Abstract
Yersinia enterocolitica biovar 1B contains two type III secretion systems (TTSSs), the plasmid-encoded Ysc-Yop system and the chromosomally encoded Ysa-Ysp system. Proteins secreted from the Ysa TTSS (Ysps) have only been detected in vitro when cells are cultured at 26°C in a high-NaCl medium. However, the exact role of the Ysa TTSS is unclear. Thus, investigations into the regulation of this system may help elucidate the role of the Ysps during the life cycle of Y. enterocolitica. Here we present evidence that the AraC-like regulator YsaE acts together with the chaperone SycB to regulate transcription of the sycByspBCDA operon, a phenomenon similar to that seen in the closely related Salmonella SPI-1 and Shigella flexneri Mxi-Spa-Ipa TTSSs. Deletion of either sycB or ysaE results in a twofold reduction in the activity of a sycB-lacZ fusion compared to the wild type. In a reconstituted Escherichia coli system, transcription of sycB was activated sixfold only when both YsaE and SycB were present, demonstrating that they are necessary for activation. ysrR and ysrS are located near the ysa genes and encode a putative two-component regulatory system. Mutations in either gene indicated that both YsrR and YsrS were required for secretion of Ysps. In addition, transcription from sycB-lacZ and ysaE-lacZ fusions was decreased 6.5- and 25-fold, respectively, in the ysrS mutant compared to the wild type. Furthermore, in the absence of NaCl, the activity of ysaE-lacZ was reduced 25-fold in the wild-type and ΔysrS strains, indicating that YsrS is probably required for the salt-dependent expression of the ysa locus. These results suggest that the putative two-component system YsrRS may be a key element in the regulatory cascade for the Ysa TTSS.
The genus Yersinia has three species that are pathogenic to humans. Y. pestis is the causative agent of bubonic and pneumonic plagues, and Y. enterocolitica and Y. pseudotuberculosis primarily cause gastroenteritis. Most prevalent of the yersiniae in humans, the Y. enterocolitica infection is usually self-limiting to the gastrointestinal tract and mesenteric lymph node, causing gastroenteritis and lymphadenitis (12). However, in immunocompromised individuals, Y. enterocolitica can become systemic, and it has a 50% mortality rate in such cases (12).
Consumption of contaminated food or water is the primary source of Y. enterocolitica infection. Ingested bacteria are capable of surviving the gastric barrier and then migrate to the terminal ileum, where they attach to and subsequently invade the M cells that overlie the Peyer's patches (9, 25). Once inside the Peyer's patches, the bacteria replicate to high titers and can then disseminate to the mesenteric lymph nodes, spleen, and other organs, resulting in systemic disease (6, 8, 42, 51). The ability of Yersinia spp. to survive and replicate within the host is linked to the presence of a large virulence plasmid (43). This plasmid carries genes encoding the Ysc type III secretion apparatus as well as translocators, regulators, and effector proteins (Yops). At 26°C, the optimal growth temperature for Yersinia spp. outside the host, several copies of the secretion apparatus, called injectisomes, are detectable on the surface of the bacterium (37). At 37°C, in response to contact with target cells (in vivo) or to loss of Ca2+ ions (in vitro), the Yops are secreted. The functions of several Yops have been identified; these include translocation of effector Yops into host cells, impairment of phagocytosis, and downregulation of the host's inflammatory response (reviewed in reference 10).
While the virulence plasmid is necessary for full virulence, it is not sufficient, and several chromosomal genes have been identified as being important for the progression of disease. The genes encoding invasin, the primary invasion factor for Y. enterocolitica and Y. pseudotuberculosis, and its regulator, RovA, are located on the chromosome (26, 39, 40, 44). The highly virulent strains of biotype 1B have a high-pathogenicity island that contains genes involved in iron uptake (7). In addition, several genes have been identified through various means as having a role in virulence, but their functions are not yet understood (reviewed in reference 45).
Recently, a chromosomally encoded type III secretion system (TTSS) was discovered in Y. enterocolitica and designated Ysa, for Yersinia secretion apparatus (23). This system is only present in a subset of Y. enterocolitica strains, the highly virulent biotype 1B strains (serotypes O:4, O:8, O:13, and O:21) (19). A recent phylogenetic analysis of TTSSs revealed that the Ysa system is closely related to the Salmonella SPI-1 and Shigella flexneri Mxi/Spa TTSSs (19). Interestingly, there is a TTSS on the chromosome of Y. pestis, but it is more closely related to the Salmonella SPI-2 TTSS, indicating that the chromosomal TTSSs of Y. enterocolitica and Y. pestis were acquired after divergence of the species (19).
Several proteins, referred to as Ysps (Yersinia secreted proteins), can be detected in the supernatants of cultures grown at 26°C in the presence of high NaCl concentrations (20, 23). No proteins were detected when genes encoding putative Ysa apparatus components were disrupted or when cultures were grown at 37°C (20, 23, 55). Analysis by 50% lethal dose of a strain carrying a mutation in a putative apparatus gene, ysaV, showed no attenuation by intraperitoneal injection. However, by an oral route it was attenuated 10-fold, suggesting that the Ysa TTSS played a role in the early stages of infection (23). Seven Ysps have been identified to date. Three were identified as YopE, YopN, and YopP, which are encoded by the virulence plasmid (20, 54). The amounts of these Yops secreted under Ysa secretion conditions are significantly less than under Yop secretion conditions, and the relevance of their secretion is not understood. However, YopP, secreted only through the Ysa TTSS, was able to suppress the production of tumor necrosis factor alpha by infected macrophages (54). The other identified Ysps are encoded by the yspBCDA genes just downstream of the ysa apparatus genes (20). YspB, YspC, and YspD are homologous to proteins involved in translocation, but YspA is a unique protein (20). In addition, SycB has been demonstrated to function as a chaperone for YspB (20).
While a number of the proteins secreted by the Ysa TTSS have been identified and the functions for many genes in the locus have been inferred based on homology, nothing is known about the regulation of this system other than a requirement for growth at 26°C in high NaCl concentrations (23). The closely related Salmonella SPI-1 and S. flexneri Mxi/Spa TTSSs have an interesting regulation system that has not been observed in other TTSSs: each utilizes an AraC-like regulator and a chaperone to regulate the transcription of genes encoding secreted effectors (14, 15, 34). In Salmonella spp., it has been shown that InvF (AraC-like regulator) and SicA (chaperone) interact, and this interaction is likely to be required for the transcriptional activation because InvF alone can bind DNA but not activate transcription (15). The ysa locus has homologs of InvF and SicA, designated YsaE and SycB, respectively, and the genetic organization of these and surrounding genes is very similar (Fig. 1). Transcription of the TTS apparatus genes is regulated by HilA in Salmonella spp. (3, 31) and by VirB in S. flexneri (4), both of which are themselves regulated by various environmental conditions (reviewed in references 17 and 32). It appears that HilA and VirB serve as the focal point for transmitting the environmental signals that lead to expression of these type III secretion systems and their effectors. These regulators do not show any homology to each other, and no homologue of either protein exists in Y. enterocolitica. Therefore, identifying regulators upstream of the Ysa system is of interest and may facilitate an understanding of the role of the Ysa TTSS.
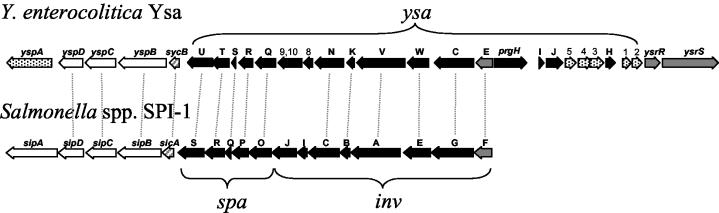
FIG. 1.
Organization of the ysa operon. Black arrows indicate putative apparatus genes, gray arrows indicate genes encoding regulators, and white arrows indicate genes encoding secreted proteins. Speckled genes and those labeled with numbers are unique. Open reading frames 8 and 9/10 are believed to encode proteins that are part of the TTSS apparatus (19) and are therefore colored black. sycB and sicA have dual functions as chaperone and regulator. Dotted lines indicate homologous genes between the two systems (only a portion of SPI-1 is shown). The intergenic region between ysaU and sycB is 96 bp; the analogous region in SPI-1 (spaS to sicA) is 137 bp. No terminator structure was predicted to exist in a 300-bp region that includes the ysaU-sycB intergenic region with Mfold (http://www.bioinfo.rpi.edu/applications/mfold/).
In this work, we investigated the transcriptional regulation of the ysa and ysp genes. We show that the AraC-like protein, YsaE, and chaperone, SycB, are both required to activate transcription of the sycByspBCDA operon, a phenomenon similar to that seen in Salmonella spp. and S. flexneri (13-15, 27, 34). In addition, we show that YsrS, the putative sensor protein of a two-component system, is required for expression of the ysaE promoter and that this activation is NaCl dependent. These results indicate that the putative two-component system YsrRS may be a key component in the regulatory cascade for the Ysa secretion apparatus.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this work are listed in Table 1 and described in detail below. Overnight cultures were typically grown in Luria broth (LB) (170 mM NaCl; Difco) at 26°C for Y. enterocolitica or 37°C for Escherichia coli, unless otherwise stated. For examination of secreted proteins, cultures were grown overnight in L-broth (1% tryptone, 0.5% yeast extract; referred to hereafter as LB-0) and subcultured into L-broth containing 290 mM NaCl (referred to hereafter as LB-290). Antibiotics were added as needed at the following concentrations: ampicillin, 100 μg/ml; kanamycin, 100 μg/ml; nalidixic acid, 20 μg/ml; chloramphenicol, 12.5 μg/ml; spectinomycin, 50 μg/ml; streptomycin, 50 μg/ml; and tetracycline, 7.5 μg/ml. 5-Bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) was added to a final concentration of 40 μg/ml.
TABLE 1.
Strains and plasmids used in this work
| Strain or plasmid | Relevant genotype | Reference or source |
|---|---|---|
| E. coli | ||
| DH5α | F−φ 80ΔlacZM15 Δ(lacZYA-argF)U169 deoP recA1 endA1 hsdR17 (rK− mK−) | Invitrogen |
| S17-1λpir | Tpr StrrrecA thi pro hsdR hsdM+ RP4::2-Tc::Mu::Km Tn7 λpir lysogen | 39 |
| CC118λpir | araD139 Δ(ara leu)7697 ΔlacX74 phoA20 galE galK thi rpsE rpoB argE(Am) recA1 λpir lysogen | 24 |
| VM1264 | CC118λpir carrying pRW50, pWKS130, pHG329 | This work |
| VM1265 | CC118λpir carrying pKW21, pKW22, pKW23 | This work |
| VM1266 | CC118λpir carrying pKW21, pWKS130, pKW23 | This work |
| VM1267 | CC118λpir carrying pKW21, pKW22, pHG329 | This work |
| VM1272 | CC118λpir carrying pKW21, pWKS130, pHG329 | This work |
| Y. enterocoliticaa | ||
| JB580v | 8081v (r− m+ Nalr) | 28 |
| JB580c | JB580v cured of pYVe8081 | Lab strain |
| YVM1063 | JB580c carrying pKW22 | This work |
| YVM1064 | JB580c carrying pWKS130 | This work |
| YVM886 | JB580v ysaC::pEP185.2 | This work |
| YVM932 | JB580v ΔysaE | This work |
| YVM981 | JB580v ΔsycB | This work |
| YVM997 | JB580v ΔysaE ΔsycB | This work |
| YVM969 | JB580v ΔysrS | This work |
| YVM1006 | JB580v ΔysrR | This work |
| YVM917 | YVM886c | This work |
| YVM971 | YVM932c | This work |
| YVM1025 | YVM971 carrying pKW22 | This work |
| YVM1042 | YVM971 carrying pWKS130 | This work |
| YVM996 | YVM981c | This work |
| YVM1035 | YVM996 carrying pKW28 | This work |
| YVM1052 | YVM996 carrying pKW27 | This work |
| YVM972 | YVM969c | This work |
| YVM1081 | YVM972 carrying pKW31 | This work |
| YVM1043 | YVM972 carrying pWKS130 | This work |
| YVM1089 | YVM1006c | This work |
| YVM1102 | YVM1089 carrying pKW24 | This work |
| YVM1103 | YVM1089 carrying pWKS130 | This work |
| YVM987 | JB580v sycB-lacZYA | This work |
| YVM988 | YVM981 sycB-lacZYA | This work |
| YVM989 | YVM932 sycB-lacZYA | This work |
| YVM1002 | YVM997 sycB-lacZYA | This work |
| YVM990 | YVM969 sycB-lacZYA | This work |
| YVM1054 | YVM988 carrying pKW28 | This work |
| YVM1054 | YVM988 carrying pKW27 | This work |
| YVM1019 | YVM989 carrying pKW22 | This work |
| YVM1044 | YVM989 carrying pWKS130 | This work |
| YVM1093 | YVM1002 carrying pKW28 | This work |
| YVM1094 | YVM1002 carrying pKW27 | This work |
| YVM1020 | YVM1002 carrying pKW22 | This work |
| YVM1045 | YVM1002 carrying pWKS130 | This work |
| YVM1062 | YVM1002 carrying pKW28 and pKW22 | This work |
| YVM1087 | YVM1002 carrying pKW27 and pWKS130 | This work |
| YVM1073 | YVM990 carrying pKW31 | This work |
| YVM1056 | YVM990 carrying pWKS130 | This work |
| YVM1061 | YVM990 carrying pKW28 and pKW22 | This work |
| YVM1060 | YVM990 carrying pKW27 and pWKS130 | This work |
| YVM1074 | YVM995 carrying pKW31 | This work |
| YVM1057 | YVM995 carrying pWKS130 | This work |
| YVM925 | JB580v ysaE-lacZYA | This work |
| YVM970 | YVM932 ysaE-lacZYA | This work |
| YVM995 | YVM969 ysaE-lacZYA | This work |
| Plasmids | ||
| pRW50 | Tetr; low-copy-number transcriptional reporter vector | 30 |
| pSR47S | Kanr; MobRP4 oriR6K, cloning vector | 36 |
| pWKS130 | Kanr; low-copy-number cloning vector | 52 |
| pHG329 | Ampr; medium-copy-number cloning vector | 48 |
| pEP185.2 | Cmr; MobRP4 oriR6K, cloning vector | 28 |
| pGEX-6P-1 | Ampr; cloning vector for generation of GST fusion proteins | Amersham |
| pKN8 | Cmr; MobRP4 oriR6K, transcriptional reporter vector | 18 |
| pKW27 | Cmr Strr Spr; pACYC184 with Str/Sp cassette | This work |
| pKW6 | ysrS lacking codons for amino acids 196-589 cloned into pSR47S | This work |
| pKW7 | ysaE lacking codons for amino acids 91-178 cloned into pSR47S | This work |
| pKW10 | sycB lacking codons for amino acids 68-140 cloned into pSR47S | This work |
| pKW16 | ysrR lacking codons for amino acids 25-207 cloned into pSR47S | This work |
| pSAH1 | ysaC codons 240-447 cloned into pEP185.2 | This work |
| pKW11 | sycB promoter region cloned into pKN8 | This work |
| pKW5 | ysaE promoter region cloned into pKN8 | This work |
| pKW21 | sycB promoter region cloned into pRW50 | This work |
| pKW22 (pYsaE) | ysaE coding sequence and promoter cloned into pWKS130 | This work |
| pKW23 | sycB coding sequence and promoter cloned into pHG329 | This work |
| pKW24 (pYsrR) | ysrR coding sequence and promoter cloned into pWKS130 | This work |
| pKW28 (pSycB) | sycB coding sequence and promoter cloned into pKW27 | This work |
| pKW31 (pYsrS) | ysrS coding sequence cloned into pWKS130 | This work |
| pYW2 | yspC coding sequence cloned into pGEX-6P-1 | This work |
The suffix v denotes strains carrying the pYVe8081 virulence plasmid, and the suffix c denotes strains that have been cured of this plasmid as described in the text.
Construction of plasmids.
The plasmids used to generate in-frame deletion mutants of sycB, ysrR, ysaE, and ysrS were constructed as follows. Primers sycB-delA and -delB were used to amplify a ≈500-bp region containing a few N-terminal codons and upstream sequence. This fragment was digested with SalI and BamHI and cloned into the same sites of pSR47S. Primers sycB-delC and -delD were used to generate a similarly sized fragment containing a few C-terminal codons and additional downstream sequence. This product was digested with BamHI and NotI and cloned into those sites of pSR47S containing the upstream fragment, resulting in plasmid pKW10. Plasmid pKW16 was constructed in an identical fashion with primer pairs ysrR-delA/delB and ysrR-delC/delD.
The PCR products generated with ysaE-delA/delB and ysaE-delC/delD were digested with ClaI and BglII and with BglII and XbaI, respectively. They were sequentially cloned into pEP185.2, generating pKW4. The SalI-NotI fragment from pKW4, which contained the ysaE inserts, was subcloned into pSR47S to make pKW6. PCR products generated with ysrS-delA/delB and ysrS-delC/delD were digested with KpnI and XbaI and with XbaI and SacI, respectively, and sequentially cloned into pEP185.2 to make pKW3. Primers ysrS-delA and ysrS-delD were used to amplify the ysrS inserts from pKW3, and the product was cloned into pCR2.1 TOPO. The SalI-NotI fragment from this plasmid was then subcloned into pSR47S, generating pKW7. Following ligation, each plasmid was transformed into E. coli strain S17-1λpir by electroporation. All constructs were confirmed by restriction digestion and sequenced to ensure that no errors were generated during amplification.
For generating a disruption in the ysaC gene, an internal region containing approximately 500 bp of the ysaC gene was amplified with primers ysaC-F1 and ysaC-R1 (Table 2) and cloned into the pCR2.1-TOPO vector (Invitrogen). The KpnI-XhoI fragment was cleaved out of that plasmid and cloned into the same sites of pEP185.2, resulting in pSAH1. This plasmid was transformed into E. coli strain S17-1λpir by electroporation and confirmed by restriction digestion.
TABLE 2.
Primers used in this work
| Primer | Sequencea (5′→3′) |
|---|---|
| ysaC-F1 | GGGTGAACCGACGATCGAA |
| ysaC-R1 | CAAGTTTGCCCGAGTTGTCA |
| ysaE-delA | CCATCGATCGATTCGATGGCTACCCGCTTTGAG |
| ysaE-delB | GAAGATCTTGCAGCATCAATCGTTGCGAGAGTTTCG |
| ysaE-delC | GAAGATCTGGCGTCTCTGCGGCCTACTTCAGGC |
| ysaE-delD | GCTCTAGACGGCTTCTCCAGCCGTTCAGCGACG |
| ysaE-FP1 | GCTCTAGACGATTCGATGGCTACCCGCTTTGAG |
| ysaE-FP4 | CGGGATCCGATTCGATGGCTACCCGCTTTGAG |
| ysaE-RP2 | CCCAAGCTTATGCAGCATCAATCGTTGCGAGAG |
| sycB-delA | ACGCGTCGACGGCTGGTACGCGTTGAGCTGG |
| sycB-delB | CGGGATCCGCGAAAGAACGTTTCGGCTTC |
| sycB-delC | CGGGATCCGGGAGTGATGATTTGGAGTTG |
| sycB-delD | ATAAGAATGCGGCCGCGCCAACGACCCCATCAACGATG |
| sycB-FP1 | GCTCTAGACCGGTAGCACGGCAGCTATGGCGG |
| sycB-RP1 | GAAGATCTGCTGATAAACAGCTGCCAACCCC |
| sycB-FP2 | CGGAATTCCCGGTAGCACGGCAGCTATGGCGG |
| sycB-FP3 | CCCAAGCTTCCGGTAGCACGGCAGCTATGGCGG |
| ysrS-delA | GGGGTACCTCACCGCAAGAGCTGG |
| ysrS-delB | GCTCTAGACGCAGCTTCAGCCTGCCG |
| ysrS-delC | GCTCTAGACCTGCTGCGGCTCGTGGG |
| ysrS-delD | GCGAGCTCACGGGCGCGCTGCGCATC |
| ysrS-OEF1 | GCGTCGACGGGCTTACTTCAAACACTGATTTC |
| ysrS-RP3 | ATAAGAATGCGGCCGCTCAGTCATGTTCTTTTTCCTTAG |
| ysrR-de1A | ACGCGTCGACGCAGGATAATCCGATGAAATCTCG |
| ysrR-de1B | CGGGATCCCATCAGCGCAAGGCGACTGAAAGG |
| ysrR-de1C | CGGGATCCGTATCGAACACGAAAACGCGTGCC |
| ysrR-de1D | ATAAGAATGCGGCCGCGCTTGGTAAACCACTCAATCAGCG |
| ysrR-RP1 | GGGGTACCTGGCCTCGGCAGCATAAACAGCCG |
| yspC-1.1 | ACGCGTCGACTCATGACCACTATTCAACAAGCCACGCAC |
| yspC-2.2 | ATATTGAATGCGGCCGCTTAACCCTTAACAATGGCCTGATTG |
| KW114 | TAGCACGGCAGCTATGGC |
| KW115 | AGCTGATAAACAGCTGCCAAC |
Restriction enzyme sites are underlined.
Transcriptional lacZ fusions were constructed by cloning putative promoter regions into pKN8 (18). For the ysaE and sycB promoters, approximately 300 bp of promoter sequence and 250 bp of coding sequence were amplified, digested with XbaI and BglII, and cloned into those sites of pKN8, resulting in plasmids pKW5 and pKW11, respectively. The plasmids were transformed into S17-1λpir by electroporation, confirmed by restriction digestion, and sequenced to ensure that no errors were generated during amplification. Plasmid pKW21 was made by digesting a PCR-generated fragment of the sycB promoter region with EcoRI and BamHI and ligating it into the same sites of pRW50. The ligated plasmid was transformed into E. coli strain DH5α, confirmed by restriction digestion, and sequenced. The primer sequences and pairs used for these constructs are listed in Tables 2 and 3, respectively.
TABLE 3.
Primer pairs used for transcriptional fusions and complementing clones
| Gene | Plasmid | 5′ primer | 3′ primer | Region amplified (bp)a |
|---|---|---|---|---|
| ysaE | pKW5 | ysaE-FP1 | ysaE-de1B | −291 to +249 |
| sycB | pKW11 | sycB-FP1 | sycB-RP1 | −311 to +264 |
| sycB | pKW21 | sycB-FP2 | sycB-de1B | −311 to +200 |
| sycB | pKW23 | sycB-FP3 | sycB-RP2 | −311 to +574 |
| sycB | pKW28 | sycB-FP1 | sycB-RP2 | −311 to +574 |
| ysaE | pKW22 | ysaE-de1A | ysaE-de1D | −291 to +963 |
| ysrS | pKW31 | ysrS-OEF1 | ysrS-RP3 | −31 to +2376 |
| ysrR | pKW24 | ysrR-de1A | ysrR-RP1 | −476 to +772 |
Base pairs amplified relative to the putative start codon (+1).
Two plasmids carrying the sycB coding region were constructed. In the first, primers sycB-FP3 and -RP2 were used to amplify the sycB gene and promoter region. The product was digested with EcoRI and HindIII, cloned into those sites of pHG329, and transformed into DH5α, giving pKW23. The second PCR product, generated with primers sycB-FP1 and -RP2, was digested with BglII and HindIII and cloned into those sites in pKW27, giving pKW28. Plasmid pKW27 is pACYC184 with a streptomycin-spectinomycin resistance cassette from p34-Sm (16) cloned into the BamHI site; the tetracycline resistance cassette was disrupted by this insertion. The ysaE-complementing clone was made by digesting a PCR product from primers ysaE-delA and -delD with SalI and NotI and cloning it into those sites of pWKS130 to give pKW22. The ysrR-complementing clone was made by digesting a PCR product from primers ysrR-delA and -RP1 with SalI and KpnI and cloning it into those sites of pWKS130 to give pKW24. For the ysrS-complementing clone, a PCR product generated by primers ysrS-OEF1 and -RP3 was digested with SalI and NotI and cloned into those sites of pWKS130 to give pKW31. All plasmids were transformed into DH5α by electroporation, confirmed by restriction digestion, and sequenced to ensure that no errors occurred during amplification. Primer sequences and pairs used for these constructs are given in Tables 2 and 3, respectively.
Primers yspC 1.1 and yspC 2.2 (Table 2) were used to amplify the entire coding sequence of YspC. The PCR product was digested with SalI and NotI and cloned into the same sites of pGEX-6P-1 (Amersham). The resulting plasmid, pYW2, encodes a glutathione S-transferase (GST)-YspC fusion protein.
Strain construction. (i) In-frame deletions.
Strains YVM969, YVM932, YVM981, and YVM1006 containing chromosomal in-frame deletions in ysrS, ysaE, sycB, and ysrR, respectively, were made by conjugation as follows. Equal volumes of saturated cultures of E. coli carrying the desired plasmid (pKW6, -7, -10, or -16) and Y. enterocolitica JB580v were mixed, plated on LB agar, and allowed to incubate at 26°C overnight. The resulting lawn of cells was scraped into 1 ml of 1× PBS, diluted 1:100, and plated on LB agar plates containing nalidixic acid to select against E. coli and kanamycin to select against Y. enterocolitica lacking the plasmid. Replication of pSR47S requires the pir protein; all Y. enterocolitica strains used lack pir, and thus survivors would have undergone site-specific recombination. Several transconjugants were streaked onto LB agar plates containing nalidixic acid and 5% sucrose for selection of colonies that had undergone a second recombination step and lost the vector. Cells retaining a functional sacB gene should not grow in the presence of sucrose. Several of these colonies were then screened for kanamycin sensitivity, of which 10 to 20 were picked for confirmation of the in-frame deletion by colony PCR (see below).
To ensure that recombination occurred in the proper location, strains selected for experiments were analyzed by Southern blotting. Strains used for analysis of Ysa-dependent secretion were subsequently cured of the virulence plasmid by inoculation on LB agar containing 20 mM MgCl2 and 20 mM NaC2O4 and grown at 37°C. Loss of the virulence plasmid was verified by visualization of plasmid DNA preparations on 0.8% agarose gels. The test strains were always compared to plasmid preparations from JB580v and JB580c, which served as positive and negative controls, respectively. The cured strains of YVM969, YVM932, YVM981, and YVM1006 are designated YVM972, YVM971, YVM996, and YVM1089, respectively.
(ii) Plasmid integrations.
Strain YVM886 was generated by conjugating pSAH1 into JB580v as described above but with selection on LB agar containing nalidixic acid and chloramphenicol. Proper insertion of the plasmid was confirmed by Southern blotting. YVM886 was then cured of pYVe8081 as described above to yield YVM917. Strains carrying chromosomal promoter-lacZ fusions were constructed by conjugating either pKW5 or pKW11 into the desired Y. enterocolitica strain, followed by selection on LB agar containing nalidixic acid and chloramphenicol, giving YVM925 and YVM987, respectively. Because of the region that was cloned, merodiploid strains were generated by the recombination event. Proper integration of the plasmid was confirmed by Southern blotting prior to analysis. These strains were not subsequently cured of pYVe8081.
(iii) Plasmid transformation.
For Y. enterocolitica, the strain to be transformed was inoculated into LB broth containing 1% glucose and grown overnight. Approximately 500 μl of the saturated culture was washed twice with an equal volume of ice-cold distilled H2O and resuspended in 40 μl of 10% glycerol; 1 to 2 μl of plasmid DNA was added to the cells and electroporated by standard procedures. Ten percent of the recovered culture was plated on LB agar with appropriate antibiotics. When two plasmids were needed, both were transformed simultaneously. For E. coli strains VM1265, VM1266, VM1267, and VM1272, electrocompetent CC118 λpir cells were simultaneously transformed with all three desired plasmids. Ten percent of the recovered culture was plated on LB agar with appropriate antibiotics.
PCR and DNA sequencing.
Standard methods for PCR were conducted under the conditions specified by the supplier with either Pfu polymerase (Stratagene, La Jolla, Calif.) or Taq polymerase (Qiagen, Valencia, Calif.). Colony PCR was performed in a standard reaction in 50 μl. A single colony was resuspended in 50 μl of distilled H2O and vortexed, and 5 μl was used as the template. A 5-min incubation at 95°C preceded the cycling reactions to ensure cell lysis. DNA sequencing was performed with the Big Dye termination cycle sequencing ready reaction system under the conditions specified by the supplier (PE Applied Biosystems, Foster City, Calif.). Reactions were analyzed at the Protein and Nucleic Acid Chemistry Laboratory at the Washington University School of Medicine.
β-Galactosidase assays.
Saturated cultures grown overnight in LB-0 were diluted into fresh LB-290 to an initial optical density at 600 nm (OD600) of 0.2 and grown for 4 h at 26°C on a roller drum. Antibiotics were added as necessary to retain plasmids. Assays were performed as described before (38).
Total RNA extraction and RT-PCR.
A saturated culture of JB580v grown overnight in LB-0 was diluted into fresh LB-290 to an initial OD600 of 0.2 and grown for 4 h at 26°C on a roller drum. RNAprotect bacterial reagent (Qiagen) was added to the cell sample as described by the manufacturer. The cells were collected by centrifugation, and total RNA was extracted with the MasterPure RNA extraction kit from Epicenter. DNA was removed from 20 μg of sample with DNA-free (Ambion) following the manufacturer's protocol. For cDNA synthesis, 2 μg of RNA was used as a template with 200 U of Superscript III as described by the supplier (Invitrogen). PCR was performed with the cDNA synthesis products as the template with primers KW114 and KW115 (Table 2) and Taq polymerase (Qiagen) in a 50-μl reaction volume. For controls, PCR was performed without cDNA template as well as with genomic DNA to show the expected size of the product generated with these primers. One fifth of the reaction was separated on a 1.2% agarose gel and stained with ethidium bromide.
Preparation of secreted proteins, SDS-PAGE, and Western blot analysis.
Extracellular proteins were collected as described before (55). Briefly, saturated cultures grown in LB-0 were diluted into fresh LB-290 to an initial OD600 of 0.2 and grown for 6 h at 26°C on a roller drum. The longer culture time for protein preparations than for expression studies was chosen because the preparations were cleaner, making detection of individual bands easier. Antibiotics were added as necessary to retain plasmids. The cells were removed from 4.5 ml of culture by centrifugation in microcentrifuge tubes for 1 min at 13,000 rpm. The supernatant was centrifuged a second time, followed by passage through a 0.22-μm syringe filter. Ice-cold trichloroacetic acid was added to a final concentration of 10% (vol/vol) and incubated on ice for 10 to 20 min. The samples were centrifuged at 4°C for 10 min at 13,000 rpm, washed once with ice-cold acetone, and resuspended in 1 M Tris-HCl, pH 9.0. The proteins were boiled for 5 min in 1× sample buffer (46), and OD600 equivalents were loaded onto sodium dodecyl sulfate (SDS)-polyacrylamide gels. Proteins were visualized by staining with silver nitrate (Bio-Rad) or transferred to nitrocellulose for Western analysis with a Bio-Rad Trans-Blot SD semidry transfer apparatus as specified by the supplier. Blots were blocked in 1× phosphate-buffered saline (PBS) with 0.1% Tween 20 and 5% skim milk (PBST-milk) for 1 h at room temperature. Primary antibody directed against YspC was diluted 1:1,000 in PBST-milk and allowed to react overnight at 4°C. The membranes were washed several times with PBST-milk and then incubated with goat anti-rabbit immunoglobulin G-horseradish peroxidase at 1:25,000 in PBST-milk for 1 h at room temperature. The membranes were washed again in PBST-milk, and proteins were detected by chemiluminescence (ECL; Amersham).
To generate the anti-YspC antibody, a GST-YspC fusion protein was purified from E. coli carrying pYW2 with the bulk GST purification module as specified by the supplier (Amersham). Approximately 1 mg of GST-YspC was sent to Covance Research Products for immunization of a New Zealand White rabbit.
RESULTS
Regulation of the sycB-yspBCDA operon.
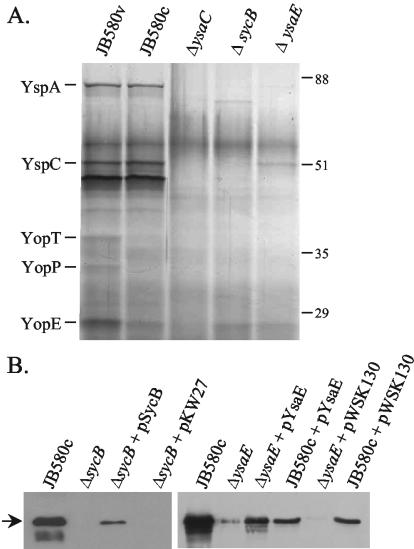
The organization of the ysa-syc-ysp genes in Yersinia spp. is quite similar to that in Salmonella. SPI-1 and to a lesser degree to the S. flexneri mxi-spa-ipa genes. Given that both the Salmonella and Shigella systems employ an AraC-like regulator (InvF and MxiE, respectively) and a TTSS chaperone (SicA and IpgC, respectively) to regulate the expression of genes encoding secreted proteins, it was of interest to investigate if the analogous proteins, YsaE and SycB, functioned similarly in the Ysa system. To test this hypothesis, in-frame deletions of ysaE and sycB were constructed, and the strains were examined for their ability to secrete proteins after growth in L-broth with 290 mM NaCl (LB-290). Both strains showed a loss of most protein bands (Fig. 2A). This indicates that YsaE and SycB are required for wild-type levels of secretion.
FIG. 2.
Strains lacking sycB or ysaE do not secrete Ysps. Proteins were precipitated from culture supernatants and separated by SDS-10% polyacrylamide gel electrophoresis as described in the text. (A) Silver-stained gel showing loss of Ysps from the culture supernatants of ΔsycB and ΔysaE strains. ΔysaC carries a disruption in the ysaC gene and is thought to have a defective apparatus. The culture equivalent of 2 OD units was loaded in each lane. (B) Complementation of the mutant strains as determined by Western blotting with anti-YspC antibody. YspC is indicated by the arrow. The culture equivalent of 1 OD unit was loaded in each lane.
Western blots probed with anti-YspC antibody, which recognizes the secreted protein YspC, showed that secretion was restored when the wild-type gene was provided in trans, demonstrating that the reduced secretion was due to the loss of the deleted gene (Fig. 2B). Curiously, the complemented ΔysaE strain consistently appeared to secrete less YspC than the wild-type strain. The presence of kanamycin in the medium did not significantly impair the growth of the cultures (not shown). However, the antibiotic (or plasmid carriage) may somehow interfere with secretion or precipitation of Ysps, since the wild-type strain carrying pWKS130 or pKW22 secreted less YspC than the strain without either plasmid (Fig. 2B). A similar phenomenon was observed with the sycB-complementing clone (not shown).
In related TTSSs, InvF/SicA and MxiE/IpgC act by stimulating the transcription of genes encoding secreted proteins (13-15, 27, 34). To test the hypothesis that YsaE and SycB were acting as transcriptional regulators, a sycB-lacZ fusion was introduced into the ΔsycB and ΔysaE strains, and β-galactosidase activities were determined. The activity of the sycB promoter decreased about twofold in the ΔysaE and ΔsycB strains, suggesting that both proteins play a role in the transcription of the sycByspBCDA operon (Table 4). The ysaE and sycB mutants could be fully complemented for sycB expression when the respective wild-type gene was provided on a plasmid; strains carrying the vector alone showed no change in activity from the mutants. A similar twofold reduction in transcription was observed in the ysaE sycB double mutant, and activity of sycB-lacZ was only restored when both ysaE and sycB were provided in trans. Promoter activity in each of the complemented strains was much higher than in the wild type, indicating that YsaE or SycB may be limiting in the wild type under the conditions examined. Expression in the wild-type strain carrying plasmid pSycB, pYsaE, or both was also increased, but no difference was observed with the vectors (not shown). The observed effects of SycB and YsaE were independent of genes encoded by the virulence plasmid. Loss of the virulence plasmid had no effect on sycB-lacZ transcription in a wild-type strain (not shown).
TABLE 4.
Regulation of sycB promoter by SycB and YsaE
| Relevant phenotype | Strain | Plasmida | Mean β-galactosidase activity (Miller units)b ± SD |
|---|---|---|---|
| SycB+, YsaE+ | YVM987 | None | 512 ± 58 |
| SycB−, YsaE+ | YVM988 | None | 282 ± 33 |
| YVM1054 | pSycB | 2,851 ± 426 | |
| YVM1055 | pKW27 | 295 ± 4 | |
| SycB+, YsaE− | YVM989 | None | 297 ± 13 |
| YVM1019 | pYsaE | 1,784 ± 286 | |
| YVM1044 | pWKS130 | 263 ± 9 | |
| SycB−, YsaE− | YVM1002 | None | 331 ± 26 |
| YVM1093 | pSycB | 267 ± 20 | |
| YVM1094 | pKW27 | 232 ± 12 | |
| YVM1020 | pYsaE | 279 ± 3 | |
| YVM1045 | pWKS130 | 292 ± 23 | |
| YVM1062 | pSycB, pYsaE | 3,867 ± 132 | |
| YVM1087 | pKW27, pWKS130 | 297 ± 52 |
Plasmid pSycB is pKW28, and pYsaE is pKW22; pKW27 is the vector for pSycB, and pWSK130 is the vector for pYsaE.
Miller units represent the mean of at least three independent assays with standard deviations.
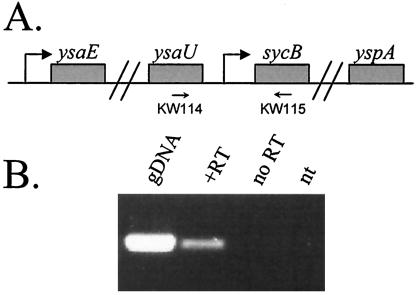
The possibility existed that transcription initiating at the ysaE promoter may also transcribe the sycByspBCDA genes, as is thought to be the case in SPI-1 (13). If this is so, then the decrease in transcription of the sycB promoter observed in the ΔysaE and ΔsycB strains may not be an accurate measure of the contribution of these two proteins on transcription initiating at the sycB promoter. To determine if a transcript existed that initiated upstream of ysaU, RT-PCR was used to amplify a region that encompassed the ysaU-sycB intergenic region. A product was detected only when genomic DNA or cDNA was added to the reaction, but not from reactions containing no template or template from a cDNA synthesis reaction lacking reverse transcriptase (Fig. 3). These data demonstrate that transcription of sycByspBCDA genes can indeed initiate at a promoter upstream of ysaU and contributes to the transcription of this operon.
FIG. 3.
Transcription of sycB originates at a promoter upstream of ysaU. (A) Schematic of the ysa locus encompassing the sycB promoter region. The approximate locations of primers KW114 and KW115 are indicated. (B) RT-PCR was performed with primers KW114 and KW115 and cDNA generated from 2 μg of total RNA that was isolated from JB580v as described in the text; 20% of the reaction was loaded on a 1.2% agarose gel and stained with ethidium bromide. Templates for the PCR are listed above each lane and were as follows: gDNA, genomic DNA; +RT, products from cDNA synthesis reaction with Superscript III added; no RT, products from cDNA synthesis reaction with no Superscript III; nt, no DNA or cDNA added.
In order to analyze the effects of SycB and YsaE on sycB-lacZ expression without the contributions from the upstream promoter, a plasmid-based system was reconstituted in E. coli. Transcription from sycB-lacZ was at background levels if only SycB or YsaE was present (Table 5). However, when both YsaE and SycB were present, the sycB promoter was activated about sixfold. This suggests that both proteins are necessary and probably sufficient to stimulate transcription from the sycB promoter, although we cannot exclude the existence of additional regulators.
TABLE 5.
YsaE and SycB are sufficient to activate the sycB promoter in E. coli CC118 λpir
| Strain | Plasmida
|
Mean β-galactosidase activity (Miller units)b ± SD | ||
|---|---|---|---|---|
| sycB-lacZ | YsaE | SycB | ||
| VM1265 | + | + | + | 57.8 ± 2 |
| VM1266 | + | − | + | 9.9 ± 0 |
| VM1267 | + | + | − | 9.1 ± 1 |
| VM1272 | + | − | − | 10.0 ± 1 |
Plasmid constructs are as follows: sycB-lacZ, pKW21; YsaE, pKW22; SycB, pKW23; +, presence of construct; −, vector only.
Miller units represent the mean of at least three independent assays with standard deviations.
Furthermore, these data, combined with the RT-PCR data, indicate that at least two promoters are transcribing the sycByspBCDA genes.
Regulation of the ysa operon.
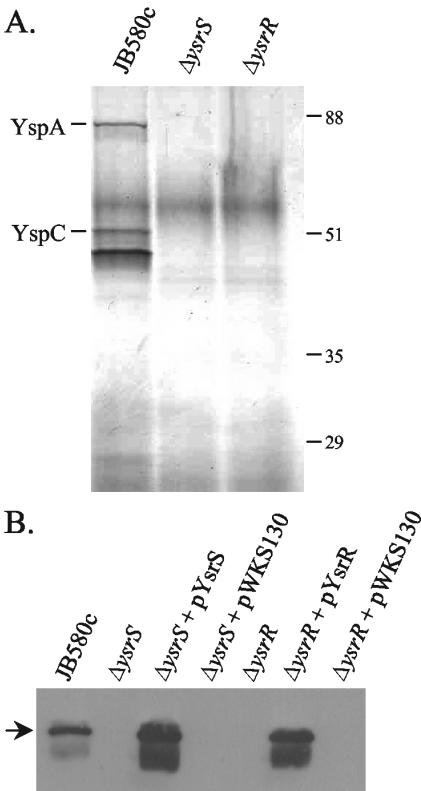
Since Y. enterocolitica does not appear to have a homologue to known key regulators of the SPI-1 and Mxi-Spa TTSSs, there is no obvious regulator for the expression of the ysa genes. However, proximal to the ysa locus are two genes that encode a putative two-component regulatory system, ysrR and ysrS (23) (Fig. 1). These genes are closely related to the rcsB and rcsC genes, respectively, which encode a two-component regulatory system that regulates gene transcription in a number of enteric organisms, often in response to osmotic shock (2, 47). YsrS is the putative hybrid sensor component and has regions similar to the conserved His- and Asp-containing domains of sensor proteins but, like RcsC, lacks the conserved His residue in the phosphotransfer (HPt) domain. YsrR has regions similar to the Asp-containing receiver domain and a putative helix-turn-helix DNA binding domain. To see if YsrRS is involved in regulating the ysa genes, in-frame deletions were constructed in the ysrS and ysrR genes. Examination of proteins in culture supernatants after growth in LB-290 indicated that both the ΔysrS and ΔysrR strains were defective in the secretion of all of the Ysps (Fig. 4A). These mutant phenotypes could be complemented by providing the wild-type gene in trans, indicating that the loss of secretion was due to the absence of functional YsrR or YsrS (Fig. 4B).
FIG. 4.
Strains lacking ysrS or ysrR do not secrete Ysps. Proteins were precipitated from culture supernatants and separated by SDS-10% polyacrylamide gel electrophoresis as described in the text. (A) Silver-stained gel showing the loss of all Ysps from the culture supernatants of the ΔysrS and ΔysrR strains. The culture equivalent of 2 OD units was loaded in each lane. (B) Complementation of the mutant strains as determined by Western blotting with anti-YspC antibody. YspC is indicated by the arrow. The culture equivalent of 1 OD unit was loaded in each lane.
To determine if the defect in Ysp secretion observed in the ΔysrS and ΔysrR strains was due to decreased transcription, the activity of the ysaE-lacZ and sycB-lacZ fusions was examined in the ΔysrS strain. The β-galactosidase activities of the sycB and ysaE promoters were down 6.5- and 25-fold, respectively, compared to the wild-type strain, indicating that YsrS is required for full activity of both promoters (Table 6). Addition of ysrS in trans restored promoter activity, demonstrating that the observed decrease was indeed due to loss of ysrS; no change in activity was observed in strains carrying the vector. However, the YsrS regulatory pathway could be acting indirectly at the sycB promoter by activating expression of the upstream ysaE promoter and thus production of YsaE and SycB. To determine if the reduced sycB-lacZ activity resulted from a direct loss of activation at the sycB promoter, the ΔysrS strain was transformed with pSycB and pYsaE. If the ysrS defect can be overcome by expressing ysaE and sycB in trans, this would suggest that the YsrRS system acts indirectly at the sycB promoter. Indeed, the activity of the sycB-lacZ fusion was restored in the ΔysrS strain carrying pYsaE and pSycB, indicating that the observed decrease in activity at this promoter in the ΔysrS strain is probably a downstream effect from the loss of activation at the ysaE promoter.
TABLE 6.
Regulation by YsrS in Y. enterocolitica
| Relevant phenotype | Plasmida | Strain | β-Galactosidase activity with sycB-lacZb | Strain | β-Galactosidase activity with ysaE-lacZb |
|---|---|---|---|---|---|
| YsrS+ | None | YVM987 | 512 ± 58 | YVM925 | 398 ± 6 |
| YsrS− | None | YVM990 | 78 ± 3 | YVM995 | 16 ± 1 |
| pYsrS | YVM1073 | 3,770 ± 369 | YVM1074 | 1,688 ± 70 | |
| pWKS130 | YVM1056 | 75 ± 5 | YVM1057 | 16 ± 1 | |
| pSycB, pYsaE | YVM1061 | 1,622 ± 251 | ND | ||
| pKW27, pWKS130 | YVM1060 | 41 ± 2 | ND |
Plasmid pYsrS is pKW31, pSycB is pKW28, and pYsaE is pKW22. pKW27 is the vector for pSycB, and pWSK130 is the vector for pYsaE and pYsrS.
Values are expressed as Miller units and represent the mean of at least three independent assays with standard deviations. ND, not determined.
Regulation by YsrS requires NaCl.
Because the function of sensor proteins is generally to detect environmental cues and secretion of Ysps is only observed when cells are cultured in high NaCl, we hypothesized that YsrS responds to the NaCl in the growth medium. To test the idea that YsrS senses NaCl, wild-type and ΔysrS strains containing the ysaE-lacZ fusion were grown in LB-0 and LB-290 and assayed for β-galactosidase activity (Table 7). In LB-0, β-galactosidase activity in the wild-type strain was the same as in the ΔysrS strain. In addition, no induction by NaCl was observed in the ΔysrS strain, yet the wild-type strain showed 25-fold induction. Furthermore, induction of ysaE by NaCl could be restored in the ΔysrS strain by providing ysrS in trans. This revealed that, in the absence of YsrS or in the absence of NaCl, there is no activation of the ysaE promoter.
TABLE 7.
Regulation of the ysaE promoter by YsrS requires NaCl
| Relevant phenotype | Strain | Plasmida | β-Galactosidase activityb (% of wild-type level) with:
|
|
|---|---|---|---|---|
| 290 mM NaCl | 0 mM NaCl | |||
| YsrS+ | YVM925 | None | 398 ± 6 (100) | 16 ± 1 (4) |
| YsrS− | YVM995 | None | 16 ± 1 (4) | 20 ± 2 (5) |
| YVM1074 | pYsrS | 1,688 ± 70 (425) | 30 ± 7 (7) | |
| YsaE− | YVM970 | None | 309 ± 13 (78) | 16 ± 1 (4) |
pYsrS is pKW31.
Values are expressed as Miller units and represent the mean of at least three independent assays with standard deviations. Percent activity for each promoter relative to the wild-type strain grown in 290 mM NaCl is shown in parentheses.
To confirm that YsrS was required for the NaCl-dependent activation, the ysaE-lacZ fusion was similarly tested in the ΔysaE strain. Consistent with previous studies on InvF (22), loss of YsaE did not lead to a significant reduction in activity of its own promoter when cultured in LB-290. Loss of SycB also did not affect the expression of ysaE-lacZ (not shown). However, just as in the wild-type strain, activity from the ysaE promoter was reduced when it was cultured in LB-0. This indicates that the NaCl-dependent activation of the ysa operon requires YsrS but not YsaE. This effect could be a response to changes in osmolarity rather than a specific NaCl-dependent effect, but this has not been fully explored.
Intriguingly, the growth rate of the ΔysrS and ΔysrR strains was slightly but reproducibly faster than that of the wild-type strain when grown in LB-290. Doubling times for the ΔysrS and ΔysrR strains was typically about 10 min faster than for the wild type (≈83 min for the ΔysrS and ΔysrR strains and ≈93 min for the wild type). This increased doubling time was not observed for these strains grown in LB-0, nor was it observed for the ΔysaE or ΔysaC strains grown in LB-290, indicating that it is specific to the ΔysrS and ΔysrR strains when cultured in the presence of NaCl. This also suggests that it is not related to the secretion of Ysps.
DISCUSSION
In this work, we showed that the AraC-like regulator YsaE and the chaperone SycB are involved in the regulation of a subset of the Ysps. Further examination of this phenomenon showed that the sycByspBCDA operon is transcriptionally regulated by YsaE and SycB. Loss of either activator resulted in a reduction in sycB-lacZ activity in Y. enterocolitica. Similarly, a reconstituted system in E. coli showed a sixfold activation of sycB-lacZ only in the presence of both regulators. These data indicate that YsaE and SycB are necessary and likely sufficient to activate transcription from this promoter, although the existence of additional regulators cannot be excluded. The increased activation observed in the isolated E. coli system compared to that observed in Y. enterocolitica is probably a consequence of transcription of sycByspBCDA initiating at the upstream promoter (ysaE) that is not subject to regulation by YsaE and SycB. This is also probably the case in Salmonella SPI-1, where transcription of the sicAsipBCDA genes can occur through initiation at an upstream promoter (possibly the invF promoter), as well as at the sicA promoter (13).
A similar mechanism of type III effector gene regulation by MxiE and IpgC exists in S. flexneri. Here, it is not the translocator operon that is affected but a set of proteins whose secretion is only observed under conditions of active secretion (27, 34). By incrementally overexpressing IpgC, Mavris et al. showed that the expression of these proteins increased as the concentration of IpgC increased (34). The authors concluded that the level of free IpgC, which would be found when its cognate proteins had been secreted, is the signal that leads to increased transcription of the secreted proteins. This report marks the third example of an AraC-like regulator acting with a type III chaperone to stimulate transcription of secreted proteins. Thus, it is likely that this is a conserved mechanism by which the cell monitors its secretion state and links it to transcriptional regulation.
The mechanism(s) behind this activation is not well understood. It has been shown that InvF can bind DNA in the absence of SicA, but SicA by itself does not bind DNA (15). The DNA binding sites for InvF and MxiE have been identified and are strikingly similar; in the center of each site, there is a T-rich region (15, 35), which may facilitate bending of the DNA by the protein. These promoters also lack an obvious −35 consensus sequence. In the region thought to contain the sycB promoter, a T-rich region is in roughly the same location as in the InvF- and MxiE-regulated genes, and there is no definable −35 region. However, much work remains to determine if this is indeed a YsaE binding site.
Many bacteria, both gram-negative and gram-positive, use two-component regulatory systems to regulate virulence genes. For example, RcsBC is required for the expression of capsule genes in E. coli (49), Erwinia amylovora (5), and Klebsiella pneumoniae (1) as well as Vi antigen expression in Salmonella enterica serovar Typhi (2). In S. enterica serovar Typhimurium, the type III secretion systems encoded on both SPI-1 and SPI-2 are regulated by multiple two-component systems (29, 32, 33, 41). In this work, we present data suggesting that expression of the ysaE promoter requires YsrS and most likely YsrR. YsrS and YsrR are encoded by genes adjacent to the ysa locus and comprise a putative two-component regulatory system (19, 23). Analysis of secreted proteins as well as the lacZ fusion to ysaE in the ysrS mutant revealed that YsrS is a key component in the expression of the ysa locus. Loss of YsrS also resulted in lower sycB promoter activity. However, providing ysaE and sycB in trans in the ΔysrS strain complemented sycB activity. Together, these results indicate that the ysaE promoter is regulated by YsrS and also suggests that initiation at ysaE can lead to transcription of sycByspBCDA. YsrR is also a critical component, as an in-frame deletion of ysrR similarly resulted in complete loss of all secreted proteins in culture supernatants. Secretion of Ysps has not been observed in the absence of NaCl in the culture medium, and stimulation of ysaE transcription by YsrS requires NaCl. Thus, YsrS may be functioning as an environmental sensor of NaCl or osmolarity. Furthermore, since YsrS is probably a membrane-bound protein, it is more likely that the actual transcriptional regulator is YsrR; however, this has yet to be experimentally tested.
BLAST searches with YsrS revealed homology to RcsC in E. coli and other enteric bacteria. While much of the similarity was limited to the conserved domains, YsrS is also similar to RcsC in that they both lack a histidine phosphotransferase (HPt) domain. This domain contains the second His site that transfers the phosphate to the Asp residue on the response regulator (53). In E. coli, the HPt-containing protein YojN has been identified as the intermediate between RcsC and RcsB (50). Interestingly, YojN lacks the other necessary His and Asp residues typically found in sensors, suggesting that the only functional domain is the HPt (50). Another example of a third partner in phosphorelay is with the LuxN-LuxU-LuxO system in Vibrio harveyi. LuxN lacks an HPt domain, while LuxU contains the appropriate His residue (21). There are several open reading frames that contain HPt domains within the Y. enterocolitica genome, suggesting that such an intermediate protein providing this domain for the YsrS-YsrR phosphorelay does indeed exist. YsrR has homology to a number of response regulators, including RcsB. However, most of the conserved residues are in the LuxR-type helix-turn-helix DNA-binding motif.
Although the activity of YsrR and YsrS appears to require NaCl, they probably do not have a role in osmoprotection, as is suggested for the RcsC-YojN-RcsB system (56). This is evidenced by the increased growth rate observed in the ΔysrS and ΔysrR strains in the presence of high NaCl concentrations. In fact, the Y. enterocolitica genome contains genes that are probably the true RcsBC orthologues, based on amino acid similarity and genetic organization (http://www.sanger.ac.uk/Projects/Y_enterocolitica/). Thus, YsrRS represent a new and uncharacterized two-component system. In virulence plasmid-containing Yersinia strains, growth of the bacterium slows when Yop secretion is induced (11 and references therein). The ΔysaC strain, which is defective in secretion but not transcription, does not display altered growth. Therefore, the faster growth of the ΔysrS and ΔysrR strains might be a consequence of not using metabolic resources for expressing the ysa and ysp genes.
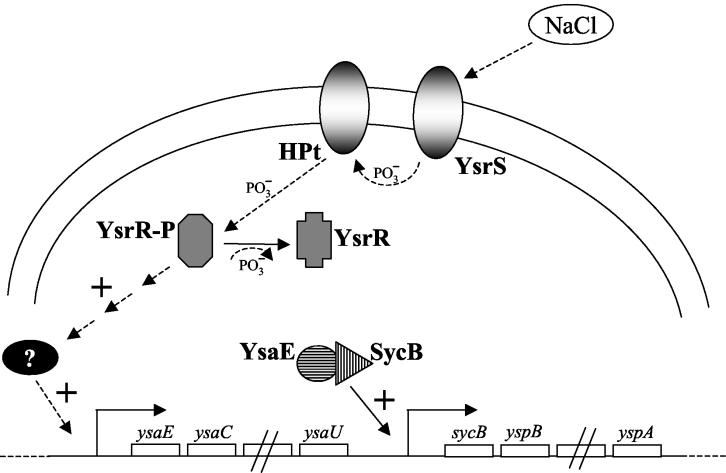
Based on the results presented here, a schematic model for Ysa TTSS regulation is proposed in Fig. 5. We hypothesize that YsrS senses an environmental cue (NaCl), transfers phosphate to an HPt-containing intermediate (HPt), which then transfers the phosphate to YsrR. The activated phospho-YsrR then stimulates transcription of the ysaE promoter, either directly or indirectly. Once levels of YsaE and free SycB are sufficient, they stimulate transcription of the sycB promoter and possibly others. From the experiments conducted with InvF and MxiE (15, 35), we can infer that YsaE probably binds DNA in the absence of SycB. It will be of interest to investigate if SycB enhances the DNA binding affinity of YsaE, if SycB makes any contact with the DNA, and if either protein contacts RNA polymerase. Similar questions arise surrounding the regulation by YsrRS. It remains to be demonstrated that YsrR directly regulates promoter activity and if it is indeed involved in a phosphorylation cascade with YsrS. It also remains to be demonstrated whether or not YsrS directly senses NaCl and other environmental cues for activating this system.
FIG. 5.
Model for activation of the ysaE and sycB promoters. YsrS senses NaCl in the culture medium by an unknown mechanism and initiates a phosphorelay that leads to phosphorylation of YsrR. The activated YsrR then stimulates transcription of the ysaE promoter, either directly or indirectly. Once sufficient levels of YsaE and SycB have accumulated, they stimulate transcription of the sycB promoter.
Acknowledgments
We are indebted to Yanli Wu for the cloning and purification of GST-YspC and for arranging the preparation of antibody with Covance Research Products. We thank Scott Handley for the construction and initial characterization of YVM886 (ΔysaC). We thank Damon Ellison for critical review of the manuscript.
This research was supported by National Institutes of Health grant AI42736 awarded to V. L. Miller.
REFERENCES
- 1.Allen, P., C. A. Hart, and J. R. Saunders. 1987. Isolation from Klebsiella and characterization of two rcs genes that activate colanic acid capsular biosynthesis in Escherichia coli. J. Gen. Microbiol. 133:331-340. [DOI] [PubMed] [Google Scholar]
- 2.Arricau, N., D. Hermant, H. Waxin, C. Ecobichon, P. S. Duffey, and M. Y. Popoff. 1998. The RcsB-RcsC regulatory system of Salmonella typhi differentially modulates the expression of invasion proteins, flagellin and Vi antigen in response to osmolarity. Mol. Microbiol. 29:835-850. [DOI] [PubMed] [Google Scholar]
- 3.Bajaj, V., R. L. Lucas, C. Hwang, and C. A. Lee. 1996. Co-ordinate regulation of Salmonella typhimurium invasion genes by environmental and regulatory factors is mediated by control of hilA expression. Mol. Microbiol. 22:703-714. [DOI] [PubMed] [Google Scholar]
- 4.Beloin, C., S. McKenna, and C. J. Dorman. 2002. Molecular dissection of VirB, a key regulator of the virulence cascade of Shigella flexneri. J. Biol. Chem. 277:15333-15344. [DOI] [PubMed] [Google Scholar]
- 5.Bereswill, S., and K. Geider. 1997. Characterization of the rcsB gene from Erwinia amylovora and its influence on exoploysaccharide synthesis and virulence of the fire blight pathogen. J. Bacteriol. 179:1354-1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bradford, W. D., P. S. Noce, and L. T. Gutman. 1974. Pathologic features of enteric infection with Yersinia enterocolitica. Arch. Pathol. 98:17-22. [PubMed] [Google Scholar]
- 7.Carniel, E. 1999. The Yersinia high-pathogenicity island. Int. Microbiol. 2:161-167. [PubMed] [Google Scholar]
- 8.Carter, P. B. 1975. Pathogenicity of Yersinia enterocolitica for mice. Infect. Immun. 11:164-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clark, M. A., B. H. Hirst, and M. A. Jepson. 1998. M-cell surface beta1 integrin expression and invasin-mediated targeting of Yersinia pseudotuberculosis to mouse Peyer's patch M cells. Infect. Immun. 66:1237-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cornelis, G. R. 2002. The Yersinia Ysc-Yop ′type III' weaponry. Nat. Rev. Mol. Cell. Biol. 3:742-752. [DOI] [PubMed] [Google Scholar]
- 11.Cornelis, G. R., A. Boland, A. P. Boyd, C. Geuijen, M. Iriarte, C. Neyt, M. P. Sory, and I. Stainier. 1998. The virulence plasmid of Yersinia, an antihost genome. Microbiol. Mol. Biol. Rev. 62:1315-1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cover, T. L., and R. C. Aber. 1989. Yersinia enterocolitica. N. Engl. J. Med. 321:16-24. [DOI] [PubMed] [Google Scholar]
- 13.Darwin, K. H., and V. L. Miller. 1999. InvF is required for expression of genes encoding proteins secreted by the SPI1 type III secretion apparatus in Salmonella typhimurium. J. Bacteriol. 181:4949-4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Darwin, K. H., and V. L. Miller. 2000. The putative invasion protein chaperone SicA acts together with InvF to activate the expression of Salmonella typhimurium virulence genes. Mol. Microbiol. 35:949-960. [DOI] [PubMed] [Google Scholar]
- 15.Darwin, K. H., and V. L. Miller. 2001. Type III secretion chaperone-dependent regulation: activation of virulence genes by SicA and InvF in Salmonella typhimurium. EMBO J. 20:1850-1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dennis, J. J., and G. J. Zylstra. 1998. Plasposons: modular self-cloning minitransposon derivatives for rapid genetic analysis of gram-negative bacterial genomes. Appl. Environ. Microbiol. 64:2710-2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dorman, C. J., and M. E. Porter. 1998. The Shigella virulence gene regulatory cascade: a paradigm of bacterial gene control mechanisms. Mol. Microbiol. 29:677-684. [DOI] [PubMed] [Google Scholar]
- 18.Ellison, D. W., B. Young, K. Nelson, and V. L. Miller. 2003. YmoA negatively regulates expression of invasin from Yersinia enterocolitica. J. Bacteriol. 185:7153-7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Foultier, B., P. Troisfontaines, S. Muller, F. R. Opperdoes, and G. R. Cornelis. 2002. Characterization of the ysa pathogenicity locus in the chromosome of Yersinia enterocolitica and phylogeny analysis of type III secretion systems. J. Mol. Evol. 55:37-51. [DOI] [PubMed] [Google Scholar]
- 20.Foultier, B., P. Troisfontaines, D. Vertommen, M. N. Marenne, M. Rider, C. Parsot, and G. R. Cornelis. 2003. Identification of substrates and chaperone from the Yersinia enterocolitica 1B Ysa type III secretion system. Infect. Immun. 71:242-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Freeman, J. A., and B. L. Bassler. 1999. Sequence and function of LuxU: a two-component phosphorelay protein that regulates quorum sensing in Vibrio harveyi. J. Bacteriol. 181:899-906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Galan, J. E., and R. Curtiss 3rd. 1989. Cloning and molecular characterization of genes whose products allow Salmonella typhimurium to penetrate tissue culture cells. Proc. Natl. Acad. Sci. USA 86:6383-6387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haller, J. C., S. Carlson, K. J. Pederson, and D. E. Pierson. 2000. A chromosomally encoded type III secretion pathway in Yersinia enterocolitica is important in virulence. Mol. Microbiol. 36:1436-1446. [DOI] [PubMed] [Google Scholar]
- 24.Herrero, M., V. de Lorenzo, and K. N. Timmis. 1990. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 172:6557-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Isberg, R. R., and J. M. Leong. 1990. Multiple beta 1 chain integrins are receptors for invasin, a protein that promotes bacterial penetration into mammalian cells. Cell 60:861-871. [DOI] [PubMed] [Google Scholar]
- 26.Isberg, R. R., D. L. Voorhis, and S. Falkow. 1987. Identification of invasin: a protein that allows enteric bacteria to penetrate cultured mammalian cells. Cell 50:769-778. [DOI] [PubMed] [Google Scholar]
- 27.Kane, C. D., R. Schuch, W. A. Day, Jr., and A. T. Maurelli. 2002. MxiE regulates intracellular expression of factors secreted by the Shigella flexneri 2a type III secretion system. J. Bacteriol. 184:4409-4419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kinder, S. A., J. L. Badger, G. O. Bryant, J. C. Pepe, and V. L. Miller. 1993. Cloning of the YenI restriction endonuclease and methyltransferase from Yersinia enterocolitica serotype O8 and construction of a transformable R-M+ mutant. Gene 136:271-275. [DOI] [PubMed] [Google Scholar]
- 29.Lee, A. K., C. S. Detweiler, and S. Falkow. 2000. OmpR regulates the two-component system SsrA-SsrB in Salmonella pathogenicity island 2. J. Bacteriol. 182:771-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lodge, J., J. Fear, S. Busby, P. Gunasekaran, and N. R. Kamini. 1992. Broad host range plasmids carrying the Escherichia coli lactose and galactose operons. FEMS Microbiol. Lett. 74:271-276. [DOI] [PubMed] [Google Scholar]
- 31.Lostroh, C. P., V. Bajaj, and C. A. Lee. 2000. The cis requirements for transcriptional activation by HilA, a virulence determinant encoded on SPI-1. Mol. Microbiol. 37:300-315. [DOI] [PubMed] [Google Scholar]
- 32.Lostroh, C. P., and C. A. Lee. 2001. The Salmonella pathogenicity island-1 type III secretion system. Microbes Infect. 3:1281-1291. [DOI] [PubMed] [Google Scholar]
- 33.Lucas, R. L., and C. A. Lee. 2000. Unravelling the mysteries of virulence gene regulation in Salmonella typhimurium. Mol. Microbiol. 36:1024-1033. [DOI] [PubMed] [Google Scholar]
- 34.Mavris, M., A. L. Page, R. Tournebize, B. Demers, P. Sansonetti, and C. Parsot. 2002. Regulation of transcription by the activity of the Shigella flexneri type III secretion apparatus. Mol. Microbiol. 43:1543-1553. [DOI] [PubMed] [Google Scholar]
- 35.Mavris, M., P. J. Sansonetti, and C. Parsot. 2002. Identification of the cis-acting site involved in activation of promoters regulated by activity of the type III secretion apparatus in Shigella flexneri. J. Bacteriol. 184:6751-6759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Merriam, J. J., R. Mathur, R. Maxfield-Boumil, and R. R. Isberg. 1997. Analysis of the Legionella pneumophila fliI gene: intracellular growth of a defined mutant defective for flagellum biosynthesis. Infect. Immun. 65:2497-2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Michiels, T., J. C. Vanooteghem, C. Lambert de Rouvroit, B. China, A. Gustin, P. Boudry, and G. R. Cornelis. 1991. Analysis of virC, an operon involved in the secretion of Yop proteins by Yersinia enterocolitica. J. Bacteriol. 173:4994-5009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miller, J. H. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 39.Miller, V. L., and S. Falkow. 1988. Evidence for two genetic loci in Yersinia enterocolitica that can promote invasion of epithelial cells. Infect. Immun. 56:1242-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nagel, G., A. Lahrz, and P. Dersch. 2001. Environmental control of invasin expression in Yersinia pseudotuberculosis is mediated by regulation of RovA, a transcriptional activator of the SlyA/Hor family. Mol. Microbiol. 41:1249-1269. [DOI] [PubMed] [Google Scholar]
- 41.Ochman, H., F. C. Soncini, F. Solomon, and E. A. Groisman. 1996. Identification of a pathogenicity island required for Salmonella survival in host cells. Proc. Natl. Acad. Sci. USA 93:7800-7804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pepe, J. C., and V. L. Miller. 1993. Yersinia enterocolitica invasin: a primary role in the initiation of infection. Proc. Natl. Acad. Sci. USA 90:6473-6477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Portnoy, D. A., and R. J. Martinez. 1985. Role of a plasmid in the pathogenicity of Yersinia species. Curr. Top. Microbiol. Immunol. 118:29-51. [DOI] [PubMed] [Google Scholar]
- 44.Revell, P. A., and V. L. Miller. 2000. A chromosomally encoded regulator is required for expression of the Yersinia enterocolitica inv gene and for virulence. Mol. Microbiol. 35:677-685. [DOI] [PubMed] [Google Scholar]
- 45.Revell, P. A., and V. L. Miller. 2001. Yersinia virulence: more than a plasmid. FEMS Microbiol. Lett. 205:159-164. [DOI] [PubMed] [Google Scholar]
- 46.Sambrook, J. E., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 47.Sledjeski, D. D., and S. Gottesman. 1996. Osmotic shock induction of capsule synthesis in Escherichia coli K-12. J. Bacteriol. 178:1204-1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stewart, G. S., S. Lubinsky-Mink, C. G. Jackson, A. Cassel, and J. Kuhn. 1986. pHG165: a pBR322 copy number derivative of pUC8 for cloning and expression. Plasmid 15:172-181. [DOI] [PubMed] [Google Scholar]
- 49.Stout, V., and S. Gottesman. 1990. RcsB and RcsC: a two-component regulator of capsule synthesis in Escherichia coli. J. Bacteriol. 172:659-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Takeda, S., Y. Fujisawa, M. Matsubara, H. Aiba, and T. Mizuno. 2001. A novel feature of the multistep phosphorelay in Escherichia coli: a revised model of the RcsC -> YojN -> RcsB signalling pathway implicated in capsular synthesis and swarming behaviour. Mol. Microbiol. 40:440-450. [DOI] [PubMed] [Google Scholar]
- 51.Vantrappen, G., E. Ponette, K. Geboes, and P. Bertrand. 1977. Yersinia enteritis and enterocolitis: gastroenterological aspects. Gastroenterology 72:220-227. [PubMed] [Google Scholar]
- 52.Wang, R. F., and S. R. Kushner. 1991. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100:195-199. [PubMed] [Google Scholar]
- 53.West, A. H., and A. M. Stock. 2001. Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem. Sci. 26:369-376. [DOI] [PubMed] [Google Scholar]
- 54.Young, B. M., and G. M. Young. 2002. Evidence for targeting of Yop effectors by the chromosomally encoded Ysa type III secretion system of Yersinia enterocolitica. J. Bacteriol. 184:5563-5571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Young, B. M., and G. M. Young. 2002. YplA is exported by the Ysc, Ysa, and flagellar type III secretion systems of Yersinia enterocolitica. J. Bacteriol. 184:1324-1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou, L., X. H. Lei, B. R. Bochner, and B. L. Wanner. 2003. Phenotype microarray analysis of Escherichia coli K-12 mutants with deletions of all two-component systems. J. Bacteriol. 185:4956-4972. [DOI] [PMC free article] [PubMed] [Google Scholar]