I gene was isolated from the Xylanase-Dockerin Type I construct21 with primers 3 and 4. Codon optimized sequences were purchased from Geneart/Invitrogen. The genes of interest were designed in such a way that they already contained overlapping sequences with their neighboring partners (pET28a and Dockerin Type I). In the case of the Spectrin, two domains were linked with a flexible Glycine-Serine (x6)-linker. For Fibronectin, four type III domains were fused separated by Glycine-Serine (x6)-linkers. The expression vector in all cases was a pET28a plasmid with a modified multiple cloning site (sequence attached). After construction, clones were verified via sequencing and amplified in NEB5alpha E. coli cells. Following plasmid preparation, samples were concentrated up to 500 ng/μl prior to microspotting.
DNA microspotting
A 24×60 mm #1 thickness coverslip (Thermo scientific) was silanized with 3-Aminopropyldimethyl-ethoxysilane (ABCR) following literature protocols30.
The DNA solution containing 1 % (w/v) nuclease-free Bovine Serum Albumin (Carl Roth) in nuclease-free water, was micro-spotted under humid atmosphere onto the silanized coverslip using the GIX Microplotter II (Sonoplot) and a glass capillary with a 30 μm tip diameter (World Precision Instruments) according to the manufacturer’s instructions in a rectangular 40×16 pattern with 320 μm column pitch and 678 μm row pitch. Alignment of the DNA array and the microfluidic chip was done manually using a stereomicroscope. Bonding between the glass cover slip and microfluidic device was achieved by thermal bonding for 5 h at 80 °C on a hot plate.
Details on device operation and force spectroscopy measurements can be found in the supplementary information.
Supplementary Material
Acknowledgements
M.O. is grateful to the Elite Network of Bavaria (IDK-NBT) for a doctoral fellowship. M.A.N. gratefully acknowledges support from Society in Science – The Branco Weiss Fellowship administered by the ETH Zürich. The authors acknowledge support from the European Research Council, DFG Sonderforschungsbereich 1032, and the Center for Integrated Protein Science Munich (CIPSM) Excellence Cluster. The authors thank Prof. Edward Bayer at the Weizmann Institute for donation of starting genetic materials used for Doc and Coh modules.
DNA Sequence Accession Codes
Plasmids of all constructs are deposited in the Addgene database:
| Addgene ID | Construct |
|---|---|
| 58708 | pET28a-ybbR-HIS-sfGFP-DocI |
| 58709 | pET28a-ybbR-HIS-CBM-CohI |
| 58710 | pET28a-StrepII-TagRFP-CohI |
| 58711 | pET28a-ybbR-HIS-Xyl-DocI |
| 58712 | pET28a-ybbR-HIS-10FNIII(x4)-DocI |
| 58713 | pET28a-ybbR-HIS-Spec(x2)-DocI |
Footnotes
Competing financial interests statement: The authors declare no competing financial interests.
References
- 1.Thomas WE, Trintchina E, Forero M, Vogel V, Sokurenko EV. Cell. 2002;109:913–923. doi: 10.1016/s0092-8674(02)00796-1. [DOI] [PubMed] [Google Scholar]
- 2.Li C, Xu Q. Cellular signalling. 2000;12:435–445. doi: 10.1016/s0898-6568(00)00096-6. [DOI] [PubMed] [Google Scholar]
- 3.Engler AJ, Sen S, Sweeney HL, Discher DE. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 4.Muller D, Helenius J, Alsteens D, Dufrêne YF. Nature Methods. 2009;5:383–390. doi: 10.1038/nchembio.181. [DOI] [PubMed] [Google Scholar]
- 5.Florin E-L, Moy VT, Gaub HE. Science. 1994;264:415–417. doi: 10.1126/science.8153628. [DOI] [PubMed] [Google Scholar]
- 6.Rief M, Gautel M, Oesterhelt F, Fernandez J, Gaub H. Science. 1997;276:1109–1112. doi: 10.1126/science.276.5315.1109. [DOI] [PubMed] [Google Scholar]
- 7.Fernandez J, Li H. Science. 2004;303:1674–1678. doi: 10.1126/science.1092497. [DOI] [PubMed] [Google Scholar]
- 8.Oesterhelt F, et al. Science. 2000;288:143–146. doi: 10.1126/science.288.5463.143. [DOI] [PubMed] [Google Scholar]
- 9.Abbondanzieri EA, Greenleaf WJ, Shaevitz JW, Landick R, Block SM. Nature. 2005;438:460–465. doi: 10.1038/nature04268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bryant Z, et al. Nature. 2003;424:338–341. doi: 10.1038/nature01810. [DOI] [PubMed] [Google Scholar]
- 11.Linke WA. Cardiovascular Research. 2008;77:637–648. doi: 10.1016/j.cardiores.2007.03.029. [DOI] [PubMed] [Google Scholar]
- 12.Yin J, et al. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15815–15820. doi: 10.1073/pnas.0507705102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maerkl SJ, Quake SR. Science. 2007;315:233–237. doi: 10.1126/science.1131007. [DOI] [PubMed] [Google Scholar]
- 14.Rockel S, Geertz M, Hens K, Deplancke B, Maerkl SJ. Nucleic Acids Research. 2013;41:e52. doi: 10.1093/nar/gks1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gerber D, Maerkl SJ, Quake SR. Nature Methods. 2008;6:71–74. doi: 10.1038/nmeth.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Otten M, Wolf P, Gaub HE. Lab on a Chip. 2013;13:4198. doi: 10.1039/c3lc50830g. [DOI] [PubMed] [Google Scholar]
- 17.Thorsen T, Maerkl SJ, Quake SR. Science. 2002;298:580–584. doi: 10.1126/science.1076996. [DOI] [PubMed] [Google Scholar]
- 18.Garcia-Cordero JL, Maerkl SJ. Chemical Communications. 2013;49:1264. doi: 10.1039/c2cc37740c. [DOI] [PubMed] [Google Scholar]
- 19.Gumpp H, Stahl SW, Strackharn M, Puchner EM, Gaub HE. Review of Scientific Instruments. 2009;80:063704. doi: 10.1063/1.3148224. [DOI] [PubMed] [Google Scholar]
- 20.Bayer EA, Belaich J-P, Shoham Y, Lamed R. Annual Review of Microbiology. 2004;58:521–554. doi: 10.1146/annurev.micro.57.030502.091022. [DOI] [PubMed] [Google Scholar]
- 21.Stahl SW, et al. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:20431–20436. doi: 10.1073/pnas.1211929109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li L, Huang HH-L, Badilla CL, Fernandez JM. Journal of Molecular Biology. 2005;345:817–826. doi: 10.1016/j.jmb.2004.11.021. [DOI] [PubMed] [Google Scholar]
- 23.Rief M, Pascual J, Saraste M, Gaub HE. Journal of Molecular Biology. 1999;286:553–561. doi: 10.1006/jmbi.1998.2466. [DOI] [PubMed] [Google Scholar]
- 24.Dietz H, Rief M. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:1244–1247. doi: 10.1073/pnas.0509217103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Greene DN, et al. Biophysical Journal. 2008;95:1360–1370. doi: 10.1529/biophysj.108.130237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Puchner EM, Franzen G, Gautel M, Gaub HE. Biophysical Journal. 2008;95:426–434. doi: 10.1529/biophysj.108.129999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jobst MA, Schoeler C, Malinowska K, Nash MA. JoVE. 2013 doi: 10.3791/50950. doi:10.3791/50950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gibson CT, Smith DA, Roberts CJ. Nanotechnology. 2005;16:234–238. doi: 10.1088/0957-4484/16/2/009. [DOI] [PubMed] [Google Scholar]
- 29.Gibson DG, et al. Nature Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 30.Zimmermann JL, Nicolaus T, Neuert G, Blank K. Nat Protoc. 2010;5:975–985. doi: 10.1038/nprot.2010.49. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.