Abstract
Dps, the nonspecific DNA-binding protein from starved cells, is the most abundant protein in stationary-phase Escherichia coli. Dps homologs are found throughout the bacteria and in at least one archaeal species. Dps has been shown to protect cells from oxidative stress during exponential-phase growth. During stationary phase, Dps organizes the chromosome into a highly ordered, stable nucleoprotein complex called the biocrystal. We show here that Dps is required for long-term stationary-phase viability under competitive conditions and that dps mutants have altered lag phases compared to wild-type cells. We also show that during stationary phase Dps protects the cell not only from oxidative stress but also from UV and gamma irradiation, iron and copper toxicity, thermal stress, and acid and base shock. The protective roles of Dps are most likely achieved through a combination of functions associated with the protein-DNA binding and chromosome compaction, metal chelation, ferroxidase activity, and regulation of gene expression.
Dps, the DNA-binding protein from starved cells, is among the most abundant proteins in stationary-phase Escherichia coli. As cells transition from exponential-phase growth to stationary phase, Dps is induced in an RpoS-dependent manner, reaching up to 200,000 molecules per cell (2). Dps is also regulated by OxyR and σ70 in response to oxidative stress during exponential phase (3). The protein has nonspecific DNA-binding activity, affects the expression of at least three dozen genes, and plays a role in various stress responses, including protection from oxidative damage (2, 18). The protein's active form is a hollow, spherical, dodecamer with an outer diameter of ∼90 Å and an interior core diameter of ∼45 Å (2, 27). During stationary phase, Dps binds the chromosome, forming a highly ordered and stable nucleoprotein complex called a biocrystal. This biocrystalline form is distinct from the exponential-phase configuration of the nucleoid and is unique to stationary-phase cells (12, 19, 27). Biocrystal formation contributes to the ability of Dps to affect gene expression and protects chromosomal DNA. Dps shows significant structural homology to the ferritins, a highly conserved family of proteins found in virtually all organisms, and possesses ferroxidase activity and the ability to sequester iron (15). This iron-binding activity very likely plays a significant role in the ability of Dps to protect the chromosome from oxidative damage.
For E. coli, most studies of Dps have focused on its role in oxidative damage protection, particularly against peroxides, during exponential phase (2, 18). Here we expand the repertoire of potential stressors to include long-term stationary-phase incubation under conditions of nutrient stress and competition, the presence of oxidizing agents and heavy metals, thermal stress, ionizing and UV radiation, and extremes of pH. Our working hypothesis is that this multifunctional protein is involved in many cellular processes and probably functions via several different mechanisms. First, through direct Dps-DNA interactions, the protein protects DNA by sequestering the chromosome into the highly stable biocrystal complex. Dps may also directly protect DNA by serving as an alternative target for reactive agents. Second, the protein reduces the production of oxidative radicals that can damage DNA and protein by sequestering and mineralizing metal ions, especially ferrous iron, that can engage in the Fenton reaction (9, 26). Third, Dps can neutralize toxic peroxides through its ferroxidase activity. Finally, Dps may regulate the expression of genes that are required for the long-term survival and stress responses of stationary-phase cells. We chose to test this hypothesis by determining the relative sensitivities of wild-type and dps mutant cells to a wide variety of environmental stresses.
MATERIALS AND METHODS
Bacterial strains and plasmids.
All experiments were performed with strains derived from E. coli K-12 strain ZK126 (28) and are listed in Table 1. The plasmid pJE106 includes the dps gene with its own regulatory region (2).
TABLE 1.
Bacterial strains
Culture conditions, media, and titration assays.
All strains were cultured in 5 ml of Luria-Bertani (LB) broth (Difco) and incubated at 37°C with aeration in a test tube roller unless otherwise noted. Viable-cell counts were determined by serial dilution of cells removed periodically from a culture, followed by plating on LB agar containing the appropriate antibiotic(s). Antibiotics were used at the following concentrations: nalidixic acid, 20 μg/ml; streptomycin, 25 μg/ml; chloramphenicol, 30 μg/ml; kanamycin, 50 μg/ml; ampicillin, 150 μg/ml. All chemicals were obtained from Sigma-Aldrich.
Long-term growth, lag phase, and stationary-phase competition assays.
To measure long-term survival of individual strains, 5-ml LB broth cultures were inoculated 1:1,000 (vol:vol) from fresh overnight cultures started directly from frozen LB broth-glycerol stocks. These monocultures were incubated, and viable counts were periodically determined. To determine the length of lag phase of cells exiting long-term stationary-phase incubation, cultures were initially inoculated as described above and allowed to incubate for 10 days. Every 24 h, 10 μl of cells was removed and used to inoculate new 5-ml LB broth cultures. Viable counts and optical density at 600 nm were then determined every 30 min for 5 h. The lag time was calculated as the length of time from inoculation until exponential growth begins, which was determined by interpolating a line through the exponential-phase time points and marking the point of inflection with respect to the horizontal line representing the initial density. All growth phase and competition experiments were performed at least three times. Lag-phase determinations were performed four or five times.
To determine relative fitness, wild-type, mutant, and/or plasmid-transformed strains carrying different antibiotic resistance markers were coinoculated 1:1,000 (vol:vol) from fresh overnight cultures. Individual subpopulation titers were determined after periodic sampling, serial dilution, and plating on medium containing appropriate antibiotics. For experiments where the relative fitness of strains carrying dps-expressing plasmids was determined, at each time point serial dilutions were plated on agar containing either kanamycin alone, kanamycin and ampicillin, chloramphenicol alone, or chloramphenicol and ampicillin. Plating cells on medium containing a single drug compared to two drugs allows the extent of plasmid loss to be determined. For all experiments, the limit of detection is <1,000 CFU/ml.
Oxidative-stress assays.
Overnight stationary-phase cultures of wild-type or dps mutant cells were incubated at 37°C with aeration in the presence or absence of hydrogen peroxide at concentrations up to 400 mM. After the addition of H2O2 (which becomes time zero in the experiment), viable-cell counts were periodically determined. All assays were performed at least three times.
UV and gamma irradiation.
Overnight stationary-phase cultures of wild-type or dps mutant cells were exposed to UV light. Five-milliliter cultures were transferred to a sterile 10-cm-diameter petri dish, placed under a germicidal lamp (254 nm, 30 J/m2), and sampled periodically for up to 5 min. After exposure, samples were kept on ice until viable-cell counts were determined. All UV experiments were performed five times.
Overnight stationary-phase cultures of wild-type or dps mutant cells were exposed to gamma radiation, up to 700 Gy, at the Jet Propulsion Laboratory, Pasadena, Calif. One milliliter of each culture was transferred to microcentrifuge tubes and exposed without shaking at ambient temperature. The amount of irradiation was proportional to the exposure time. After exposure, samples were kept on ice until viable-cell counts were determined. Gamma irradiation experiments were performed in duplicate.
Metal stress assays.
Exponential-phase or stationary-phase cultures of wild-type or dps mutant cells were incubated at 37°C with aeration in the presence or absence of different metals at the following concentrations: FeSO4, 5 to 10 mM in log phase and 20 to 200 mM in stationary phase; CuSO4, 5 to 10 mM; 50 to 100 mM; MnCl2, 50 to 100 mM and 100 mM; SnCl2, 50 to 100 mM and 50 to 100 mM; K2Cr2O7, 5 to 10 mM and 100 mM; Pb(NO3)2, 50 to 100 mM and 50 to 100 mM; Na2SeO3, 5 to 10 mM and 50 to 100 mM; CdCl2, 5 to 500 mM and 250 to 500 mM; NiSO4, 5 to 10 mM and 250 to 500 mM; HgCl2, 5 to 10 mM and 50 to 100 mM. After addition of the metal solution (which becomes time zero in the experiment), viable-cell counts were periodically determined. All metal stress assays were performed in duplicate.
To determine the metal sensitivity of cells during log-phase growth, overnight cultures were inoculated 1:500 (vol:vol) into fresh LB broth and grown for 2 h. Cultures were then treated with metals, and viable counts were periodically determined.
Temperature stress.
Wild-type or dps mutant cells were incubated overnight at 37°C. Cultures were then shifted to a shaking water bath at a temperature of 45 to 60, 4, or 18°C. Viable-cell counts were periodically determined. All assays were performed four times.
For thermal adaptation experiments, cells were initially grown as described above. Wild-type or dps mutant cultures were then shifted to 53°C for 10 min, followed by a further shift to 55°C. Viable-cell counts were periodically determined. Experiments were performed in duplicate.
pH stress.
Overnight stationary-phase cultures of wild-type or dps mutant cells were incubated at 37°C with aeration in medium adjusted to pH 2 with 10% HCl or adjusted to pH 12 with 5 N NaOH. After the pH shift (which becomes time zero in the experiment), viable-cell counts were periodically determined. All pH stress assays were performed in duplicate.
RESULTS
dps is required for long-term stationary-phase survival during competition in coculture, but not in monoculture.
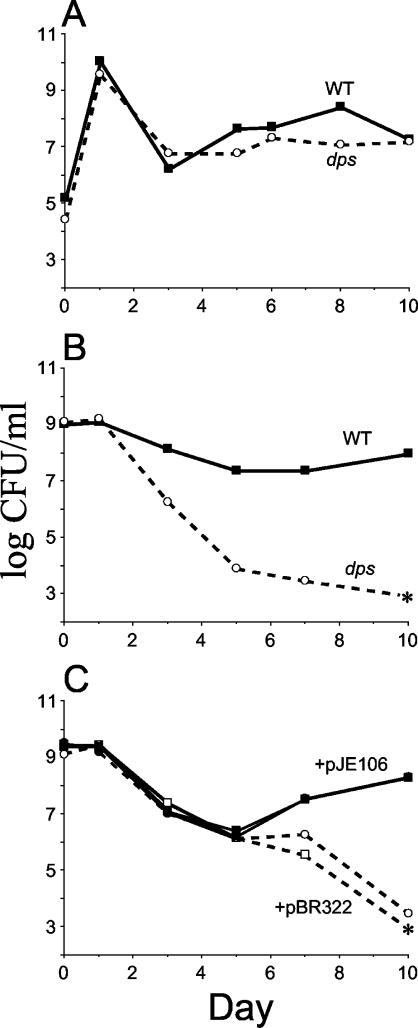
Since Dps is maximally expressed during stationary phase, we wished to determine if the protein is required for stationary-phase survival. Wild-type or dps mutant cells were inoculated in monoculture into LB broth and incubated at 37°C (Fig. 1A). Viable counts of both wild-type and mutant cells are comparable over 10 days of incubation, initially reaching ∼109 CFU/ml upon entry into stationary phase and dropping to ∼107 CFU/ml after passing through death phase. The viable counts remain at ∼107 CFU/ml for more than 15 days of continuous batch culture incubation, without the addition of nutrients (data not shown).
FIG. 1.
Long-term survival and competition patterns. (A) Superimposition of growth curves of cells grown separately in monoculture. WT, wild type. (B) Competition assay of cells grown in coculture. (C) Competition of dps null mutants containing dps-expressing (pJE106) or control (pBR322) plasmids. Solid symbols, SF2049(pJE106-Dps+) plated on kanamycin only (squares) or on kanamycin and ampicillin (circles). Open symbols, SF2043(pBR322-Dps−) plated on chloramphenicol only (squares) or on chloramphenicol and ampicillin (circles). Asterisks indicate no detectable cells (limit of detection, <1,000 CFU/ml). Representative data are shown.
In contrast, when wild-type and dps mutant cells are coinoculated into the same culture, dps mutants show a stationary-phase-specific competition defect (9; R. Yalamanchili and S. E. Finkel, unpublished data). This inability to compete is observed in two types of competition experiments. First, when overnight, stationary-phase cultures of both wild-type and mutant cells are mixed 1:1 and then allowed to continue incubating in stationary phase, the mutant population begins to decline compared to the wild type after 1 day of coincubation (Fig. 1B). After 3 days, dps mutant titers are <1% of maximum cell density, and, after 7 days, dps mutants are no longer detectable in the culture; wild-type cells maintain ∼107 CFU/ml. Essentially the same results are observed when a fresh LB broth culture is inoculated 1:1,000 with equal numbers of wild-type and dps mutant cells. Titers of both subpopulations initially reach the same density of ∼109 CFU/ml in stationary phase, and then the dps mutant population gradually declines until no cells are detectable after 7 to 10 days (data not shown).
The stationary-phase-specific competition defect of dps mutants can be eliminated by expression of Dps from a plasmid. When long-term incubation experiments are performed using monocultures of dps mutants carrying either a Dps-expressing plasmid or a control plasmid, neither strain shows a difference in stationary-phase survival compared to cells without the plasmid (data not shown). However, when a dps mutant carrying a plasmid with a dps gene (pJE106) is competed in coculture with a dps strain carrying a control plasmid (pBR322), the strain with the control vector is outcompeted by cells expressing dps (Fig. 1C). dps mutants carrying the control plasmid not only fail in competition but also lose the vector plasmid over time (when experiments are performed in the absence of drug selection), suggesting that Dps expression helps to maintain the plasmid (data not shown). These results demonstrate that dps mutants survive in monoculture but cannot compete with wild-type strains in nutritionally restricted medium.
Lag-phase differences between dps mutants and wild-type cells after stationary-phase incubation.
The nucleoid structures of wild-type and dps mutant cells are known to differ considerably during stationary phase (10). Cells expressing Dps form the highly ordered biocrystal, while the nucleoid in dps mutants transitions into a highly stable, gel-like cholesteric phase during long-term culture (12, 17). As previously reported, when spread onto fresh nutrient agar, dps mutants from 6-day-old cultures form colonies more slowly than wild-type cells (12). This observation led us to study the effect of Dps during exit from long-term stationary phase by determining the length of the lag phase.
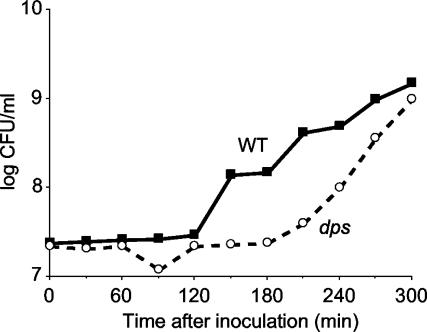
Wild type or dps mutant cells were inoculated into LB broth and incubated at 37°C for 10 days. Each day, 10 μl was removed from each culture and inoculated into fresh medium. The lag time and exponential phase growth rate were then determined. Dps mutants have an increased lag period compared to wild-type cells during the first 2 days of long-term stationary-phase incubation, approximately double that observed for the wild-type (Fig. 2; Table 2). Surprisingly, after 3 days of incubation, lag times are similar through day 6. After this time, as the age of the inoculum increases, lag times of dps mutant cultures again increase relative to those of the wild type until day 9 (Table 2). Although the lengths of lag phase differ, once cells enter exponential phase, growth rates and final cell densities are comparable to those for wild-type cells and densities reach ∼109 CFU/ml after overnight incubation.
FIG. 2.
Comparison of lag phases of wild-type (WT) and dps mutant cells from 1-day-old stationary-phase cultures. Shown is a superimposition of growth curves of cells grown separately in monoculture.
TABLE 2.
Lag times of wild-type and dps strains
| Days in stationary phase | Length of lag time (min) ± SD
|
Lag time ratio (dps mutant/wild type) | |
|---|---|---|---|
| Wild type | dps | ||
| 1 | 83 ± 23 | 152 ± 30 | 1.8 |
| 2 | 88 ± 19 | 175 ± 50 | 2.0 |
| 3 | 81 ± 31 | 100 ± 20 | 1.2 |
| 4 | 105 ± 11 | 105 ± 11 | 1 |
| 5 | 98 ± 4 | 98 ± 4 | 1 |
| 6 | 100 ± 21 | 100 ± 21 | 1 |
| 7 | 90 ± 42 | 140 ± 55 | 1.6 |
| 8 | 100 ± 0 | 110 ± 14 | 1.1 |
| 9 | 125 ± 45 | 125 ± 45 | 1 |
| 10 | 95 ± 6 | 95 ± 6 | 1 |
dps mutants are sensitive to oxidative stress during stationary phase.
Most studies of the sensitivity to oxidative stress of dps mutants have focused on exponential-phase cells. Here we examine stationary-phase cultures. Overnight cultures of wild-type or dps mutant cells were treated with hydrogen peroxide, and survival patterns were determined. Unlike exponential-phase cells, which are killed by H2O2 at concentrations above 50 mM, stationary-phase wild-type and dps mutant cells are resistant to hydrogen peroxide for more than 3 h at concentrations up to 200 mM. Above these concentrations, dps mutants start to show increased sensitivity to hydrogen peroxide. dps mutants treated with 400 mM H2O2 lose viability after 30 min of treatment, whereas wild-type cells maintain viability at ∼106 CFU/ml for ∼90 min (Fig. 3).
FIG. 3.
Hydrogen peroxide stress. Overnight cultures of wild-type (WT; squares) or dps (circles) cells were treated with 400 mM H2O2. Solid symbols, untreated cells; open symbols, peroxide-treated cells. Asterisks indicate no detectable cells (limit of detection, <1,000 CFU/ml). Representative data are shown.
dps mutants are sensitive to UV irradiation.
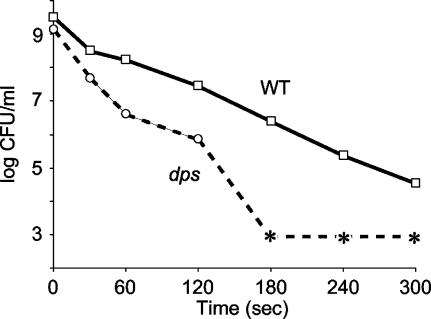
Stationary-phase cultures of wild-type or dps mutant cells were exposed to UV irradiation, and survival patterns were determined. dps mutants are more sensitive to UV light than wild-type cells (Fig. 4). After 30 s of exposure, more than 99% of dps cells are killed, compared to 90% of wild-type cells. After 60 s of exposure, mutant-cell viability is further reduced compared to the wild-type cell viability. After 3 min, wild-type cell counts are ∼106 CFU/ml, while dps mutants are undetectable.
FIG. 4.
UV stress. Overnight cultures of wild-type (WT) or dps cells were exposed to UV radiation. Asterisks indicate no detectable cells (limit of detection, <1,000 CFU/ml). Representative data are shown.
dps mutants are sensitive to gamma irradiation.
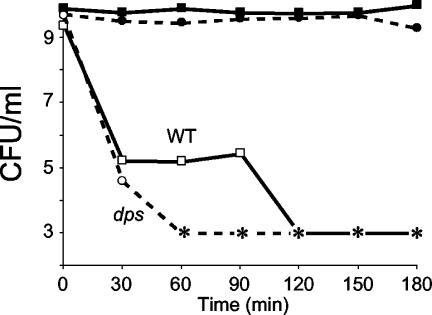
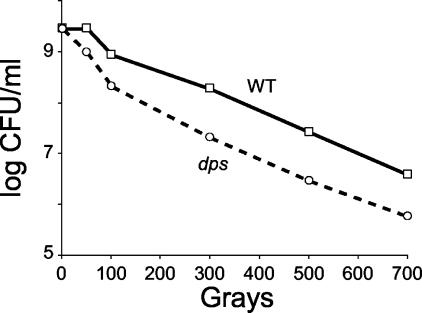
Stationary-phase cultures of dps mutant or wild-type cells were exposed to gamma radiation. The lethal radiation dose for E. coli during exponential phase is approximately 30 to 70 Gy (10). During stationary phase, wild-type cells completely lose viability above ∼700 to 1,000 Gy. dps mutants are more sensitive to gamma irradiation at all levels of exposure, consistently showing a 10-fold-lower rate of survival at exposures of 300 Gy and above (Fig. 5).
FIG. 5.
Gamma radiation stress. Overnight cultures of wild-type (WT) or dps cells were exposed to gamma irradiation. Representative data are shown.
dps mutants show differential sensitivity to certain metals during stationary phase and exponential phase.
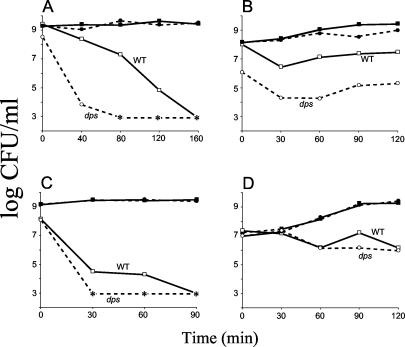
Stationary-phase cultures of wild-type and dps mutant cells were treated with various metals. Treatment with cadmium, chromium, lead, mercury, manganese, nickel, selenium, and tin reveals no Dps-specific effect; wild-type and dps mutant cells show similar sensitivities (data not shown). A significant Dps-specific effect is observed with iron or copper treatment during stationary phase. After 45 min of treatment with 100 mM iron, wild-type cell densities are 107 CFU/ml, while mutant titers are reduced to ∼103 CFU/ml (Fig. 6A). After 80 min of treatment dps mutants are undetectable; it takes wild-type cells twice as long to completely lose viability. A Dps-specific effect was observed in log phase as well. During early log phase, dps mutant cells are more sensitive than wild-type cells when treated with 5 to 10 mM Fe2+ (Fig. 6B).
FIG. 6.
Metal stress during stationary phase and log phase. Wild type (WT; squares) or dps (circles) cells were treated with either iron (A and B) or copper (C and D) during stationary phase (A and C) or log phase (B and D). Open symbols, metal treatment; filled symbols, no-treatment controls. Asterisks indicate no detectable cells (limit of detection, <1,000 CFU/ml). Representative data are shown.
When treated with 50 mM copper, stationary-phase cultures of dps mutants completely lose viability after 30 min of exposure, whereas wild-type cells take ∼90 min to lose viability (Fig. 6C). However, unlike what is found for iron treatment, dps mutants show no significant increase in sensitivity to copper during exponential-phase growth (Fig. 6D).
Zinc treatment shows a dps-specific effect only during the transition from late exponential phase into stationary phase. Upon treatment with 100 mM zinc, mutant-cell titers are reduced over 100-fold (from 106 to 104 CFU/ml) whereas wild-type titers are reduced only 10-fold (from 107 to 106 CFU/ml). This Dps-specific difference in survival is not observed in early-exponential-phase or stationary-phase cultures (data not shown).
dps mutants are sensitive to thermal stress and do not show temperature-dependent thermal adaptation.
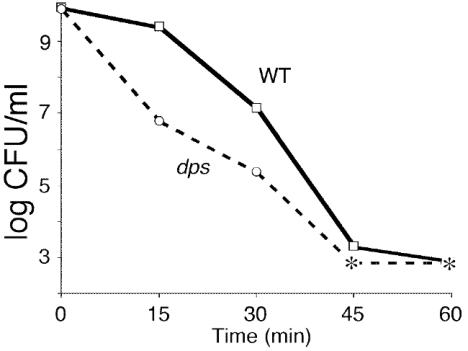
Overnight stationary-phase cultures of wild-type or dps mutant cells were shifted to incubation temperatures ranging from 45 to 60°C, after which cell viability was determined every 5 min for 1 h. At 45°C no difference in viability between wild-type and dps mutant cells was observed. At 50 and 53°C viability of wild-type cells is 10-fold higher than that of mutants after 90 min of exposure. After 15 min at 55°C, a 100-fold difference in survival between wild-type cells and dps mutants is observed; mutant titers drop to 107 CFU/ml, while wild-type titers remain at 109 CFU/ml (Fig. 7). After an additional 15 min, dps cell titers decrease to 105 CFU/ml, and at 45 min titers are undetectable; wild-type cells survive for ∼60 min. After 5 min at 58°C, wild-type cells show titers of 104 CFU/ml, whereas dps mutants are inviable (data not shown).
FIG. 7.
Temperature stress. Overnight cultures of wild-type (WT) or dps cells were incubated at 55°C. Asterisks indicate no detectable cells (limit of detection, <1,000 CFU/ml). Representative data are shown.
Given the increased sensitivity of dps mutants to thermal stress, we determined if Dps plays a role in thermal adaptation. Thermal adaptation is a phenomenon in which cells that are initially exposed to an elevated, sublethal temperature become resistant to higher temperatures that are usually lethal (16, 23, 24). To observe Dps-specific effects on thermal adaptation, stationary-phase cells of both the mutant and wild-type strains were pretreated at 53°C for 10 min, followed by incubation at 55°C. dps mutant cell titers dropped to 106 CFU/ml within 15 min at 55°C, whereas wild-type cells apparently underwent thermal adaptation, remaining at 109 CFU/ml. Wild-type-cell titers remain high for 30 min (data not shown).
Similar survival experiments were done for cold shock. Stationary-phase cultures of both wild-type and dps mutant cells were shifted to either 4 or 18°C. There was no observed difference in cell viability between the wild-type and mutant strains after 2 h of incubation at either temperature or after 10 days of incubation at 4°C (data not shown).
dps mutants are sensitive to alkaline and acid pH.
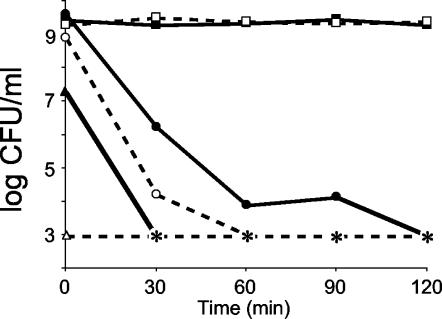
The pH of stationary-phase cultures of wild-type or dps mutant cells was shifted over a range of 2 to 12 by addition of HCl or NaOH, respectively. Under conditions of acid stress, dps mutants show 100-fold-greater sensitivity after 30 min at pH 2 and 104-fold-greater sensitivity after 60 min of exposure, the time at which mutants become undetectable (Fig. 8). It takes twice as long for wild-type cells to completely lose viability.
FIG. 8.
pH stress. Overnight cultures of wild-type (solid symbols) or dps (open symbols) cells were incubated at pH 7 (squares), 2 (circles), or 12 (triangles). Asterisks indicate no detectable cells (limit of detection, <1,000 CFU/ml). Representative data are shown.
Under alkaline conditions (pH 12) dps mutants instantly lose viability upon addition of NaOH (Fig. 8). Though wild-type cells show an immediate 100-fold decrease in cell titer, it takes 30 min for these cells to completely lose viability.
DISCUSSION
We have identified Dps homologs in over 130 bacterial species, and one member of the archaea, representing a wide variety of prokaryotic organisms (S. Nair and S. E. Finkel, unpublished data). The ubiquity across species, high abundance, DNA-binding activity, and protective functions of Dps led us to study the role of this protein in long-term stationary-phase survival. This study shows that cells expressing Dps are able to endure many environmental stresses to a greater degree than dps mutants. There are a number of mechanisms by which Dps can protect cells against these stresses based on four intrinsic properties of the protein: DNA binding, metal binding and sequestration, ferroxidase activity, and ability to affect gene regulation. These are each discussed below.
Dps monomers assemble into a dodecameric functional unit, and these dodecamers pack (like oranges stacked in a crate) with DNA into a three-dimensional hexagonal structure (10, 11, 16, 24). In the biocrystal, the positively charged DNA backbone most likely interacts with Dps through basic residues on the dodecamer surfaces (13). During stationary phase, Dps condenses the chromosome such that most other macromolecules are excluded. By directly binding the chromosome, Dps can shield DNA from damage caused by peroxides and other agents. This model is supported by studies showing that Dps protects plasmid DNA against peroxide and DNase I attack (12). The protection against gamma irradiation provided to DNA by HU, another bacterial histone-like protein, is similar to the protection by Dps shown here (6). HU mutants are five times more sensitive to gamma radiation than wild-type cells. We propose that Dps protects DNA against UV and gamma radiation by a mechanism similar to that for HU: prevention of the single- and double-strand breaks associated with irradiation. It may also serve as a competitive substrate for the oxidative radicals generated by various solvents, metals, and radiation.
It is also possible that Dps increases the efficiency and accuracy of the repair process by recruiting repair enzymes or other proteins to the chromosome. For example, Dps may modulate interactions with proteins involved in maintaining the supercoiled state of the DNA. Since, as shown here, dps mutants are temperature sensitive, by forming the DNA-bound biocrystal, Dps may prevent relaxation of DNA during heat shock (1). This may be similar to the observed interactions between DnaK and DNA gyrase. During heat shock, the DnaK chaperone associates with the chromosome, facilitating DNA gyrase activity (20). It will be interesting to determine whether Dps interacts similarly with any such proteins.
We also can see similarities between the activity of Dps and the SASP (small acid soluble) proteins of spores of Bacillus subtilis (25). SASP proteins bind to DNA nonspecifically and change its conformation from B form to A form. During UV irradiation, SASP proteins prevent formation of bipyrimidine photoproducts, such as the T-T photodimer. Instead, photoproducts called SP lesions form and are repaired by a spore-specific repair system (22). The SASP proteins also change the chemical reactivity of DNA during heat stress, slowing the rate of depurination. Dps, by forming the biocrystal, may similarly change the reactivity of the DNA so that it is less sensitive to heat stress and UV irradiation. It will be useful to determine the conformation of DNA bound by Dps and compare the relative number of depurination sites after heat stress in wild-type and dps null strains.
Dps is a structural homolog of ferritin, the primary iron storage protein of virtually all organisms (13, 14). Functionally, ferritins are defined by the ability to bind iron and the presence of a ferroxidase activity. The fundamental difference between Dps and the ferritins is that Dps binds DNA. Dps and ferritin monomers have the same four-helix-bundle tertiary structure and form multiprotein spheres with hollow cores, functioning as 12-mers and 24-mers, respectively. In both multimer spheres, three monomer subunits form trimers (four in Dps and eight for the ferritins) with a pore of ∼20 Å in diameter. These pores are lined with acidic residues that can accommodate positively charged ions, such as iron (13). Dps homologs in other organisms, such as Dps from Listeria innocua (7) and DpsA from Halobacterium salinarium (22), have also been shown to be ferritins. DpsA from Synechococcus is a bacterioferritin, as it contains a heme moiety (21). It was recently shown that Dps acts as an iron storage protein, participating in its binding, oxidation, and mineralization (15). Each dodecamer can accommodate ∼500 iron atoms. Metal chelation protects organisms from assault by various reactive metals, including Fe(II), which engages in the Fenton reaction: Fe2+ + H2O2 → Fe3+ + OH− + OḢ. This reaction yields hydroxyl radicals that can damage macromolecules, including proteins, membrane lipids, and DNA. In ferritins, Fe(II) is oxidized at the ferroxidase center, precipitating as insoluble ferric oxyhydroxide, Fe(III), and is stored in the protein's hollow core. The following reaction summarizes the reactions that take place during iron oxidation and mineralization when dioxygen is the oxidant (14): 4Fe2+ + O2 + 6H2O → 4FeOOH + 8H+. In contrast, iron oxidation by Dps occurs slowly when O2 is the oxidant. However, in the presence of hydrogen peroxide, iron oxidization and mineralization are rapid (29). The rate of oxidation is more than 100 times faster in the presence of H2O2 than O2. This difference may be due to different amino acids forming the ferroxidase active site compared to those for the ferritins (13, 15). Dps does not have the conserved residues associated with the ferroxidase center of the ferritin family. The Dps ferroxidase consumes H2O2 in two ways, both for oxidation and for mineralization (29), summarized by the reaction 2Fe2+ + H2O2 + 2H2O → 2FeOOH + 4H+. In this reaction Fe(II) is oxidized to Fe(III) by H2O2 and oxidized Fe(III) is mineralized at the core as insoluble ferric(III) oxohydroxide at the core of the dodecamer (29). One molecule of H2O2 is consumed for every two molecules of Fe(II) oxidized.
Thus by oxidizing and mineralizing iron, Dps serves two purposes: (i) both processes lead to quenching of H2O2 and prevent it from reacting with ferrous iron in the Fenton reaction and (ii) through mineralization, Fe(III) is stored in the Dps core. The trapped iron is sequestered from the cytosol, a reducing environment that can convert it back into reactive Fe2+. This prevents it from interacting with H2O2 formed during various metabolic activities. Dps also has been shown to have a weak catalase activity, further neutralizing hydrogen peroxide (29). This results in two potentially deleterious reactants being converted to harmless products.
Dps is known to affect gene expression both positively and negatively. When two-dimensional gel electrophoresis of cell extracts prepared from wild-type and dps mutant cells is performed, differences in protein spot patterns are observed (2). Some spots are present in wild-type cells but not in the mutant, and vice versa. During long-term stationary-phase incubation (up to 10 days), differential protein expression has been observed (S. E. Finkel and R. Kolter, unpublished results). In eukaryotes, histones undergo reversible covalent modifications to make DNA accessible or inaccessible to the transcriptional machinery, thereby regulating gene expression (4). It will be interesting to determine whether Dps plays a similar role in differential gene expression by undergoing covalent modifications. Dps may regulate gene expression both by modulating the structure of the DNA and by interacting with the transcription machinery, possibly recruiting transcription factors. Also of interest will be a comparison of gene expression patterns under various stress conditions, such as those described here.
The presence of Dps homologs in a wide variety of organisms, though with functional divergence, suggests its importance in stress survival in many environments. In most organisms it is likely a DNA binding protein and protects the organism against various stresses. However, in some pathogenic organisms, such as Treponema pallidum and Haemophilus ducreyi, it is a major virulence determinant required for pathogenicity (5, 8, 11). Studies of stationary-phase bacteria may serve as a better approximation of the natural environments where these organisms reside, and such studies will help us to better understand the myriad roles of this highly abundant protein.
Acknowledgments
We thank Michael Farrell, Vyacheslav Palchevskiy, Evan Pepper, Julie Badger, George O'Toole, and Dianne Newman for helpful discussions and comments on the manuscript. We thank Pamela Conrad for assistance with gamma irradiation experiments.
This work was supported in part by the USC/Norris Comprehensive Cancer Center and the W. M. Keck Foundation.
REFERENCES
- 1.Adamcik, J., V. Viglasky, F. Valle, M. Antalik, D. Podhradsky, and G. Dietler. 2002. Effect of bacteria growth temperature on the distribution of supercoiled DNA and its thermal stability. Electrophoresis 23:3300-3309. [DOI] [PubMed] [Google Scholar]
- 2.Almiron, M., A. J. Link, D. Furlong, and R. Kolter. 1992. A novel DNA-binding protein with regulatory and protective roles in starved Escherichia coli. Genes Dev. 6:2646-2654. [DOI] [PubMed] [Google Scholar]
- 3.Altuvia, S., M. Almiron, G. Huisman, R. Kolter, and G. Storz. 1994. The dps promoter is activated by OxyR during growth and by IHF and sigma S in stationary phase. Mol. Microbiol. 13:265-272. [DOI] [PubMed] [Google Scholar]
- 4.Berger, S. L. 1999. Gene activation by histone and factor acetyltransferases. Curr. Opin. Cell Biol. 11:336-341. [DOI] [PubMed] [Google Scholar]
- 5.Borenstein, L. A., J. D. Radolf, T. E. Fehniger, D. R. Blanco, J. N. Miller, and M. A. Lovett. 1988. Immunization of rabbits with recombinant Treponema pallidum surface antigen 4D alters the course of experimental syphilis. J. Immunol. 140:2415-2421. [PubMed] [Google Scholar]
- 6.Boubrik, F., and J. Rouviere-Yaniv. 1995. Increased sensitivity to gamma irradiation in bacteria lacking protein HU. Proc. Natl. Acad. Sci. USA 92:3958-3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bozzi, M., G. Mignogna, S. Stefanini, D. Barra, C. Longhi, P. Valenti, and E. Chiancone. 1997. A novel non-heme iron-binding ferritin related to the DNA-binding proteins of the Dps family in Listeria innocua. J. Biol. Chem. 272:3259-3265. [DOI] [PubMed] [Google Scholar]
- 8.Brentjens, R. J., M. Ketterer, M. A. Apicella, and S. M. Spinola. 1996. Fine tangled pili expressed by Haemophilus ducreyi are a novel class of pili. J. Bacteriol. 178:808-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buda, F., B. Ensing, M. C. Gribnau, and E. J. Baerends. 2003. O2 evolution in the Fenton reaction. Chemistry 9:3436-3444. [DOI] [PubMed] [Google Scholar]
- 10.Byun, M. W., O. J. Kwon, H. S. Yook, and K. S. Kim. 1998. Gamma irradiation and ozone treatment for inactivation of Escherichia coli O157:H7 in culture media. J. Food Prot. 61:728-730. [DOI] [PubMed] [Google Scholar]
- 11.Fehniger, T. E., J. D. Radolf, and M. A. Lovett. 1986. Properties of an ordered ring structure formed by recombinant Treponema pallidum surface antigen 4D. J. Bacteriol. 165:732-739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frenkiel-Krispin, D., S. Levin-Zaidman, E. Shimoni, S. G. Wolf, E. J. Wachtel, T. Arad, S. E. Finkel, R. Kolter, and A. Minsky. 2001. Regulated phase transitions of bacterial chromatin: a non-enzymatic pathway for generic DNA protection. EMBO J. 20:1184-1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grant, R. A., D. J. Filman, S. E. Finkel, R. Kolter, and J. M. Hogle. 1998. The crystal structure of Dps, a ferritin homolog that binds and protects DNA. Nat. Struct. Biol. 5:294-303. [DOI] [PubMed] [Google Scholar]
- 14.Harrison, P. M., and P. Arosio. 1996. The ferritins: molecular properties, iron storage function and cellular regulation. Biochim. Biophys. Acta 1275:161-203. [DOI] [PubMed] [Google Scholar]
- 15.Ilari, A., P. Ceci, D. Ferrari, G. L. Rossi, and E. Chiancone. 2002. Iron incorporation into Escherichia coli Dps gives rise to a ferritin-like microcrystalline core. J. Biol. Chem. 277:37619-37623. [DOI] [PubMed] [Google Scholar]
- 16.Jenkins, D. E., J. E. Schultz, and A. Matin. 1988. Starvation-induced cross protection against heat or H2O2 challenge in Escherichia coli. J. Bacteriol. 170:3910-3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leforestier, A., and F. Livolant. 1993. Supramolecular ordering of DNA in the cholesteric liquid crystalline phase: an ultrastructural study. Biophys. J. 65:56-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martinez, A., and R. Kolter. 1997. Protection of DNA during oxidative stress by the nonspecific DNA-binding protein Dps. J. Bacteriol. 179:5188-5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Minsky, A., E. Shimoni, and D. Frenkiel-Krispin. 2002. Stress, order and survival. Nat. Rev. Mol. Cell Biol. 3:50-60. [DOI] [PubMed] [Google Scholar]
- 20.Ogata, Y., T. Mizushima, K. Kataoka, K. Kita, T. Miki, and K. Sekimizu. 1996. DnaK heat shock protein of Escherichia coli maintains the negative supercoiling of DNA against thermal stress. J. Biol. Chem. 271:29407-29414. [DOI] [PubMed] [Google Scholar]
- 21.Pena, M. M., and G. S. Bullerjahn. 1995. The DpsA protein of Synechococcus sp. strain PCC7942 is a DNA-binding hemoprotein. Linkage of the Dps and bacterioferritin protein families. J. Biol. Chem. 270:22478-22482. [DOI] [PubMed] [Google Scholar]
- 22.Reindel, S., S. Anemuller, A. Sawaryn, and B. F. Matzanke. 2002. The DpsA-homologue of the archaeon Halobacterium salinarum is a ferritin. Biochim. Biophys. Acta. 1598:140-146. [DOI] [PubMed] [Google Scholar]
- 23.Riehle, M. M., A. F. Bennett, and A. D. Long. 2001. Genetic architecture of thermal adaptation in Escherichia coli. Proc. Natl. Acad. Sci. USA 98:525-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schultz, J. E., G. I. Latter, and A. Matin. 1988. Differential regulation by cyclic AMP of starvation protein synthesis in Escherichia coli. J. Bacteriol. 170:3903-3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Setlow, P. 1995. Mechanisms for the prevention of damage to DNA in spores of Bacillus species. Annu. Rev. Microbiol. 49:29-54. [DOI] [PubMed] [Google Scholar]
- 26.Stadtman, E. R., and B. S. Berlett. 1991. Fenton chemistry. Amino acid oxidation. J. Biol. Chem. 266:17201-17211. [PubMed] [Google Scholar]
- 27.Wolf, S. G., D. Frenkiel, T. Arad, S. E. Finkel, R. Kolter, and A. Minsky. 1999. DNA protection by stress-induced biocrystallization. Nature 400:83-85. [DOI] [PubMed] [Google Scholar]
- 28.Zambrano, M. M., D. A. Siegele, M. Almiron, A. Tormo, and R. Kolter. 1993. Microbial competition: Escherichia coli mutants that take over stationary phase cultures. Science 259:1757-1760. [DOI] [PubMed] [Google Scholar]
- 29.Zhao, G., P. Ceci, A. Ilari, L. Giangiacomo, T. M. Laue, E. Chiancone, and N. D. Chasteen. 2002. Iron and hydrogen peroxide detoxification properties of DNA-binding protein from starved cells. A ferritin-like DNA-binding protein of Escherichia coli. J. Biol. Chem. 277:27689-27696. [DOI] [PubMed] [Google Scholar]