Abstract
Cell type specific delivery of RNAi to T cells has remained to be a challenge. Here we describe an aptamer mediated delivery of shRNA to CD4+ T cells targeting RORγt to suppress Th17 cells. A cDNA encoding CD4 aptamer and RORγt shRNA was constructed and the chimeric CD4 aptamer-RORγt shRNA (CD4-AshR-RORγt) was generated using in vitro T7 RNA transcription. 2′-F-dCTP and 2′-F-dUTP were incorporated into CD4-AshR-RORγt for RNase resistance. CD4-AshR-RORγt was specifically uptaken by CD4+ Karpas 299 cells and primary human CD4+ T cells. The RORγt shRNA moiety of CD4-AshR-RORγt chimera was cleaved and released by Dicer. Furthermore, CD4-AshR-RORγt suppressed RORγt gene expression in Karpas 299 cells and CD4+ T cells and consequently inhibited Th17 cell differentiation and IL-17 production. These results demonstrate that aptamer-facilitated cell specific delivery of shRNA represents a novel approach for efficient RNAi delivery and is potentially to be developed for therapeutics targeting specific T cells subtypes.
Introduction
RNA interfering (RNAi)-mediated gene silencing holds great promise for manipulating T cells to study basic T cell biology and for developing potential T cell targeted therapeutics. However, efficient delivery of small interfering RNA (siRNA) into primary T cells represents a major hurdle to the widely use of RNAi technology [1]. T cells are known to be “hard to transfect”. Several methods of transfection have been applied to T cells with satisfactory efficiency in primary T cells but with several caveats [1]. Electroporation and nucleofection suffer excessive cell loss and may require pre-activation of T cells [2,3]. It was reported that chemically modified synthetic siRNA with Acell agents can also be used to transfect siRNA into primary T cells but these require prolonged pre-incubation with T cells and works in only a small number of cells [4]. The most notable disadvantage of these methods is that they are not suitable for in vivo use. Retroviral vectors are effective methods to transfect siRNA into T cells [5,6] since the viral vectors integrate into the host genome and thus the siRNA is stably expressed for the lifetime of the cell. The same reason limits the viral vector transfection for potential therapeutics due to the concern about malignant transformation [1]. Nanoparticles are effective vehicles for siRNA delivery to T cells and an in vivo system has been reported, but the delivery is not T cell specific [7]. Peptides, including polyarginine with cell-penetrating properties, have been studied to deliver siRNA to cells [8]. Using an anti-CD7 single chain antibody conjugated to a 9-arginine peptide we have attempted to deliver siRNA to T cells but achieved inefficiency due to precipitation of anti-CD7 single chain antibody-arginine-siRNA complex.
Aptamers are single stranded oligonucleotides selected from random sequence libraries with high affinity and specificity to the target molecules [9,10]. Besides being effective therapeutic agents, aptamers have been actively exploited for targeted delivery of drugs including siRNA [11]. In theory, due to their high specificity and affinity, aptamers can deliver siRNA into any cell type provided the cells express the ligand for aptamer to bind. The aptamer-siRNA chimera, first described in 2006 by McNamara et al [12] has been exploited to deliver siRNA into prostate cancer cells. Zhou et al [13] modified the aptamer-siRNA chimera with aptamer specific to HIV envelope protein expressed by viral infected T cells and siRNA to viral genes and successfully suppressed HIV replication in HIV infected human CD4+ T cells. Wheeler et al [14,15] developed a CD4 aptamer-siRNA chimera that targeted CCR5, gag and vif and delivered to infected human CD4+ T cells and suppressed the targeted gene expression and killed HIV.
Here we describe a CD4 aptamer-shRNA chimera specific to RORγt to suppress T helper 17 (Th17) cells with potential to develop for a Th17 specific therapeutic agent in Th17 mediated inflammatory diseases. Increasing evidence indicates that Th17 cells and their released cytokines play a critical role in the pathogenesis of autoimmune and inflammatory diseases [16]. Th17 cells preferentially express and produce its signature cytokine IL-17A, and IL-17F, IL-21 and IL-22 as well. Th17 cells and their secreted cytokines are considered to account for initiation and maintenance of several autoimmune and inflammatory disorders [16,17,18,19]. Blocking IL-17A activity has been proven to be highly effective to treat immune mediated inflammatory disease models and clinical trials with blocking IL-17 are ongoing with promising results to treat inflammatory diseases [20,21,22]. However, IL-17A and IL-17F are also produced by many other innate immune cells and are important cytokines in host defense [23]. Moreover, it is Th17 cells that are detrimental and are to be blocked for therapeutic purpose. Therefore, it is highly desirable to narrow the target to Th17 cells and spare IL-17 cytokines produced by innate immune cells from being blocked.
Materials and Methods
Synthesis of CD4 aptamer-RORγt shRNA chimera
Chimera synthesis was modified from previously described methods [14,24,25,26]. DNA oligos used for PCR (supplementory Table 1) were commercially synthesized (Integrated DNA Technologies). cDNA Template containing T7 promoter used for synthesis of chimera was synthesized with Pfu DNA polymerase (Thermo Fisher Scientific) and purified with QIAquick Gel purification kit (Qiagen). The sequence of cDNA was verified by sequencing. The RNA CD4 aptamer-shRNA chimera was transcribed using T7 polymerase in vitro using DuraScribe kit (Illumina). 2′-F-dCTP and 2′-F-dUTP were incorporated to enhance RNase resistance andCy3-CTP (GE) was incorporated (Cy3-CTP/2′-F-dCTP ratio = 1/9) for visualization and resolved on 6% dPAGE gel for Cy3 scanning and then ethidium bromide staining prior to purification with G25 column (GE) following phenol extraction and sodium acetate/ethanol precipitation. The sequences of the chimeras of CD4 or mock CD4 aptamer-shRNAs against retinoic related orphan receptor (ROR)γt and CCR5 or scrambled shRNA are shown in supplementary Table 2. Additionally, in order to investigate if the CD4 aptamer shRNA chimera transcribed in vitro is the substrate for the endoribonuclease Dicer that processes longer endogenous RNA precursors into short RNA as an intracellular step of the RNAi pathway, Dicer cleavage of the chimera was assayed in vitro with recombinant human Dicer kit (Genlantis) in accordance to the manufactory’s instruction.
T lymphocyte cell lines and T-enriched PBMCs
Karpas 299 cell line was obtained from Dr. Zu (Houston Methodist Hospital Research Institute) [27] and maintained in RPMI1640 containing 10% FBS. For evaluation of Cy3-labeled chimera internalization, Karpas 299 cells were incubated with 200 nM chimera overnight. For analysis of the function of chimeras in silencing RORγt and IL-17 production, Karpas 299 cells were incubated with the chimera for 72h. Fresh PMBCs from healthy donors were isolated by Ficoll (GE) density centrifugation and cultured in RPMI 1640 medium containing 10% human AB serum. T enriched PBMCs were prepared by adding anti-CD11c, CD11b, CD19, CD56 and immnunomagnetic beads to PBMCs (BD Bioscience) and purified CD4+ primary T cells are derived by removing CD8+ T cell from T enriched PBMCs with anti-CD8 and immnunomagnetic beads.
Fluorescent microscopy and flow cytometry
Internalization of the synthesized chimera was determined by incubating 200 nM or 1 μM Cy3-labeled chimera with Karpas 299 cells or T-cell enriched PBMC overnight. The cells were stained with FITC-anti-CD4 (BioLegend) and analyzed by confocal microscopy. T-cell enriched PBMCs were stimulated with anti-CD3/CD28 conjugated to MACS beads for 5 days. For Th1 cells, IL-12 (10 ng/ml) was added; for Th2 cells, IL-4 (10 ng/ml) and anti-human IFN-γ (10 μg/ml) were added; for Th17 cells, LPS (100 ng/ml) was added in the culture. PMA (50 ng/ml) and Ionomycin (500 ng/ml) were added 5 h prior to harvest for intracellular staining. Intracellular staining for RORγt and IL-17A was performed with PE-anti-mouse/human RORγt and PE-anti human IL-17A (eBioscience); staining for IFN-γ and IL-4 was performed with PE-anti-human IFN-γ and PE-anti-human IL-4 (BioLegend) and analyzed by flow cytometry.
Real-time PCR
Real-time PCR was performed as previously described [28]. The probe and primers mixes for RORC2 (Hs01076112), TBX21 (Hs00203436), GATA3 (Hs00231122) and GUSB (Hs9999908) were purchased from Thermo Fisher Scientific. mRNA levels for RORC, TBX21 and GATA3 were normalized by GUSB.
Quantification of cytokines
IL-17A levels in the supernatant were quantified by ELISA (eBiosciences) as previously described [29]. Karpas 299 cells were incubated with 50 ng/ml PMA and additional 40 mM sodium chloride for 48 h prior to harvesting the supernatant. T-cell enriched PBMCs were activated with biotinylated antibodies against human CD3 and CD28, conjugated to anti-biotin MACS beads (Miltenyi Biotec Inc.) and 100 ng/ml lipopolysaccharide (LPS) 48h prior to collecting the supernatant.
Statistics
Data are presented as mean ± SD. Data of real-time PCR, ELISA and flow cytometry were analyzed by one-way ANOVA followed by Dunnett comparison test. P value < 0.05 was considered significant.
Results and Discussion
CD4 aptamer-RORγt shRNA chimera was specifically internalized into human CD4+ T cells
RNA aptamer that is identified by SELEX can specifically bind cellular membrane proteins with high-affinity due to its complex and functional secondary and probably tertiary structures determined by its unique nucleotide sequences [30]. Furthermore, it is demonstrated that CD4 RNA aptamer can conjugate and deliver siRNAs/shRNAs targeting CCR5 and HIV gp120 gene into the T cells that express CD4 [14,15]. We constructed a cDNA template by PCR to encode a CD4 aptamer, RORγt shRNA sense chain, loop and RORγt shRNA antisense chain. The cDNA sequence was verified by sequencing. The RNA of CD4 aptamer-RORγt shRNA chimera (CD4-AshR-RORγt) was transcribed as a single molecule in vitro by T7 polymerase transcription. A mock CD4 aptamer was created using scrambled sequence. Both CD4-AshR-RORγt and mock-CD4-AshR-RORγt chimeras consist of 133 nucleotides in length (Figure 1A). The predicted secondary structures of CD4-AshR-RORγt and mock CD4-AshRRORγt chimeras that were generated by the computational method (Rochester University) were shown in Figure 1B and C. Similarly, we generated CD4 aptamer-CCR5 shRNA (CD4-AshR-CCR5) and CD4 aptamer-scrambled shRNA (CD4-AshR-scrambled) chimeras as negative controls for RORγt shRNA. All of the chimeras incorporated 2′-F-CTP and 2′-F-UTP for enhanced resistance to RNase. In order to track internalization of the chimeras, two Cy3-CTPs were incorporated into each chimera as determined by spectophotometric analysis. A strong fluorochrome signal was readily detected by fluorescent gel scanning (Figure 2A). Consistent with the characteristic of specifically and effectively delivering, the Cy3-labeled CD4-AshR-RORγt entered human CD4+ T cell line, Karpas 299 cells and CD4+ T cells in PBMC, as assessed with fluorescent confocal microscope and flow cytometric analysis (Figure 2B, 2C and 2D). In contrast, Cy3-labeled mock CD4-AshR-RORγt, in which the sequence of CD4 aptamer was scrambled, was unable to be internalized into Karpas 299, nor in CD4+ T cells (Figure 2D). Consistent with the findings in previous studies, our results showed that only negligible amount of CD4 aptamers or CD4 aptamer conjugated to siRNAs is up-taken by CD8+ T cells (Figure 2D) [14]. These suggest that the synthesized CD4 aptamer-shRNA chimera can be uniquely and sufficiently transferred into the CD4+ human T cells.
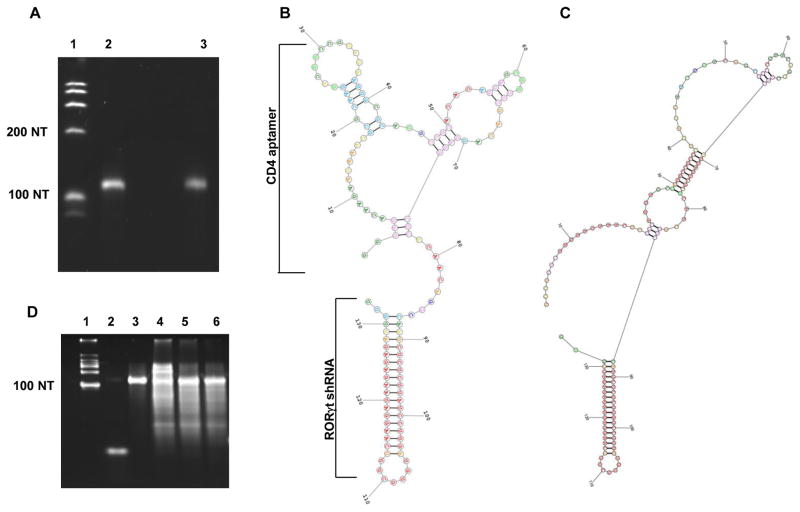
Figure 1. CD4-AshR-RORγt chimera.
(A) Chimera in vitro transcribed by T7 RNA polymerase was analyzed by denatured PAGE and ethidium bromide staining. Lane 1, ssRNA ladder; Lane 2, CD4-AshR-RORγt chimera; Lane 3, mock-CD4-AshR-RORγt chimera. (B and C) Predicted secondary structure of CD4-AshR-RORγt chimera (B) and mock-CD4-AshR-RORγt chimera (C). The region of the CD4 aptamer (clone 9 [26]) responsible for binding to CD4 is outlined. The shRNA portion of the chimera consists of targeted RORγt siRNA with 2 overhang nucleotides at its 3′end and a 7 nucleotide loop. (D) Cleavage analysis of synthesized chimeras by Dicer. Lane 1, ssRNA ladder; Lane 2, antisense siRNA to RORγt; Lane 3, intact CD4-AshR-RORγt chimera; Lane 4–6, chimeras were digested with Dicer: Lane 4, Mock CD4-AshR-RORγt chimera; Lane 5, CD4-AshR-RORγt chimera; Lane 6, CD4-AshR-scrambled control chimera (representative of two experiments).
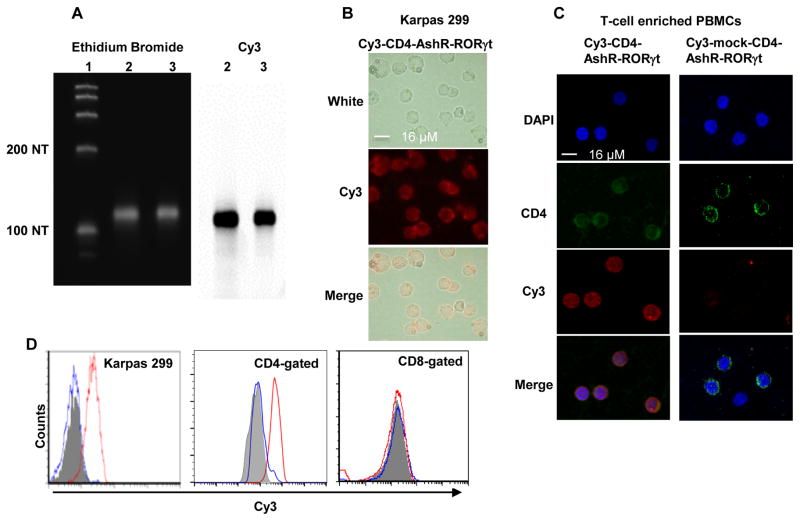
Figure 2. CD4-AshR-RORγt chimera efficiently entered CD4+ human T cells.
(A) CD4- and mock-CD4-AshR-RORγt chimeras were labeled by incorporating Cy3-CTP during in vitro transcription. Cy3 scanning (right panel) showed strong Cy3-signaling bands that were at an appropriate size of transcripts shown in ethidium bromide imaging (left panel). Lane 1, ssRNA ladder; Lane 2, CD4-AshR-RORγt chimera; Lane 3, mock-CD4-AshR-RORγt chimera. (B and C) Uptake of Cy3-labeld CD4-AshR-RORγt chimera by CD4+ human T cell line Karpas 299 cells and CD4+ T cells in PBMCs. (D) Flow cytometric analysis showed that Cy3-labeled CD4-AshR-RORγt chimera was significantly internalized in CD4+ Karpas 299 cells and CD4+ T cells but not in CD8+ T cells. There is no uptake of Cy3-labeled mock-CD4-AshR-RORγt chimera by Karpas 299 cells or T-cell enriched PBMCs. Gray: PBS; Red line: Cy3-labeled CD4-AshR-RORγt chimera; Blue line: Cy3-labeled mock-CD4-AshR-RORγt chimera (representative of 2–5 experiments).
Several strategies have been exploited to link an siRNA to an aptamer [31]. Aptamer-siRNA chimera linking an aptamer with an siRNA directly without using a linker sequence provides effective and specific delivery of siRNA into target cells [12]. To make an aptamer-siRNA chimera, an apteamer-siRNA-sense strand is transcribed then is annealed to the separately synthesized antisense of siRNA. We found that the annealing efficiency of antisense to sense strand linked to the aptamer is not consistent. Whereas, aptamer-shRNA chimera has a unique advantage being synthesized as a single RNA strand which does not require annealing with other RNAs. High yield production of aptamer-shRNA as a single molecule can be consistently achieved. This is particularly important for large scale of production of aptamer-shRNA for in vivo use. Moreover, the siRNA moiety of apatamer-shRNA chimera folds into a short hairpin structure (Figure 1B and C) which closely resembles endogenous microRNA. This has been demonstrated to be more readily processed by the RNAi machinery [31].
CD4-AshR-RORγt chimera significantly silenced RORγt expression in human CD4+ T cells
Intracellular small hairpin RNA should be cleaved into 21–25 nucleotide double strand RNA by Dicer and then the guide strand of the resulting duplexes are processed to the RNA-induced silencing complex (RISC) to degrade the complementary mRNA [25]. Consistent with this, as shown in the Figure 1A and D, the size of CD4-AshR-RORγt chimera produced in vitro by T7 RNA polymerase transcription was originally 133 nucleotides in length. The shRNA moiety of CD4-AshR-RORγt chimera was released into short paired double stranded RNA after cleavage by Dicer (Figure 1D). To confirm the silencing effect on specific gene expression, the level of RORγt mRNA was reduced by CD4-AshR-RORγt in a concentration-dependent fashion in the CD4+ Karpas 299 cells and T cell-enriched PBMCs, but not by mock CD4-AshR-RORγt, CD4-AshR-scrambled control, or CD4-AshR-CCR5 (Figure 3A–3C), as assayed by quantitative real-time PCR. This was further demonstrated by intracellular RORγt staining with flow cytometrey (Figure 3D–F). The suppressive effect of CD4-AshR-RORγt delivered specific shRNA on RORγt expression is consistent with specific siRNAs transfected by lipid transfection agents [32]. In contrast, expression of TBX21 and GATA3 was not altered by CD4-AshR-RORγt (Figure 3C). These data demonstrated that CD4-AshR-RORγtspecifically suppressed RORγt gene expression.
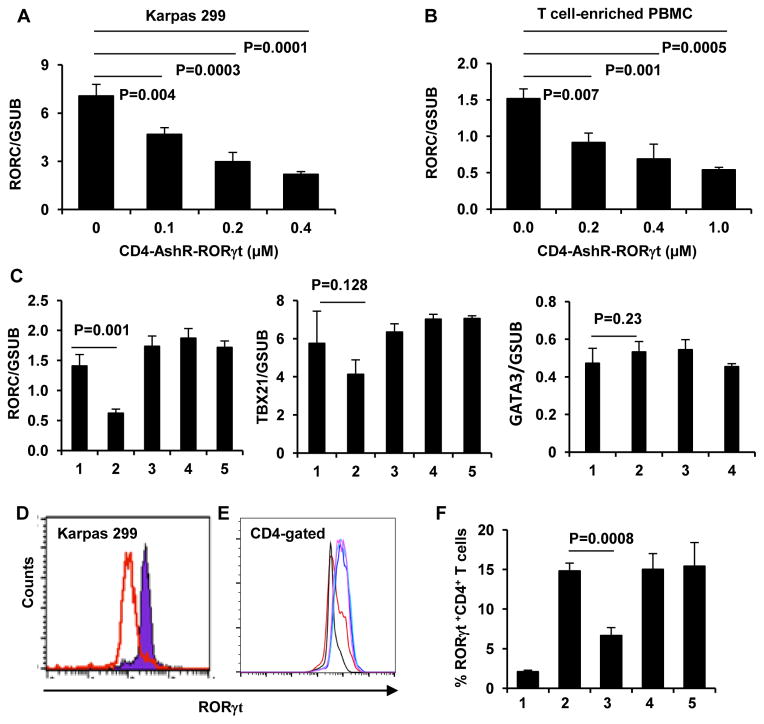
Figure 3. Specific silencing of RORγt in human CD4+ T cells by CD4-AshR-RORγt chimera.
Karpas 299 cells and PBMCs were treated as described in methods. (A through C) Quantitative real-time PCR assay for RORγt gene expression. RORγt gene expression was significantly reduced by CD4-AshR-RORγt chimera in a concentration-dependent manner in Kapas 299 cells (A) and T-cell enriched PBMCs (B). Mock-CD4-AshR-RORγt chimera, CD4-AshR-scrambled control or CD4-AshR-CCR5 chimera had no effect on RORγt gene expression in T-cell enriched PBMCs (C). Additionally, all the chimeras lacked a significant inhibition on TBX21or GATA3 in T-cell enriched PBMCs (C). (Data are presented as mean ± SD of three experiments). 1, PBS; 2, CD4-AshR-RORγt chimera; 3, mock-CD4-AshR-RORγt chimera; 4, CD4-AshR-scrambled control chimera; 5, CD4-AshR-CCR5 chimera. (D and E) RORγt protein expression was analysed by flow cytometry. (D) Karpas 299 cells were stimulated with PMA 50 ng/ml for 24 h. Red line, CD4-AshR-RORγt chimera; Blue line, Mock-CD4-AshR-RORγt chimera (representative of three experiments). (E) PBMCs were stimulated with anti-CD3/CD28 and LPS for 48 h. RORγt expression was reduced by CD4-AshR-RORγt chimera (red line), but not by mock-CD4-AshR-RORγt chimeras (blue line) or CD4-AshR-scrambled control chimera (purple line). Black line, PBMCs without stimulation; green line, PBMCs with stimulation but without chimeras (representative of three experiments). (F) The percentage of RORγt+ cells in stimulated T-cell enriched PBMCs was reduced by CD4-AshR-RORγt chimeras, but not by mock-CD4-AshR-RORγt chimera or CD4-AshR-scrambled control chimera. 1, PBMCs without stimulation; 2, PBMCs with stimulation but without chimeras; 3, Stimulated PBMCs were treated with CD4-AshR-RORγt chimeras; 4, Stimulated PBMCs were treated with mock-CD4-AshR-RORγt chimera; 5, Stimulated PBMCs were treated with CD4-AshR-scrambled control chimera (Data are presented as mean ± SD of three experiments).
CD4-AshR-RORγt chimera significantly inhibited IL17 production by CD4+ human T cells
Down-regulation of RORγt function by its antagonists like digoxin derivatives could result in decrease of both Th17 cells and IL17 production [33,34]. As shown in Figure 4A–D, consistent with decreased RORγt, CD4-AshR-RORγt exerted a concentration-dependent suppression of IL-17A production in CD4+ Karpas 299 cells and T cell-enriched PBMC. In parallel with altered secretion of IL-17A, intracellular IL-17A staining is significantly impaired by CD4-AshR-RORγt, whereas mock CD4-AshR-RORγt, CD4-AshR-scrambled control or CD4-AshR-CCR5 showed no effect. As shown in the Figure 4E–H, the intracellular staining for IFN-γ and IL-4 was not changed by CD4-AshR-RORγt, suggesting it did not affect the synthesis of Th1 or Th2 cytokines. This further confirmed that RORγt is a valid target for regulating Th17 cell differentiation and IL-17 production.
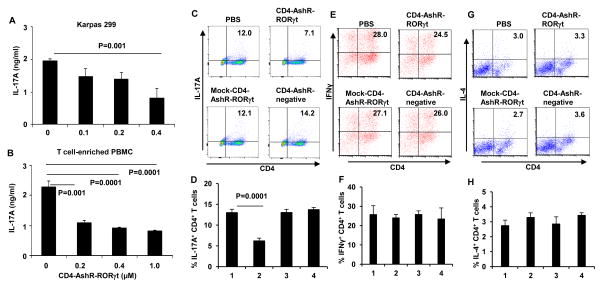
Figure 4. Reduction of IL-17A synthesis in CD4+ T cells by CD4-AshR-RORγt chimera.
Karpas 299 cells were stimulated with PMA and T-cell enriched PBMCs were stimulated with anti-CD3/CD28 and cytokines with anti-cytokine antibodies or LPS for Th1, Th2 or Th17 polarization respectively. (A and B) IL-17A in the supernatant was measured by ELISA. IL-17A production was significantly decreased by CD4-AshR-RORγt chimera in a concentration-dependent fashion (Data are presented as mean ± SD of three experiments). (C and D) Reduction of IL-17A-producing CD4+ T cells by CD4-AshR-RORγt chimera, but not by mock- CD4-AshR-RORγt or CD4-AshR-scrambled control chimeras. (E, F, G and H) CD4-AshR-RORγt chimera had no significant impacts on IFN-γ- and IL-4-producing CD4+ T cells. 1, PBS; 2; CD4-AshR-RORγt chimera; 3, mock-CD4-AshR-RORγt chimera; 4, CD4-AshR-scrambled control chimera (Data are presented as mean ± SD of three experiments).
The present data in our study revealed that CD4 aptamer can serve as a delivery vehicle for shRNA that targets a specific gene in CD4+ human T cells. The internalized RORγt shRNA via CD4 aptamer can be cleaved and released by Dicer and then specifically silenced the targeted RORγt gene expression and finally led to marked decrease of Th17 differentiation and IL-17 production. This particular CD4 aptamer does not alter the cell surface levels of CD4 or other activation markers of the host CD4+ T cells [14]. By substituting the shRNA for targeted genes, this CD4 aptamer may be used as a universal vehicle to introduce RNAi into CD4+ T cells. Compared with other vehicles for siRNA delivery into T cells, aptamers have many advantages. First, the size of aptamers is relatively smaller and less likely to be immunogenic. This is particularly critical for in vivo use as therapeutics. Aptamers can be chemically synthesized and it is relatively less expensive to generate aptamer-shRNA/siRNA. Thus, it is of great interest to evaluate the use of this CD4-AshR-RORγt chimera in treatment of Th17 mediated inflammatory disorders.
Supplementary Material
Highlights.
CD4 aptamer-RORγt shRNA chimera was generated using in vitro T7 RNA transcription.
CD4 aptamer-RORγt shRNA chimera was specifically delivered into CD4+ T cells.
The RORγt shRNA moiety of the chimera was cleaved and released by Dicer.
CD4 aptamer-RORγt shRNA chimera suppressed RORγt gene expression.
CD4 aptamer-RORγt shRNA specifically inhibited Th17 cell differentiation.
Acknowledgments
This work was supported by a grant from NIH (AR055254), ACR Rheumatology Research Foundation and National Psoriasis Foundation.
We would like to thank the following individuals for technical assistance: Drs. Jiehua Zhou and John Rossi (City of Hope), Dr. James McNamara (University of Iowa), Lee Adams Wheeler and Dr. Judy Lieberman (Harvard University).
Footnotes
Disclosures: The authors have no financial conflicts of interest.
The online version of this article contains supplemental material.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Freeley M, Long A. Advances in siRNA delivery to T-cells: potential clinical applications for inflammatory disease, cancer and infection. Biochem J. 2013;455:133–147. doi: 10.1042/BJ20130950. [DOI] [PubMed] [Google Scholar]
- 2.Gehl J. Electroporation: theory and methods, perspectives for drug delivery, gene therapy and research. Acta Physiologica Scandinavica. 2003;177:437–447. doi: 10.1046/j.1365-201X.2003.01093.x. [DOI] [PubMed] [Google Scholar]
- 3.Lai W, Chang CH, Farber DL. Gene transfection and expression in resting and activated murine CD4 T cell subsets. J Immunol Methods. 2003;282:93–102. doi: 10.1016/j.jim.2003.07.015. [DOI] [PubMed] [Google Scholar]
- 4.Gomez-Valades AG, Llamas M, Blanch S, Perales JC, Roman J, Gomez-Casajus L, Mascaro C. Specific Jak3 Downregulation in Lymphocytes Impairs gammac Cytokine Signal Transduction and Alleviates Antigen-driven Inflammation In Vivo. Mol Ther Nucleic Acids. 2012;1:e42. doi: 10.1038/mtna.2012.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rangachari M, Zhu C, Sakuishi K, Xiao S, Karman J, Chen A, Angin M, Wakeham A, Greenfield EA, Sobel RA, Okada H, McKinnon PJ, Mak TW, Addo MM, Anderson AC, Kuchroo VK. Bat3 promotes T cell responses and autoimmunity by repressing Tim-3-mediated cell death and exhaustion. Nat Med. 2012;18:1394–1400. doi: 10.1038/nm.2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jin HS, Liao L, Park Y, Liu YC. Neddylation pathway regulates T-cell function by targeting an adaptor protein Shc and a protein kinase Erk signaling. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:624–629. doi: 10.1073/pnas.1213819110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu Z, Winters M, Holodniy M, Dai H. siRNA delivery into human T cells and primary cells with carbon-nanotube transporters. Angewandte Chemie-International Edition. 2007;46:2023–2027. doi: 10.1002/anie.200604295. [DOI] [PubMed] [Google Scholar]
- 8.Eguchi A, Meade BR, Chang YC, Fredrickson CT, Willert K, Puri N, Dowdy SF. Efficient siRNA delivery into primary cells by a peptide transduction domain-dsRNA binding domain fusion protein. Nat Biotechnol. 2009;27:567–571. doi: 10.1038/nbt.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou J, Neff CP, Swiderski P, Li H, Smith DD, Aboellail T, Remling-Mulder L, Akkina R, Rossi JJ. Functional in vivo delivery of multiplexed anti-HIV-1 siRNAs via a chemically synthesized aptamer with a sticky bridge. Mol Ther. 2013;21:192–200. doi: 10.1038/mt.2012.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ni X, Castanares M, Mukherjee A, Lupold SE. Nucleic acid aptamers: clinical applications and promising new horizons. Curr Med Chem. 2011;18:4206–4214. doi: 10.2174/092986711797189600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou J, Bobbin ML, Burnett JC, Rossi JJ. Current progress of RNA aptamer-based therapeutics. Front Genet. 2012;3:234. doi: 10.3389/fgene.2012.00234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McNamara JO, 2nd, Andrechek ER, Wang Y, Viles KD, Rempel RE, Gilboa E, Sullenger BA, Giangrande PH. Cell type-specific delivery of siRNAs with aptamer-siRNA chimeras. Nat Biotechnol. 2006;24:1005–1015. doi: 10.1038/nbt1223. [DOI] [PubMed] [Google Scholar]
- 13.Zhou J, Li H, Li S, Zaia J, Rossi JJ. Novel dual inhibitory function aptamer-siRNA delivery system for HIV-1 therapy. Mol Ther. 2008;16:1481–1489. doi: 10.1038/mt.2008.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wheeler LA, Trifonova R, Vrbanac V, Basar E, McKernan S, Xu Z, Seung E, Deruaz M, Dudek T, Einarsson JI, Yang L, Allen TM, Luster AD, Tager AM, Dykxhoorn DM, Lieberman J. Inhibition of HIV transmission in human cervicovaginal explants and humanized mice using CD4 aptamer-siRNA chimeras. J Clin Invest. 2011;121:2401–2412. doi: 10.1172/JCI45876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wheeler LA, Vrbanac V, Trifonova R, Brehm MA, Gilboa-Geffen A, Tanno S, Greiner DL, Luster AD, Tager AM, Lieberman J. Durable knockdown and protection from HIV transmission in humanized mice treated with gel-formulated CD4 aptamer-siRNA chimeras. Mol Ther. 2013;21:1378–1389. doi: 10.1038/mt.2013.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miossec P, Kolls JK. Targeting IL-17 and TH17 cells in chronic inflammation. Nat Rev Drug Discov. 2012;11:763–776. doi: 10.1038/nrd3794. [DOI] [PubMed] [Google Scholar]
- 17.van Hamburg JP, Corneth OB, Paulissen SM, Davelaar N, Asmawidjaja PS, Mus AM, Lubberts E. IL-17/Th17 mediated synovial inflammation is IL-22 independent. Ann Rheum Dis. 2013;72:1700–1707. doi: 10.1136/annrheumdis-2012-202373. [DOI] [PubMed] [Google Scholar]
- 18.van Hamburg JP, Asmawidjaja PS, Davelaar N, Mus AM, Colin EM, Hazes JM, Dolhain RJ, Lubberts E. Th17 cells, but not Th1 cells, from patients with early rheumatoid arthritis are potent inducers of matrix metalloproteinases and proinflammatory cytokines upon synovial fibroblast interaction, including autocrine interleukin-17A production. Arthritis & Rheumatism. 2011;63:73–83. doi: 10.1002/art.30093. [DOI] [PubMed] [Google Scholar]
- 19.Cascao R, Moura RA, Perpetuo I, Canhao H, Vieira-Sousa E, Mourao AF, Rodrigues AM, Polido-Pereira J, Queiroz MV, Rosario HS, Souto-Carneiro MM, Graca L, Fonseca JE. Identification of a cytokine network sustaining neutrophil and Th17 activation in untreated early rheumatoid arthritis. Arthritis Res Ther. 2010;12:R196. doi: 10.1186/ar3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chu CQ, Song Z, Mayton L, Wu B, Wooley PH. IFNgamma deficient C57BL/6 (H-2b) mice develop collagen induced arthritis with predominant usage of T cell receptor Vbeta6 and Vbeta8 in arthritic joints. Ann Rheum Dis. 2003;62:983–990. doi: 10.1136/ard.62.10.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lubberts E. IL-17/Th17 targeting: on the road to prevent chronic destructive arthritis? Cytokine. 2008;41:84–91. doi: 10.1016/j.cyto.2007.09.014. [DOI] [PubMed] [Google Scholar]
- 22.Genovese MC, Van den Bosch F, Roberson SA, Bojin S, Biagini IM, Ryan P, Sloan-Lancaster J. LY2439821, a humanized anti-interleukin-17 monoclonal antibody, in the treatment of patients with rheumatoid arthritis: A phase I randomized, double-blind, placebo-controlled, proof-of-concept study. Arthritis & Rheumatism. 2010;62:929–939. doi: 10.1002/art.27334. [DOI] [PubMed] [Google Scholar]
- 23.Korn T, Bettelli E, Oukka M, Kuchroo VK. IL-17 and Th17 Cells. Annu Rev Immunol. 2009;27:485–517. doi: 10.1146/annurev.immunol.021908.132710. [DOI] [PubMed] [Google Scholar]
- 24.Zhou J, Rossi JJ. Aptamer-targeted RNAi for HIV-1 therapy. Methods Mol Biol. 2011;721:355–371. doi: 10.1007/978-1-61779-037-9_22. [DOI] [PubMed] [Google Scholar]
- 25.Ni X, Zhang Y, Ribas J, Chowdhury WH, Castanares M, Zhang Z, Laiho M, DeWeese TL, Lupold SE. Prostate-targeted radiosensitization via aptamer-shRNA chimeras in human tumor xenografts. J Clin Invest. 2011;121:2383–2390. doi: 10.1172/JCI45109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Davis KA, Lin Y, Abrams B, Jayasena SD. Staining of cell surface human CD4 with 2′-F-pyrimidine-containing RNA aptamers for flow cytometry. Nucleic Acids Res. 1998;26:3915–3924. doi: 10.1093/nar/26.17.3915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang P, Zhao N, Zeng Z, Feng Y, Tung CH, Chang CC, Zu Y. Using an RNA aptamer probe for flow cytometry detection of CD30-expressing lymphoma cells. Lab Invest. 2009;89:1423–1432. doi: 10.1038/labinvest.2009.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yomogida K, Chou Y, Pang J, Baravati B, Maniaci BJ, Wu S, Zhu Y, Chu CQ. Streptavidin suppresses T cell activation and inhibits IL-2 production and CD25 expression. Cytokine. 2012;58:431–436. doi: 10.1016/j.cyto.2012.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yomogida K, Chou YK, Chu CQ. Superantigens induce IL-17 production from polarized Th1 clones. Cytokine. 2013;63:6–9. doi: 10.1016/j.cyto.2013.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bouchard PR, Hutabarat RM, Thompson KM. Discovery and development of therapeutic aptamers. Annu Rev Pharmacol Toxicol. 2010;50:237–257. doi: 10.1146/annurev.pharmtox.010909.105547. [DOI] [PubMed] [Google Scholar]
- 31.Dassie JP, Giangrande PH. Current progress on aptamer-targeted oligonucleotide therapeutics. Ther Deliv. 2013;4:1527–1546. doi: 10.4155/tde.13.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burgler S, Mantel PY, Bassin C, Ouaked N, Akdis CA, Schmidt-Weber CB. RORC2 is involved in T cell polarization through interaction with the FOXP3 promoter. Journal of Immunology. 2010;184:6161–6169. doi: 10.4049/jimmunol.0903243. [DOI] [PubMed] [Google Scholar]
- 33.Huh JR, Leung MW, Huang P, Ryan DA, Krout MR, Malapaka RR, Chow J, Manel N, Ciofani M, Kim SV, Cuesta A, Santori FR, Lafaille JJ, Xu HE, Gin DY, Rastinejad F, Littman DR. Digoxin and its derivatives suppress TH17 cell differentiation by antagonizing RORgammat activity. Nature. 2011;472:486–490. doi: 10.1038/nature09978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huh JR, Littman DR. Small molecule inhibitors of RORgammat: targeting Th17 cells and other applications. Eur J Immunol. 2012;42:2232–2237. doi: 10.1002/eji.201242740. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.