Abstract
Mycoplasma mobile is a flask-shaped bacteria that binds to a substrate and glides towards its tapered end, the so-called “head-like protrusion,” by an unknown mechanism. To search for cellular structures underlying this motility, the cell-substrate interface of actively gliding cells was visualized by rapid-freeze-and-freeze-fracture rotary-shadow electron microscopy. Novel structures, called “spikes,” were observed to protrude from the cell membrane and attach to the glass surface at their distal end. The spikes were on average 50 nm in length and 4 nm in diameter, most abundant around the head, and not observed in a nonbinding mutant. The spikes may be involved in the mechanism of binding, gliding, or both.
Mycoplasma gliding.
Mycoplasmas are bacteria that lack a cell wall and are parasitic or commensal to many kinds of hosts, including humans, animals, and plants (25). Several mycoplasma species have a membrane protrusion at one end, called the “head-like protrusion,” which creates the organism's characteristic flask-shaped cell morphology. These bacteria are capable of gliding motility, enabling them to translocate smoothly on solid surfaces, always in the direction of the head-like protrusion (3, 13). Although gliding motility of mycoplasmas is believed to be involved in pathogenicity, the mechanism of gliding motility has not been well investigated. Mycoplasmas do not have any homologs of genes that encode pili, flagella, genes related to other bacterial motility, or motor proteins that are common in eukaryotic motility, such as myosins (5, 7, 9, 19). These facts suggest that mycoplasmas glide by an entirely unknown mechanism, but this mechanism has been difficult to isolate because many species of mycoplasma travel at low speed and with interrupted motility.
The species Mycoplasma mobile provides an opportunity to study mycoplasma gliding motility, because this species is the fastest gliding and, unlike other species, glides without interruption (13-15, 20, 21, 27). At all stages of growth, M. mobile glides smoothly and continuously on glass at an average speed of 2.0 to 4.5 μm/s, or about 3 to 7 times the length of the cell per s (27), exerting a force of up to 27 piconewtons (pN) (20, 21). These distinct characteristics enabled detailed analyses of gliding (6, 20-22, 26-28) and isolation of gliding mutants that were characterized by reduced or deficient gliding or enhanced speed (23, 28).
Attempts to visualize machinery.
Since mycoplasmas bind to and crawl on solid surfaces, a specialized structure may be responsible for this function. However, previous attempts to visualize such structures by various electron microscopy (EM) techniques have been unsuccessful. In this study, we applied rapid-freeze-and-freeze-fracture rotary-shadow EM to M. mobile and visualized novel structures, which we call “spikes,” that are possibly involved in the mechanism of gliding motility. This study is the first to use both rapid freezing and freeze-fracture to visualize adhered and gliding mycoplasmas. While other studies used freeze-fracture (18), they used nongliding species of mycoplasma (2, 24, 29), a suspension of mycoplasma cells that were not adhered to or gliding on a surface (17, 29), or a slower freezing method that was less effective at preserving cell structure (4, 17, 29).
Rapid-freeze-and-freeze-fracture rotary-shadow EM.
The rapid-freeze method described previously was applied to mycoplasma with slight modifications (11, 12). M. mobile strain 163K (ATCC 43663) and its mutants (23) were grown as described previously (28). The mycoplasma cells were collected by centrifugation (28) and suspended in fresh Aluotto medium at a 25-fold-higher concentration. A droplet (20 μl) of cell suspension was placed on a 5-mm2 square nitric acid-cleaned coverslip at room temperature for 10 min to let mycoplasmas bind to and glide on the glass surface (Fig. 1). Unattached cells were rinsed away, while attached cells continued to glide normally, by placement of the coverslip in a washing buffer that consisted of 70 mM NaCl, 5 mM MgCl2, 3 mM EGTA, and 30 mM HEPES buffer (pH 7.5). All cells on the glass surface were confirmed to be gliding under this condition by phase-contrast microscopy. After 30 s of rinsing, the coverslip was transferred to a freezing stage, excess rinsing buffer was wicked away with filter paper, and the coverslip was slammed against a super-cooled copper block in a Life Cell freezing machine. For freeze-fracture/deep-etch preparations, rapidly frozen specimens were knife-fractured at nominally −108°C in a Balzers 301 freeze-fracture apparatus (Lichtenstein, Austria) equipped with a high-speed rotary stage. Platinum was rotary applied to the fractured sample from 20°, and then a carbon backing was rotary applied from above. Replicas were quickly removed from the glass coverslip with hydrofluoric acid, cleaned in commercial bleach, rinsed in distilled water, and picked up on 400-mesh copper EM grids.
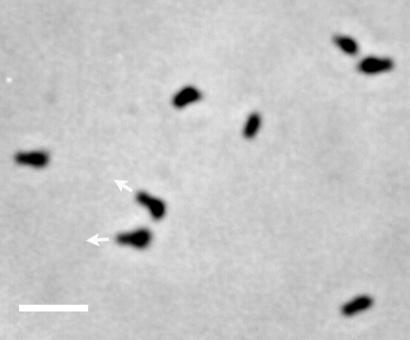
FIG. 1.
Phase-contrast microscopy of M. mobile gliding on glass surface. Mycoplasma cells gliding on glass were fixed and observed as described previously (28). The gliding direction is indicated by arrows for two cells. Bar, 2 μm.
Replicas were viewed in a JEOL 200CX electron microscope and photographed at tilt angles of ±5°. Electron micrographs were scanned directly using an Agfa DuoScan HiD scanner, and tilt pair images were merged for stereo view analysis.
Fracturing pattern.
By the technique applied here, the cells were frozen with millisecond time resolution (8) while attached and gliding on the glass substrate and then fractured, creating the fracture faces schematized in Fig. 2. Most frequently, the fracture plane traveled between lipid bilayers rather than traversing cell cytoplasm, because the binding between leaflets is relatively weak compared to the force required to fracture through cytoplasm. Thus, the most commonly viewed fracture faces are P and E faces, representing the interior of the plasma membrane viewed from the outside or inside, respectively.
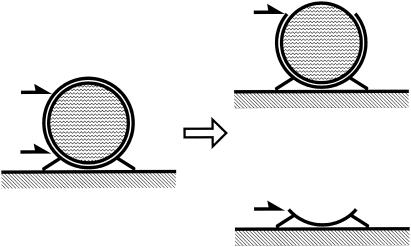
FIG. 2.
Schematic illustration of freeze-fracture faces. (Left) Mycoplasma cell attached to glass surface prior to fracture. The outer and inner leaflets of cell membrane, cytoplasm, glass, and spikes at the lower side of a cell are presented. Fracture of a cell mostly occurred in two patterns, which are presented by black half-arrows. (Right top) P face. Shown is the outer view of the inner leaflet of the cell membrane. (Right bottom) E face. Shown is the inner view of the outer leaflet of the membrane.
Spikes at mycoplasma-glass interface.
To reveal structures involved in the adherence and gliding of a mycoplasma cell on the glass surface, we focused on the cell-glass interface. This perspective was most readily visualized in E-face views (Fig. 2, bottom right) in which most of the cell was removed during fracture, leaving behind only the portion of the outer bilayer leaflet, closest to the glass surface. In all of such images, spikes that stuck out from the membrane were clearly visible (Fig. 3). Three-dimensional stereo observation showed that the spikes contacted the glass surface at their distal end. The angles of the spikes relative to the cell axis and to the glass surface were variable. Spikes were most abundant around the head-like protrusion (Fig. 3), concentrated mainly around the neck, and slightly distributed into the body. Sometimes, structures resembling spikes were observed also at the cell's posterior; however, they were not identical in appearance to the spikes around the head. Further studies are necessary to conclude if these structures are spikes.
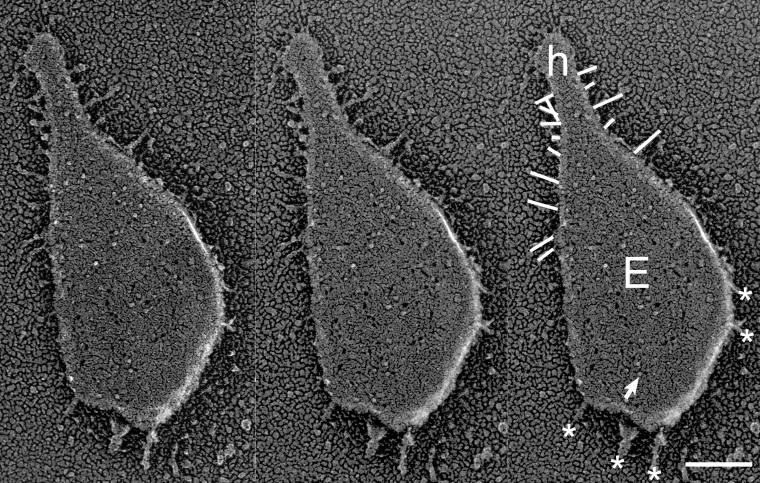
FIG. 3.
E-face image of gliding mycoplasma. The left and middle images are a stereo pair. Stereo view shows a typical E-face image in which the remaining outer leaflet of lower membrane forms a bowl-like shape containing transmembrane protein molecules and holes from which membrane proteins were pulled out with inner leaflet. The right image is identical to the middle one but contains the following indications: the spike, transmembrane protein, head-like protrusion, and protrusions at cell body, which are marked by lines, arrow, the letter “h,” and asterisks, respectively. Bar, 100 nm.
Previously, we studied the movement of cells attached with beads on surfaces (20, 21) and cells having an elongated head-like protrusion (22). Based on these studies, we predicted that the force of movement is generated around the head-like protrusion, and indeed, the observed distribution of spikes is consistent with this prediction. We previously isolated several mutants of M. mobile that are defective in gliding motility (23). We looked for spikes on the mutant called m12, which does not bind to glass and thus does not exhibit gliding motility. We failed to find evidence of any spikes on the m12 mutant cells (data not shown), supporting the notion that spikes are related to binding or gliding.
To ensure accurate measurements of spike dimensions, we found small pieces of membrane containing spikes that were aligned at very low angles to the glass surface (Fig. 4) and whose entire length was clearly visible. The spikes had rather uniform dimensions, if we measure the diameter of straight area, averaging 51.2 ± 10.0 nm in length and 4.3 ± 1.6 nm in diameter (n = 18). It is possible that the spikes observed in this study may be integral membrane proteins that were deformed by the force of the fracture, producing the spike-shaped structures. However, the uniformity of spike length and diameter argues against this possibility. A deformity produced by the force of the fracture would produce a range of spike lengths and diameters, since the amount of force experienced by each cell, and its sides, would vary. If the spikes were deformed membrane proteins, one would not expect such consistency in spike dimension.
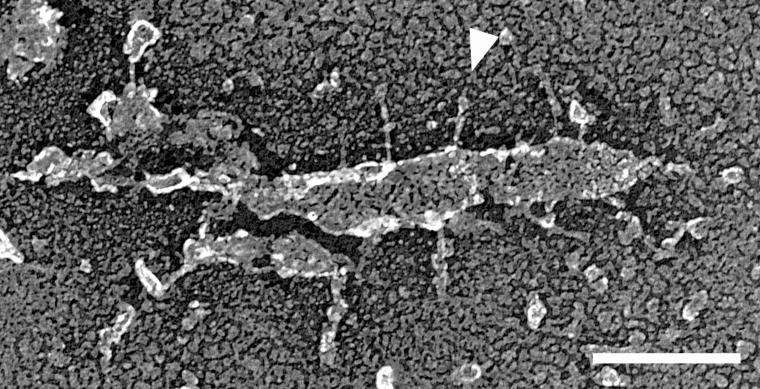
FIG. 4.
Spikes protruding from a piece of membrane on glass. Virtually the entire cell was removed by fracture, leaving a fragment of outer bilayer leaflet. A spike is marked by an arrowhead. Bar, 100 nm.
Although the spikes were easily found around the head-like protrusion at the cell-glass interface, it was difficult to find them in P-face images, in which the outer bilayer leaflet was removed by fracture. This fact suggests that the spikes may have been bound mainly to the outer leaflet of the cell membrane and removed with the outer leaflet during fracture. Alternatively, spikes may have been absent in areas of the cell not adjacent to the substrate. Currently, the mycoplasma cell is divided into the head-tail axis. If the spikes are present only on the surface of the mycoplasma that contacts the substrate, a substrate-attached versus nonattached axis should be added to the mycoplasma cell.
Previously, EM studies revealed two components of the head-like protrusion in the gliding species M. pneumoniae that may be involved in binding and gliding and may be similar to the spikes observed on the head-like protrusion of M. mobile. The head-like protrusion of M. pneumoniae is called the “attachment organelle,” and it contains an electron-dense core that is thought to function as a cytoskeletal support for this attachment organelle. The first structure, revealed by thin-section EM of M. pneumoniae bound to a tracheal organ culture (30), is a 200-nm-long, fibrillar element that is approximately 2 nm in diameter and extends from the extracellular surface of the attachment organelle to the tissue. The fibrillar element was well stained with ruthenium red, which specifically stains mucopolysaccharides. The second structure, observed by negative staining of M. pneumoniae, is a peplomer-like structure, called “nap” (1, 10). The localization of nap is also restricted to the extracellular surface of the attachment organelle, similar to the distribution of spikes on the head-like protrusion of M. mobile. While nap has also been found in other gliding species (Mycoplasma genitalium, Mycoplasma pulmonis, and Mycoplasma gallisepticum), it has not been observed in M. mobile (16). Whether a functional commonality exists between the spike of M. mobile and the fibrillar element or nap remains to be clarified.
Other structures.
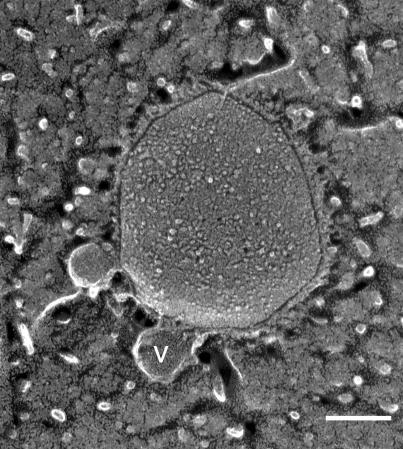
We found small vesicles attached to the mycoplasma cell exterior, which have not been previously reported (Fig. 5). These vesicles had a rather regular diameter of 57.6 ± 9.5 nm (n = 12). Possibly, in previous studies that did not utilize the rapid-freeze technique, such vesicles were not preserved. The vesicles were found more frequently in the nonbinding mutant, m12, we examined (data not shown). Occurrence of vesicles may be related to the maintenance of cell morphology, which is disordered in the nonbinding and nongliding strains (23).
FIG. 5.
Vesicles in the P face. A vesicle is marked by the letter “v.” Bar, 100 nm.
An individual M. mobile cell generates a rather large force of up to 27 pN, which has raised speculation that the cell's binding and gliding machinery is anchored to an stationary structure rather than floating freely in the membrane (20). Indeed, it has been proposed that the electron-dense core observed in M. pneumoniae may support its attachment organelle and serve as scaffolding for that species' binding and gliding machinery (16a, 25a). Like the attachment organelle in M. pneumoniae, the head-like protrusion of M. mobile may also contain cytoskeletal scaffolding that may anchor components of the binding or gliding machinery, such as the spikes, which may be necessary for binding or gliding. However, such a cytoskeletal structure has not yet been detected in the head-like protrusion of M. mobile.
Acknowledgments
We are grateful to Thomas S. Reese, Laboratory of Neurobiology, National Institutes of Health, for critical reading of the manuscript. We thank Atsuko Uenoyama, Osaka City University, for technical assistance.
This work was partly supported by Rowland Institute for Science; Grants-in-Aid for Scientific Research (C) to M.M.; and Science Research on Priority Area (“motor proteins,” “genome science,” and “infection and host response”) from the Ministry of Education, Science, Sports and Culture, and Technology to M.M.
REFERENCES
- 1.Baseman, J. B., R. M. Cole, D. C. Krause, and D. K. Leith. 1982. Molecular basis for cytadsorption of Mycoplasma pneumoniae. J. Bacteriol. 151:1514-1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bittman, R., S. Clejan, and S. W. Hui. 1990. Increased rates of lipid exchange between Mycoplasma capricolum membranes and vesicles in relation to the propensity of forming nonbilayer lipid structures. J. Biol. Chem. 265:15110-15117. [PubMed] [Google Scholar]
- 3.Bredt, W. 1979. Motility, p. 141-145. In M. F. Barile, S. Razin, J. G. Tully, and R. F. Whitcomb (ed.), The mycoplasmas, vol. 1. Academic Press, New York, N.Y. [Google Scholar]
- 4.Carson, J. L., and A. M. Collier. 1982. Host-pathogen interactions in experimental Mycoplasma pneumoniae disease studied by the freeze-fracture technique. Rev. Infect. Dis. 4(Suppl.):S173-S178. [DOI] [PubMed] [Google Scholar]
- 5.Chambaud, I., R. Heilig, S. Ferris, V. Barbe, D. Samson, F. Galisson, I. Moszer, K. Dybvig, H. Wroblewski, A. Viari, E. P. Rocha, and A. Blanchard. 2001. The complete genome sequence of the murine respiratory pathogen Mycoplasma pulmonis. Nucleic Acids Res. 29:2145-2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fischer, M., H. Kirchhoff, R. Rosengarten, G. Kerlen, and K.-H. Seack. 1987. Gliding movement of Mycoplasma sp. nov. strain 163K on erythrocytes. FEMS Microbiol. Lett. 40:321-324. [Google Scholar]
- 7.Fraser, C. M., J. D. Gocayne, O. White, M. D. Adams, R. A. Clayton, R. D. Fleischmann, C. J. Bult, A. R. Kerlavage, G. Sutton, J. M. Kelley, R. D. Fritchman, J. F. Weidman, K. V. Small, M. Sandusky, J. Fuhrmann, D. Nguyen, R. Utterback, D. M. Saudek, C. A. Phillips, J. M. Merrick, J.-F. Tomb, B. A. Dougherty, K. F. Bott, P.-C. Hu, T. S. Lucier, S. N. Peterson, H. O. Smith, C. A. Hutchison III, and J. C. Venter. 1995. The minimal gene complement of Mycoplasma genitalium. Science 270:397-403. [DOI] [PubMed] [Google Scholar]
- 8.Heuser, J. E., T. S. Reese, M. J. Dennis, Y. Jan, L. Jan, and L. Evans. 1979. Synaptic vesicle exocytosis captured by quick freezing and correlated with quantal transmitter release. J. Cell Biol. 81:275-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Himmelreich, R., H. Hilbert, H. Plagens, E. Pirkl, B.-C. Li, and R. Herrmann. 1996. Complete sequence analysis of the genome of the bacterium Mycoplasma pneumoniae. Nucleic Acids Res. 24:4420-4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu, P. C., R. M. Cole, Y. S. Huang, J. A. Graham, D. E. Gardner, A. M. Collier, and W. A. Clyde, Jr. 1982. Mycoplasma pneumoniae infection: role of a surface protein in the attachment organelle. Science 216:313-315. [DOI] [PubMed] [Google Scholar]
- 11.Khan, S., M. Dapice, and T. S. Reese. 1988. Effects of mot gene expression on the structure of the flagellar motor. J. Mol. Biol. 202:575-584. [DOI] [PubMed] [Google Scholar]
- 12.Khan, S., I. H. Khan, and T. S. Reese. 1991. New structural features of the flagellar base in Salmonella typhimurium revealed by rapid-freeze electron microscopy. J. Bacteriol. 173:2888-2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kirchhoff, H. 1992. Motility, p. 289-306. In J. Maniloff, R. N. McElhaney, L. R. Finch, and J. B. Baseman (ed.), Mycoplasmas: molecular biology and pathogenesis. American Society for Microbiology, Washington, D.C.
- 14.Kirchhoff, H., P. Beyene, M. Fischer, J. Flossdorf, J. Heitmann, B. Khattab, D. Lopatta, R. Rosengarten, G. Seidel, and C. Yousef. 1987. Mycoplasma mobile sp. nov., a new species from fish. Int. J. Syst. Bacteriol. 37:192-197. [Google Scholar]
- 15.Kirchhoff, H., and R. Rosengarten. 1984. Isolation of a motile mycoplasma from fish. J. Gen. Microbiol. 130:2439-2445. [DOI] [PubMed] [Google Scholar]
- 16.Kirchhoff, H., R. Rosengarten, W. Lotz, M. Fischer, and D. Lopatta. 1984. Flask-shaped mycoplasmas: properties and pathogenicity for man and animals. Isr. J. Med. Sci. 20:848-853. [PubMed] [Google Scholar]
- 16a.Krause, D. C. 1996. Mycoplasma pneumoniae cytadherence: unravelling the tie that binds. Mol. Microbiol. 20:247-253. [DOI] [PubMed] [Google Scholar]
- 17.Maniloff, J. 1972. Cytology of mycoplasmas. Associated Scientific Publishers, Amsterdam, The Netherlands.
- 18.Maniloff, J., and H. J. Morowitz. 1972. Cell biology of the mycoplasmas. Bacteriol. Rev. 36:263-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McBride, M. J. 2001. Bacterial gliding motility: multiple mechanisms for cell movement over surfaces. Annu. Rev. Microbiol. 55:49-75. [DOI] [PubMed] [Google Scholar]
- 20.Miyata, M., W. S. Ryu, and H. C. Berg. 2002. Force and velocity of Mycoplasma mobile gliding. J. Bacteriol. 184:1827-1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miyata, M., W. S. Ryu, and H. C. Berg. 2001. Force-velocity relationship of mycoplasma gliding. Biophys. J. 80:70. [Google Scholar]
- 22.Miyata, M., and A. Uenoyama. 2002. Movement on the cell surface of gliding bacterium, Mycoplasma mobile, is limited to its head-like structure. FEMS Microbiol. Lett. 215:285-289. [DOI] [PubMed] [Google Scholar]
- 23.Miyata, M., H. Yamamoto, T. Shimizu, A. Uenoyama, C. Citti, and R. Rosengarten. 2000. Gliding mutants of Mycoplasma mobile: relationships between motility and cell morphology, cell adhesion and microcolony formation. Microbiology 146:1311-1320. [DOI] [PubMed] [Google Scholar]
- 24.Razin, S., M. Hasin, Z. Ne'eman, and S. Rottem. 1973. Isolation, chemical composition, and ultrastructural features of the cell membrane of the mycoplasma-like organism Spiroplasma citri. J. Bacteriol. 116:1421-1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Razin, S., D. Yogev, and Y. Naot. 1998. Molecular biology and pathogenicity of mycoplasmas. Microbiol. Mol. Biol. Rev. 62:1094-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25a.Razin, S., and E. Jacobs. 1992. Mycoplasma adhesin. J. Gen. Microbiol. 138:407-422. [DOI] [PubMed] [Google Scholar]
- 26.Rosengarten, R., M. Fisher, H. Kirchhoff, G. Kerlen, and K.-H. Seack. 1988. Transport of erythrocytes by gliding cells of Mycoplasma mobile 163K. Curr. Microbiol. 16:253-257. [Google Scholar]
- 27.Rosengarten, R., and H. Kirchhoff. 1987. Gliding motility of Mycoplasma sp. nov. strain 163K. J. Bacteriol. 169:1891-1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Uenoyama, A., A. Kusumoto, and M. Miyata. 2004. Identification of a 349-kilodalton protein (Gli349) responsible for cytadherence and glass binding during gliding of Mycoplasma mobile. J. Bacteriol. 186:1537-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wall, F., R. M. Pfister, and N. L. Somerson. 1983. Freeze-fracture confirmation of the presence of a core in the specialized tip structure of Mycoplasma pneumoniae. J. Bacteriol. 154:924-929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wilson, M. H., and A. M. Collier. 1976. Ultrastructural study of Mycoplasma pneumoniae in organ culture. J. Bacteriol. 125:332-339. [DOI] [PMC free article] [PubMed] [Google Scholar]